Abstract
Cochlear sensory hair cells (HCs) are crucial for hearing as mechanoreceptors of the auditory systems. Clarification of transcriptional regulation for the cochlear sensory HC development is crucial for the improvement of cell replacement therapies for hearing loss. Transcription factor Atoh1 is the key player during HC development and regeneration. In this review, we will focus on Atoh1 and its related signaling pathways (Notch, fibroblast growth factor, and Wnt/β-catenin signaling) involved in the development of cochlear sensory HCs. We will also discuss the potential applicability of these signals for the induction of HC regeneration.
Keywords: Hair Cells, Auditory; Regeneration; Transcription Factors; Hearing Loss
INTRODUCTION
Hearing loss represents a major health problem. It affects over 5% of the world's population. The World Health Organization (WHO) has estimated that 360 million people in the world are suffering from hearing impairment (WHO, 2013). As people's life spans are extended compared to those in the past, the incidence of sensorineural hearing loss (SNHL) has also increased.1 SNHL can arise from several causes including aging, noise trauma, ototoxic drugs, genetic disorders, and infections. This disability can influence an individual's social, functional, and psychological well-being. It can cause isolation, dependence, frustration, and communication problems.2 SNHL is commonly caused by cochlear sensory hair cell (HC) defects, consequently leading to auditory spiral ganglion neuron loss.3 In mammals, hearing impairment due to sensory HC loss in the cochlea is irreversible because lost cochlear hair cells cannot be spontaneously regenerated.4,5 Current treatments for SNHL such as hearing aids, middle ear implants, and cochlear implants do provide benefits to patients. However, their outcomes are diverse.6,7 Recently, there has been a strong research focus on alternative treatments for SNHL. Understanding the transcriptional regulation mechanism involved in sensory HCs development and specification has led to a new opening of strategies for treating SNHL by cell or gene therapies.8 Here, we will review a key transcriptional factor (Atoh1) and its related signaling pathway (Notch, fibroblast growth factor, and Wnt/β-catenin signaling) involved in the development of cochlear sensory HCs. We will also discuss the potential applicability of these signals for the induction of HC regeneration.
THE SENSORY EPITHELIUM OF THE COCHLEA
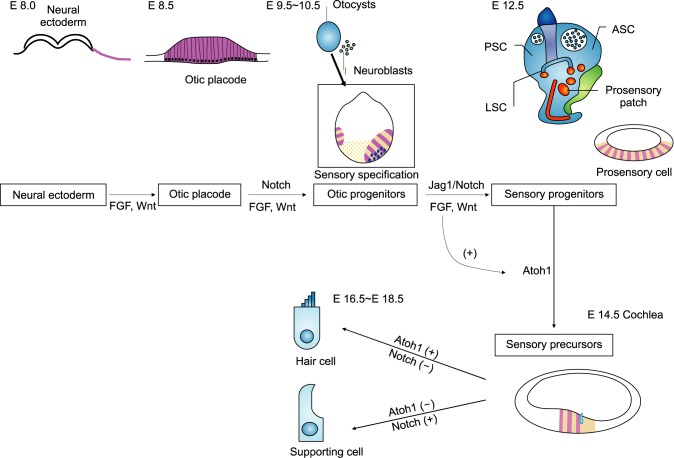
In mammals, hearing and balance are regulated by sensory HCs located in the cochlear duct and vestibular organs of the inner ear. Cochlear sensory hair cells are mechanosensory receptors. They convert sound into electrical signals.9 There are approximately 16,000 sensory HCs located in the organ of Corti. The inner HCs are arranged in a single row and outer HCs are arranged in three rows (Fig. 1). Inner HCs excite type I spiral ganglion neurons. A single inner HC makes synaptic connection with numerous type I afferent nerve fibers. Outer HCs enhance the responsiveness of the sensory epithelium to specific frequencies and detect low intensity sound.10 They are innervated by type II afferent nerve fibers. A single afferent nerve fiber can innervate many outer HCs.11 These complex cochlear sensory systems are formed via delicate cell fate decision and differentiation processes tightly controlled by a combination of numerous intrinsic and extrinsic signaling pathways.12 The specification of prosensory regions in the otocyst is the initial step during the development of sensory epithelia in the inner ear. During the development of the prosensory region, numerous intrinsic and extrinsic signaling pathways are required. These singling pathways are highly ordered to generate proper functioning HCs.13,14 Numerous genes are related to sensory inner ear HC specification/differentiation, such as Atonal homolog 1 (Atoh1; Math1), growth factor independent 1 (Gfi1), Pou4f3, miRNA-183,15 and connexin26 gene (GJB2).16,17 Among these potential genes for HC specification and regeneration, Atoh1 is a ‘key gene’ in HC development because its activity at different developmental periods can sufficiently induce cochlear HC differentiation.2
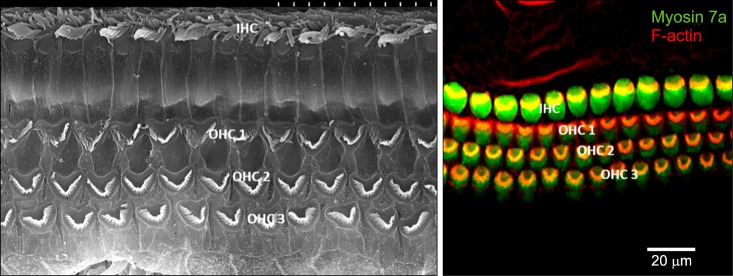
FIG. 1. Sensory hair cells in organ of Corti. Inner hair cells are arranged in one row on the medial side of the organ of Corti. Outer hair cells are arranged in three rows on the lateral side. Left side figure is scanning electron microscopy image and right side figure is immunofluorescent image. IHC: inner hair cell, OHC: outer hair cell.
