ABSTRACT
Opsonophagocytic assays (OPAs) are routinely used for assessing the immunogenicity of pneumococcal vaccines, with OPA data often being utilized for licensure of new vaccine formulations. However, no reference serum for pneumococcal OPAs is available, making evaluation of data among different laboratories difficult. This international collaboration was initiated to (i) assign consensus opsonic indexes (OIs) to FDA pneumococcal reference serum lot 007sp (here referred to as 007sp) and a panel of serum samples used for calibration of the OPA and (ii) determine if the normalization of the OPA results obtained with test samples to those obtained with 007sp decreases the variability in OPA results among laboratories. To meet these goals, six participating laboratories tested a panel of serum samples in five runs for 13 serotypes. For each serum sample, consensus OIs were obtained using a mixed-effects analysis of variance model. For the calibration serum samples, normalized consensus values were also determined on the basis of the results obtained with 007sp. For each serotype, the overall reduction in interlaboratory variability was calculated by comparing the coefficients of variation of the unadjusted and the normalized values. Normalization of the results substantially reduced the interlaboratory variability, ranging from a 15% reduction in variability for serotype 9V to a 64% reduction for serotype 7F. Normalization also increased the proportion of data within 2-fold of the consensus value from approximately 70% (average for all serotypes) to >90%. On the basis of the data obtained in this study, pneumococcal reference standard lot 007sp will likely be a useful reagent for the normalization of pneumococcal OPA results from different laboratories. The data also support the use of the 16 FDA serum samples used for calibration of the OPA as part of the initial evaluation of new assays or periodic assessment of established assays.
KEYWORDS: 007sp, OPA, opsonic, pneumococcus, standardization
INTRODUCTION
Conjugate vaccines targeting the capsular polysaccharides (PSs) of Streptococcus pneumoniae have been largely successful in reducing the incidence of invasive pneumococcal disease caused by vaccine serotypes in both children and adults in various countries throughout the world (reviewed in reference 1). Although large clinical trials were performed to demonstrate the efficacy of the first pneumococcal conjugate vaccine, such efficacy trials are not practical for the evaluation of new vaccines with increased serotypic coverage. Therefore, surrogate markers of protection have been used to evaluate new vaccine formulations.
Comparison of the immunogenicity of a prospective vaccine to that of the current vaccine in terms of serotype-specific IgG concentrations using enzyme-linked immunosorbent assay (ELISA) has been utilized in assessing the efficacy of candidate vaccines in pediatric populations. Since a correlation between serum antibody concentrations and vaccine efficacy has been established only for this age group (2), the use of ELISA for the evaluation of vaccine efficacy is restricted to pediatric populations. Although the development of a third-generation ELISA (reviewed in reference 3) has increased the specificity of the assay, serum antibody concentrations measured in an ELISA may not always reflect their functional capacity, especially in adults (4, 5). Therefore, ELISA data are not accepted for licensure of pneumococcal vaccines for use in adults.
In vivo, antibodies against pneumococcal capsular PSs are thought to function by opsonizing the bacteria for subsequent phagocytosis and killing by granulocytes (6). Thus, in vitro opsonophagocytic assays (OPAs) were developed to mimic this mechanism (7–9). Multiple improvements have since been made to the assay, resulting in various assay protocols (10, 11), including an OPA with a multiplexed format (MOPA) (12), that are suitable for use in vaccine studies. Consequently, the OPA has become an important tool for evaluating the immunogenicity of new pneumococcal vaccines, particularly in adults.
A previous international interlaboratory study of OPAs (13) showed a reasonably good correlation between results obtained by different laboratories that utilized different assay formats. However, the absolute agreement of the results was less than desirable, with values with a 2-fold range above and below the consensus values (i.e., a 4-fold range from the upper limit to the lower limit) encompassing approximately 68% of the results. Values with a 4-fold range above and below the consensus value (i.e., a 16-fold range from the upper limit to the lower limit) were needed to capture approximately 90% of the results. The study authors concluded that OPAs may need additional control and absolute OPA results were not necessarily comparable among different laboratories.
When the new pneumococcal ELISA reference serum, pneumococcal reference serum lot 007sp (here referred to as 007sp), was produced, it was intended to serve as a reference serum for both ELISAs and OPAs (14). When sera were pooled to create 007sp, 16 serum samples from single donors were prepared for OPA calibration purposes (14). In 2012, an international collaborative study involving 6 laboratories familiar with OPAs was devised to (i) assign consensus values to 007sp and a panel of 16 serum samples to be used for calibrating OPAs and (ii) determine if the normalization of the results obtained with 007sp had any impact on the interlaboratory agreement of OPA results. The results of the collaboration are described here.
RESULTS
Consensus values for 007sp.
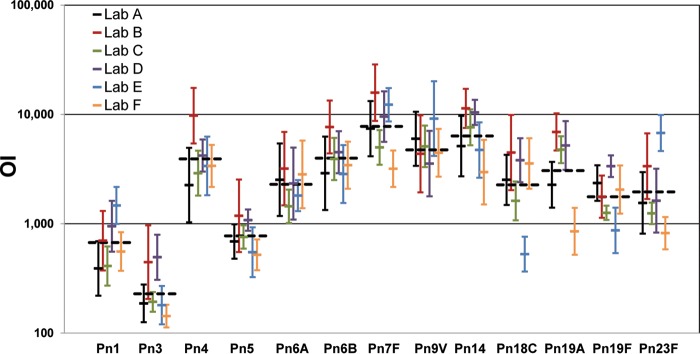
In order to estimate the consensus values for 007sp, the 007sp opsonic indexes (OIs) were fit using a mixed-effects analysis of variance (ANOVA) model. For each of the 13 target serotypes, the individual laboratory geometric mean OIs (GMOIs) as well as the consensus value for 007sp are shown in Table 1 and Fig. 1. The consensus values ranged from 229 (for serotype 3) to 7,776 (for serotype 7F). The width of the 95% confidence intervals (CIs) ranged from 1.9-fold for serotype 6A to >10-fold for serotype 19A. The confidence intervals for the other serotypes were generally in the 2- to 3-fold range except for serotypes 18C and 23F, which had confidence intervals of 5.2- and 4.8-fold, respectively.
TABLE 1.
