Abstract
Membrane proteins are substantially more challenging than natively soluble proteins as subjects for structural analysis. Thus, membrane proteins are greatly under-represented in structural databases. Recently, as a consequence of focused attention by consortium efforts and advances in methodology, the pace has accelerated for atomic-level structure determination of membrane proteins. Enabling advances have come in methods for protein production, for crystallographic analysis, and for cryo-EM analysis.
Proteins in membranes provide the portals through which cells, and membrane-delimited organelles within cells, communicate with their external environments, including other cells. Thereby, membrane proteins are crucial components in cellular physiology and biochemistry. Membrane proteins are also involved in processes of disease, where they are the molecular targets of over 40% of all FDA-approved drugs1. As for soluble proteins, atomic-level structure informs us greatly about the biochemistry and physiology of processes that these molecules affect. Moreover, new principles of structure and connections to function are also developing that are special to the proteins in lipid bilayers. We can expect further fundamental understanding from more comprehensive structural information on membrane proteins.
Integral membrane proteins present formidable, but not insurmountable challenges for biochemical and structural characterization. Problems arise in the recombinant expression of membrane proteins, especially so for those from eukaryotes; biochemical purification and characterization is more challenging for membrane proteins than for natively soluble proteins; and many membrane proteins are intrinsically flexible. Typically, one must isolate membrane proteins by detergent extraction from cellular lipid bilayers for purification even if they are to be reconstituted into lipidic environments for biochemical characterization or structural analysis, which further complicates structural analysis by crystallography or single-particle cryogenic electron microscopy (cryo-EM).
Progress in atomic-level structure determination
Despite the challenges, impressive progress has been made in the production and analysis of membrane proteins, particularly of late. The triumphs of membrane protein structure are legendary, starting with bacteriorhodopsin at a helix-resolving resolution in 19752 and the photosynthetic reaction center at an atomic-level in 19853, and on through potassium channels4 and G-protein coupled receptor (GPCR) complexes5. Hundreds of others have come along the way and since. The vast majority of atomic-level membrane protein structures have been determined by x-ray crystallography, but some have come from NMR spectroscopy and, recently, cryo-EM has become an exciting contributor.
The rate of production of atomic-level membrane protein structures is accelerating (Figure 1); 80% of all unique membrane proteins were reported in the past decade and over half came in the last five years. Nevertheless, the structural output on membrane proteins remains at a very low level when compared with that for soluble proteins. Whereas the Protein Data Bank (PDB) contained a total of 108,902 active ‘Protein Only’ depositions through 22 March 2016, White’s compilation of known structures for membrane protein6 records only 1,897 coordinate files (604 unique) through 17 March 2016. Thus, while membrane proteins comprise 20–30% of all proteins in both prokaryotic and eukaryotic organisms7, they presently constitute just 1.7% of known atomic-level structures. Although this fraction is up from the 1.0% of six years ago, membrane proteins still remain grossly under-represented. It is less easy to quantify the progress on functional characterization of membrane proteins, but many complications that affect structural analysis inevitably also affect biochemical characterization.
Figure 1.