THE ROLE OF ATOH1 IN INNER EAR HC DEVELOPMENT
Atoh1 is a proneural basic helix-loop-helix (bHLH) transcription factor. It is the first gene discovered in inner ear development.18 Several studies have revealed that Atoh1 regulates not only the development of various kinds of neural cells such as the spinal cord,19 inner ear HCs,20,21,22 cerebellar neurons,23 and proprioceptive system neurons,24,25 but also several types of nonneural cells such as intestinal secretory cells.26,27 During inner ear development, Atoh1 has a key role in HC differentiation. Atoh1 expression represents the prosensory region in the development of cochlea. Atoh1 positive cells can generate sensory HCs.28 In mice, the organ of Corti develops from a postmitotic prosensory domain in the cochlear duct formed between embryonic days (E) 12.5 and E14.29,30 Atoh1 is first expressed in the base of the cochlea at E 12.5. It is up-regulated in primitive HCs in the mid-basal region between E13.5 and E 14.5 in mice. During inner ear development in mice, it spreads apically along the prosensory domain until modelling of organ of Corti is complete around E17.5.31 In addition, Atoh1 is essential for HC survival shortly after differentiation between E15.5 to E17.5. It is required for HC maturation, including the formation of stereociliary bundles (Fig. 2).32,33 On the other hand, Atoh1 ectopic expression could generate diverse phenotypes of cochlea. In the embryonic stage, overexpressed Atoh1 can form supernumerary cochlear sensory HCs.34 In neonatal mice, Atoh1 overexpression can form HCs from supporting cells.35,36,37,38,39,40 However, such ectopic HC generation ability of Atoh1 is significantly reduced in mature or undamaged cochlea.36,41 Many molecules including Pou4f3, Gfi1, Myosin 7a were suggested to be down-stream genes of Atoh1 by transcriptome analysis. However, the concise role of these genes in the context of Atoh1 work is yet to be defined.
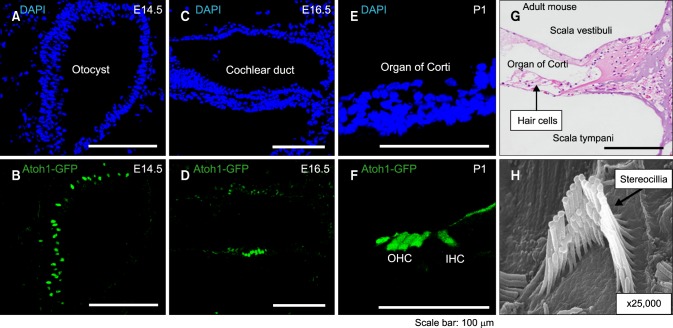
FIG. 2. Atoh1 gene expression during inner ear development. Atoh1 is expressed in the proneuronal cells at E14.5 (A, B) and expressed in sensory hair cells in the cochlea at E16.5 and P1 (C-F). In adult mammals, hair cells are located in the organ of Corti (G, H&E staining) and have stereocillia (H, scanning electro-microscopic image). OHC: outer hair cell, IHC: inner hair cell.
Because Atoh1 has a crucial role in the generation of cochlear HCs, many cochlear HC regeneration studies have focused on Atoh15,42,43 However, these studies have limitations in that the efficacy of Atoh1 might be variable and regenerated HC did not function properly.42,43 Although Atoh1 itself is insufficient to regenerate functional HCs, these studies have served as a foundation for other studies to determine candidate signaling pathways that can control Atoh1 expression and HC differentiation. Those signaling pathways are discussed below.
ATOH1 RELATED SIGNALING PATHWAYS INVOLVED IN HC DEVELOPMENT AND REGENERATION
1. Notch signaling (Fig. 3)
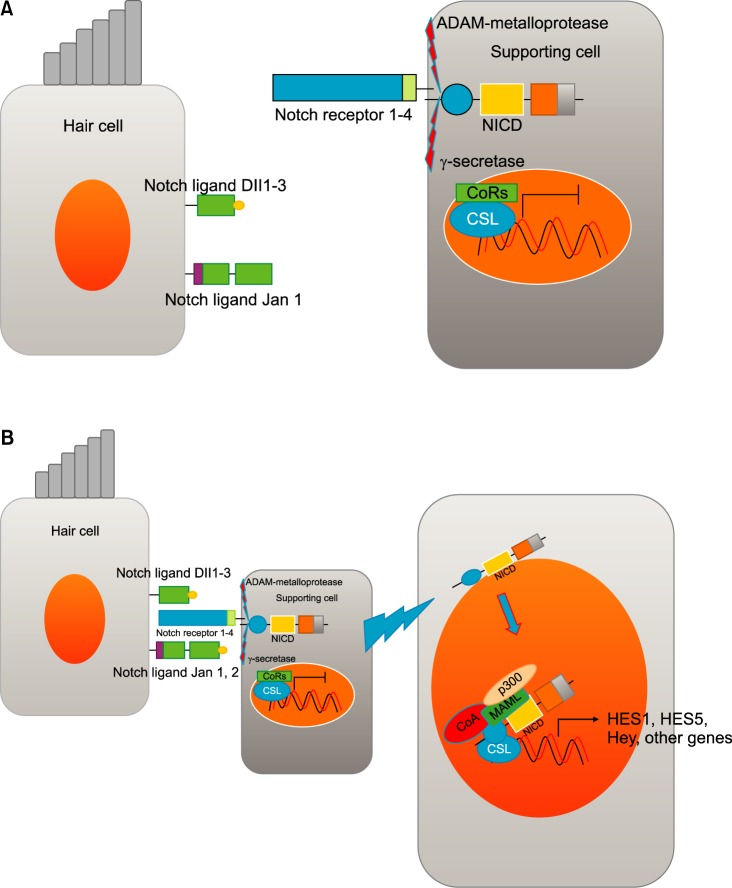
FIG. 3. Notch signaling during mammalian hair cell development. (A) There are 5 notch ligands (Dll1, Dl12, Dll3, jag 1, jag2) in hair cell and 4 notch receptors in the supporting cells. γ-secretase is an internal protease that cleaves the NICD. (B) Notch signaling is activated upon cell-to-cell contact. Downstream genes (Hes1, 5, Hey, other genes) are produced by Notch signaling. These produced inhibitory basic helix-loop-helix proteins (HES1, HES5) blocking the effects of prosenosry genes (Atoh1) that leads to inhibition of the hair cell fate. These inhibited cells differentiate into supporting cells. Dll: delta-like ligand, NICD: notch intracellular domain, CoRs: co-repressor, CSL: CBF1, Su(H) and LAG-1,CoA: co-activator, MAML: mastermind ligand.
