Abstract
A 4-year-old boy with severe intellectual disability (ID) and characteristics of autism was found to have a de novo 1.9-Mb microdeletion in 7q31.33q32.1, in which LRRC4, GRM8, and 11 other genes were included. GRM8 is associated with attention deficit hyperactivity disorder. LRRC4 is related to synaptic cell adhesion molecules, some of which are associated with autism. The deletion of LRRC4 may be responsible for the severe ID and characteristics of autism observed in the present patient.
Interstitial deletions encompassing the 7q31 region are rare, and there are few reports of affected patients.1,2 We identified a de novo 1.9-Mb deletion in a patient with severe intellectual disability and characteristics of autism. Here, we report the patient’s clinical characteristics and discuss the genotype–phenotype correlation.
The boy, who was 4 years and 7 months old, was born to non-consanguineous healthy Japanese parents. There were no remarkable episodes during pregnancy. There was no family history of neurodevelopmental disorders. His two older sisters are healthy. He was born at 38 weeks and 5 days of gestation by cesarean section and weighed 4,180 g (>97th percentile). Just after birth, he showed poor sucking. Developmental delays had been noted since early infancy; head control occurred at 7 months, sitting occurred at 18 months, and walking alone occurred at 2 years and 3 months. On the basis of the Enjoji developmental test, his developmental quotient was evaluated as 19 at 3 years of age, thus indicating severe intellectual disability. Behaviors related to attention deficit and/or hyperkinetic disorder was rarely observed. Instead, he exhibited stereotyped motions, with head swinging, turning around in circles in the same place, and self-injuring by banging his head on the floor. He often exhibited a panicked state when he did not get things in the way that he wanted. He was not shy, but he showed poor eye contact. Sleep disturbance was frequently observed. These findings were considered to be characteristics of autism (a formal examination could not be performed, owing to severe intellectual disability). He experienced an episode of afebrile seizure with generalized tonic convulsion at the age of 3 years and 4 months.
At present, his height is 102.1 cm (25–50th percentile), his weight is 16.2 kg (25–50th percentile), and his occipitofrontal circumference is 50.0 cm (25–50th percentile), thus indicating proportional stature. His facial features are not distinctive. He still cannot speak any meaningful words. He uses diapers because of incontinence. Assistance in his daily life activities, including getting dressed and eating, is required.
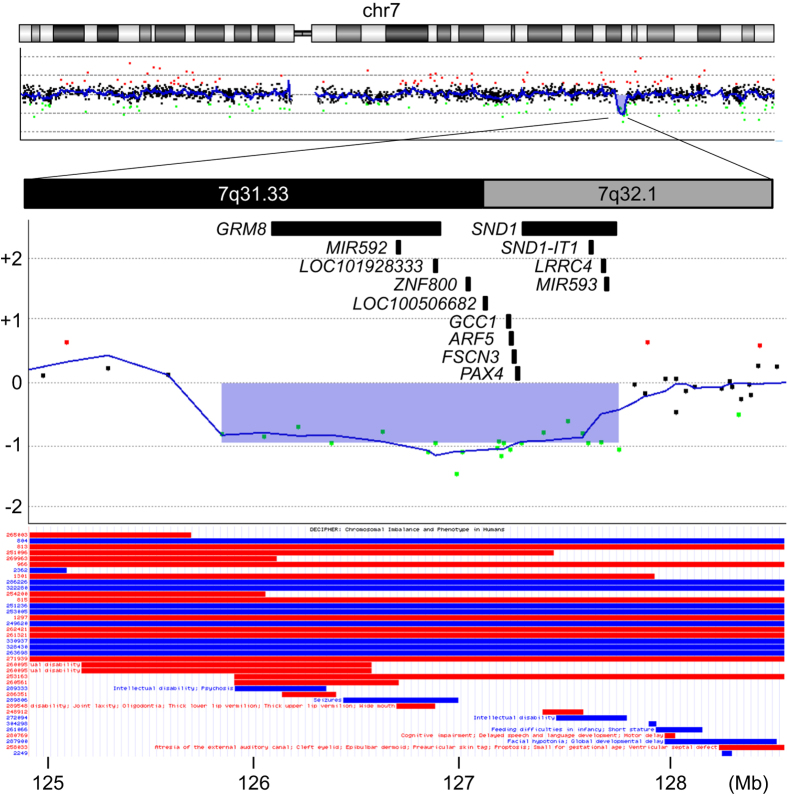
This study was approved by the ethics committee of Tokyo Women’s Medical University. After written informed consent had been obtained from the patient’s family, peripheral blood samples were obtained from the patient and his parents. Genomic DNA was extracted with a QIAquick DNA extraction kit (Qiagen, Hilden, Germany). Chromosomal microarray testing was performed with an Agilent 60 K Human Genome CGH Microarray platform (Agilent Technologies, Santa Clara, CA, USA), as previously described.3 Next, a genomic copy number loss at the 7q31.33 region [arr 7q31.33q32.1(125,875,286–127,816,631)×1] was identified (Figure 1). The deletion length was 1.9 Mb, and 13 RefSeq genes were included in the deletion (Table 1, Figure 1). The same deletion was not detected in the parental samples through microarray testing, thus indicating a de novo occurrence. These data have been registered as #332875 in the DECIPHER database (https://decipher.sanger.ac.uk/). The data presented by the UCSC genome browser are depicted in Figure 1 for comparison.
