Summary
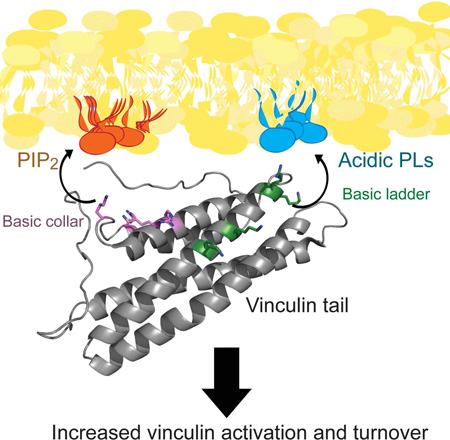
Vinculin, a scaffolding protein that localizes to focal adhesions (FA) and adherens junctions, links the actin cytoskeleton to the adhesive super-structure. While vinculin binds to a number of cytoskeletal proteins, it can also associate with phosphatidylinositol 4,5-bisphosphate (PIP2) to drive membrane association. To generate a structural model for PIP2-dependent interaction of vinculin with the lipid bilayer, we conducted lipid-association, nuclear magnetic resonance, and computational modeling experiments. We find that two basic patches on the vinculin tail drive membrane association: the basic collar specifically recognizes PIP2, while the basic ladder drives association with the lipid bilayer. Vinculin mutants with defects in PIP2-dependent liposome association were then expressed in vinculin knockout murine embryonic fibroblasts. Results from these analyses indicate that PIP2-binding is not required for localization of vinculin to FAs or FA strengthening but is required for vinculin activation and turnover at FAs to promote its association with the force-transduction FA nanodomain.
Graphical abstract
Introduction
Vinculin (Vcl) is a large (117 kDa), ubiquitously expressed and highly conserved scaffolding protein present in higher eukaryotes. It localizes to focal adhesions (FAs) and adherens junctions (AJs) (Johnson et al., 1998), and has been implicated in regulating cell morphology, cell migration (Coll et al., 1995), cell stiffness (le Duc et al., 2010; Shen et al., 2011), adhesion strength (Mierke et al., 2008), FA turnover, and FA morphology (Xu et al., 1998a; Xu et al., 1998b).
Vcl contains an N-terminal head domain (Vh) and a C-terminal tail domain (Vt) connected by a proline-rich linker (Bakolitsa et al., 2004). Vh and Vt bind each other and maintain Vcl in an autoinhibited conformation (Bakolitsa et al., 2004; Cohen et al., 2005; Johnson and Craig, 1995b). Release of autoinhibition occurs via binding of ligands and possibly phosphorylation (Chen et al., 2006; Golji et al., 2012). While Vcl has been shown to associate with ~20 binding partners, the tail domain alone binds, filamentous actin (F-actin) (Huttelmaier et al., 1997) and acidic phospholipids (Johnson and Craig, 1995a), specifically phosphatidylinositol 4,5-bisphosphate (PIP2) (Palmer et al., 2009). These tail domain interactions are believed to coordinate with Vh interactions to facilitate Vcl activation (Weekes et al., 1996).
In FAs, PIP2 is generated by phosphatidylinositol phosphate kinase type 1 gamma (PIPKIγ) (van den Bout and Divecha, 2009) and through PIP2 production, PIPKIγ regulates FA dynamics (Li et al., 2013; Wu et al., 2011). PIP2 is also required for FA formation (Sun et al., 2007) and promotes recruitment and activation of Vcl at FAs (Legate et al., 2011; Wu et al., 2011). However, the consequences of the Vcl/PIP2 interaction are less clear. While association of Vcl with PIP2 has been implicated in numerous processes including cell spreading, cell migration, FA turnover, and force transduction (Chandrasekar et al., 2005; Chinthalapudi et al., 2014; Diez et al., 2009; Saunders et al., 2006), the Vcl constructs used in these studies may not be selective in their disruption of PIP2-binding. Additionally, PIP2 has been reported to regulate Vcl phosphorylation (Ziegler et al., 2002) and trafficking (Halstead et al., 2010; Marquez et al., 2009).
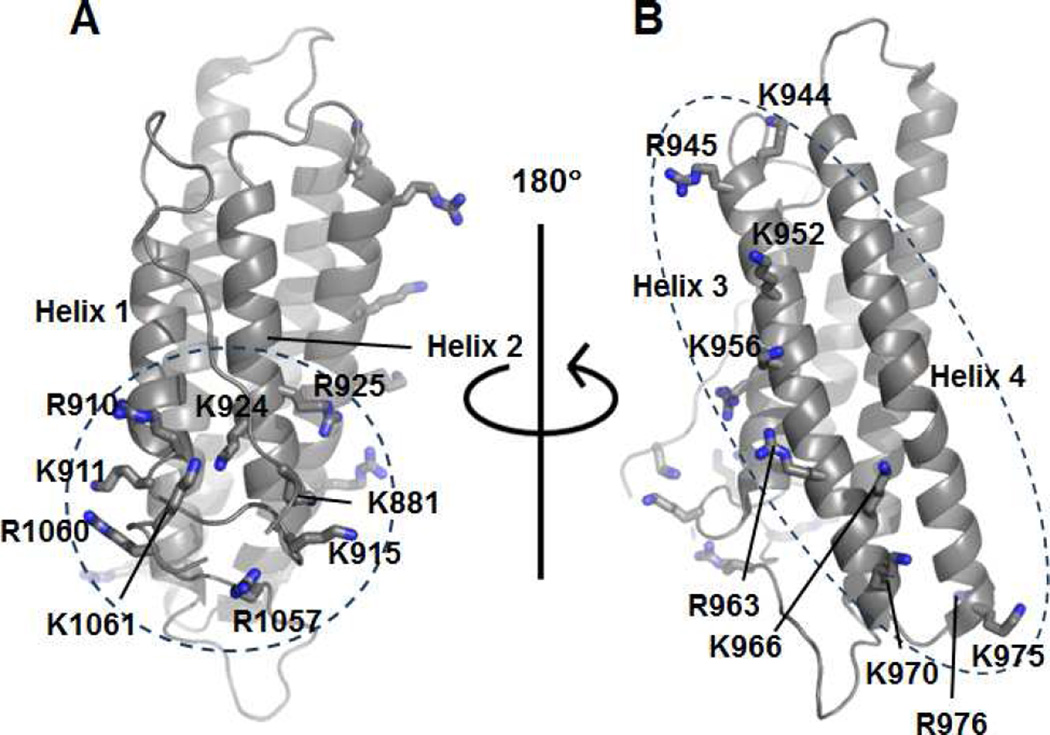
Vt contains two strongly basic surfaces, the basic ladder and the basic collar (Figure 1) (Bakolitsa et al., 1999). Mutation of the basic collar (Bakolitsa et al., 2004; Chinthalapudi et al., 2014; Saunders et al., 2006; Ziegler et al., 2002), the basic ladder (Chandrasekar et al., 2005; Chinthalapudi et al., 2014), or deletion of the C-terminus (Chandrasekar et al., 2005; Diez et al., 2009; Saunders et al., 2006; Wirth et al., 2010; Ziegler et al., 2002) impairs PIP2 binding. However, the use of multiple mutations or deletions within Vt can alter its structure and perturb other Vt functions (Palmer et al., 2009). For example, two separate Vcl variants with reported PIP2 defects of comparable severity, Vcl K952Q/K956Q/R963Q/R966Q and Vcl R1060Q/K1061Q, exhibit different phenotypes in exerting force on a substrate (Diez et al., 2009).
Figure 1. Vt contains two basic regions predicted to bind PIP2.
(A) Ribbon diagram of Vt (PDB 1ST6) highlighting basic collar residues within the N-terminal strap, helices 1 and 2, and the C-terminus.
(B) Ribbon diagram of Vt (PDB 1ST6) highlighting basic ladder residues within helices 3 and 4.
Recently, a crystal structure of a Vt mutant bound to a soluble, short-chain PIP2 was solved, providing the first structural model for how PIP2 binds to Vt (PDB 4PR9) (Chinthalapudi et al., 2014). In the structure, Vt R1060A forms a trimeric oligomer when bound to 1,2-dioctanoyl-sn-glycero-3-phospho-(1’-myo-inositol-4’,5’-bisphosphate) (PIP2-C8). Two of the Vt molecules recognize the PIP2 head group via the basic collar, whereas the third Vt molecule interacts with the PIP2 head group through residues K944 and R945 in the basic ladder. Mutations to the basic collar (Vt K1061Q) or the basic ladder (K944Q/R945Q) disrupt PIP2-dependent lipid co-sedimentation (Chinthalapudi et al., 2014). Later work confirmed that Vt dimerizes in the presence of PIP2 (Chinthalapudi et al., 2015). However, these models fail to explain how PIP2 association with Vt promotes membrane insertion. Additionally, the two lipid-disrupting mutants produced distinct phenotypes when expressed in Vcl-null mouse embryonic fibroblasts (Vcl −/− MEFs). Whereas Vcl K944Q/R945Q prevents exchange of Vcl at FAs, Vcl K1061Q only mildly disrupts exchange of Vcl at FAs (Chinthalapudi et al., 2014).
Using computational modeling, site-directed mutagenesis and lipid co-sedimentation of Vt in membrane mimetics, we confirm that both the basic ladder and collar are involved in binding to acidic phospholipids. However, our data suggest an alternative structural model: the basic collar promotes PIP2-specific binding while the basic ladder drives membrane insertion. Cells expressing Vcl variants specifically deficient in lipid-binding retain the ability to reinforce cell stiffness upon mechanical deformation but show altered FA turnover, perturbed nano-scale Vcl localization within FAs, and reduced Vcl activation at FAs.