007sp consensus values and statisticsa
| Pn | GMOI for laboratory: |
Consensus OI (95% CI) | Value for variance component: |
Total % CV | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | Lab | Run(Lab) | Plate(Run × Lab) | |||
| 1 | 390 | 700 | 411 | 949 | 1,468 | 556 | 672 (396; 1,143) | 0.2496 | 0.0003 | 0.0622 | 75 |
| 3 | 187 | 445 | 193 | 494 | 180 | 143 | 229 (131; 398) | 0.2710 | 0.0013 | 0.0408 | 75 |
| 4 | 2,259 | 9,738 | 2,899 | 4,204 | 3,375 | 3,381 | 3,912 (2,351; 6,511) | 0.2264 | 0.0006 | 0.0721 | 73 |
| 5 | 688 | 1,181 | 757 | 1,080 | 548 | 519 | 774 (535; 1,119) | 0.1180 | 0.0006 | 0.0372 | 48 |
| 6A | 2,522 | 3,191 | 1,443 | 2,331 | 1,811 | 2,825 | 2,293 (1,683; 3,123) | 0.0782 | 0.0001 | 0.1205 | 56 |
| 6B | 2,893 | 7,668 | 3,915 | 4,515 | 2,849 | 3,430 | 3,976 (2,707; 5,841) | 0.1280 | 0.0001 | 0.0804 | 58 |
| 7F | 7,402 | 15,789 | 4,991 | 9,552 | 12,246 | 3,182 | 7,776 (4,207; 14,371) | 0.3360 | 0.0004 | 0.0572 | 87 |
| 9V | 5,989 | 4,356 | 5,095 | 3,551 | 9,149 | 4,451 | 4,733 (3,083; 7,268) | 0.1507 | 0.0031 | 0.0802 | 62 |
| 14 | 5,130 | 11,345 | 7,618 | 10,441 | 4,722 | 2,965 | 6,349 (3,680; 10,951) | 0.2647 | 0.0000 | 0.0743 | 79 |
| 18C | 2,518 | 4,475 | 1,618 | 3,806 | 527 | 3,559 | 2,264 (996; 5,145) | 0.5996 | 0.0014 | 0.0693 | 127 |
| 19A | 2,271 | 6,909 | 4,755 | 5,192 | NT | 852 | 3,059 (948; 9,867) | 0.8827 | 0.0025 | 0.0463 | 163 |
| 19F | 2,357 | 1,768 | 1,258 | 3,362 | 869 | 2,055 | 1,766 (1,093; 2,855) | 0.2032 | 0.0007 | 0.0369 | 63 |
| 23F | 1,552 | 3,355 | 1,244 | 1,627 | 6,757 | 820 | 1,952 (895; 4,259) | 0.5370 | 0.0036 | 0.0611 | 117 |
For each serotype, the laboratory-specific 007sp GMOI and the consensus 007sp OI are shown. Estimates of various variance components and the total CV, determined by ANOVA, are also indicated. Pn, pneumococcal serotype; OI, opsonic index; GMOI, geometric mean opsonic index; CI, confidence interval; CV, coefficient of variation (expressed as a percent); NT, not tested; Lab, variability among the laboratories; Run(Lab), variability among runs within each laboratory; Plate(Run × Lab), variability among plates within each combination of run and laboratory.
FIG 1.
007sp results. For each serotype (x axis), the line representing each laboratory (see the key) indicates the laboratory-specific GMOI (y axis, center tick) ± 2 standard deviations (y axis, terminal ticks). The consensus OI for each serotype is indicated by the black, dashed horizontal line.
The total coefficient of variation (CV; expressed as a percentage) and various variance components [Lab, which is the variability among the laboratories; Run(Lab), which is the variability among runs within each laboratory; Plate(Run × Lab), which is the variability among plates within each combination of run and laboratory] estimated from the ANOVA are also shown in Table 1. Serotype 5 had the lowest CV (48%), and serotype 19A had the highest CV (163%). Serotypes 18C and 23F also had relatively high total CVs (127% and 117%, respectively). The total CVs of the remaining serotypes were generally in the range of 60% to 80%. Analysis of the variance components showed that the laboratory (Lab in Table 1) was the major source of variation for all serotypes except serotype 6A, where the variation was mostly associated with the Plate(Run × Lab) variance component.
FDA calibration serum sample-normalized consensus values.
To estimate the consensus OIs for each of the FDA calibration serum samples, OIs were normalized on the basis of the overall consensus value for 007sp and the value for 007sp estimated in each corresponding run. The normalized consensus values (and the corresponding 95% CIs) for each of the FDA calibration serum samples were calculated and are shown in Table 2 for the 13 target serotypes. These consensus values were determined after removing outliers (20 outliers were identified among the calibration serum samples; see Table 7). The laboratory-specific GMOIs (unadjusted and normalized) for each of the calibration serum samples are included in Table S1 in the supplemental material, with the 20 results identified as outliers being indicated in red. Generally, the confidence intervals for the normalized consensus values are quite small, with most being less than 2-fold.
TABLE 2.
Normalized consensus OIs for calibration serum samplesa
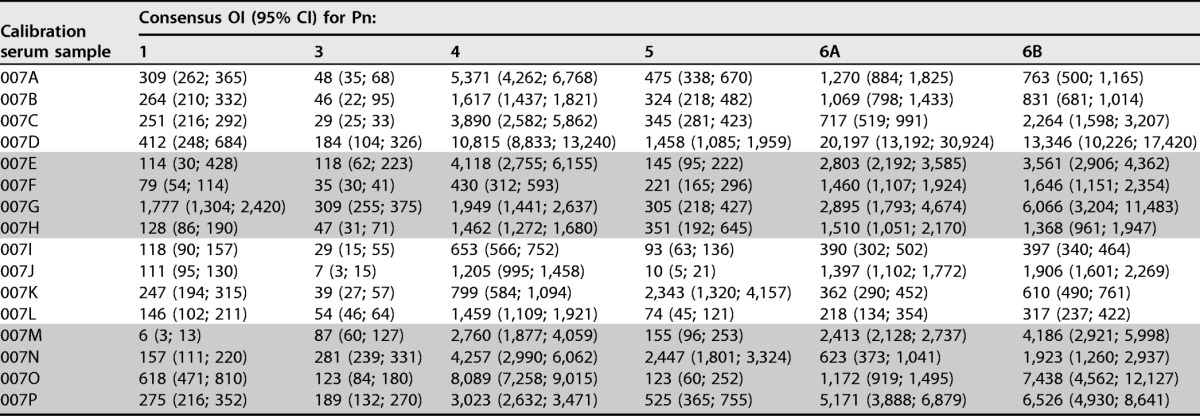
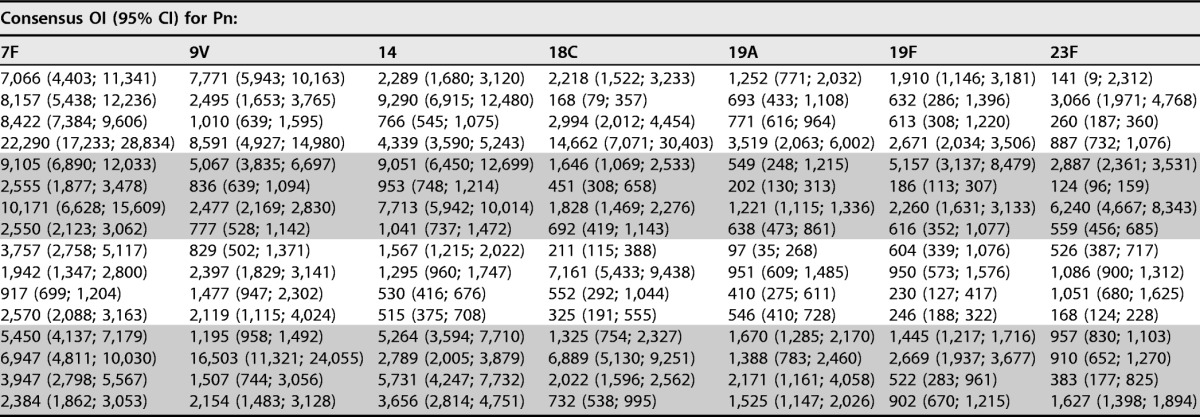
For each calibration serum sample, the normalized consensus OIs (95% CIs) for the indicated serotypes are shown. Outliers were excluded from these analyses. Pn, pneumococcal serotype; OI, opsonic index; CI, confidence interval.