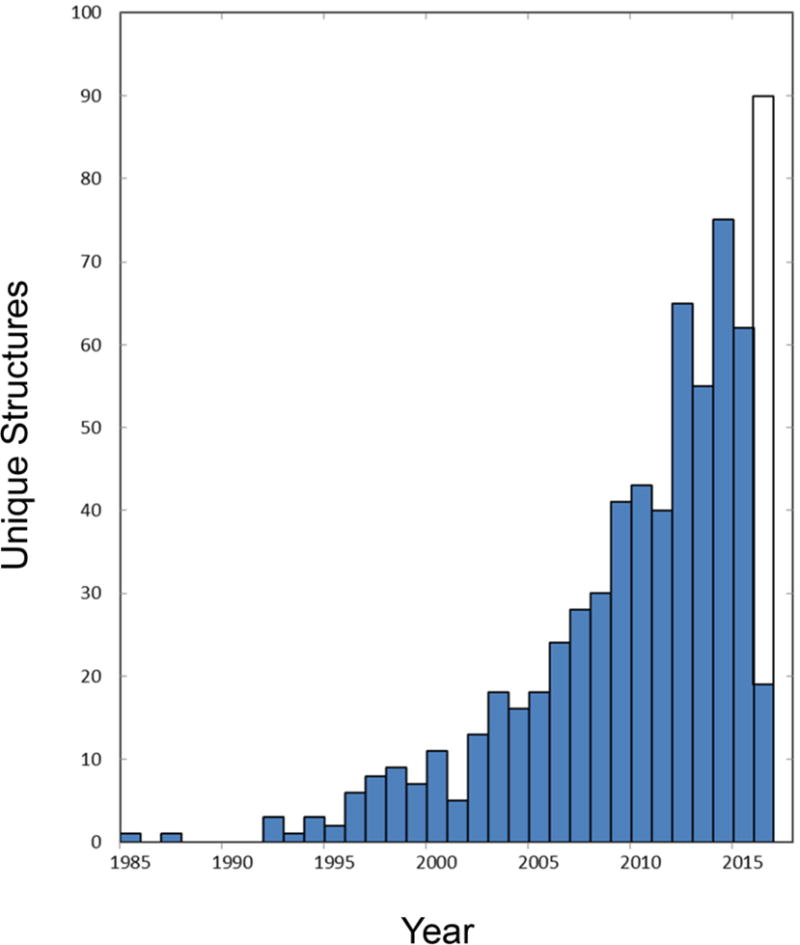
Growth in the production of membrane protein structures. Annual numbers of unique structures are plotted as a function of time. The numbers of unique structures are taken as defined and recorded through 17 March 2016 in the White database of membrane proteins of known structure (http://blanco.biomol.uci.edu/mpstruc/). The open portion of the bar for 2016 projects for production continuing at the same rate as for this year to date.
The situation for membrane proteins also differs from that for soluble proteins with respect to structural novelty. For soluble proteins, the number of solved structures has reached the point that novelty is approaching saturation for the numbers of families, superfamilies, and folds8. This is in keeping with the common observation that a protein with a sequence unlike that of any known structure proves unexpectedly to have a three-dimensional (3D) structure clearly like something already known. This was seen especially frequently in the Protein Structure Initiative (PSI) where attention was placed on sequence families without prior structural representation. A similarly careful analysis8 has not been made for membrane proteins; however, albeit anecdotally, we know from the structural genomics experience of the New York Consortium on Membrane Protein Structure (NYCOMPS) that such novelty is still commonplace. For 25 structures obtained by NYCOMPS from putatively novel starting points, we judge that 16 had new folds, three belonged to new superfamilies, and five were in new structural families at the time of structure solution. Since membrane proteins are under-represented in the PDB at present, it follows that newly solved membrane protein structures are relatively likely to be novel.
Advances in expression and purification
Many factors are furthering progress in structural analysis of membrane proteins, and certainly improved proficiency in recombinant expression and purification is a major contributor. Efficient procedures have been developed for screening for the expression of membrane proteins in bacterial systems9, although their effectiveness for eukaryotic proteins has been limited. Various alternatives are in common use for the recombinant production of eukaryotic proteins, usually from synthetic genes, with a focus on baculovirus-infected insect cells and on cultures of mammalian cells, typically human embryonic kidney cells, HEK293. Procedures have also been developed to couple these two aspects in baculovirus transduction of mammalian cells10.
Size exclusion chromatography (SEC) has proved to be highly effective for the identification of suitable constructs and appropriate detergents for solubilization, with high yields in monodisperse elution profiles predictive of readiness for structure analysis. SEC can be performed after partial purification based on an affinity tag, typically a polyhistidine fusion peptide; however, through the use of GFP fusions, such candidates can be identified as well by fluorescence-detection SEC (FSEC) without purification11. Most laboratories have settled on dodecylmaltoside (DDM, n-dodecyl β-D-maltoside) as the detergent of choice for initial solubilization of proteins from membranes, but exchange can then be made into alternative detergents with SEC assessment. Experience shows that harsher detergents, particularly so for phosphocholines (Fos-choline), may be disruptive of 3D structure and this has led to development of detergents that better mimic the lipids of membranes. Notable success has come from maltose-neopentyl glycols (MNGs)12. In addition, of particular relevance for cryo-EM, synthetic polymers known as amphipols can be used to displace detergents13.