In many tissues, notch signaling is one of the most important pathways that regulates cell differentiation, proliferation, and cell death by lateral inhibition. Notch receptors and ligands Delta and Serrate (Jagged in mammals) are trans-membrane receptor proteins highly expressed in many tissues.44 The Notch trans-membrane receptor molecule is activated via direct interaction with an adjacent cell surface trans-membrane ligand. This interaction promotes cleavage of Notch receptors to release notch intracellular domain (NICD). The released NICD is translocated to the nucleus to bind to RBP-JK family and act as a transcriptional coactivator. This transcriptional coactivator activates the expression of target genes such as Hes1, leading to repression of downstream target genes.
During the differentiation of HCs and supporting cells, Notch signaling plays an important role by lateral inhibition. In mammals, there are four notch receptors (Notch 1-4) and five canonical notch ligands (Delta-like 1, 3, 4 and Jagged 1, 2).45 Many studies have demonstrated that Notch ligands are expressed in developing HCs and that Notch receptors are expressed more broadly than Notch ligands.46,47,48,49 Notch signaling is activated upon cell-to-cell contact, which is an initial step of lateral inhibition. Notch signaling produces inhibitory bHLH proteins (HES1 and HES5) that block the effect of Atoh1, leading to inhibition of the HC fate. These inhibited cells subsequently differentiate into supporting cells. Individual deletion of notch ligand Delta-like (Dl1) or Jagged 2 results in a significant increase in sensory HCs.49,50
Downstream molecules of Notch signaling during hair cell differentiation are products of Hairy and Enhancer- of-Split genes in Drosophila. In vertebrates, bHLH transcription factor family genes, Hes/Hey are homologs of Hairy and Enhancer-of-split genes.51 Several studies have revealed that Hes/Hey family members are expressed in supporting cells and that Hes/Hey deletions promote hair cell differentiation.52,53,54,55 Interestingly, Hes/Hey family members are individually expressed in different part of the cochlea. They may control different supporting cell populations. For example, overproduction of inner HCs has occurred via Hes1 inactivation whereas overproduction of outer HCs can be caused by Hes5 deletion.53 Several studies have also revealed that many Hes/Hey family members and other downstream factors of Notch are effectors in lateral inhibition.55,56
Numerous studies have shown that Notch inhibition can affect hair cell regeneration. The most commonly studied drugs used to block Notch signaling are γ-secretase inhibitors.57,58 These studies have demonstrated that γ-secretase inhibitors can increase the number of regenerated HCs in damaged zebrafish lateral line57 or damaged chick cochlea.58 In addition, Notch inhibition could change supporting cells to new nascent HCs in undamaged neonatal mouse cochlea.59,60 In mammalian vestibular organs, Notch signaling inhibition could significantly induce more supporting cells to activate Atoh1 gene expression and change supporting cells to mature myosin VIIa-expressing HCs after vestibular HCs damage.61,62,63 However, in damaged, mature mammalian cochlea, pharmacological trials using Notch signaling inhibitors have been unsatisfactory.64,65 Taken together, these results imply that Notch inhibition is a promising strategy for HC regeneration in damaged cochlea.
2. Fibroblast growth factor (FGF)
Currently, 22 FGF ligand genes and 4 FGF receptor genes have been identified in mammals. They play important roles during the development of CNS66 and multiple sensory organs.67,68 During inner ear development, FGF signaling regulates otic placode induction69 and early otocyst formation.68 In the later stages during inner ear development, FGF family also regulates the formation of auditory HCs. In particular, FGF20 signals through FGF receptor 1 to promote early HC specification in mice developing cochlea and treatment with FGF signaling inhibitors decreases Atoh1 expression at E14.70,71 After initiating HC differentiation, interaction between FGF8 in nascent inner HCs and FGF receptor 3 in supporting cells is essential for the development of pillar cells (which are found in the organ of Corti and act as supporting cells for HCs) and inhibition of such interaction leads to the formation of excess ectopic HCs and a decrease of supporting cells.72,73,74,75 Interestingly, Notch inhibition could not convert pillar cells into HCs because they are specified by notch independent Hey2 expression which is activated by FGF signaling, unlike other supporting cell subtypes. Therefore, FGF receptor blockage allows pillar cells to be more responsive to Notch inhibition.76
Furthermore, FGF signaling pathway is related to Atoh1 during inner ear development.77 For example, in the presence of FGF2, several neurotropic factors (NGF, BDNF and NT-3) can significantly increase Atoh1 expressions.78 In zebrafish, FGF3 and FGF8 signals are upstream activators of Atoh1 genes during the development of preotic placode and otic vesicle.79 FGF20 also indirectly activates Atoh1 expression through FGF receptor 1 and activates Ets-domain transcription factors PEA3 and ERM, which then can activate Atoh1 expression.70
Collectively, these studies reveal that FGF signaling could control the specification of prosensory cells and subsequent differentiation into HCs and supporting cells during inner ear development. In addition, the FGF signaling pathway interacts with the Notch signaling pathway during cochlear development and controls Atoh1 expression.
3. Wnt/β-catenin signaling
The Wnt (Wingless-related integration site) signaling pathway is composed of a set of secreted factors (Wnts) and Frizzled receptors. There are 19 different Wnt genes in murine and human genomes.80 The Wnt signaling pathway is a hallmark of many embryonic stages with multiple different roles at each time point. In the inner ear, it participates in otic specification, vestibular organ formation, and development of the cochlea.