TABLE 7.
Data excluded from analysesa
| Pn | Indeterminable results |
Intralaboratory outliers |
Interlaboratory outliers |
|||
|---|---|---|---|---|---|---|
| Calibration serum sample | Source (no. of serum samples) | Calibration serum sample | Source (no. of serum samples) | Calibration serum sample | Source (no. of serum samples) | |
| 1 | 007E | Lab C (1) | 007E | Lab A (1) | 007E | Lab A (1) |
| 007M | Lab D (1) | 007M | Lab E (1) | |||
| 007N | Lab A (1) | |||||
| 3 | 007C | Lab E (1) | 007B | Lab F (1) | 007A | Lab E (1) |
| 007E | Lab F (3) | 007B | Lab E (1) | |||
| 007E | Lab F (1) | |||||
| 007J | Lab E (1) | |||||
| 4 | 007F | Lab F (1) | 007E | Lab A (1) | ||
| 007J | Lab F (2) | 007M | Lab A (2) | |||
| 007P | Lab E (1) | |||||
| 5 | 007F | Lab E (1) | ||||
| 6A | 007C | Lab F (1) | 007L | Lab E (1) | ||
| 6B | 007I | Lab E (3) | 007L | Lab E (1) | 007L | Lab E (1) |
| 007L | Lab E (2) | |||||
| 7F | 007M | Lab A (2) | ||||
| 14 | 007P | Lab E (1) | ||||
| 18C | 007I | Lab E (2) | 007I | Lab E (1) | 007I | Lab E (1) |
| 007M | Lab E (2) | 007M | Lab E (1) | 007M | Lab E (1) | |
| 19A | 007I | Lab A (1) | 007E | Lab F (1) | ||
| 007F | Lab F (1) | |||||
| 19F | 007A | Lab E (1), F (1) | 007F | Lab E (1) | ||
| 007C | Lab E (4) | 007L | Lab E (1) | |||
| 007D | Lab E (1) | |||||
| 007O | Lab E (2) | |||||
| 23F | 007A | Lab A (1), C (5) | 007A | Lab D (1) | 007A | Lab F (1) |
| 007B | Lab E (1) | 007C | Lab F (1) | 007C | Lab F (1) | |
| 007C | Lab E (4), F (1) | 007F | Lab F (2) | 007F | Lab E (1) | |
| 007F | Lab E (1) | 007I | Lab A (1) | 007I | Lab F (1) | |
| 007H | Lab F (1) | 007O | Lab A (1) | 007L | Lab E (1) | |
| 007J | Lab F (1) | 007O | Lab D (1) | 007O | Lab F (1) | |
| 007L | Lab E (3) | |||||
For each serotype, the results for the calibration serum samples identified as indeterminable, intralaboratory outliers, and interlaboratory outliers are indicated. Indeterminable results are defined as failing to meet the testing laboratory's assay system suitability criteria, intralaboratory outliers are defined as an OI within a laboratory differing by more than 4-fold from the corresponding laboratory's median OI, and interlaboratory outliers are defined as a laboratory's geometric mean OI differing by more than 4-fold from the corresponding consensus OI. Pn, pneumococcal serotype.
Effect of normalization on interlaboratory variation.
To determine the effect of normalization on assay variation, the variability of the unadjusted results (including interlaboratory outliers) from the 20 test serum samples was compared to the variability of the normalized results (also including outliers). For each of the 13 target serotypes, the variability (expressed as percent CV) of the unadjusted and normalized results is presented in Table 3. The percent reduction in variability due to normalization, as well as the various variance components estimated by the ANOVA, are also indicated in Table 3 (see Table S2 in the supplemental material for the effect of normalization on individual samples). A noticeable improvement in interlaboratory agreement with normalization based on the performance with 007sp was seen, ranging from a 15% reduction in variability for serotype 9V to a 64% reduction for serotype 7F. Although the CVs were still high for some serotypes after normalization (for example, for serotype 23F, the CV decreased from 244% to 180% with normalization), the four artificially created low-OI serum samples are responsible for the high CVs in many instances.
TABLE 3.