Results
Computational modeling predicts distinct roles for the basic collar and basic ladder
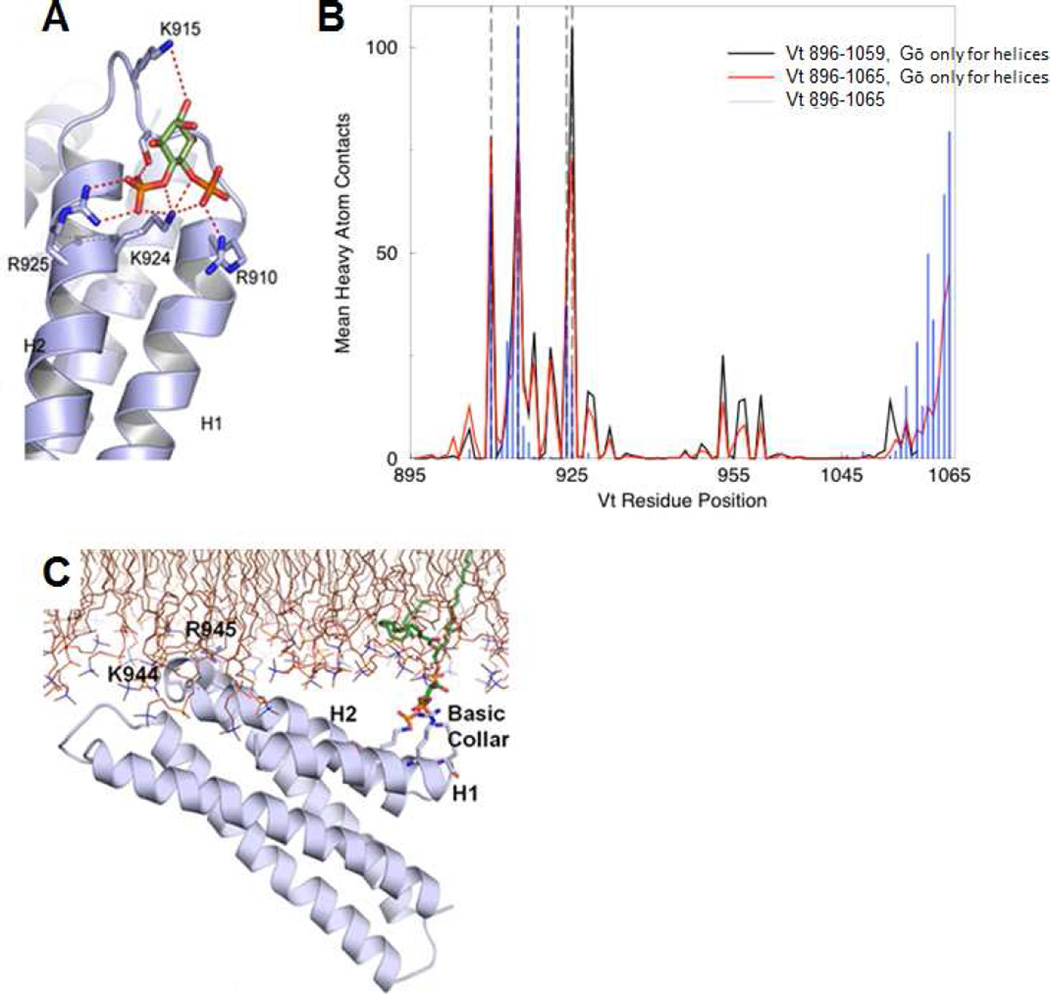
To better understand how Vt binds PIP2, we modeled the interaction of the PIP2 head group with Vt. The head group, D-inositol 4,5-bisphosphate, was docked to Vt residues 895–1065 using MedusaDock (Ding et al., 2010; Yin et al., 2008). The N-terminal strap was omitted from Vt for docking, as its removal retains the Vt fold and actin binding, but increases association with PIP2 (Palmer et al., 2009). MedusaDock identified the basic collar, specifically residues R910, K915, K924, and R925, as the Vt site with the highest specificity for the PIP2 head group. The side chain of R910 interacts primarily with the hydroxyl of C4, K915 with the hydroxyl of C6 (which would place it near the phosphoryl group on C1 in the full PIP2 molecule), K924 with the hydroxyl of C5, and R925 with the phosphoryl group on C4 (Figure 2A).
Figure 2. Simulations of PIP2-mediated Vt membrane association identify distinct roles for the basic collar and basic ladder.
(A) MedusaDock results displaying PIP2 head group (green) interactions with the Vt basic collar.
(B) Results from DMD simulations of 1-stearoyl-2-arachidonoyl-sn-glycero-3-phospho-(1’-myo-inositol-4’,5’-bisphosphate) with Vt. The average number of heavy atom Vt contacts with PIP2 are mapped per residue for simulations with Gō constraints for helices and Vt 896–1059 (black), Vt 896–1065 (red), and Gō constraints for all Vt residues 896–1065 (blue). R910, K915, K924 and R925 are denoted with dashed vertical lines. (C) GROMACS simulation results (final pose) of Vt, in the presence of POPC and one molecule of PIP2 (green carbons). The basic collar, helices 1 and 2, and K944 and R945 are labeled. See also Figures S1, S2 and Movies S1, S2, S3.
Next, discrete molecular dynamics (DMD) simulations (Ding et al., 2008; Dokholyan et al., 1998; Shirvanyants et al., 2012) were performed with a full-length PIP2, 1-stearoyl-2-arachidonoyl-sn-glycero-3-phospho-(1’-myo-inositol-4’,5’-bisphosphate), constraining the PIP2 head group to the basic collar and using Gō constraints on C(3/Ca contacts (with the interaction square-well depth 0.5e, where e is the DMD energy unit (Dokholyan et al., 1998; Proctor et al., 2011)) for all residues in Vt (residues 896–1065) or Gō constraints only for residues in alpha-helices (using Vt constructs 896–1065 or 896–1059). After selecting 2000 snapshots with a binding energy less than −10 kcal/mol (Ding et al., 2010), the residues in contact with PIP2 were tallied (Figure 2B). In all cases, the contacts at the basic collar remained high. When using Gō constraints only for helical residues, some contacts were observed at the basic ladder, though contacts at the basic collar were more frequent (Figure 2B). For more complete sampling of full-length PIP2 binding Vt, we performed DMD simulations in which the PIP2 head group was unrestrained. In these simulations, we see PIP2 sampling all regions of Vt which argues against the head group binding the basic collar being a kinetically trapped state. We clustered the DMD snapshots. Four of the seven clusters feature the head group interacting with the basic collar (Figure S1A). Furthermore, the largest cluster of conformations recapitulates head group interactions with the basic collar that were seen in the docking simulations (Figure S1B).
While both MedusaDock and DMD simulations identify the basic collar as the primary site for PIP2 head group recognition, these simulations were performed in the absence of a lipid bilayer. To better mimic the physiological interaction at the membrane, all-atom, explicit solvent molecular dynamics simulations of Vt bound to PIP2 in a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayer were conducted. At the start of the simulation, Vt (residues 896–1055) was bound to PIP2 as in the docking pose (Figure 2A) and the Vt helix bundle was oriented roughly parallel to the lipid bilayer. We performed three simulations of 100 ns each. In all three simulations, the contacts between basic collar and PIP2 head group were maintained, even as the PIP2 molecule diffused around in the bilayer (Figure S2A). Furthermore, helix 3 of Vt was found to contact the bilayer in all three simulations. The side chains of basic ladder residues K944, R945, K952, and K956 inserted into the lipid bilayer (Figure 2C, Movies S1, S2). We also performed three 100 ns simulations of Vt in the same starting configuration in a POPC-only bilayer. In these control simulations, basic collar interactions with the membrane were not observed and interactions of helix 3 with the membrane were inconsistent. In one of the three simulations, Vt diffuses away from the membrane (Movie S3). The control simulations confirm that PIP2 anchors Vt to the membrane through the basic collar, which facilitates basic ladder interactions with the bilayer.
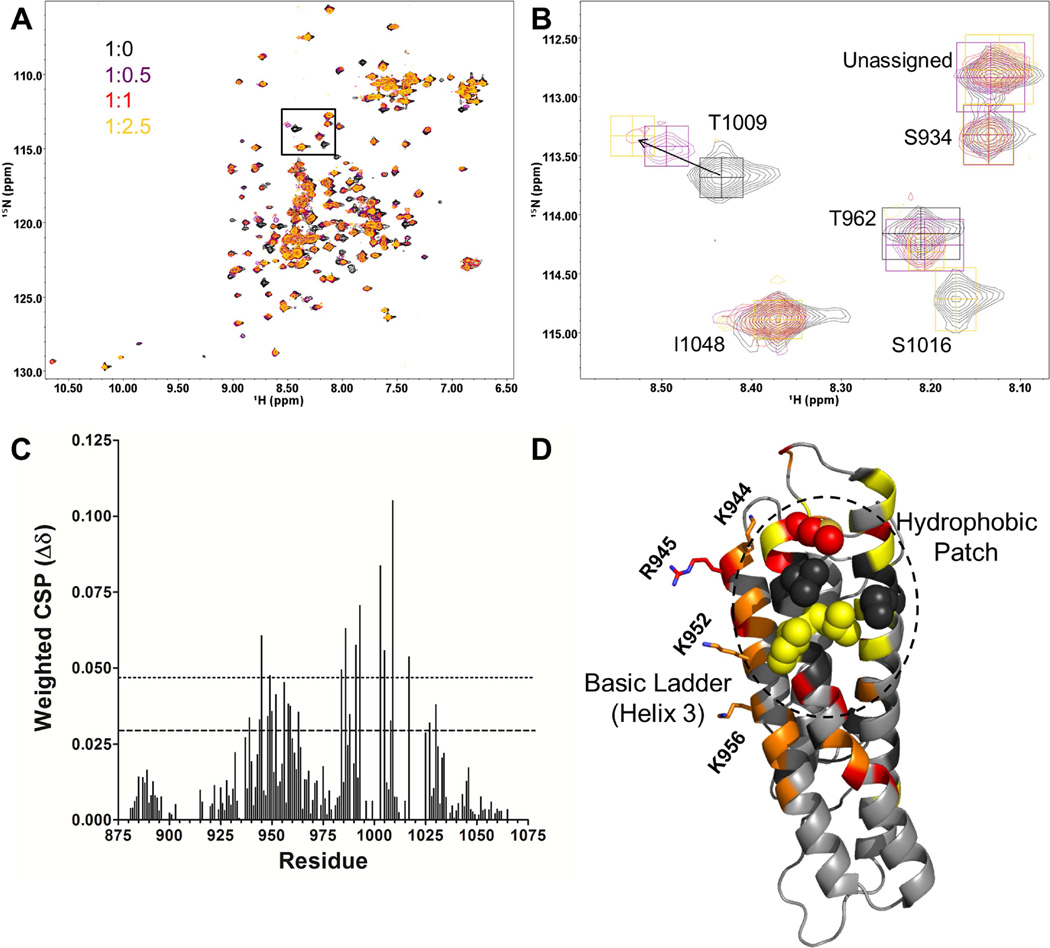
Short-chain PIP2 binds to Vt at a hydrophobic patch on helices 4 and 5 in solution
The mode of binding of Vt to membrane-associated PIP2 from our simulations (Figure 2C) is significantly different from the recently published crystal structure of Vt R1060A complexed to a short-chain PIP2 lipid (PIP2-C8) (Chinthalapudi et al., 2014) and the model docked to a lipid bilayer (Chinthalapudi et al., 2015). Chinthalapudi et al show the sidechain of K1061 binds to the PIP2 head group, but not R910 and R925, which interact in our model (Chinthalapudi et al., 2015; Chinthalapudi et al., 2014). Additionally, the proposed orientations of membrane-associated Vt molecules within a dimer do not match our modeled orientation. A possible explanation for this discrepancy is the use of PIP2-C8 instead of PIP2 contained in a lipid bilayer. We titrated 15N-enriched Vt with PIP2-C8 at various ratios and monitored NMR spectral changes by 2D 1H-15N heteronuclear single quantum coherence (HSQC) analyses (Figure 3A). In doing so, we hoped to avoid potential artifacts from crystallization and use of the R1060A mutant. At Vt:PIP2-C8 ratios of 1:2.5 and 1:6 a white precipitate formed, as previously reported (Chinthalapudi et al., 2015), suggesting Vt was saturated with PIP2-C8 at a ratio of 1:2.5. Perturbations in chemical shift or line width map to a site on helices 3, 4, and 5, comprised primarily by a hydrophobic patch. Peaks corresponding to these residues significantly shift or broaden, suggesting a strong interaction (Figure 3B–D). In contrast, peaks corresponding to residues in the basic collar do not significantly shift or broaden with increasing PIP2-C8 (Figure 3B–D). Weaker shifts are found in peaks corresponding to the basic ladder. These results suggest that the fatty acid tails of PIP2-C8 drive binding in solution, and that the PIP2 head group associates with the basic ladder because of its proximity to the hydrophobic patch. An interaction between the acyl chains and the hydrophobic patch was not observed in the crystal structure of the Vt:PIP2-C8 complex (Chinthalapudi et al., 2014).