Many advances in methods for the handling and analysis of membrane proteins have come from individual laboratories in response to the challenges of difficult problems. Considerable development has also come from larger initiatives, however. The NIH Protein Structure Initiative included a particular emphasis on this area, with nine centers devoted to membrane proteins in its PSI:Biology phase, including the NYCOMPS14 project mentioned above (led by myself at NYSBC), the Center for Membrane Protein Structures (led by Bob Stroud at UCSF), and the GPCR Network15 (led by Ray Stevens, then at Scripps). In addition, other consortium efforts are in place including the Structural Genomics Consortium (led by Aled Edwards overall with membrane proteins led by Liz Carpenter at Oxford), the Membrane Protein Structural Dynamics Consortium (led by Eduardo Perozo at Chicago), and the Membrane Protein Laboratory at the Diamond Light Source and Research Complex at Harwell (established by So Iwata and now coordinated by Isabel Moraes). These centers have brought robotics and high-throughput methods to bear on membrane protein production and analysis, and they have contributed substantially in the generation of structures.
Advances in crystallographic analysis
Improvements in methods for analyzing membrane protein structure by x-ray crystallography are also contributing to the growth in structural results. Part of this is strongly connected to the advances in expression and purification, notably the ability to test multiple constructs, including insertions of stabilizing fusion domains16. Another part is due to the ready availability of crystallization screens, crystallization robotics, and automated visualization of crystallization trials. A special boost for membrane proteins crystallization has come from advances in lipidic cubic phase (LCP) crystallization17. LCP-grown crystals form in lipidic bilayers where they make direct protein-protein contacts laterally, often mediated by monooleins of the LCP medium, and other contacts between the layers (Figure 2). The resulting network has sufficient rigidity to produce exceptional diffraction patterns in many cases. Typically, LCP-grown crystals are quite small (order of 10 μm), a property that has been met with parallel developments in fabricated mounts.
Figure 2.

Latticework in a lipidic cubic phase (LCP) crystal of a membrane protein. The crystal structure of TSPO in the apo type 2 LCP lattice is drawn with protein molecules as ribbon diagrams, interstitial monoolein chains in stick representation, and water molecules as red dots.
Structural analysis of membrane proteins has also benefited from advances in x-ray sources and diffraction methods. Synchrotron beamlines have been developed to deliver x-ray beams matched in size to microcrystals18, which is especially advantageous for LCP-grown crystals of membrane proteins. X-ray free electron laser (XFEL) experiments are having particular impact for membrane proteins. An LCP injector has been developed for efficient delivery of LCP-grown microcrystals19. Various devices have been developed for the deployment of crystals, including microfluidics, acoustic droplet ejection and grid-based systems.
Since many membrane proteins of interest have novel structures, de novo phase evaluations are required for crystal structure determination. As for other biological crystals, anomalous diffraction methods dominate for de novo phasing of membrane proteins; however, anomalous signals are often weak because of poor diffraction overall. This can be true even when selenomethionine substitution provides the phasing element, but it is all the more so for analyses based on native elements such as sulfur. By using high multiplicity from multiple crystals20 and from varied crystal orientations21, as well as lower than usual x-ray energies, excellent progress is being made in solving structures from weak anomalous signals22. Because LCP-grown crystals often diffract particularly well, such structures can truly be at an atomic level (Figure 3a).
Figure 3.