Model-based assessment of effect of normalizationa
| Pn | Unadjusted |
Normalized |
Variability reduction (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Result for variance component: |
% CV | Result for variance component: |
% CV | ||||||||
| Lab | Sample × Lab | Run(Lab) | Sample × Run(Lab) | Lab | Sample × Lab | Run(Lab) | Sample × Run(Lab) | ||||
| 1 | 0.2456 | 0.3424 | 0.0103 | 0.0631 | 126 | 0.0000 | 0.3099 | 0.0196 | 0.0646 | 87 | 40 |
| 3 | 0.2848 | 0.2032 | 0.0197 | 0.0687 | 114 | 0.0457 | 0.1729 | 0.0180 | 0.0735 | 75 | 46 |
| 4 | 0.3046 | 0.3975 | 0.0218 | 0.0632 | 143 | 0.0290 | 0.4374 | 0.0135 | 0.0696 | 110 | 30 |
| 5 | 0.1437 | 0.1511 | 0.0096 | 0.0480 | 81 | 0.0200 | 0.1236 | 0.0135 | 0.0572 | 59 | 39 |
| 6A | 0.1088 | 0.2606 | 0.0304 | 0.0900 | 101 | 0.0185 | 0.2810 | 0.0096 | 0.0941 | 89 | 18 |
| 6B | 0.2177 | 0.3247 | 0.0244 | 0.0578 | 120 | 0.0251 | 0.3774 | 0.0290 | 0.0657 | 102 | 20 |
| 7F | 0.6062 | 0.0750 | 0.0208 | 0.0781 | 142 | 0.0574 | 0.0897 | 0.0198 | 0.1113 | 69 | 64 |
| 9V | 0.0702 | 0.3027 | 0.0501 | 0.0771 | 103 | 0.0310 | 0.2594 | 0.0315 | 0.1010 | 92 | 15 |
| 14 | 0.1515 | 0.0451 | 0.0286 | 0.0465 | 68 | 0.0275 | 0.0427 | 0.0073 | 0.0491 | 43 | 53 |
| 18C | 0.9021 | 0.3757 | 0.0398 | 0.1014 | 229 | 0.1726 | 0.5400 | 0.0232 | 0.0999 | 149 | 41 |
| 19A | 1.1479 | 0.1970 | 0.0282 | 0.0501 | 230 | 0.1144 | 0.3457 | 0.0113 | 0.0940 | 112 | 60 |
| 19F | 0.4291 | 0.1988 | 0.0297 | 0.0681 | 134 | 0.1736 | 0.1790 | 0.0221 | 0.0840 | 97 | 37 |
| 23F | 0.4649 | 0.9584 | 0.0299 | 0.0739 | 244 | 0.0673 | 0.9215 | 0.0072 | 0.0666 | 180 | 30 |
The overall reduction in variability due to normalization is shown for each serotype. Estimates of the various variance components and CVs of the unadjusted and normalized results from the ANOVA are also shown. Pn, pneumococcal serotype; CV, coefficient of variation (expressed as a percent); Lab, variability among the laboratories; Run(Lab), variability among runs within each laboratory; Sample × Lab, variability associated with the interaction between test sample and laboratory; Sample × Run(Lab), variability associated with the interaction between test sample and runs within a laboratory.
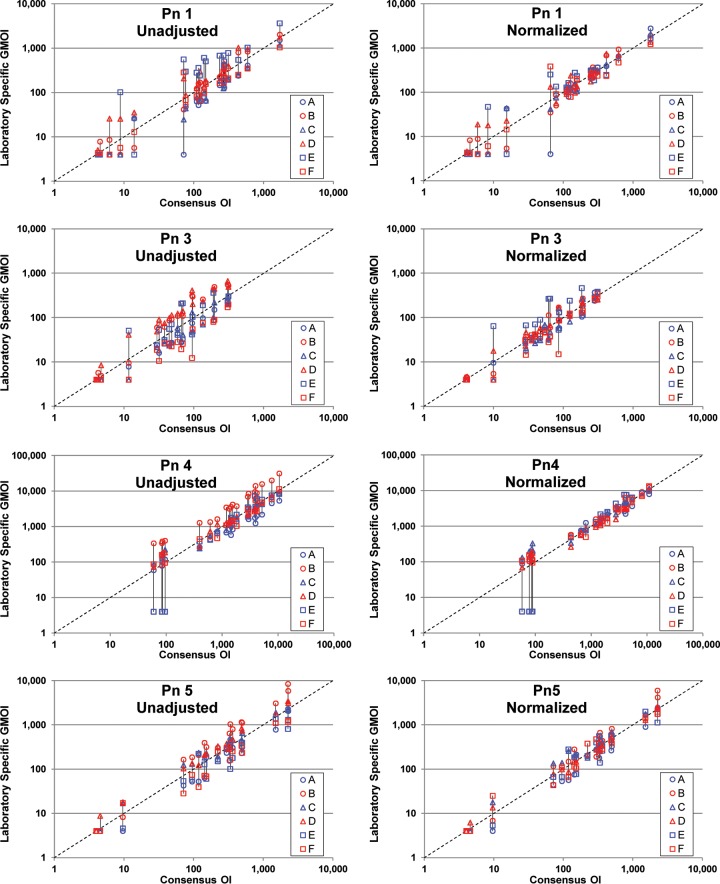
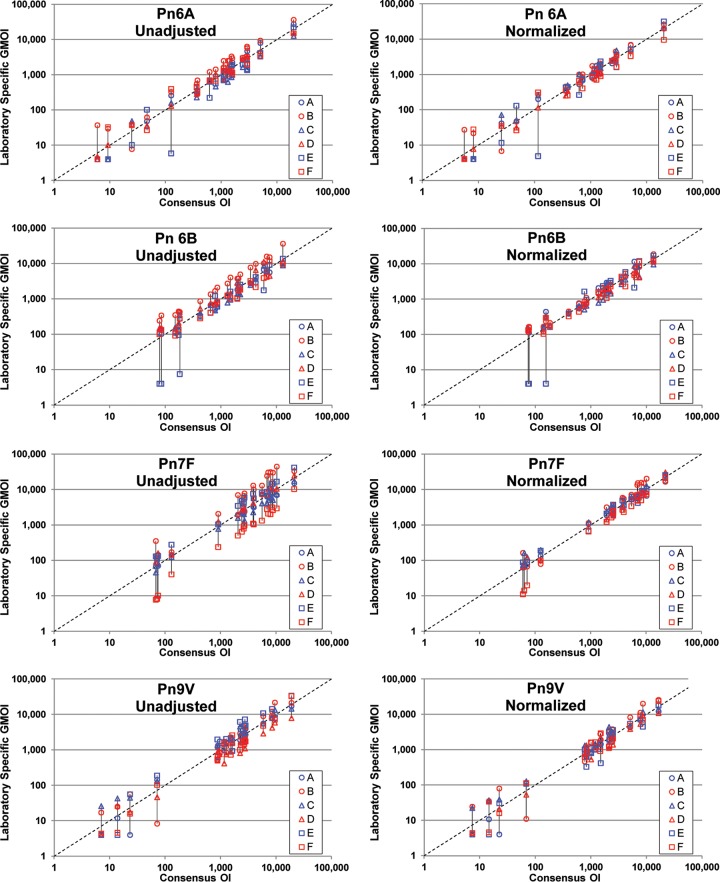
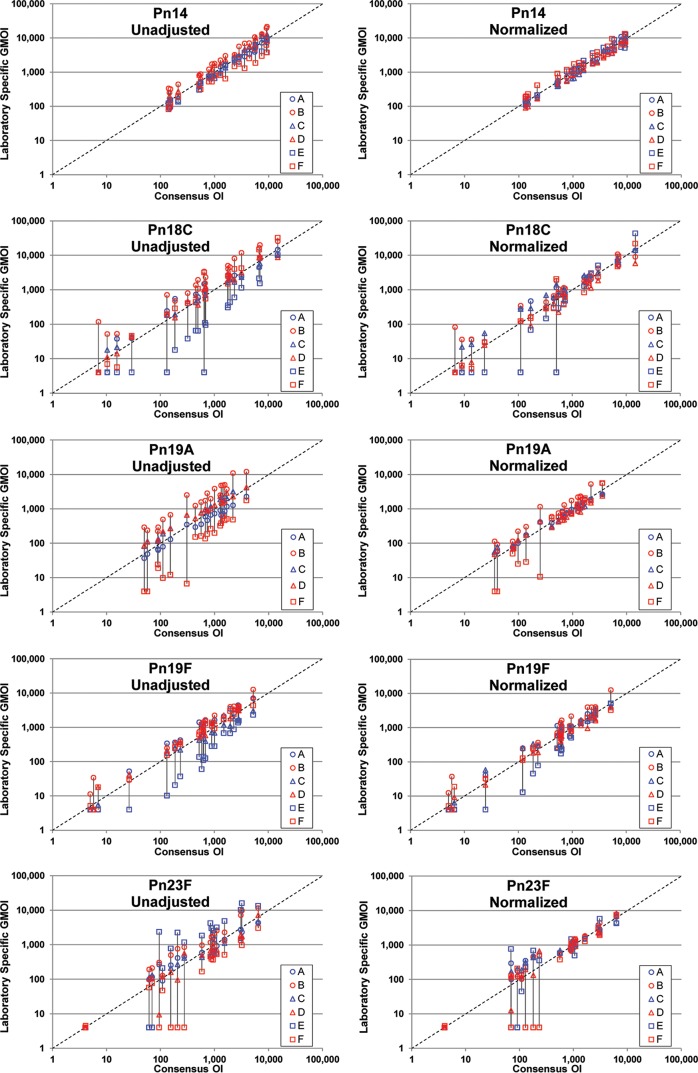
As a visual depiction of the effect of normalization, unadjusted and normalized results are presented graphically in Fig. 2. For the indicated serotype, each panel shows the laboratory-specific GMOIs (y axis; see the Fig. 2 legend) as a function of the overall consensus OI (x axis) for each of the 20 serum samples tested. The left panels display the unadjusted results, and the right panels display the normalized results. The results for each sample are connected by a solid black line for clarity. The effect of normalization can be seen by comparing the length of the vertical lines connecting the data. For most of the serum samples, normalization resulted in shorter lines, indicating better agreement among the laboratories.
FIG 2.