Figure 3. Titration of 15N-enriched Vt with PIP2-C8.
(A, B) 2D NMR 1H-15N HSQC titration of PIP2-C8 into 15N-enriched Vt (50 µM) at the listed ratios. The inset (B) shows an expanded view of the T1009, S1016 and I1048 resonances. The T1009 amide peak undergoes chemical shift changes and the S1016 peak shows line width broadening; the I10148 resonance is unperturbed.
(C) The weighted chemical shift perturbation (CSP) is shown for each assigned residue. The dashed line corresponds to the average CSP plus one standard deviation, while the dotted line corresponds to the average CSP plus two standard deviations.
(D) CSP and line broadening changes mapped to the Vt structure. Residues with CSP greater than twice the standard deviation from the mean are shown in red. Those with CSP greater than one standard deviation from the mean but less than two are shown in orange. Residues in yellow broaden significantly and are difficult to track. Residues in black are unassigned. K944, R945, K952, and K956, which reside in the basic ladder are shown as sticks. Aliphatic residues in the hydrophobic patch on helices 4 and 5 are shown as spheres.
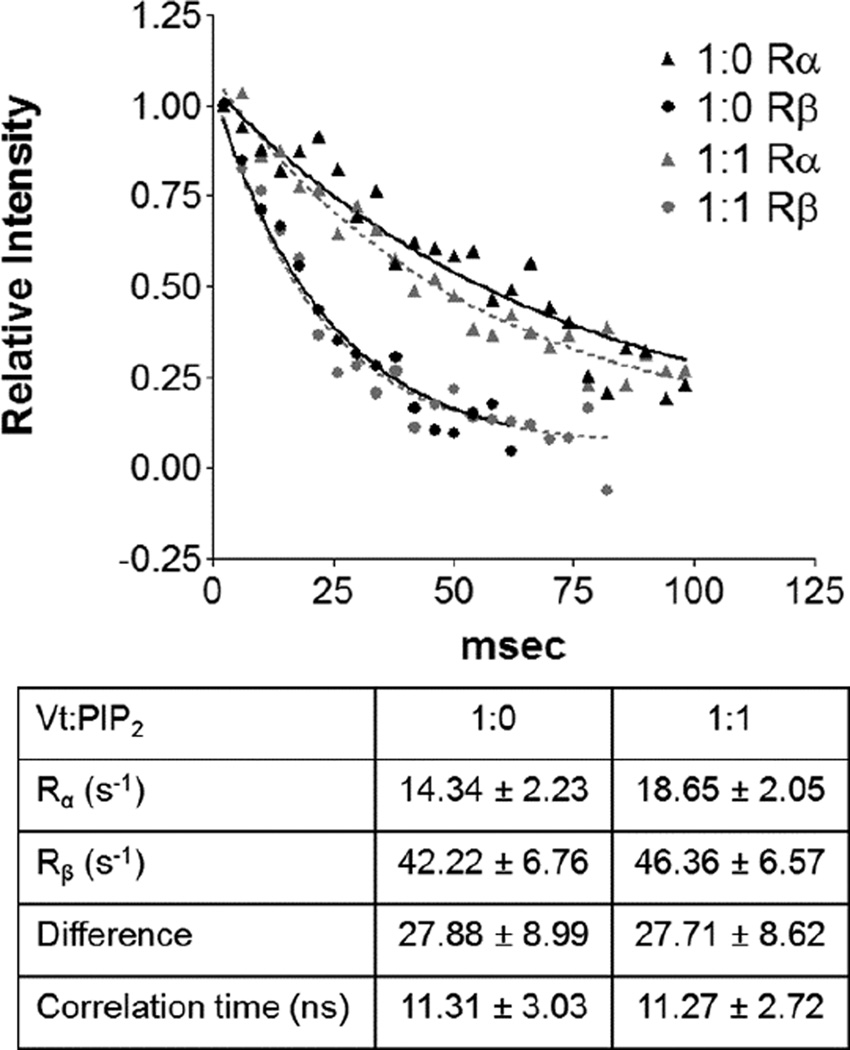
PIP2 was also deemed sufficient for the generation of Vt oligomers (Chinthalapudi et al., 2015; Chinthalapudi et al., 2014). To test the ability PIP2-C8 to induce Vt oligomerization in solution we used the TRACT NMR experiment (Lee et al., 2006) to calculate the effective rotational correlation time of Vt (Figure 4). Oligomerization of Vt would slow tumbling and significantly increase the rotational correlation time. However, the rotational correlation time of Vt did not increase at a 1:1 ratio of Vt:PIP2-C8, suggesting that either Vt does not oligomerize in the presence of PIP2-C8 or that Vt oligomers induced by binding PIP2-C8 are unobservable by solution NMR. Such oligomers would either be insoluble or large multimers with very fast magnetic relaxation. These explanations are inconsistent with a functional Vt dimer or trimer, suggesting that PIP2-C8 is insufficient to appropriately study the Vt:PIP2 interaction.
Figure 4. Rotational correlation time of Vt is not altered upon binding of PIP2-C8.
A 1D plot of amide resonance intensity determined from the TRACT experiment at two different Vt:PIP2 ratios. The curves were analyzed for different relaxation rates and the effective rotational correlation time was calculated. At both ratios, Vt exhibited a correlation time of roughly 11.3 nanoseconds.
The R1060Q/K1061Q PIP2-deficient Vt variant is a poor actin crosslinker
The Vcl variant, LD-CT (Vcl R1060Q/K1061Q), has been used in multiple studies as a lipid-binding deficient Vcl construct (Chandrasekar et al., 2005; Diez et al., 2009; Ziegler et al., 2002). High-speed co-sedimentation assays revealed that Vt LD-CT retains actin binding (Figure S3), while low-speed actin co-sedimentation assays show that Vt LD-CT is deficient in F-actin crosslinking (Figure S3).