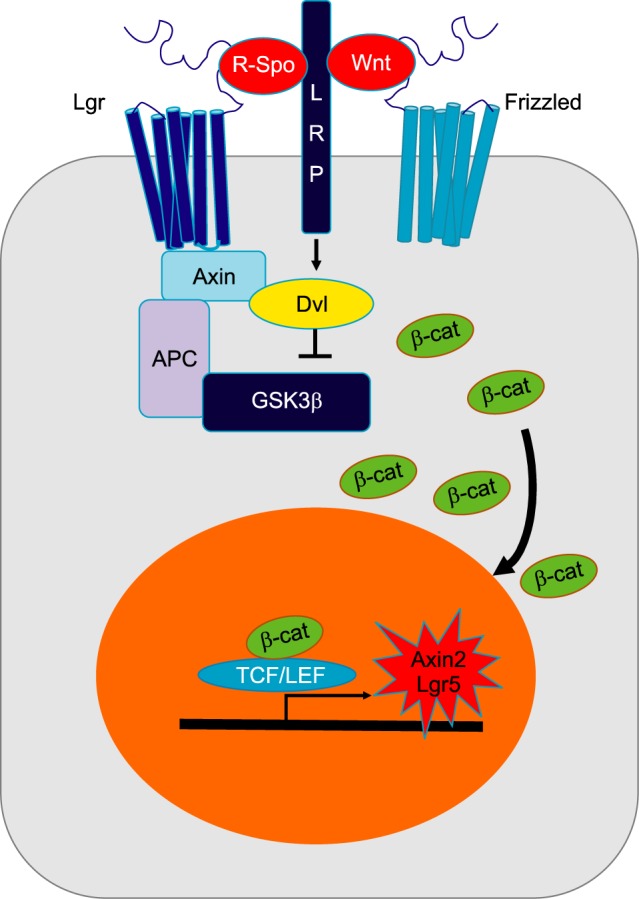
In cochlear development, active canonical Wnt signaling is mediated by β-catenin translocated into the nucleus. It can bind the T cell factor/lymphoid enhancer-binding factor (TCF/LEF) family of transcription factors to initiate transcription (Fig. 4). This canonical Wnt/β-catenin signaling is known to control both development and homeostasis in many tissues.81 Canonical Wnt/β-catenin signaling has been proposed to be upstream of both Atoh1 and Sox2 during cochlear HC and retina cell development.82,83 During otic placode development, a gradient of Wnt signaling determines the size of otic placode. The Wnt pathway also interacts with the notch pathway.84 Recently, it has been reported that canonical Wnt/β-catenin signaling is upregulated in cochlear prosensory cells and that inhibition of Wnt/β-catenin signaling blocks the development of prosensory cells during early cochlear development.85 Furthermore, Wnt signaling activator LiCl can expand Sox2-expressing prosensory cells and increase sensory HC number. Another experiment using transgenic mice revealed that β-catenin deletion at different developmental stages resulted in different phenotypes. When β-catenin is deleted during the prosensory period, differentiation of both HCs and pillar cells was prevented. However, when β-catenin was deleted after sensory HC differentiation in the cochlea, it failed to affect HC maturation.86 These results suggest that Wnt/β-catenin signaling is indispensable for prosensory cell proliferation and sensory HC differentiation but not for sensory HC maturation. In addition, Wnt/β-catenin activation can drive G-protein-coupled receptor 5 (Lgr5)-expressing cells to act as HC progenitors in neonatal cochlea.87,88
FIG. 4. Wnt/β-catenin signaling. Without Wnt ligands, GSK3β complex destructs β-catenin (β-cat) by phosphorylation. This signaling is activated when Wnt ligands bind to Fizzled receptors and their co-receptor lipoprotein receptor-related protein (LRP) 5/6. Subsequently, this lead to sequestration of Axin, recruitment of Disheveled (Dvl), and the release of β-cat. Frizzled receptor is stabilized by attachment of R-spondins (R-spo) to Lgr4/5/6 receptor. Released β-cat is translocated into the nucleus. Ant it will bind to T cell factor/lymphoid enhancer-binding factor (TCF/LEF) family to release Wnt target genes, such as Axin2 and Lgr5.
Although many studies have determined the roles of canonical Wnt signaling during mammalian cochlea development, only a few studies have begun to elucidate its potential for regenerating HCs in damaged cochlea. As previously described, the Lgr5 positive cell is one candidate cell for HC regeneration in damaged cochlea. It requires a Wnt signaling agonist to acquire proliferative behavior. Over-expression of β-catenin can also induce ectopic HC formation in the cochlea.85,86 HC damage can stimulate Lgr5 positive supporting cells to proliferate and regenerate HCs or directly transdifferentiate into HCs.89 During HC regeneration, Wnt/β-catenin signaling can promote HC regeneration by increasing cell proliferation and Atoh1 expression (Fig. 5).87,88,90 More recently, Kuo et al. have reported that β-catenin and Atoh1 have synergistic effects on both cochlear HC generation and survival in neonatal mice.91 However, there have been no mitogenic or prosensory effects after damage in the supporting cells of adult mouse cochlea.87
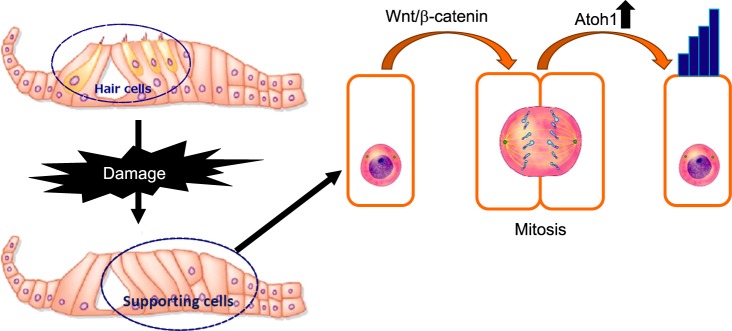
FIG. 5. The proposed Role of Wnt/β-catenin signaling in hair cell regeneration. Wnt/β-catenin signaling can increase hair cell regeneration by stimulating cell mitoic proliferation and up-regulating Atoh1 expression.
In summary, these studies demonstrate that activation of Wnt/β-catenin signaling can force neonatal cochlear supporting cell proliferation and lead to HC regeneration. Therefore, it is reasonable to establish a regenerative strategy by initial Wnt activation to enhance supporting cell proliferation, followed by prosensory regenerating signals such as notch inhibition and forced Atoh1 expression to induce HC differentiation.92 Because the interaction between Notch and Wnt pathways in inner ear is limited,59 more studies are required to further reveal the mechanisms involved in cochlear HC regeneration.
4. Transcriptional profiling of mESCs after forced Atoh1 expression
Several studies have attempted to find down-stream molecules of Atoh1. We also conducted this work using Atoh1 overexpressing (doxycycline inducible system) embryonic stem cells (ESCs) as follows. ESCs were spontaneously differentiated for 2 days followed by Atoh1 over-expression for another 2 days. Cells were collected and transcriptome changes were evaluated by RNA-sequencing. Gene ontology analysis showed several up-regulated pathways such as Notch signaling, neuron migration, glutamate receptor, action potential regulation, and neurotransmitter regulation (unpublished data). On the other hand, BMP signaling, Wnt signaling, and mesoderm formation related genes were down-regulated by Atoh1 overexpression. In similar contexts with previous transcriptome studies,93,94 these pathways might play a role in the hair cell development.