Results of normalization. For the 20 serum samples tested, each plot shows the six laboratory-specific GMOIs (y axis) as a function of the consensus OI (x axis). Unadjusted data are displayed in the left plots, and normalized data are shown in the right plots. The laboratory-specific GMOIs for each sample are connected by a vertical line for visualization. Each plot also has a dashed line indicating identity. Outliers were included in these analyses.
The data in Fig. 2 also indicate that the benefit of normalization may be dependent on the OI. For serum samples with low consensus values, normalization provided little benefit. In fact, in several instances the variability increased for serum samples with low OIs.
The effect of normalization can also be seen by comparing the breadth of the confidence intervals for the calibration serum samples in Table S2, where normalization reduced the confidence interval for most samples and most serotypes. For some serotypes (serotype 19A, for example), the effect is quite striking.
The effect of normalization for each serotype was also assessed by determining the percentage of laboratory-specific GMOIs (with and without normalization) within 2- and 3-fold of the consensus OI for the 16 FDA calibration sera. The resulting percentages combined over the set of laboratories are provided in Table 4. Table 4 also includes the fold range (above and below the overall consensus OIs) needed to encompass 90% of the results. Normalization resulted in consistently higher percentages of results within 2- and 3-fold of the consensus values, with the increase in results within 2-fold being the most pronounced. For example, for serotype 19A, the percentage of estimates within 2-fold increased from 56% to 91%.
TABLE 4.
Frequency-based assessment of effect of normalizationa
| Pn | Unadjusted |
Normalized |
||||
|---|---|---|---|---|---|---|
| % laboratory-specific OIs within 2- and 3-fold of overall consensus value |
Fold (90%) | % laboratory-specific OIs within 2- and 3-fold of overall consensus value |
Fold (90%) | |||
| 2-fold | 3-fold | 2-fold | 3-fold | |||
| 1 | 73 | 92 | 2.90 | 91 | 96 | 1.95 |
| 3 | 60 | 90 | 3.00 | 88 | 96 | 2.13 |
| 4 | 79 | 96 | 2.49 | 99 | 100 | 1.49 |
| 5 | 73 | 97 | 2.40 | 92 | 100 | 1.96 |
| 6A | 89 | 98 | 2.04 | 94 | 99 | 1.76 |
| 6B | 83 | 98 | 2.25 | 96 | 99 | 1.70 |
| 7F | 67 | 81 | 3.53 | 99 | 100 | 1.60 |
| 9V | 86 | 100 | 2.10 | 94 | 99 | 1.90 |
| 14 | 91 | 99 | 1.95 | 100 | 100 | 1.49 |
| 18C | 63 | 77 | 5.30 | 80 | 97 | 2.18 |
| 19A | 56 | 71 | 4.50 | 91 | 95 | 1.80 |
| 19F | 75 | 90 | 3.00 | 82 | 97 | 2.23 |
| 23F | 56 | 73 | 5.11 | 75 | 87 | 3.20 |
The percentage of laboratory-specific OIs within 2- and 3-fold of the overall consensus value is shown. The fold range from the overall consensus needed to include 90% of the values [identified as Fold (90%)] is also shown. Outlier results are included in this analysis. Pn, pneumococcal serotype.
A similar assessment was also performed for each individual laboratory. The percentages for the individual laboratory assessments are presented in Table 5. For each serotype, 80% of the results were within 2-fold of the consensus values with the following exceptions: laboratory B for serotype 19F; laboratory E for serotypes 3, 18C, 19F, and 23F; and laboratory F for serotype 23F.
TABLE 5.