The basic collar is required for lipid binding and F-actin crosslinking
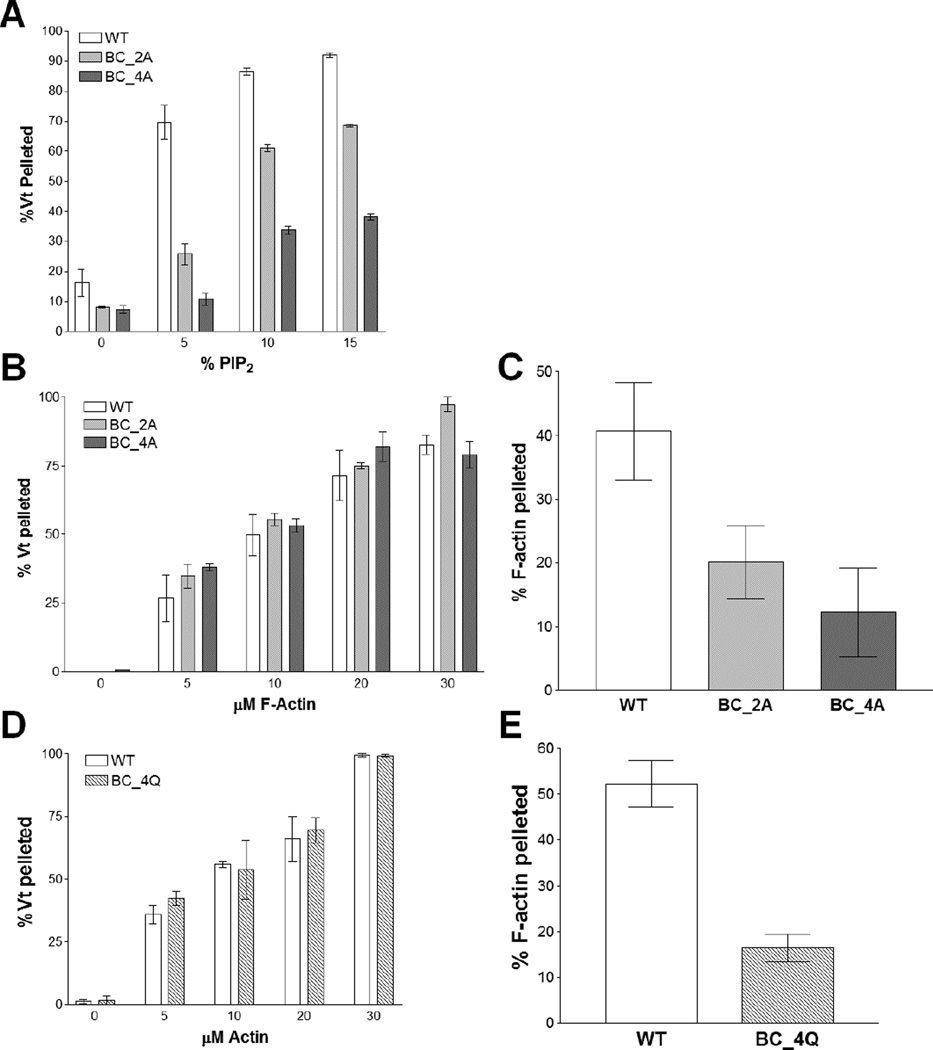
As our molecular dynamics simulations indicate that R910, K915, K924, and R925 play key roles in binding the PIP2 head group, we measured PIP2-binding of Vt K924A/R925A (Vt BC_2A) and Vt R910A/K915A/K924A/R925A (Vt BC_4A) using a PIP2 co-sedimentation assay (Palmer et al., 2009). While Vt BC_2A exhibited a modest decrease in binding to PIP2, with roughly half as much Vt binding at 5% PIP2 (Figure 5A), Vt BC_4A showed an even greater drop in PIP2 binding, with a 6-fold decrease in binding at 5% PIP2. These findings suggest that the basic collar residues R910, K915, K924, and R925 are important for PIP2 binding.
Figure 5. Mutations within the Vt basic collar disrupt PIP2 binding and F-actin crosslinking.
(A) Liposome co-sedimentation assay. Mutations in the Vt basic collar disrupt binding to PIP2-containing liposomes.
(B–E) Actin co-sedimentation assays. At high-speed co-sedimentation (B, D) the Vt basic collar mutants retain actin binding, but at low-speed co-sedimentation (C, E) a decrease in F-actin crosslinking is observed. N ≥ 3, error bars are ± SEM. See also Figure S3.
We found that both Vt BC_2A and Vt BC_4A retain actin binding (Figure 5B), consistent with the current structural model of the Vt/actin complex (Kim et al., 2014). However, these variants exhibited a significant decrease in the ability of Vt to bundle F-actin filaments, as measured by a low-speed co-sedimentation assay (Figure 5C). This decrease was not rescued by the more conservative Vt variant, Vt BC_4Q (R910Q/K915Q/K924Q/R925Q) (Figure 5E), suggesting that retention of the basic collar positive charge is important for bundling of F-actin by Vcl.
The basic ladder is required for lipid association
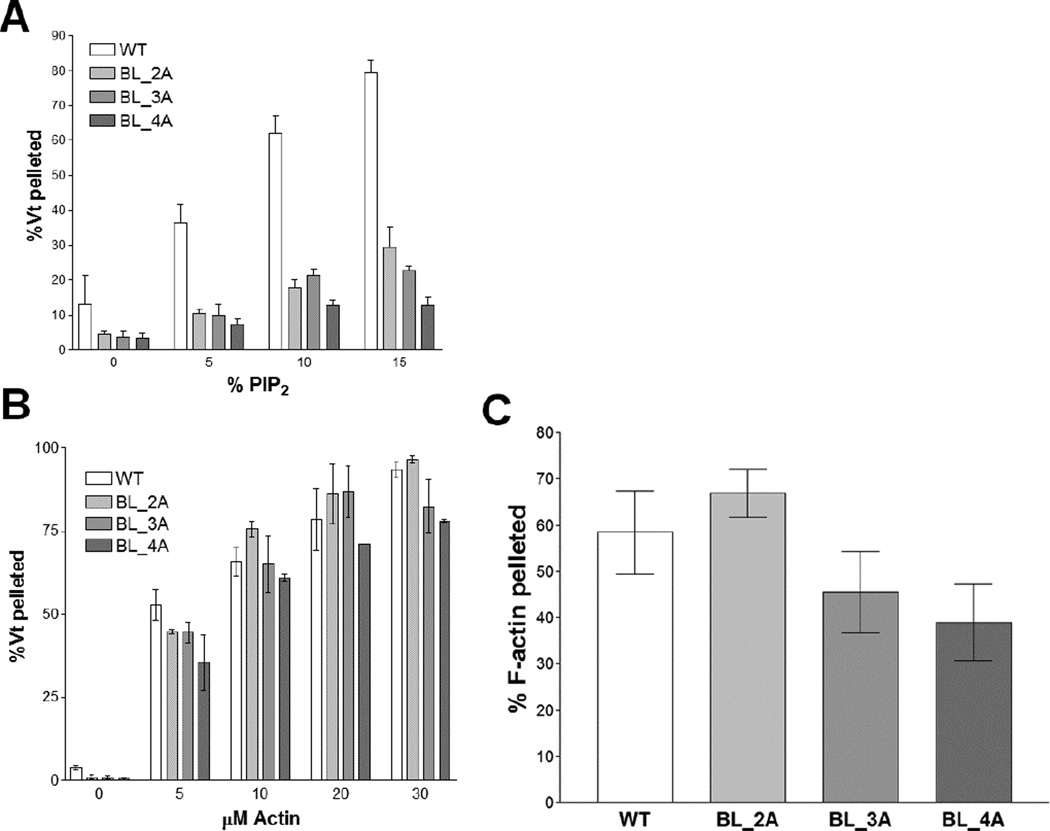
Since mutations to the basic collar disrupt the ability of Vt to bundle F-actin, Vcl variants with mutations in the basic collar cannot be used to specifically report on the role of lipid binding by Vcl in cells. Mutation of basic residues within the Vt basic ladder can also disrupt binding to PIP2-containing liposomes (Chandrasekar et al., 2005; Chinthalapudi et al., 2014), suggesting that these residues may be good targets for disrupting PIP2-binding. Three constructs were generated to test the importance of the basic ladder in binding to PIP2-containing liposomes: Vt K944A/R945A (BL_2A), Vt K944A/R945A/K952A (BL_3A), and Vt K944A/R945A/K952A/K956A (BL_4A). All three constructs all show decreased binding to PIP2-containing liposomes (Figure 6A), with Vt BL_4A exhibiting the most severe defect.
Figure 6. Mutations to the basic ladder disrupt PIP2 binding but do not significantly impair F-actin binding or crosslinking.
(A) Liposome co-sedimentation assay. Mutation of the basic ladder disrupts binding to PIP2-containing liposomes.
(B, C) Actin co-sedimentation assays. Basic ladder mutants retain F-actin binding and crosslinking (bundling), respectively, as determined by high-speed co-sedimentation (B) and low-speed co-sedimentation (C). N ≥ 3, error bars are ± SEM.
Actin binding and crosslinking activities for these variants were evaluated to test the specificity of the PIP2-binding defect. Vt BL_2A, Vt BL_3A, and Vt BL_4A all retain actin binding (Figure 6B) and crosslinking (Figure 6C). These data support the use of these basic ladder variants in studying the biological function of the Vcl/PIP2 interaction.
The Vt basic ladder drives association with acidic liposomes
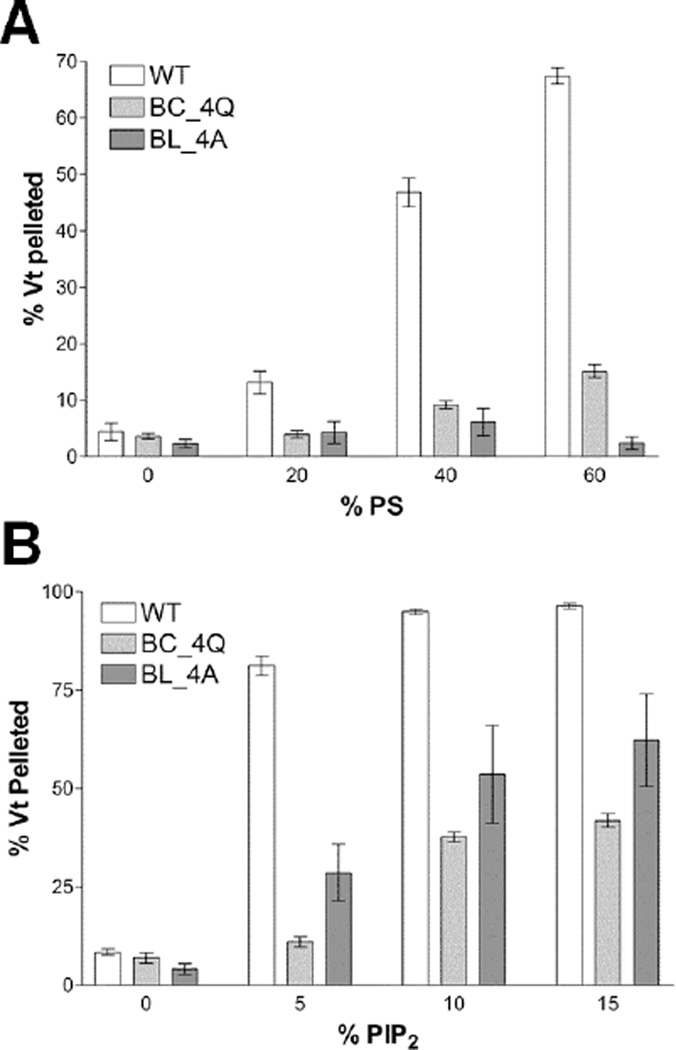
As mutations in both the basic collar and basic ladder impair Vt binding to PIP2-containing liposomes, it is difficult to determine which site is responsible for PIP2 specificity. Co-sedimentation experiments conducted with Vt BC_4Q or Vt BL_4A and increasing amounts of PIP2 showed similar association profiles (Figure 7A). We tested the hypothesis that the basic ladder is critical for membrane insertion by performing lipid co-sedimentation assays in the absence of PIP2, but with increasing concentrations of phosphatidylserine (PS). WT Vt bound to these liposomes in a PS-dependent manner, with nearly 70% of Vt bound when the liposomes contained 60% PS (Figure 7B). Vt BC_4Q retained some binding, with 15% of Vt BC_4Q associated with 60% PS-containing liposomes. However, Vt BL_4A did not show significant binding, even at high levels of PS, suggesting that the basic ladder drives interactions with negatively-charged lipid membranes. As mutations within the N-terminal half of helix 3 impair lipid association yet retain actin interactions, these variants are reasonable tools to study the effect of lipid binding by Vcl in cells.