Figure 1.
Results of the chromosomal microarray testing superimposed with genes located in this region. Chromosome view (top) and gene view (middle) created by Agilent Genomic Workbench (Agilent Technologies), showing an interstitial 1.9-Mb deletion at 7q31.33q32.1. The web image of the UCSC genome browser (https://genome.ucsc.edu/ (visited 20 September 2016)), in which the DECIPHER data in the region of interest are shown, was captured and pasted (bottom). The X and Y axes indicate genomic location and signal log2 ratio, respectively. The deleted region (shown by blue translucent rectangle) includes 13 RefSeq genes, which are depicted with gene symbols. Black bars indicate the gene locations. Red and blue bars in DECIPHER data indicate the loss and the gain of genomic copy numbers, respectively.
Table 1. List of genes included in the deletion region.
| Gene symbol | Description |
Positiona |
Gene/locus MIM number | Phenotype | Phenotype MIM number | Inheritance trait | |
|---|---|---|---|---|---|---|---|
| Start | End | ||||||
| GRM8 | Glutamate receptor, metabotropic 8 | 126,078,652 | 126,883,569 | #601116 | ADHD | ||
| MIR592 | MicroRNA 592 | 126,698,142 | 126,698,238 | ||||
| LOC101928333 | 126,855,181 | 126,869,975 | |||||
| ZNF800 | Zinc-finger protein 800 | 127,010,354 | 127,032,323 | ||||
| LOC100506682 | 127,116,937 | 127125858 | |||||
| GCC1 | GRIP and coiled-coil domain containing 1 | 127,220,682 | 127,225,654 | #607418 | |||
| ARF5 | ADP-ribosylation factor 5 | 127,228,406 | 127,231,759 | #103188 | |||
| FSCN3 | Fascin homolog 3, actin-bundling protein, testicular | 127,231,463 | 127,236,057 | #615800 | |||
| PAX4 | Paired box 4 | 127,250,346 | 127,255,780 | #167413 | Diabetes mellitus, type 2 | #125853 | AD |
| Maturity-onset diabetes of the young, type IX | #612225 | ||||||
| SND1 | Staphylococcal nuclease and tudor domain containing 1 | 127,292,202 | 127,732,659 | #602181 | |||
| SND1-IT1 | SND1 intronic transcript 1 (non-protein coding) | 127,637,562 | 127,640,130 | ||||
| LRRC4 | Leucine-rich repeat containing 4 | 127,667,124 | 127,671,002 | #610486 | |||
| MIR593 | MicroRNA 593 | 127,721,913 | 127,722,012 | ||||
Abbreviations: AD, autosomal dominant; ADP, adenosine diphosphate; ADHD, attention deficit/hyperkinetic disorder; MIM, Mendelian inheritance in man.
Genomic positions are referred to build19.
As shown in Figure 1, there were some overlapping deletions in the DECIPHER database; however, informed consent and detailed clinical information of the registered patients were unavailable. Therefore, we were not able to perform a genotype–phenotype correlation study using the data included in the DECIPHER database. In addition, we found only two reports of overlapping deletions in the literature. One of the deletions was derived from complex chromosomal translocations;1 the deletion was large and included the deletion region identified in the present patient. The other overlapping deletion coexisted with a 17q21.31 microduplication.2 The phenotypic features of these patients with complicated chromosomal abnormalities were inappropriate for use in a genotype–phenotype correlation study.
The deletion identified in this study included 13 RefSeq genes. According to RefSeq gene information (Table 1), only the paired box 4 gene (PAX4) is related to the OMIM phenotype (http://omim.org/); the PAX4 mutation is related to autosomal dominant diabetes mellitus type 2 (MIM #125853).4 At present, there have been no symptoms of diabetes mellitus in this patient. Careful observation is required to identify these symptoms.
In addition to PAX4, the leucine-rich repeat containing 4 gene (LRRC4) and the glutamate receptor metabotropic 8 gene (GRM8) showed some functional relevance to neurological features. Elia et al.5 have performed a large cohort study on patients with attention deficit and/or hyperkinetic disorder, and have identified statistically significant findings of copy number variations, including genes related to glutamate receptor gene networks and G-protein-coupled receptors involved in modulating excitatory synaptic transmission. Some reports have also suggested a relationship between copy number variations in glutamate receptor gene regions and attention deficit and/or hyperkinetic disorder.6,7 A partial duplication of GRM8 has been identified in an individual with autism.8 On the basis of these findings, copy number changes in GRM8 may be related to neurodevelopmental disorders. More recently, GRM8 has been reported to be associated with psychiatric disorders.9–11 However, the clinical features of the present patient are more severe than those observed in patients with such neuropsychiatric disorders.