Frequency-based assessment of normalization for individual laboratoriesa
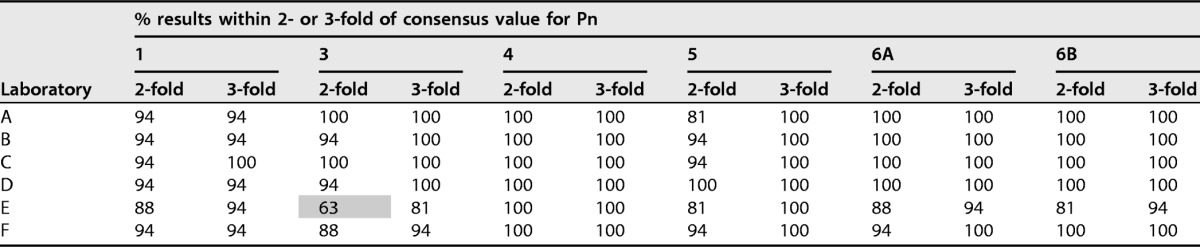
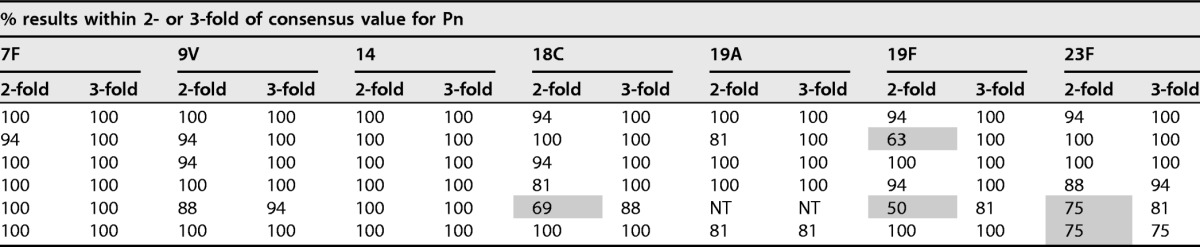
For each laboratory, the percentages of results within 2- and 3-fold of the consensus values listed in Table 2 are indicated. Outliers were included in these analyses. Results of less than 80% for 2-fold are indicated by gray shading. Pn, pneumococcal serotype; NT, not tested.
Effect of normalization on intralaboratory variation.
As a secondary goal of this study, the impact of standardization on intralaboratory variation was also examined. The results for the calibration serum samples from each run were normalized using the results for 007sp from the corresponding run and the consensus OI for 007sp. For each combination of laboratory and serotype, the overall intralaboratory percent CV was calculated with and without normalization, and the reduction in variability due to normalization was determined (Table S3). The results varied considerably, ranging from a 71% reduction in variation (laboratory D, serotype 9V) to a 69% increase in variation (laboratory E, serotype 9V), with no obvious trend for a particular laboratory or a particular serotype being found.
DISCUSSION
In this study, we describe the results of an international collaborative study with two primary goals: (i) to assign consensus values for a pneumococcal OPA standard and the FDA calibration serum samples and (ii) to determine the impact of normalization of the results with the results for the OPA standard.
Although different assay formats and lots of reagents were used in this study, we were able to assign the consensus values for 007sp with relatively narrow confidence intervals, suggesting that pneumococcal OPAs produce consistent results despite variations in assay formats and materials among laboratories. For 007sp, there was more variability associated with the consensus values for serotypes 18C, 19A, and 23F. For serotypes 18C and 19A, the increased variability was mostly driven by one laboratory (laboratory E for 18C and laboratory F for 19A; Table 1 and Fig. 1). The source of the higher variability for serotype 23F is not clear.
Generally, the confidence intervals for the unadjusted results for the FDA calibration serum samples were comparable to those for 007sp. However, normalization substantively reduced the size of the confidence intervals, with most being less than 2-fold. Both model-based and frequency-based approaches suggested a considerable benefit to normalization in regard to interlaboratory agreement. The results of the ANOVA indicated at least a 15% reduction in variability due to normalization with the results for 007sp. For the frequency-based approach, the percentage of results within 2- and 3-fold of the consensus values for the 16 serum samples used for calibration increased substantially (especially within 2-fold of the consensus values) after normalization (Table 4).
However, the data in Fig. 2 indicate that the benefit of normalization may be dependent on the magnitude of the OI. Normalization did not improve the agreement for samples with low OIs, as some laboratories were not able to detect OIs for these samples. Since undetectable values cannot be normalized, normalization provides little benefit for these serum samples with low OIs. Moreover, the confidence intervals of unadjusted estimates for these low-titer serum samples were higher than those for the calibration serum samples. The fact that the serum samples with low OIs are not naturally incurred (i.e., they were created by spiking serum samples with high OIs into a negative matrix) may contribute to the increase in variability. Also, the high degree of variability in these serum samples may be due to the differences in the sensitivity of the assays used by different laboratories.
Applying the frequency-based method to the unadjusted data, the percentage of results within 2-fold of the consensus values varied by serotype, ranging from 56% (serotypes 19A and 23F) to 91% (serotype 14) (Table 4). Although differences among individual serotypes were seen, the average for the 13 serotypes (∼73%) was quite consistent with that found in a previously published study examining the interlaboratory agreement of OPA data (13), which found that approximately 68% of the results were within 2-fold of the consensus values. On the basis of the results of the previously published study and this study, approximately 70% of the results can be expected to be within 2-fold of the consensus value when unadjusted OPA results from different laboratories are compared, especially when multiple assay formats and/or protocols are utilized. However, our data suggest that normalization can improve the overall agreement, with more than 90% (average across all serotypes) of the results being within 2-fold of the consensus value. In the previous study (13), a 4-fold range above and below (16-fold overall) the consensus value was needed to encompass 90% of the results.
As noted in the Materials and Methods section, normalization of the sample data was based on the performance of 007sp on the same assay plate. In order to maximize assay throughput and minimize the consumption of 007sp, two alternate analytical approaches were also evaluated: normalization of the data using the average result for 007sp from all plates in a run and normalization of the data using the result for 007sp from the first plate of a run. Normalization using either the OI for 007sp from the same assay plate or the average OI for the run for 007sp performed similarly in terms of reducing the variation between laboratories (data not shown). However, normalization on the basis of the performance of 007sp on only the first assay plate was less effective in reducing the variation (data not shown). Therefore, ideally, 007sp would be included 2 to 3 times in each run.
As a secondary objective of the study, the effect of normalization on intralaboratory variation was examined. As shown in Table S3 in the supplemental material, the effect of normalization ranged from a 71% decrease in variability to a 69% increase in variability. Although there was no obvious advantage to this approach, the data for this study were collected over a relatively short time frame. Normalization may be useful for improving long-term assay stability when implemented properly. However, normalization should be used cautiously when qualifying new reagents or operators to ensure that it does not mask potential shifts in assay performance.
We see two primary limitations of this study. First, although each participating laboratory was quite experienced with OPAs, the degree and depth of assay validation varied among the participating laboratories. Simply using 007sp as a reference standard does not guarantee that every laboratory will generate comparable results. Second, the range of OIs of the calibration serum samples was limited for some serotypes and generally lacked low OIs.
Based on the data generated in this study, we believe that 007sp may be a useful reference to normalize data across laboratories. We found the most efficient method for normalization to be the use of the consensus value for 007sp and the average of the within-run estimates using the following formula: normalized OI = unadjusted OI × (consensus OI for 007sp/geometric mean OI for 007sp from the run), where the consensus OI for 007sp is from Table 1 and the geometric mean OI is for when 007sp is included 2 to 3 times in each run.