Figure 7. Comparative lipid co-sedimentation assays for Vt BC_4Q and Vt BL_4A.
(A) Lipid co-sedimentation assay with PIP2-containing liposomes. Mutations within the Vt basic collar (Vt BC-4Q) or basic ladder (Vt BL-4A) disrupt binding to PIP2-containing liposomes.
(B) Lipid co-sedimentation assay with PS-containing liposomes. Mutations within the Vt basic collar (Vt BC-4Q) or basic ladder (Vt BL-4A) disrupt binding to PS-containing liposomes. Larger defects in PS-dependent lipid binding are found when the basic ladder is mutated compared to the basic collar. N=3, bars are ± SEM.
Lipid-binding regulates Vcl nano-scale localization, activation, and turnover in FAs
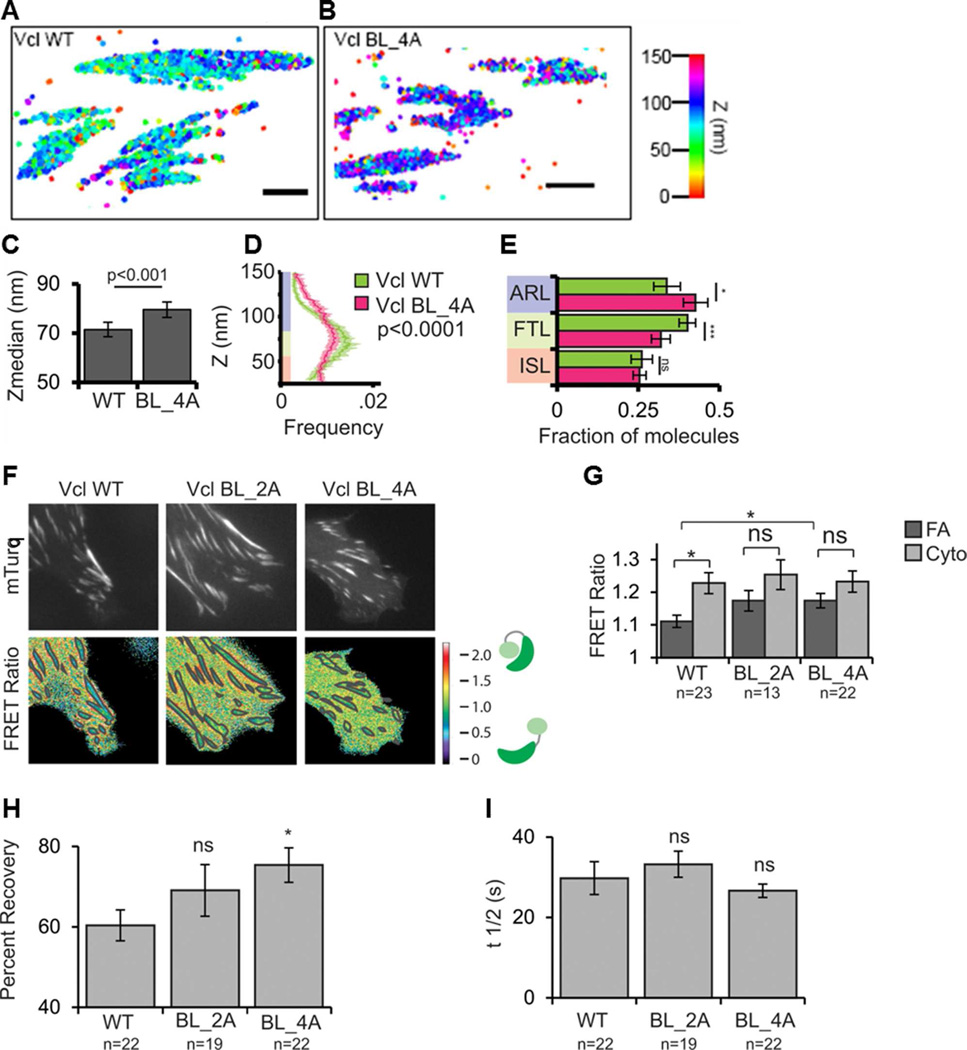
Our structural and biochemical characterization of Vt basic collar and ladder mutants identified a Vcl variant with a specific lipid-binding deficiency to facilitate studies of Vcl lipid association in a cellular context. We expressed Vcl WT, Vcl BL_2A, or Vcl BL_4A in Vcl−/− MEFs and plated them on fibronectin (FN)-coated coverslips. Tagging with GFP revealed that Vcl BL_2A and Vcl BL_4A still localize to FAs (Figure S4A). Because Vcl nano-scale localization within FAs has been shown to correlate with Vcl function (Case et al., 2015), we utilized iPALM, a super-resolution fluorescence microscopy technique that provides nanometer (10–20 nm) localization accuracy of individual proteins (Case et al., 2015; Kanchanawong et al., 2010) to determine the nano-scale position of tandem-Eos Vcl BL_4A in FAs. Previous studies have established protein localization to three nano-domains organized along the Z-axis in FAs: the membrane proximal integrin signaling layer (25–54 nm from the extracellular matrix (ECM) on the coverslip surface) where Vcl is initially recruited and activated; the force transduction layer (55–84 nm from the ECM) where active Vcl interacts with talin and strengthens FAs; and the actin regulatory layer (85–150 nm from the ECM) where Vcl binds actin (Case et al., 2015; Case and Waterman, 2015). iPALM analysis showed that while Vcl WT molecules had a median localization of 70 nm above the ECM, as previously reported (Case et al., 2015; Kanchanawong et al., 2010; Liu et al., 2015), Vcl BL_4A molecules localized significantly deeper at a median distance of 80 nm above the ECM. This change in distribution of Vcl BL_4A relative to Vcl-WT was driven by a loss of molecules from the force transduction layer and re-localization to the actin regulatory layer (Figure 8E). These results suggest that lipid binding plays a role in regulating Vcl’s vertical position in the FA, promoting its association with the force transduction layer and likely F-actin.
Figure 8. PIP2-binding regulates Vcl nano-scale localization, activation, and turnover in FAs.
(A, B) Representative iPALM renderings for MEFs expressing Vcl WT-tdEos (A) or Vcl BL_4A–tdEos (B) are shown. The color scale represents Z-position (nm), FAs oriented with the distal tip facing right, scale bar = 1 micron.
(C) Mean of Z-median measurements from individual FAs.
(D) Averaged Z-position frequency histograms of molecules within FAs. Solid line, mean frequency; Shaded region, bootstrapped 95% confidence about the mean.
(E) Mean fraction of molecules localized to each of the three FA layers in FAs. Coloring in (D,E) is used to highlight the three FA layers. ISL: integrin signaling layer (red, 25–54 nm above the coverslip), FTL: force transduction layer (green, 55–84 nm above the coverslip), ARL: actin regulatory layer (purple, 85–150 nm above the coverslip). Graphs in (C–E) represent measurements of n=59 FAs from 3 Vcl WT-tdEos expressing cells and n=58 FAs from 6 Vcl BL_4A–tdEos expressing cells.
(F) Localization and FRET signal of Vcl constructs. mTurquoise (mTurq, top) and processed mTurqouise/NeonGreen FRET ratio image (bottom) of Vcl−/− MEFs expressing either WT, BL_2A, or BL_4A Vcl FRET biosensor. The FA mask (grey lines) was created from the mTurq image and superimposed onto the FRET ratio image.
(G) Quantification of the mean FRET ratio value inside FAs (FA) and outside FAs (Cyto). (H,I) FRAP measurements of Vcl constructs. Mean percent recovery (H) and t1/2 (I) of Vcl obtained from single exponential fits of FRAP recovery curves. The fraction of Vcl that turns over at FAs (mobile fraction) increases upon disruption of lipid binding. Data in all bar graphs are represented as mean ± 95% confidence intervals. In (G–I), n = number of cells measured and significance is tested with ANOVA test followed by Tukey test post-hoc analysis. (* difference is significant at p<0.05 cutoff, ns: not significant). See also Figures S4, S5, S6.
As PIP2-binding contributes to Vcl activation in vitro (Bakolitsa et al., 2004; Bakolitsa et al., 1999; Izard et al., 2004; Weekes et al., 1996), and Vcl activation promotes a shift of Vcl from the force transduction layer to the actin regulatory layer of the FA (Case et al., 2015), we employed a Förster resonance energy transfer (FRET)-based biosensor designed to monitor Vcl’s activation state (Case et al., 2015; Chen et al., 2005) in Vcl−/− MEFs, to determine if disruption of PIP2-binding alters Vcl activation. The biosensor exhibits a low FRET ratio when Vcl is active and a high FRET ratio when Vcl is inactive (Case et al., 2015; Chen et al., 2005). The FRET ratio for Vcl WT was significantly lower in FAs than the ratio in the cytoplasm, indicating that a substantial fraction of Vcl WT is activated at FAs (Case et al., 2015; Ling et al., 2002). In contrast, Vcl variants Vcl BL_2A and Vcl BL_4A exhibited higher FRET ratios in FAs, closer to values obtained in the cytoplasm. Additionally, the FRET ratio in FAs for Vcl WT was significantly lower than the FRET ratio in FAs for Vcl BL_4A. These results suggest that Vcl membrane association plays an important role in activating Vcl at FAs.