Another candidate gene, LRRC4, consists of only one exon, and encodes a 653-amino-acid-long protein. LRRC4 is a member of the synaptic cell adhesion molecule family. The netrin-G ligands (NGLs) belong to the superfamily of leucine-rich repeat (LRR) proteins. Three known members of the NGL family, NGL-1 (LRRC4C), NGL-2 (LRRC4), and NGL-3 (LRRC4B), localize mainly to the postsynaptic side of the excitatory synapse and interact with the presynaptic ligands including netrin-G1, netrin-G2, and leukocyte antigen-related.12,13 The NGL-dependent adhesion system is important for the development of axons, dendrites, and synapses. Consistently with these functions, defects in NGLs and their ligands are associated with impaired learning/memory and hyperactivity. Many synaptic proteins and receptors are defective in patients with autism, thus strongly suggesting that the heterozygous deletion of LRRC4 is a possible explanation for the severe intellectual disability and characteristics of autism observed in the present patient.14
In conclusion, the identified 7q31.33 deletion was considered to be a pathogenic copy number loss in the present patient.
Acknowledgments
We would like to express our gratitude to the patient and his parents for their cooperation. We also acknowledge the DECIPHER database for checking overlapping deletions. We are grateful to the technicians from our laboratories, including Ms Yumiko Ondo for her skillful help. This research was supported by the Practical Research Project for Rare/Intractable Diseases from the Japan Agency for Medical Research and Development (AMED), and the Japan Society for the Promotion of Science (JSPS) KAKENHI grant number 15K09631 (TY).
Footnotes
The authors declare no conflict of interest.
References
- Tyson C, McGillivray B, Chijiwa C, Rajcan-Separovic E. Elucidation of a cryptic interstitial 7q31.3 deletion in a patient with a language disorder and mild mental retardation by array-CGH. Am J Med Genet A 2004; 129A: 254–260. [DOI] [PubMed] [Google Scholar]
- Mc Cormack A, Taylor J, Te Weehi L, Love DR, George AM. A case of 17q21.31 microduplication and 7q31.33 microdeletion, associated with developmental delay, microcephaly, and mild dysmorphic features. Case Rep Genet 2014; 2014: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangu N, Okamoto N, Shimojima K, Ondo Y, Nishikawa M, Yamamoto T. A de novo microdeletion in a patient with inner ear abnormalities suggests that the 10q26.13 region contains the responsible gene. Hum Genome Var 2016; 3: 16008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvais-Jarvis F, Smith SB, Le May C, Leal SM, Gautier JF, Molokhia M et al. PAX4 gene variations predispose to ketosis-prone diabetes. Hum Mol Genet 2004; 13: 3151–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia J, Glessner JT, Wang K, Takahashi N, Shtir CJ, Hadley D et al. Genome-wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nat Genet 2012; 44: 78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akutagava-Martins GC, Salatino-Oliveira A, Genro JP, Contini V, Polanczyk G, Zeni C et al. Glutamatergic copy number variants and their role in attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet 2014; 165B: 502–509. [DOI] [PubMed] [Google Scholar]
- Asadollahi R, Oneda B, Joset P, Azzarello-Burri S, Bartholdi D, Steindl K et al. The clinical significance of small copy number variants in neurodevelopmental disorders. J Med Genet 2014; 51: 677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serajee FJ, Zhong H, Nabi R, Huq AH. The metabotropic glutamate receptor 8 gene at 7q31: partial duplication and possible association with autism. J Med Genet 2003; 40: e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ju K, Li Z, He K, Chen J, Wang Q et al. Significant association of GRM7 and GRM8 genes with schizophrenia and major depressive disorder in the Han Chinese population. Eur Neuropsychopharmacol 2016; 26: 136–146. [DOI] [PubMed] [Google Scholar]
- Long EC, Aliev F, Wang JC, Edenberg HJ, Nurnberger J Jr, Hesselbrock V et al. Further analyses of genetic association between GRM8 and alcohol dependence symptoms among young adults. J Stud Alcohol Drugs 2015; 76: 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhong X, An Z, Han S, Luo X, Shi Y et al. Association analysis of the GRM8 gene with schizophrenia in the Uygur Chinese population. Hereditas 2014; 151: 140–144. [DOI] [PubMed] [Google Scholar]
- Woo J, Kwon SK, Kim E. The NGL family of leucine-rich repeat-containing synaptic adhesion molecules. Mol Cell Neurosci 2009; 42: 1–10. [DOI] [PubMed] [Google Scholar]
- Xu G, Wang R, Wang Z, Lei Q, Yu Z, Liu C et al. NGL-2 is a new partner of PAR complex in axon differentiation. J Neurosci 2015; 35: 7153–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yu S, Fu Y, Li X. Synaptic proteins and receptors defects in autism spectrum disorders. Front Cell Neurosci 2014; 8: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Data Citations
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.