The data also support the use of the 16 FDA calibration sera as part of the initial evaluation of new assays or periodic assessment of established assays. The level of agreement between the laboratory values and the consensus values reported here may help a laboratory evaluate the performance of its assays.
MATERIALS AND METHODS
Participating laboratories.
The locations and the formats used by each of the six laboratories selected to participate in this study are indicated in Table 6. Note that the laboratories are listed alphabetically in Table 6 and this order is not associated with the letter designations (A to F) utilized throughout the article. Participation in the study was limited to laboratories with substantial experience with pneumococcal OPAs that routinely performed the assays.
TABLE 6.
Participating laboratories and assay formatsa
| Institution name | Location | OPA format | Assay reference |
|---|---|---|---|
| Ewha Womans University | Seoul, South Korea | MOPA | 12 |
| GSK Vaccines | Rixensart, Belgium | Singleplex | 11 |
| Lanzhou Institute of Biological Products | Lanzhou, China | MOPA | 12 |
| Pfizer Vaccine Research | Pearl River, NY, USA | Singleplex | 17 |
| UCL Institute of Child Health | London, England | MOPA | 12 |
| University of Alabama at Birmingham | Birmingham, AL, USA | MOPA | 12 |
The affiliation and location of each participating laboratory are indicated. The assay format (including reference) utilized is also indicated. Note that the laboratories are listed here alphabetically and this order is not associated with the letter designations (A to F) utilized throughout this article.
Serum samples.
The production of 007sp and the 16 FDA calibration sera (samples 007A through 007P) have been described previously (14).
Preliminary data (data not shown) indicated that for many serotypes, the panel of serum samples used for calibration lacked samples with relatively low opsonization indexes (OIs; defined below). Four unvaccinated serum samples (15) that were included in those preliminary runs to serve as samples with low OIs produced irregular killing curves for multiple serotypes and thus were not useful for this purpose. So that samples with low OIs could be included in this evaluation, four samples with low OIs were prepared by spiking a calibration serum sample into an immunoglobulin-depleted serum sample (IDS). The IDS, purchased from BBI Solutions (Cardiff, UK), possessed some residual opsonic activity against some serotypes. Additional IgM depletion was performed using affinity chromatography (16) until the residual opsonic activity was removed. One set of four serum samples was prepared for serotypes 4, 6B, 14, and 19A (serum samples S17, S18, S19, and S20); and a second set of four serum samples was prepared for the remaining nine serotypes (serum samples S21, S22, S23, and S24).
Study design.
For the 13 target serotypes, each participating laboratory tested 21 serum samples (007sp, the 16 FDA serum samples used for calibration of the OPA, and the 4 serum samples prepared to have low OIs described above) in 5 separate runs. 007sp was included on every assay plate of every run (except in laboratory E, which included 007sp at least once in each run but not on each plate). For each sample, the OI calculated by the participating laboratory was used for statistical analyses.
OPAs.
All laboratories tested samples for serotypes 1, 3, 4, 5, 6A, 6B, 7F, 9V, 14, 18C, 19A, 19F, and 23F, except for laboratory E, which did not test for serotype 19A. The published reference for the OPA used by each participating laboratory is shown in Table 6. All participating laboratories utilized a killing-type OPA with frozen aliquots of bacteria as targets, baby rabbit serum as a complement source, and differentiated HL60 cells as effectors. The target strains were the same for all laboratories utilizing the MOPA but were different for each of the laboratories using the singleplex format.
For each result, the OI was calculated and tabulated by the participating laboratory. Although different algorithms were utilized to determine the OI, all laboratories defined an OI as the estimated dilution of serum that kills 50% of the target bacteria. Control wells (defining 0% killing) containing target bacteria, complement, and HL60 cells but no test serum were included in every assay. Results that failed to meet a laboratory's assay system suitability criteria were indicated to be indeterminable (Table 7 indicates the numbers and sources of calibration serum samples with indeterminable results).
Statistical analyses.
Within each laboratory, the results for each serum sample were screened for outliers, with outliers being defined as OIs differing by more than 4-fold from the respective laboratory-specific median OI. Results identified as intralaboratory outliers (Table 7) were removed prior to analysis.
To estimate the 007sp consensus OI for each serotype, the log-transformed OIs for 007sp were fit using a mixed-effects ANOVA model containing the random terms Lab, Run(Lab), and Plate(Run × Lab). Normalized OIs for the calibration panel were obtained by dividing the OI for the test sample by the OI for 007sp obtained on the same plate and then multiplying the resultant ratio by the consensus OI for 007sp. To estimate the nonnormalized and the normalized consensus OIs for the calibration samples, the log-transformed OIs and the log-transformed normalized OIs were fit by serotype and sample using a mixed-effects ANOVA model consisting of the random terms Lab and Run(Lab). Consensus OIs and the corresponding 95% CIs for 007sp and the calibration samples were obtained by back-transforming the intercept obtained for the model and its corresponding 95% CI.
For the purpose of estimating the consensus OIs for the calibration samples for future laboratory comparisons, the normalized individual laboratory geometric mean OIs (GMOIs) were screened for outliers relative to the corresponding consensus OI. For each sample, an individual laboratory GMOI was defined to be an interlaboratory outlier if it differed from its corresponding consensus OI by >4-fold. If, for each combination of sample and serotype, more than one laboratory OI was >4-fold from the consensus OI, then only the most extreme difference was identified as the outlier. Interlaboratory outliers are listed in Table 7 and are indicated in red font in Table S1 in the supplemental material.
For individual calibration samples, the percent reduction in interlaboratory variability due to normalization was calculated as follows:
| (1) |
where and denote the between-laboratory and run-within-laboratory variance component estimates for the nonnormalized OIs, respectively, and and , denote the corresponding variance components for the normalized OIs. In cases where the total variability for the normalized OIs exceeded that for the nonnormalized OIs, the percent reduction in interlaboratory variability due to normalization was calculated as follows:
| (2) |
To estimate the percent reduction in interlaboratory variability due to normalization across the set of calibration samples, the log-transformed OIs and normalized OIs were fit using a mixed-effects ANOVA model containing terms for Lab, Sample, Sample × Lab, Run(Lab), and Sample × Run(Lab), where Sample accounts for the differences in the average OI response level among the samples and the other terms are as defined in the footnote to Table 3. Sample was regarded as a fixed effect, and all other terms were regarded as random effects. The percent reduction in interlaboratory variability due to normalization was calculated as follows:
| (3) |
where , , , and denote the between-laboratory, sample-by-laboratory, run-within-laboratory, and sample-by-run-within-laboratory variance component estimates for the nonnormalized OIs, respectively, and , , , and denote the corresponding variance components for the normalized OIs.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by NIH contract HHSN272201200005C (to M.H.N.), 11172MFDS360 (to K.H.K.), and the UK National Institute for Health Research (to D.G.).