Previous studies have reported conflicting results regarding whether PIP2-binding contributes (Chinthalapudi et al., 2014; Saunders et al., 2006) or not (Chandrasekar et al., 2005) to Vcl binding to and dissociation from FAs, which is related to cell migration. We used fluorescence recovery after photobleaching (FRAP) experiments to monitor the relative amount of mobile and immobile GFP-tagged Vcl at FAs and to measure its dissociation from FAs (T1/2 of recovery) in MEFs. Our data showed that lipid-binding significantly increased the fraction of Vcl that remained stably bound and immobile within FAs (Figure 8H). Indeed, GFP-Vcl BL_4A fluorescence at FAs recovered by 75% after photobleaching in our FRAP assays, while only 60% of GFP-Vcl WT fluorescence recovered. An intermediate amount, 68%, of Vcl BL_2A recovered at FAs, though this was not significantly different from Vcl WT or Vcl BL_4A. Examination of the T1/2 of recovery showed that, contrary to the work from Chinthalapudi et al, lipid-binding did not affect the rate of Vcl dissociation from FAs (Figure 8). These results indicate that Vcl binding to PIP2 promotes the turnover of Vcl at FAs, but not the rate of Vcl dissociation, by decreasing the immobile fraction of Vcl. This is consistent with previous work, as constitutively active Vcl is less mobile (Cohen et al., 2006).
Vcl lipid-binding does not regulate cell spread area, FA size, FA number, or response to external forces at FAs
While the Vcl/PIP2 interaction contributes to Vcl activation, localization, and turnover at FAs, it has also been reported to have a critical role in FA number, morphology, and force sensing (Chandrasekar et al., 2005; Diez et al., 2009). To test this, we expressed N-terminally GFP-tagged Vcl WT, Vcl BL_2A, and Vcl BL_4A in Vcl −/− MEFs and monitored the number and size of FAs as well as cell spread area (Figure S4). No significant differences were observed, suggesting that lipid-binding by Vcl does not have a large impact on the general maintenance of FAs or cell spreading.
Previous work has reported that loss of lipid binding by Vcl impairs the cellular response to force (Diez et al., 2009). However, these measurements were made using Vcl LD-CT, a poor actin bundler (Figure S3B). Using three dimensional force microscopy (3DFM), we evaluated the ability of cells expressing our Vcl constructs to respond to external forces on integrins. FN-coated magnetic beads were placed in cell culture and allowed to adhere to Vcl −/− MEFs. We subsequently pulled on the bead and tracked its movement. Cells expressing Vcl BL_2A or Vcl BL_4A exhibited an expected decrease in bead displacement, though the decrease was not statistically significant for Vcl BL_2A (Figure S5). These data suggest that Vcl lipid binding does not significantly regulate the cellular response at FAs to external force.
Discussion
The lack of a strong experimentally validated structural model for the Vcl/PIP2 interaction and contradictory biochemical and cellular data has hindered our understanding how PIP2-dependent membrane association of Vcl contributes to Vcl functions. (Chandrasekar et al., 2005; Diez et al., 2009; Halstead et al., 2010; Humphries et al., 2007; Saunders et al., 2006). The use of non-specific Vcl mutants has resulted in seemingly contradictory data (Chinthalapudi et al., 2014; Diez et al., 2009). While the crystal structure from Chinthalapudi et al. provides a starting model, the studies were conducted on a Vt mutant and a soluble short-chain PIP2, which limits the extrapolation of the model to the interaction at a biological membrane (Chinthalapudi et al., 2015; Chinthalapudi et al., 2014).
Here, we provide an alternative model for how Vcl binds PIP2 at a membrane. Our model is in agreement with published biochemical data: mutation at K944 and R945 or K1061 weakens PIP2-binding by Vt (Chinthalapudi et al., 2014). However, according to our model, the basic collar and the basic ladder play unique roles in binding to PIP2-containing membranes. While the basic ladder drives insertion into acidic membranes, the basic collar specifically recognizes PIP2. The relative abilities of mutations at these sites to disrupt PIP2- or PS-dependent liposome association are consistent with this model.
Our data further highlight the importance of the C-terminus and the basic collar in F-actin crosslinking (Shen et al., 2011; Tolbert et al., 2014). The weaker actin filament crosslinking activity of Vt BC_2A and the nearly absent actin crosslinking activity Vt BC_4A and Vt BC_4Q suggest that residues R910, K915, K924, and R925 and their positive charge are integral to formation of the Vt dimer that facilitates actin bundling. As we did not see a decrease in the ability of MEFs to respond to external forces when expressing basic ladder Vcl variants, our data suggest that previous links to the Vcl/lipid interaction and mechanotransduction are due mainly to disruption of Vcl’s ability to crosslink actin (Diez et al., 2009). Indeed, it may be that the role of Vcl in mechanotransduction and cell stiffness is driven primarily by actin binding and crosslinking.
Additionally, our NMR data suggest that PIP2-C8 is a poor substitute for membrane-associated full length PIP2. Consistent with these observations, previous studies have shown that short-chain PIP2 analogues bind to the PKCα C2 domain in a distinct manner from membrane-bound PIP2 molecules (Guerrero-Valero et al., 2009; Lai et al., 2010). We find that PIP2-C8 does not associate with the basic collar, but instead binds the basic ladder and the hydrophobic patch on helices 4 and 5, consistent with previous findings that the hydrophobic PIP2-C8 acyl tails, rather than the head group, can drive binding (Pu et al., 2010). The nature of the lipid bilayer plays a significant role in the interaction of Vt with PIP2 and future studies should account for this.
Our data support the use of basic ladder mutations to study the Vcl/PIP2 interaction in cells (Cohen et al., 2005) given the actin-crosslinking defect exhibited by Vt constructs with mutations at the basic collar and the key role of the basic ladder in membrane insertion. However, care should be taken in interpretation of these results; mutation of the basic ladder appears sufficient to disrupt sustained interactions with a lipid bilayer, but may not prevent transient interactions with PIP2. This, in addition to potential redundancies in FAs, may explain the lack of a strong cellular phenotype when disrupting the Vcl/PIP2 interaction.
Our findings do provide evidence that PIP2-binding contributes to Vcl activation (Golji et al., 2012; Ziegler et al., 2002), as lipid binding increases the fraction of activated Vcl at FAs. Additionally, PIP2 association increases the immobile fraction of Vcl, contrary to previous reports that PIP2-binding recycles activated Vcl from FAs (Chinthalapudi et al., 2014). As PIP2-binding enhances Vt phosphorylation by Src and PKCα (Ziegler et al., 2002), reduced association of Vcl with the membrane would also reduce the ability of these kinases to phosphorylate the tail domain, potentially increasing the fraction of Vcl that can revert to its autoinhibited conformation. This may explain the decreased activation and broader distribution of Vcl across FA layers. We previously showed that a constitutively active mutant of Vcl (D974/K975/R976/R978A) induced a strong shift in Vcl nanoscale localization from the integrin signaling layer to the actin regulatory (Case et al., 2015); our PIP2 binding mutant induced a weaker shift from the integrin signaling layer to the force transduction layer. These findings suggest that Vcl/PIP2 binding alone is not sufficient for full activation of Vcl in FAs. Rather, it is possible that PIP2 binding “primes” Vcl for actin binding in the actin regulatory layer, but is dispensable for talin-binding in the force transduction layer. There may be multiple conformations of Vcl that do not simply correspond to “active” or “inactive,” which would correspond to the combinatorial interactions and post-translational modifications that regulate the conformation of Vcl at FAs. Our results further highlight the complex relationships between Vcl PIP2, and FAs, and demonstrate a role of PIP2 binding in Vcl activation, turnover, and localization of Vcl within FAs.
Experimental Procedures
Expression and purification of Vt
Vt expression constructs were a gift from Robert Liddington (Sanford Burnham Prebys Medical Discovery Institute). The pET15b Vt expression vector encodes a His-tag followed by a thrombin cleavage site and then chicken Vt residues 884–1066. Vt was expressed and purified as previously described (Palmer et al., 2009; Thompson et al., 2014).
Lipid co-sedimentation assays
Vt binding to PIP2 was evaluated by lipid co-sedimentation assays using small, unilamellar vesicles (SUVs) as previously reported (Palmer et al., 2009; Thompson et al., 2014). Details are provided in Supplemental Information.
Actin co-sedimentation assays
Actin co-sedimentation assays to assess actin binding and actin crosslinking (bundling) properties of Vt WT and Vt mutants were performed as previously reported (Shen et al., 2011).
Mutagenesis
Vt variants were generated using QuikChange site-directed mutagenesis (Stratagene) and the sequences were verified by DNA sequencing (Genewiz).
Computational modeling
Modeling of the interaction of the PIP2 head group with Vt (residues 896–1055) was performed with MedusaDock (Ding et al., 2010; Yin et al., 2008). MedusaDock docking simulations constrain the Vt secondary structure but allow sampling of the Vt sidechain and PIP2 head group using rotamer libraries, with the lowest energy pose reported.
Modeling of the interaction of the full PIP2 molecule, 1-stearoyl-2-arachidonoyl-sn-glycero-3-phospho-(1’-myo-inositol-4’,5’-bisphosphate), with Vt in the absence of a lipid bilayer was performed with DMD simulations (Ding et al., 2008; Dokholyan et al., 1998; Shirvanyants et al., 2012). The initial docking pose was taken from the MedusaDock simulations. In these all-atom DMD simulations, Gō constraints were used to maintain intramolecular C(3/Ca contacts between Vt residues during the simulation. These constraints were applied for all residues in Vt or only residues in alpha helices in different simulations. The Gō constraints limit the potential for Vt unfolding while the PIP2 is allowed to sample the protein surface. This combination allows for greater sampling of potential Vt:PIP2 interactions while maintaining the structural integrity of Vt.
Modeling of the interaction of PIP2 within a lipid bilayer was performed using all-atom, explicit solvent molecular dynamics simulations. A lipid bilayer of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) with a single PIP2 molecule was generated, and an initial pose of Vt (residues 896–1055) bound to PIP2 using the model generated from MedusaDock. Three simulations of Vt bound to PIP2 in a POPC bilayer and three simulations of same configuration without PIP2, each lasting 100 ns. were performed with GROMACS v5.0.2 (Hess et al., 2008) using a CHARMM27 force field (Feller and MacKerell, 2000) with the following parameters: time-step of 2 fs, temperature of 300K maintained by v-rescale thermostat with a time constant of 0.1 ps, 1 bar pressure maintained by semi-isotropic pressure coupling with 5 ps time constant, and particle mesh Ewald electrostatics.