SUMMARY
Numerous genes are related to sensory inner ear HC specification and differentiation. Among these genes, Atoh1 is a ‘key gene’ in HC development. At the otic placode stage, FGF signaling regulates its induction as well as early otocyst formation. Also, FGF20 signal promotes early HC specification via increasing Atoh1 expression at E12-E14. Wnt/β-catenin signaling has been proposed to be upstream of Atoh1 during cochlear development. It can increase mitotic HC regeneration by promoting cell proliferation and increasing Atoh1 expression during HC regeneration. During differentiation of HCs and supporting cells, Notch signaling plays an important role mainly via lateral inhibition. In addition, Notch inhibition can induce HC regeneration. Moreover, Notch inhibition can significantly increase Atoh1 expression (Fig. 6). Taken together, these results imply that Atoh1 and its related signaling pathway genes might offer promising approaches for HC regeneration in damaged cochlea.
FIG. 6. Factors that direct sensory hair cell fate in the inner ear. Beginning at otic placode stages, FGF signaling regulates otic placode development as well as early stage of otocyst formation. A gradient of Wnt signaling determines the size of otic placode. Wnt pathway interacts with notch pathway. FGF signals are upstream activators of Atoh1 genes during the development of prosensory cells. Final sensory hair cell development is regulated by the expression of Atoh1. Supporting cell development is activated by the expression of notch pathway.
ACKNOWLEDGEMENTS
This research was supported by a grant (NRF-2014-R1A1A2054755) of the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by Ministry of Education, Science and Technology. It was also supported by a grant (2013-CURIMS-DR001) from the Research Institute of Medical Science, Chonnam National University.
Footnotes
CONFLICT OF INTEREST STATEMENT: None declared.
References
- 1.Richardson RT, Atkinson PJ. Atoh1 gene therapy in the cochlea for hair cell regeneration. Expert Opin Biol Ther. 2015;15:417–430. doi: 10.1517/14712598.2015.1009889. [DOI] [PubMed] [Google Scholar]
- 2.Hongmiao R, Wei L, Bing H, Xiong DD, Jihao R. Atoh1: landscape for inner ear cell regeneration. Curr Gene Ther. 2014;14:101–111. doi: 10.2174/1566523214666140310143407. [DOI] [PubMed] [Google Scholar]
- 3.McFadden SL, Ding D, Jiang H, Salvi RJ. Time course of efferent fiber and spiral ganglion cell degeneration following complete hair cell loss in the chinchilla. Brain Res. 2004;997:40–51. doi: 10.1016/j.brainres.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 4.Groves AK. The challenge of hair cell regeneration. Exp Biol Med (Maywood) 2010;235:434–446. doi: 10.1258/ebm.2009.009281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Izumikawa M, Minoda R, Kawamoto K, Abrashkin KA, Swiderski DL, Dolan DF, et al. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med. 2005;11:271–276. doi: 10.1038/nm1193. [DOI] [PubMed] [Google Scholar]
- 6.Blamey P, Artieres F, Başkent D, Bergeron F, Beynon A, Burke E, et al. Factors affecting auditory performance of postlinguistically deaf adults using cochlear implants: an update with 2251 patients. Audiol Neurootol. 2013;18:36–47. doi: 10.1159/000343189. [DOI] [PubMed] [Google Scholar]
- 7.Blamey PJ, Dooley GJ, Parisi ES, Clark GM. Pitch comparisons of acoustically and electrically evoked auditory sensations. Hear Res. 1996;99:139–150. doi: 10.1016/s0378-5955(96)00095-0. [DOI] [PubMed] [Google Scholar]
- 8.Costa A, Sanchez-Guardado L, Juniat S, Gale JE, Daudet N, Henrique D. Generation of sensory hair cells by genetic programming with a combination of transcription factors. Development. 2015;142:1948–1959. doi: 10.1242/dev.119149. [DOI] [PubMed] [Google Scholar]
- 9.Savary E, Sabourin JC, Santo J, Hugnot JP, Chabbert C, Van De Water T, et al. Cochlear stem/progenitor cells from a postnatal cochlea respond to Jagged1 and demonstrate that notch signaling promotes sphere formation and sensory potential. Mech Dev. 2008;125:674–686. doi: 10.1016/j.mod.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Pickles JO. An Introduction to the Physiology of Hearing. London: Academic Press; 1988. [Google Scholar]
- 11.Spoendlin H. In: Basic Mechanisms in Haring. Moller AR, editor. New York: Academic Press; 1988. [Google Scholar]
- 12.Kelley MW. Regulation of cell fate in the sensory epithelia of the inner ear. Nat Rev Neurosci. 2006;7:837–849. doi: 10.1038/nrn1987. [DOI] [PubMed] [Google Scholar]
- 13.Dabdoub A, Puligilla C, Jones JM, Fritzsch B, Cheah KS, Pevny LH, et al. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci U S A. 2008;105:18396–18401. doi: 10.1073/pnas.0808175105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zine A, de Ribaupierre F. Notch/Notch ligands and Math1 expression patterns in the organ of Corti of wild-type and Hes1 and Hes5 mutant mice. Hear Res. 2002;170:22–31. doi: 10.1016/s0378-5955(02)00449-5. [DOI] [PubMed] [Google Scholar]
- 15.Pauley S, Kopecky B, Beisel K, Soukup G, Fritzsch B. Stem cells and molecular strategies to restore hearing. Panminerva Med. 2008;50:41–53. [PMC free article] [PubMed] [Google Scholar]
- 16.Denoyelle F, Marlin S, Weil D, Moatti L, Chauvin P, Garabédian EN, et al. Clinical features of the prevalent form of childhood deafness, DFNB1, due to a connexin-26 gene defect: implications for genetic counselling. Lancet. 1999;353:1298–1303. doi: 10.1016/S0140-6736(98)11071-1. [DOI] [PubMed] [Google Scholar]
- 17.Kelsell DP, Dunlop J, Stevens HP, Lench NJ, Liang JN, Parry G, et al. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature. 1997;387:80–83. doi: 10.1038/387080a0. [DOI] [PubMed] [Google Scholar]
- 18.Chen P, Johnson JE, Zoghbi HY, Segil N. The role of Math1 in inner ear development: Uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 2002;129:2495–2505. doi: 10.1242/dev.129.10.2495. [DOI] [PubMed] [Google Scholar]