The funders had no role in study design, the collection of data, interpretation of results, or the decision to publish the results.
The University of Alabama at Birmingham (UAB) has intellectual property rights to several reagents developed in M.H.N.'s laboratory, and M.H.N. and R.L.B. are UAB employees. D.W. is an employee of the GSK group of companies. D.C. is an employee of Pfizer Vaccine Research. G.L.X. is an employee of the Lanzhou Institute of Biological Products, China. We declare no other conflicts of interest.
This work is dedicated to the memory of Milan Blake, completing his vision of utilizing 007sp as a pneumococcal OPA reference serum sample.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/CVI.00457-16.
REFERENCES
- 1.Yildirim I, Shea KM, Pelton SI. 2015. Pneumococcal disease in the era of pneumococcal conjugate vaccine. Infect Dis Clin North Am 29:679–697. doi: 10.1016/j.idc.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jodar L, Butler JC, Carlone G, Dagan R, Frasch CE, Goldblatt D, Käyhty H, Klugman K, Plikaytis BD, Siber G, Kohberger R, Chang I, Cherian T. 2003. Serological criteria for evaluation and licensure of pneumococcal conjugate vaccine formulations for use in infants. Vaccine 21:3265–3272. doi: 10.1016/S0264-410X(03)00230-5. [DOI] [PubMed] [Google Scholar]
- 3.Wernette CM, Frasch CE, Madore D, Carlone G, Goldblatt D, Plikaytis B, Benjamin W, Quataert SA, Hildreth S, Sikkema DJ, Kayhty H, Jonsdottir I, Nahm MH. 2003. Enzyme-linked immunosorbent assay for quantitation of human antibodies to pneumococcal polysaccharides. Clin Diagn Lab Immunol 10:514–519. doi: 10.1128/CDLI.10.4.514-519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romero-Steiner S, Musher DM, Cetron MS, Pais LB, Groover JE, Fiore AF, Plikaytis BD, Carlone GM. 1999. Reduction in functional antibody activity against Streptococcus pneumoniae in vaccinated elderly individuals highly correlates with decreased IgG antibody avidity. Clin Infect Dis 29:281–288. doi: 10.1086/520200. [DOI] [PubMed] [Google Scholar]
- 5.Schenkein JG, Park S, Nahm MH. 2008. Pneumococcal vaccination in older adults induces antibodies with low opsonic capacity and reduced antibody potency. Vaccine 26:5521–5526. doi: 10.1016/j.vaccine.2008.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winkelstein JA. 1981. The role of complement in the host's defense against Streptococcus pneumoniae. Rev Infect Dis 3:289–298. doi: 10.1093/clinids/3.2.289. [DOI] [PubMed] [Google Scholar]
- 7.Gray BM. 1990. Opsonophagocidal activity in sera from infants and children immunized with Haemophilus influenzae type b conjugate vaccine (meningococcal protein conjugate). Pediatrics 85:694–697. [PubMed] [Google Scholar]
- 8.Nahm MH, Olander JV, Magyarlaki M. 1997. Identification of cross-reactive antibodies with low opsonophagocytic activity for Streptococcus pneumoniae. J Infect Dis 176:698–703. doi: 10.1086/514093. [DOI] [PubMed] [Google Scholar]
- 9.Romero-Steiner S, Libutti D, Pais LB, Dykes J, Anderson P, Whitin JC, Keyserling HL, Carlone GM. 1997. Standardization of an opsonophagocytic assay for the measurement of functional antibody activity against Streptococcus pneumoniae using differentiated HL-60 cells. Clin Diagn Lab Immunol 4:415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu BT, Yu X, Jones TR, Kirch C, Harris S, Hildreth SW, Madore DV, Quataert SA. 2005. Approach to validating an opsonophagocytic assay for Streptococcus pneumoniae. Clin Diagn Lab Immunol 12:287–295. doi: 10.1128/CDLI.12.2.287-295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henckaerts I, Durant N, De Grave D, Schuerman L, Poolman J. 2007. Validation of a routine opsonophagocytosis assay to predict invasive pneumococcal disease efficacy of conjugate vaccine in children. Vaccine 25:2518–2527. doi: 10.1016/j.vaccine.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 12.Burton RL, Nahm MH. 2006. Development and validation of a fourfold multiplexed opsonization assay (MOPA4) for pneumococcal antibodies. Clin Vaccine Immunol 13:1004–1009. doi: 10.1128/CVI.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rose CE, Romero-Steiner S, Burton RL, Carlone GM, Goldblatt D, Nahm MH, Ashton L, Haston M, Ekstrom N, Haikala R, Kayhty H, Henckaerts I, Durant N, Poolman JT, Fernsten P, Yu X, Hu BT, Jansen KU, Blake M, Simonetti ER, Hermans PW, Plikaytis BD. 2011. Multilaboratory comparison of Streptococcus pneumoniae opsonophagocytic killing assays and their level of agreement for the determination of functional antibody activity in human reference sera. Clin Vaccine Immunol 18:135–142. doi: 10.1128/CVI.00370-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldblatt D, Plikaytis BD, Akkoyunlu M, Antonello J, Ashton L, Blake M, Burton R, Care R, Durant N, Feavers I, Fernsten P, Fievet F, Giardina P, Jansen K, Katz L, Kierstead L, Lee L, Lin J, Maisonneuve J, Nahm MH, Raab J, Romero-Steiner S, Rose C, Schmidt D, Stapleton J, Carlone GM. 2011. Establishment of a new human pneumococcal standard reference serum, 007sp. Clin Vaccine Immunol 18:1728–1736. doi: 10.1128/CVI.05252-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plikaytis BD, Goldblatt D, Frasch CE, Blondeau C, Bybel MJ, Giebink GS, Jonsdottir I, Kayhty H, Konradsen HB, Madore DV, Nahm MH, Schulman CA, Holder PF, Lezhava T, Elie CM, Carlone GM. 2000. An analytical model applied to a multicenter pneumococcal enzyme-linked immunosorbent assay study. J Clin Microbiol 38:2043–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park S, Nahm MH. 2011. Older adults have a low capacity to opsonize pneumococci due to low IgM antibody response to pneumococcal vaccinations. Infect Immun 79:314–320. doi: 10.1128/IAI.00768-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper D, Yu X, Sidhu M, Nahm MH, Fernsten P, Jansen KU. 2011. The 13-valent pneumococcal conjugate vaccine (PCV13) elicits cross-functional opsonophagocytic killing responses in humans to Streptococcus pneumoniae serotypes 6C and 7A. Vaccine 29:7207–7211. doi: 10.1016/j.vaccine.2011.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.