NMR
NMR samples were prepared in 40 mM 2-(N-morpholino)ethanesulfonic acid, pH 5.5, 50 mM NaCl, 0.01% NaN3, 2 mM DTT, and 5% D2O. Samples contained 50 uM 15N-enriched Vt with variable amounts of short-chain PIP2 (1,2-dioctanoyl-sn-glycero-3-phospho-(1’-myo-inositol-4’,5’-bisphosphate; PIP2-C8), Avanti Polar Lipids, Inc.). NMR titrations and TROSY for rotational correlation times (TRACT) (Lee et al., 2006) experiments were collected on a 700 MHz Varian Inova magnet with Bruker cryoprobe and console. 1H-15N 2D heteronuclear single quantum coherence (HSQC) NMR experiments were performed to monitor the change in the backbone H-N signals (backbone assignments (BMRB Entry 15653) as a function of the short-chain PIP2 ligand concentration (Palmer and Campbell, 2008). Further details can be found in Supplemental Information.
Cell culture and transfection
Vinculin knockout murine embryonic fibroblasts (Vcl −/− MEFs) were provided by Dr. Eileen Adamson (Burnham Institute; La Jolla, CA) and cultured, maintained, and transfected as previously reported (Tolbert et al., 2014).
FRAP assays
Fluorescence Recovery After Photobleaching (FRAP) experiments were performed with Vcl −/− MEFs expressing enhanced Green Fluorescent Protein (EGFP)-labeled Vcl WT or Vcl variants. Time-lapse TIRF microscopy was performed at 37°C using an Apo TIRF 100× 1.49 NA oil immersion objective lens (Nikon Instruments, Melville, NY, USA) on an inverted Eclipse Ti microscope system (Nikon Instruments, Melville, NY, USA58). EGFP images were captured with a CCD (coolsnap HQ2; Photometrics, Tuscon, AZ, USA) operated in the 5 MHz readout mode using 488nm laser illumination (Coherent, Santa Clara, CA, USA). Cells were imaged for three frames at 5 sec intervals prior to photobleaching, and then the 405 laser illumination of the FRAPPA system (Andor Technology, Belfast, Northern Ireland) was used to bleach an entire FA in the protruding region of the cell. After photobleaching, cells were imaged at 2 sec intervals for 1 min, and then at 5 sec intervals for 3 min. Final images were drift corrected using the rigid body method of the StackReg ImageJ plugin (Thevenaz et al., 1998), and the mean intensity inside individual bleached FA’s was corrected for imaging-induced photobleaching using a cytoplasmic region outside of the bleached region. Fluorescence recovery was fit with a single exponential to determine the t1/2 and percent recovery of each FA. Cells that retracted after bleaching were discarded from the analysis.
FRET assays
FRET experiments were performed using widefield epifluorescence as previously described (Chen et al., 2005). Briefly, a Vcl activation FRET biosensor in which fluorophores were inserted into the Vcl protein between the head and tail (mTurquoise, donor) and that the C-terminus (mNeonGreen37, acceptor) such that FRET occurs when Vt interacts with the Vcl head in the auto-inhibited conformation and FRET decreases when auto-inhibition is relieved and Vcl is activated (Case et al., 2015). The raw mTurqouise, FRET and Neon Green images were aligned using the rigid body method of the StackReg ImageJ plugin (Thevenaz et al., 1998). The FRET ratio was calculated using MATLAB to run the Danuser Lab Biosensors processing software 2.1 (available for download: http://lccb.hms.harvard.edu/software.html). Differences between mean FRET ratios were determined by an analysis of variance (ANOVA) followed by Tukey post-hoc anlaysis and differences were considered significant at p<0.05.
iPALM
Interference photoactivation localization microscopy (iPALM) imaging of Vcl −/− MEF expressing Vcl WT or Vcl variants containing an N-terminal tdEos fluorophore was performed as described previously (Case et al., 2015). After image acquisition was complete, the raw datasets were processed to localize individual molecules and extract their X, Y, and Z coordinates, and analyzed in Matlab as previously described (Case et al., 2015). Representative iPALM images were rendered with the PALMsiever Matlab package (Pengo et al., 2015) using the 3D Hue rendering technique to represent the Z-position with color. Additional experimental procedures can be found in Supplemental Information.
Supplementary Material
Highlights.
The vinculin tail contains two basic patches with distinct lipid binding roles.
The basic collar recognizes PIP2.
The basic ladder recognizes acidic phospholipids.
Lipid binding increases vinculin activation and turnover at focal adhesions.
E-TOC Paragraph.
Vinculin a cytoskeletal protein that controls cellular adhesion and migration. Thompson et al. show that two basic patches on the tail domain cooperate to bind lipid membranes containing PIP2. These binding events regulate vinculin activation and turnover at focal adhesions.
Acknowledgments
P.M.T. was supported by the American Heart Association (12PRE11820012). Funding to S.L.C. and N.V.D. was supported by NIH (1R01GM115597). Funding to C.E.T. was supported by NIH (GM029860). The authors wish to thank Evan Nelsen, Greg Young, and Paul Sapienza for their assistance with methods as well as Keith Burridge for his feedback on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
S.L.C., N.V.D., and C.M.W. conceived and supervised the research. Computational modeling and simulations were performed by S.R. and A.T. Biochemical experiments were performed by P.M.T. and M.P. Cellular experiments were performed by C.E.T. and L.B.C. Data were analyzed by P.M.T., C.E.T., L.B.C., S.R., A.T., and M.P. P.M.T., C.E.T, and L.B.C. wrote the manuscript with discussion and improvements from all authors.
References
- Bakolitsa C, Cohen DM, Bankston LA, Bobkov AA, Cadwell GW, Jennings L, Critchley DR, Craig SW, Liddington RC. Structural basis for vinculin activation at sites of cell adhesion. Nature. 2004;430:583–586. doi: 10.1038/nature02610. [DOI] [PubMed] [Google Scholar]
- Bakolitsa C, de Pereda JM, Bagshaw CR, Critchley DR, Liddington RC. Crystal structure of the vinculin tail suggests a pathway for activation. Cell. 1999;99:603–613. doi: 10.1016/s0092-8674(00)81549-4. [DOI] [PubMed] [Google Scholar]
- Case LB, Baird MA, Shtengel G, Campbell SL, Hess HF, Davidson MW, Waterman CM. Molecular mechanism of vinculin activation and nanoscale spatial organization in focal adhesions. Nat Cell Biol. 2015;17:880–892. doi: 10.1038/ncb3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Waterman CM. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat Cell Biol. 2015;17:955–963. doi: 10.1038/ncb3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekar I, Stradal TE, Holt MR, Entschladen F, Jockusch BM, Ziegler WH. Vinculin acts as a sensor in lipid regulation of adhesion-site turnover. J Cell Sci. 2005;118:1461–1472. doi: 10.1242/jcs.01734. [DOI] [PubMed] [Google Scholar]
- Chen H, Choudhury DM, Craig SW. Coincidence of actin filaments and talin is required to activate vinculin. J Biol Chem. 2006;281:40389–40398. doi: 10.1074/jbc.M607324200. [DOI] [PubMed] [Google Scholar]
- Chen H, Cohen DM, Choudhury DM, Kioka N, Craig SW. Spatial distribution and functional significance of activated vinculin in living cells. J Cell Biol. 2005;169:459–470. doi: 10.1083/jcb.200410100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinthalapudi K, Patil DN, Rangarajan ES, Rader C, Izard T. Lipid-directed vinculin dimerization. Biochemistry. 2015;54:2758–2768. doi: 10.1021/acs.biochem.5b00015. [DOI] [PubMed] [Google Scholar]
- Chinthalapudi K, Rangarajan ES, Patil DN, George EM, Brown DT, Izard T. Lipid binding promotes oligomerization and focal adhesion activity of vinculin. J Cell Biol. 2014;207:643–656. doi: 10.1083/jcb.201404128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen DM, Chen H, Johnson RP, Choudhury B, Craig SW. Two distinct head-tail interfaces cooperate to suppress activation of vinculin by talin. J Biol Chem. 2005;280:17109–17117. doi: 10.1074/jbc.M414704200. [DOI] [PubMed] [Google Scholar]
- Cohen DM, Kutscher B, Chen H, Murphy DB, Craig SW. A conformational switch in vinculin drives formation and dynamics of a talin-vinculin complex at focal adhesions. J Biol Chem. 2006;281:16006–16015. doi: 10.1074/jbc.M600738200. [DOI] [PubMed] [Google Scholar]
- Coll JL, Ben-Ze’ev A, Ezzell RM, Rodriguez Fernandez JL, Baribault H, Oshima RG, Adamson ED. Targeted disruption of vinculin genes in F9 and embryonic stem cells changes cell morphology, adhesion, and locomotion. Proc Natl Acad Sci U S A. 1995;92:9161–9165. doi: 10.1073/pnas.92.20.9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez G, Kollmannsberger P, Mierke CT, Koch TM, Vali H, Fabry B, Goldmann WH. Anchorage of vinculin to lipid membranes influences cell mechanical properties. Biophys J. 2009;97:3105–3112. doi: 10.1016/j.bpj.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Tsao D, Nie H, Dokholyan NV. Ab initio folding of proteins with all-atom discrete molecular dynamics. Structure. 2008;16:1010–1018. doi: 10.1016/j.str.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Yin S, Dokholyan NV. Rapid flexible docking using a stochastic rotamer library of ligands. J Chem Inf Model. 2010;50:1623–1632. doi: 10.1021/ci100218t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Discrete molecular dynamics studies of the folding of a protein-like model. Fold Des. 1998;3:577–587. doi: 10.1016/S1359-0278(98)00072-8. [DOI] [PubMed] [Google Scholar]
- Feller SE, MacKerell AD. An Improved Empirical Potential Energy Function for Molecular Simulations of Phospholipids. The Journal of Physical Chemistry B. 2000;104:7510–7515. [Google Scholar]