- 19.Lai HC, Klisch TJ, Roberts R, Zoghbi HY, Johnson JE. In vivo neuronal subtype-specific targets of Atoh1 (Math1) in dorsal spinal cord. J Neurosci. 2011;31:10859–10871. doi: 10.1523/JNEUROSCI.0445-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, et al. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- 21.Mulvaney J, Dabdoub A. Atoh1, an essential transcription factor in neurogenesis and intestinal and inner ear development: function, regulation, and context dependency. J Assoc Res Otolaryngol. 2012;13:281–293. doi: 10.1007/s10162-012-0317-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stone JS, Choi YS, Woolley SM, Yamashita H, Rubel EW. Progenitor cell cycling during hair cell regeneration in the vestibular and auditory epithelia of the chick. J Neurocytol. 1999;28:863–876. doi: 10.1023/a:1007022205821. [DOI] [PubMed] [Google Scholar]
- 23.Klisch TJ, Xi Y, Flora A, Wang L, Li W, Zoghbi HY. In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proc Natl Acad Sci U S A. 2011;108:3288–3293. doi: 10.1073/pnas.1100230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helms AW, Johnson JE. Progenitors of dorsal commissural interneurons are defined by MATH1 expression. Development. 1998;125:919–928. doi: 10.1242/dev.125.5.919. [DOI] [PubMed] [Google Scholar]
- 25.Rose MF, Ahmad KA, Thaller C, Zoghbi HY. Excitatory neurons of the proprioceptive, interoceptive, and arousal hindbrain networks share a developmental requirement for Math1. Proc Natl Acad Sci U S A. 2009;106:22462–22467. doi: 10.1073/pnas.0911579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerbe F, van Es JH, Makrini L, Brulin B, Mellitzer G, Robine S, et al. Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J Cell Biol. 2011;192:767–780. doi: 10.1083/jcb.201010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 28.Driver EC, Sillers L, Coate TM, Rose MF, Kelley MW. The Atoh1-lineage gives rise to hair cells and supporting cells within the mammalian cochlea. Dev Biol. 2013;376:86–98. doi: 10.1016/j.ydbio.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YS, Liu F, Segil N. A morphogenetic wave of p27Kip1 transcription directs cell cycle exit during organ of Corti development. Development. 2006;133:2817–2826. doi: 10.1242/dev.02453. [DOI] [PubMed] [Google Scholar]
- 30.Ruben RJ. Development of the inner ear of the mouse: a radioautographic study of terminal mitoses. Acta Otolaryngol. 1967;(Suppl 220):1–44. [PubMed] [Google Scholar]
- 31.Stojanova ZP, Kwan T, Segil N. Epigenetic regulation of Atoh1 guides hair cell development in the mammalian cochlea. Development. 2015;142:3529–3536. doi: 10.1242/dev.126763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai T, Seymour ML, Zhang H, Pereira FA, Groves AK. Conditional deletion of Atoh1 reveals distinct critical periods for survival and function of hair cells in the organ of Corti. J Neurosci. 2013;33:10110–10122. doi: 10.1523/JNEUROSCI.5606-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chonko KT, Jahan I, Stone J, Wright MC, Fujiyama T, Hoshino M, et al. Atoh1 directs hair cell differentiation and survival in the late embryonic mouse inner ear. Dev Biol. 2013;381:401–410. doi: 10.1016/j.ydbio.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gubbels SP, Woessner DW, Mitchell JC, Ricci AJ, Brigande JV. Functional auditory hair cells produced in the mammalian cochlea by in utero gene transfer. Nature. 2008;455:537–541. doi: 10.1038/nature07265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly MC, Chang Q, Pan A, Lin X, Chen P. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. J Neurosci. 2012;32:6699–6710. doi: 10.1523/JNEUROSCI.5420-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Z, Dearman JA, Cox BC, Walters BJ, Zhang L, Ayrault O, et al. Age-dependent in vivo conversion of mouse cochlear pillar and Deiters’ cells to immature hair cells by Atoh1 ectopic expression. J Neurosci. 2012;32:6600–6610. doi: 10.1523/JNEUROSCI.0818-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z, Fang J, Dearman J, Zhang L, Zuo J. In vivo generation of immature inner hair cells in neonatal mouse cochleae by ectopic Atoh1 expression. PLoS One. 2014;9:e89377. doi: 10.1371/journal.pone.0089377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shou J, Zheng JL, Gao WQ. Robust generation of new hair cells in the mature mammalian inner ear by adenoviral expression of Hath1. Mol Cell Neurosci. 2003;23:169–179. doi: 10.1016/s1044-7431(03)00066-6. [DOI] [PubMed] [Google Scholar]
- 39.Yang J, Cong N, Han Z, Huang Y, Chi F. Ectopic hair cell-like cell induction by Math1 mainly involves direct transdifferentiation in neonatal mammalian cochlea. Neurosci Lett. 2013;549:7–11. doi: 10.1016/j.neulet.2013.04.053. [DOI] [PubMed] [Google Scholar]
- 40.Zheng JL, Gao WQ. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci. 2000;3:580–586. doi: 10.1038/75753. [DOI] [PubMed] [Google Scholar]
- 41.Kawamoto K, Ishimoto S, Minoda R, Brough DE, Raphael Y. Math1 gene transfer generates new cochlear hair cells in mature guinea pigs in vivo. J Neurosci. 2003;23:4395–4400. doi: 10.1523/JNEUROSCI.23-11-04395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atkinson PJ, Wise AK, Flynn BO, Nayagam BA, Richardson RT. Hair cell regeneration after ATOH1 gene therapy in the cochlea of profoundly deaf adult guinea pigs. PLoS One. 2014;9:e102077. doi: 10.1371/journal.pone.0102077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Izumikawa M, Batts SA, Miyazawa T, Swiderski DL, Raphael Y. Response of the flat cochlear epithelium to forced expression of Atoh1. Hear Res. 2008;240:52–56. doi: 10.1016/j.heares.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 45.Kiernan AE. Notch signaling during cell fate determination in the inner ear. Semin Cell Dev Biol. 2013;24:470–479. doi: 10.1016/j.semcdb.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adam J, Myat A, Le Roux I, Eddison M, Henrique D, Ish-Horowicz D, et al. Cell fate choices and the expression of Notch, Delta and Serrate homologues in the chick inner ear: parallels with Drosophila sense-organ development. Development. 1998;125:4645–4654. doi: 10.1242/dev.125.23.4645. [DOI] [PubMed] [Google Scholar]
- 47.Haddon C, Jiang YJ, Smithers L, Lewis J. Delta-Notch signalling and the patterning of sensory cell differentiation in the zebrafish ear: evidence from the mind bomb mutant. Development. 1998;125:4637–4644. doi: 10.1242/dev.125.23.4637. [DOI] [PubMed] [Google Scholar]
- 48.Morrison A, Hodgetts C, Gossler A, Hrabé de, Lewis J. Expression of delta1 and serrate1 (Jagged1) in the mouse inner ear. Mech Dev. 1999;84:169–172. doi: 10.1016/s0925-4773(99)00066-0. [DOI] [PubMed] [Google Scholar]
- 49.Lanford PJ, Lan Y, Jiang R, Lindsell C, Weinmaster G, Gridley T, et al. Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat Genet. 1999;21:289–292. doi: 10.1038/6804. [DOI] [PubMed] [Google Scholar]
- 50.Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133:1277–1286. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- 51.Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007;134:1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- 52.Zheng JL, Shou J, Guillemot F, Kageyama R, Gao WQ. Hes1 is a negative regulator of inner ear hair cell differentiation. Development. 2000;127:4551–4560. doi: 10.1242/dev.127.21.4551. [DOI] [PubMed] [Google Scholar]
- 53.Zine A, Aubert A, Qiu J, Therianos S, Guillemot F, Kageyama R, et al. Hes1 and Hes5 activities are required for the normal development of the hair cells in the mammalian inner ear. J Neurosci. 2001;21:4712–4720. doi: 10.1523/JNEUROSCI.21-13-04712.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li S, Mark S, Radde-Gallwitz K, Schlisner R, Chin MT, Chen P. Hey2 functions in parallel with Hes1 and Hes5 for mammalian auditory sensory organ development. BMC Dev Biol. 2008;8:20. doi: 10.1186/1471-213X-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tateya T, Imayoshi I, Tateya I, Ito J, Kageyama R. Cooperative functions of Hes/Hey genes in auditory hair cell and supporting cell development. Dev Biol. 2011;352:329–340. doi: 10.1016/j.ydbio.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 56.Kiernan AE, Cordes R, Kopan R, Gossler A, Gridley T. The Notch ligands DLL1 and JAG2 act synergistically to regulate hair cell development in the mammalian inner ear. Development. 2005;132:4353–4362. doi: 10.1242/dev.02002. [DOI] [PubMed] [Google Scholar]
- 57.Ma EY, Rubel EW, Raible DW. Notch signaling regulates the extent of hair cell regeneration in the zebrafish lateral line. J Neurosci. 2008;28:2261–2273. doi: 10.1523/JNEUROSCI.4372-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daudet N, Gibson R, Shang J, Bernard A, Lewis J, Stone J. Notch regulation of progenitor cell behavior in quiescent and regenerating auditory epithelium of mature birds. Dev Biol. 2009;326:86–100. doi: 10.1016/j.ydbio.2008.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li W, Wu J, Yang J, Sun S, Chai R, Chen ZY, et al. Notch inhibition induces mitotically generated hair cells in mammalian cochleae via activating the Wnt pathway. Proc Natl Acad Sci U S A. 2015;112:166–171. doi: 10.1073/pnas.1415901112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamamoto N, Tanigaki K, Tsuji M, Yabe D, Ito J, Honjo T. Inhibition of Notch/RBP-J signaling induces hair cell formation in neonate mouse cochleas. J Mol Med (Berl) 2006;84:37–45. doi: 10.1007/s00109-005-0706-9. [DOI] [PubMed] [Google Scholar]
- 61.Burns JC, Cox BC, Thiede BR, Zuo J, Corwin JT. In vivo proliferative regeneration of balance hair cells in newborn mice. J Neurosci. 2012;32:6570–6577. doi: 10.1523/JNEUROSCI.6274-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jung JY, Avenarius MR, Adamsky S, Alpert E, Feinstein E, Raphael Y. siRNA targeting Hes5 augments hair cell regeneration in aminoglycoside-damaged mouse utricle. Mol Ther. 2013;21:834–841. doi: 10.1038/mt.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin V, Golub JS, Nguyen TB, Hume CR, Oesterle EC, Stone JS. Inhibition of Notch activity promotes nonmitotic regeneration of hair cells in the adult mouse utricles. J Neurosci. 2011;31:15329–15339. doi: 10.1523/JNEUROSCI.2057-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mizutari K, Fujioka M, Hosoya M, Bramhall N, Okano HJ, Okano H, et al. Notch inhibition induces cochlear hair cell regeneration and recovery of hearing after acoustic trauma. Neuron. 2013;77:58–69. doi: 10.1016/j.neuron.2012.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tona Y, Hamaguchi K, Ishikawa M, Miyoshi T, Yamamoto N, Yamahara K, et al. Therapeutic potential of a gamma-secretase inhibitor for hearing restoration in a guinea pig model with noise-induced hearing loss. BMC Neurosci. 2014;15:66. doi: 10.1186/1471-2202-15-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hébert JM. FGFs: neurodevelopment's jack-of-all-trades - how do they do it. Front Neurosci. 2011;5:133. doi: 10.3389/fnins.2011.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pirvola U, Spencer-Dene B, Xing-Qun L, Kettunen P, Thesleff I, Fritzsch B, et al. FGF/FGFR-2(IIIb) signaling is essential for inner ear morphogenesis. J Neurosci. 2000;20:6125–6134. doi: 10.1523/JNEUROSCI.20-16-06125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schimmang T. Expression and functions of FGF ligands during early otic development. Int J Dev Biol. 2007;51:473–481. doi: 10.1387/ijdb.072334ts. [DOI] [PubMed] [Google Scholar]
- 69.Wright TJ, Mansour SL. Fgf3 and Fgf10 are required for mouse otic placode induction. Development. 2003;130:3379–3390. doi: 10.1242/dev.00555. [DOI] [PubMed] [Google Scholar]