- Golji J, Wendorff T, Mofrad MRK. Phosphorylation Primes Vinculin for Activation. Biophys J. 2012;102:2022–2030. doi: 10.1016/j.bpj.2012.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Valero M, Ferrer-Orta C, Querol-Audi J, Marin-Vicente C, Fita I, Gomez-Fernandez JC, Verdaguer N, Corbalan-Garcia S. Structural and mechanistic insights into the association of PKCalpha-C2 domain to PtdIns(4,5)P2. Proc Natl Acad Sci U S A. 2009;106:6603–6607. doi: 10.1073/pnas.0813099106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead JR, Savaskan NE, van den Bout I, Van Horck F, Hajdo-Milasinovic A, Snell M, Keune WJ, Ten Klooster JP, Hordijk PL, Divecha N. Rac controls PIP5K localisation and PtdIns(4,5)P(2) synthesis, which modulates vinculin localisation and neurite dynamics. J Cell Sci. 2010;123:3535–3546. doi: 10.1242/jcs.062679. [DOI] [PubMed] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. Journal of Chemical Theory and Computation. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Humphries JD, Wang P, Streuli C, Geiger B, Humphries MJ, Ballestrem C. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J Cell Biol. 2007;179:1043–1057. doi: 10.1083/jcb.200703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Bubeck P, Rudiger M, Jockusch BM. Characterization of two F-actin-binding and oligomerization sites in the cell-contact protein vinculin. Eur J Biochem. 1997;247:1136–1142. doi: 10.1111/j.1432-1033.1997.01136.x. [DOI] [PubMed] [Google Scholar]
- Izard T, Evans G, Borgon RA, Rush CL, Bricogne G, Bois PR. Vinculin activation by talin through helical bundle conversion. Nature. 2004;427:171–175. doi: 10.1038/nature02281. [DOI] [PubMed] [Google Scholar]
- Johnson RP, Craig SW. The carboxy-terminal tail domain of vinculin contains a cryptic binding site for acidic phospholipids. Biochem Biophys Res Commun. 1995a;210:159–164. doi: 10.1006/bbrc.1995.1641. [DOI] [PubMed] [Google Scholar]
- Johnson RP, Craig SW. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 1995b;373:261–264. doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- Johnson RP, Niggli V, Durrer P, Craig SW. A conserved motif in the tail domain of vinculin mediates association with and insertion into acidic phospholipid bilayers. Biochemistry. 1998;37:10211–10222. doi: 10.1021/bi9727242. [DOI] [PubMed] [Google Scholar]
- Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim LY, Thompson PM, Campbell SL, Alushin GM. In American Society for Cell Biology. Philadelphia: 2014. Structural basis of the vinculin–F-actin interaction. [Google Scholar]
- Lai CL, Landgraf KE, Voth GA, Falke JJ. Membrane docking geometry and target lipid stoichiometry of membrane-bound PKCalpha C2 domain: a combined molecular dynamics and experimental study. J Mol Biol. 2010;402:301–310. doi: 10.1016/j.jmb.2010.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Duc Q, Shi Q, Blonk I, Sonnenberg A, Wang N, Leckband D, de Rooij J. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J Cell Biol. 2010;189:1107–1115. doi: 10.1083/jcb.201001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Hilty C, Wider G, Wuthrich K. Effective rotational correlation times of proteins from NMR relaxation interference. J Magn Reson. 2006;178:72–76. doi: 10.1016/j.jmr.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Legate KR, Takahashi S, Bonakdar N, Fabry B, Boettiger D, Zent R, Fassler R. Integrin adhesion and force coupling are independently regulated by localized PtdIns(4,5)2 synthesis. Embo J. 2011;30:4539–4553. doi: 10.1038/emboj.2011.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhou Q, Sunkara M, Kutys ML, Wu Z, Rychahou P, Morris AJ, Zhu H, Evers BM, Huang C. Ubiquitylation of phosphatidylinositol 4-phosphate 5-kinase type I gamma by HECTD1 regulates focal adhesion dynamics and cell migration. J Cell Sci. 2013;126:2617–2628. doi: 10.1242/jcs.117044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature. 2002;420:89–93. doi: 10.1038/nature01082. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang Y, Goh WI, Goh H, Baird MA, Ruehland S, Teo S, Bate N, Critchley DR, Davidson MW, et al. Talin determines the nanoscale architecture of focal adhesions. Proc Natl Acad Sci U S A. 2015;112:E4864–E4873. doi: 10.1073/pnas.1512025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez MG, Fernandez-Tome Mdel C, Favale NO, Pescio LG, Sterin-Speziale NB. Bradykinin induces formation of vesicle-like structures containing vinculin and PtdIns(4,5)P2 in renal papillary collecting duct cells. Am J Physiol Renal Physiol. 2009;297:F1181–F1191. doi: 10.1152/ajprenal.00062.2009. [DOI] [PubMed] [Google Scholar]
- Mierke CT, Kollmannsberger P, Zitterbart DP, Smith J, Fabry B, Goldmann WH. Mechano-coupling and regulation of contractility by the vinculin tail domain. Biophys J. 2008;94:661–670. doi: 10.1529/biophysj.107.108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer SM, Campbell SL. Backbone 1H, 13C, and 15N NMR assignments of the tail domain of vinculin. Biomol NMR Assign. 2008;2:69–71. doi: 10.1007/s12104-008-9087-7. [DOI] [PubMed] [Google Scholar]
- Palmer SM, Playford MP, Craig SW, Schaller MD, Campbell SL. Lipid binding to the tail domain of vinculin: specificity and the role of the N and C termini. J Biol Chem. 2009;284:7223–7231. doi: 10.1074/jbc.M807842200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengo T, Holden SJ, Manley S. PALMsiever: a tool to turn raw data into results for single-molecule localization microscopy. Bioinformatics. 2015;31:797–798. doi: 10.1093/bioinformatics/btu720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor EA, Ding F, Dokholyan NV. Structural and thermodynamic effects of post-translational modifications in mutant and wild type Cu, Zn superoxide dismutase. J Mol Biol. 2011;408:555–567. doi: 10.1016/j.jmb.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu M, Orr A, Redfield AG, Roberts MF. Defining specific lipid binding sites for a peripheral membrane protein in situ using subtesla field-cycling NMR. J Biol Chem. 2010;285:26916–26922. doi: 10.1074/jbc.M110.123083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders RM, Holt MR, Jennings L, Sutton DH, Barsukov IL, Bobkov A, Liddington RC, Adamson EA, Dunn GA, Critchley DR. Role of vinculin in regulating focal adhesion turnover. Eur J Cell Biol. 2006;85:487–500. doi: 10.1016/j.ejcb.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Shen K, Tolbert CE, Guilluy C, Swaminathan VS, Berginski ME, Burridge K, Superfine R, Campbell SL. The vinculin C-terminal hairpin mediates F-actin bundle formation, focal adhesion, and cell mechanical properties. J Biol Chem. 2011;286:45103–45115. doi: 10.1074/jbc.M111.244293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirvanyants D, Ding F, Tsao D, Ramachandran S, Dokholyan NV. Discrete molecular dynamics: an efficient and versatile simulation method for fine protein characterization. J Phys Chem B. 2012;116:8375–8382. doi: 10.1021/jp2114576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Ling K, Wagoner MP, Anderson RA. Type I gamma phosphatidylinositol phosphate kinase is required for EGF-stimulated directional cell migration. J Cell Biol. 2007;178:297–308. doi: 10.1083/jcb.200701078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Tolbert CE, Shen K, Kota P, Palmer SM, Plevock KM, Orlova A, Galkin VE, Burridge K, Egelman EH, et al. Identification of an actin binding surface on vinculin that mediates mechanical cell and focal adhesion properties. Structure. 2014;22:697–706. doi: 10.1016/j.str.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert CE, Thompson PM, Superfine R, Burridge K, Campbell SL. Phosphorylation at Y1065 in Vinculin Mediates Actin Bundling, Cell Spreading, and Mechanical Responses to Force. Biochemistry. 2014 doi: 10.1021/bi500678x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bout I, Divecha N. PIP5K–driven PtdIns(4,5)P2 synthesis: regulation and cellular functions. J Cell Sci. 2009;122:3837–3850. doi: 10.1242/jcs.056127. [DOI] [PubMed] [Google Scholar]
- Weekes J, Barry ST, Critchley DR. Acidic phospholipids inhibit the intramolecular association between the N- and C-terminal regions of vinculin, exposing actin-binding and protein kinase C phosphorylation sites. Biochem J. 1996;314(Pt 3):827–832. doi: 10.1042/bj3140827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth VF, List F, Diez G, Goldmann WH. Vinculin’s C-terminal region facilitates phospholipid membrane insertion. Biochem Biophys Res Commun. 2010;398:433–437. doi: 10.1016/j.bbrc.2010.06.094. [DOI] [PubMed] [Google Scholar]
- Wu Z, Li X, Sunkara M, Spearman H, Morris AJ, Huang C. PIPKIgamma regulates focal adhesion dynamics and colon cancer cell invasion. PLoS One. 2011;6:e24775. doi: 10.1371/journal.pone.0024775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Baribault H, Adamson ED. Vinculin knockout results in heart and brain defects during embryonic development. Development. 1998a;125:327–337. doi: 10.1242/dev.125.2.327. [DOI] [PubMed] [Google Scholar]
- Xu W, Coll JL, Adamson ED. Rescue of the mutant phenotype by reexpression of full-length vinculin in null F9 cells; effects on cell locomotion by domain deleted vinculin. J Cell Sci. 1998b;111(Pt 11):1535–1544. doi: 10.1242/jcs.111.11.1535. [DOI] [PubMed] [Google Scholar]
- Yin S, Biedermannova L, Vondrasek J, Dokholyan NV. MedusaScore: an accurate force field-based scoring function for virtual drug screening. Journal of chemical information and modeling. 2008;48:1656–1662. doi: 10.1021/ci8001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler WH, Tigges U, Zieseniss A, Jockusch BM. A lipid-regulated docking site on vinculin for protein kinase C. J Biol Chem. 2002;277:7396–7404. doi: 10.1074/jbc.M110008200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.