- 70.Hayashi T, Ray CA, Bermingham-McDonogh O. Fgf20 is required for sensory epithelial specification in the developing cochlea. J Neurosci. 2008;28:5991–5999. doi: 10.1523/JNEUROSCI.1690-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pirvola U, Ylikoski J, Trokovic R, Hébert JM, McConnell SK, Partanen J. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 2002;35:671–680. doi: 10.1016/s0896-6273(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 72.Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 73.Hayashi T, Cunningham D, Bermingham-McDonogh O. Loss of Fgfr3 leads to excess hair cell development in the mouse organ of Corti. Dev Dyn. 2007;236:525–533. doi: 10.1002/dvdy.21026. [DOI] [PubMed] [Google Scholar]
- 74.Mueller KL, Jacques BE, Kelley MW. Fibroblast growth factor signaling regulates pillar cell development in the organ of corti. J Neurosci. 2002;22:9368–9377. doi: 10.1523/JNEUROSCI.22-21-09368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Puligilla C, Feng F, Ishikawa K, Bertuzzi S, Dabdoub A, Griffith AJ, et al. Disruption of fibroblast growth factor receptor 3 signaling results in defects in cellular differentiation, neuronal patterning, and hearing impairment. Dev Dyn. 2007;236:1905–1917. doi: 10.1002/dvdy.21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doetzlhofer A, Basch ML, Ohyama T, Gessler M, Groves AK, Segil N. Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev Cell. 2009;16:58–69. doi: 10.1016/j.devcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Su YX, Hou CC, Yang WX. Control of hair cell development by molecular pathways involving Atoh1, Hes1 and Hes5. Gene. 2015;558:6–24. doi: 10.1016/j.gene.2014.12.054. [DOI] [PubMed] [Google Scholar]
- 78.Ito H, Nakajima A, Nomoto H, Furukawa S. Neurotrophins facilitate neuronal differentiation of cultured neural stem cells via induction of mRNA expression of basic helix-loop-helix transcription factors Mash1 and Math1. J Neurosci Res. 2003;71:648–658. doi: 10.1002/jnr.10532. [DOI] [PubMed] [Google Scholar]
- 79.Millimaki BB, Sweet EM, Dhason MS, Riley BB. Zebrafish atoh1 genes: classic proneural activity in the inner ear and regulation by Fgf and Notch. Development. 2007;134:295–305. doi: 10.1242/dev.02734. [DOI] [PubMed] [Google Scholar]
- 80.Miller JR, Hocking AM, Brown JD, Moon RT. Mechanism and function of signal transduction by the Wnt/beta-catenin and Wnt/Ca2+ pathways. Oncogene. 1999;18:7860–7872. doi: 10.1038/sj.onc.1203245. [DOI] [PubMed] [Google Scholar]
- 81.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shi F, Cheng YF, Wang XL, Edge AS. Beta-catenin up-regulates Atoh1 expression in neural progenitor cells by interaction with an Atoh1 3’ enhancer. J Biol Chem. 2010;285:392–400. doi: 10.1074/jbc.M109.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Agathocleous M, Iordanova I, Willardsen MI, Xue XY, Vetter ML, Harris WA, et al. A directional Wnt/beta-catenin-Sox2-proneural pathway regulates the transition from proliferation to differentiation in the Xenopus retina. Development. 2009;136:3289–3299. doi: 10.1242/dev.040451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jayasena CS, Ohyama T, Segil N, Groves AK. Notch signaling augments the canonical Wnt pathway to specify the size of the otic placode. Development. 2008;135:2251–2261. doi: 10.1242/dev.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jacques BE, Puligilla C, Weichert RM, Ferrer-Vaquer A, Hadjantonakis AK, Kelley MW, et al. A dual function for canonical Wnt/XMLLink_XYZ-catenin signaling in the developing mammalian cochlea. Development. 2012;139:4395–4404. doi: 10.1242/dev.080358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shi F, Hu L, Jacques BE, Mulvaney JF, Dabdoub A, Edge AS. β-Catenin is required for hair-cell differentiation in the cochlea. J Neurosci. 2014;34:6470–6479. doi: 10.1523/JNEUROSCI.4305-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shi F, Hu L, Edge AS. Generation of hair cells in neonatal mice by β-catenin overexpression in Lgr5-positive cochlear progenitors. Proc Natl Acad Sci U S A. 2013;110:13851–13856. doi: 10.1073/pnas.1219952110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chai R, Kuo B, Wang T, Liaw EJ, Xia A, Jan TA, et al. Wnt signaling induces proliferation of sensory precursors in the postnatal mouse cochlea. Proc Natl Acad Sci U S A. 2012;109:8167–8172. doi: 10.1073/pnas.1202774109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jansson L, Kim GS, Cheng AG. Making sense of Wnt signaling-linking hair cell regeneration to development. Front Cell Neurosci. 2015;9:66. doi: 10.3389/fncel.2015.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shi F, Kempfle JS, Edge AS. Wnt-responsive Lgr5-expressing stem cells are hair cell progenitors in the cochlea. J Neurosci. 2012;32:9639–9648. doi: 10.1523/JNEUROSCI.1064-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kuo BR, Baldwin EM, Layman WS, Taketo MM, Zuo J. In vivo cochlear hair cell generation and survival by coactivation of β-catenin and atoh1. J Neurosci. 2015;35:10786–10798. doi: 10.1523/JNEUROSCI.0967-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Atkinson PJ, Huarcaya Najarro E, Sayyid ZN, Cheng AG. Sensory hair cell development and regeneration: similarities and differences. Development. 2015;142:1561–1571. doi: 10.1242/dev.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cai T, Jen HI, Kang H, Klisch TJ, Zoghbi HY, Groves AK. Characterization of the transcriptome of nascent hair cells and identification of direct targets of the Atoh1 transcription factor. J Neurosci. 2015;35:5870–5883. doi: 10.1523/JNEUROSCI.5083-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Krizhanovsky V, Soreq L, Kliminski V, Ben-Arie N. Math1 target genes are enriched with evolutionarily conserved clustered E-box binding sites. J Mol Neurosci. 2006;28:211–229. doi: 10.1385/JMN:28:2:211. [DOI] [PubMed] [Google Scholar]