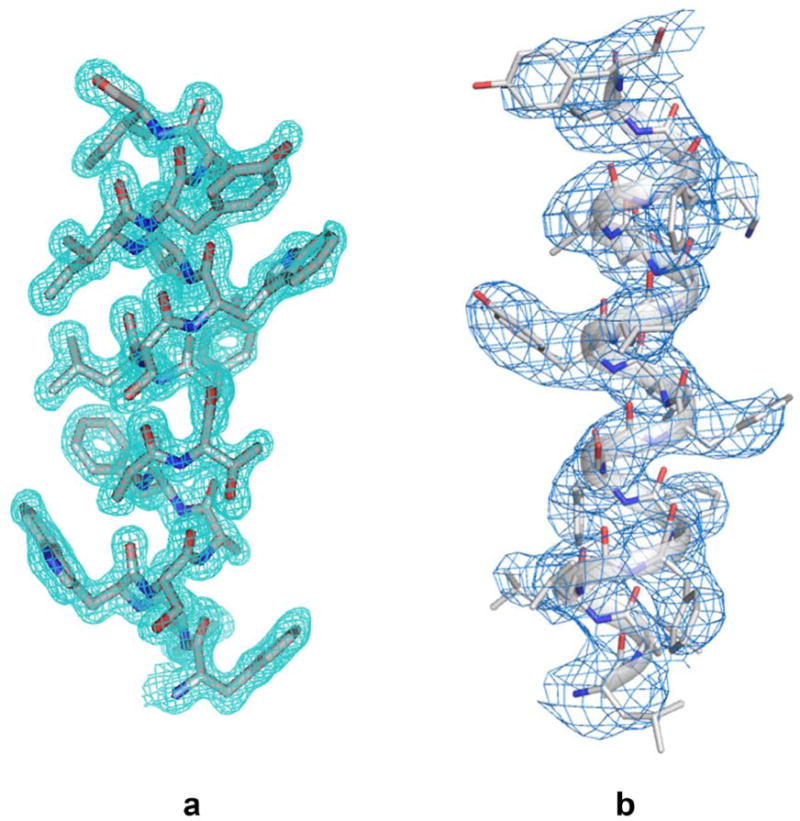
Density distributions for transmembrane helices obtained by different methods. (a) Helix TM5 (residues F136-I49) from TSPO of Bacillus cereus as determined at 1.7 Å resolution from an apo type 2 crystal grown in LCP30. Reproduced from Figure S2 of Ref. 30 with permission by Science. (b) Helix TM3 (residues L171-Y189) of presenilin from the cryo-EM structure of human γ-secretase at 3.4 Å resolution28. Adapted from Extended Data Figure 2 of Ref. 28 with permission by Nature.
Advances in cryo-EM analysis
Structural analyses by electron microscopy have been important for membrane proteins from the earliest days2, but until recently they have rarely reached an atomic-level of resolution and, if so, then only from ordered arrays or symmetric viruses. The situation changed dramatically with image corrections associated with direct-electron detectors. The advances in detector technology and computational procedures responsible for the new-found excellence of cryo-EM are beyond the scope of this commentary. Recent reviews of cryo-EM methodology nicely provide the background23–24. Suffice it to indicate here that cryo-EM is already making important contributions to membrane-protein structure.
Arguably, the resolution revolution for cryo-EM began with the structure of a membrane protein, that of the TRPV1 ion channel obtained at the ‘gold-standard’ resolution of 3.4 Å25. Since then, several other membrane protein structures have been determined at what is being called ‘near-atomic’ resolution. These include structures of the exceptionally large 2.3-MDa RYR1 calcium-release channel, determined at resolutions of 4.8 Å26 and 3.8 Å27, the much smaller and asymmetric intramembrane protease γ-secretase, determined at 3.4 Å resolution28, and the Slo2.2 potassium channel at 4.5 Å resolution29. Compared to many x-ray crystal structures these current-day cryo-EM resolutions seem modest; however, they certainly suffice for chain tracing at a poly-alanine level of analysis, and in many case side-chain definitions allow for full atomic models to be built (Figure 3b). What is remarkable, for the eyes of a crystallographer accustomed to experimental maps at comparable nominal resolutions, is the quality of cryo-EM density that comes from direct imaging without the phase evaluations required for crystallography.
Prospects
The future looks very bright for structure of membrane proteins. Advances such as those described above will remain productive, and we can expect continued technology development. New x-ray sources, such as beamlines at NSLS-II and MAX-IV, will be available soon with incredible fluxes focused into microbeams. Relevant diffraction methods will be enhancing the effectiveness of phase evaluations from native membrane proteins, without needing recourse to heavy atoms. The role of cryo-EM has just begun, and we can expect many technical advances – for specimen preparation, instrumentation, and image processing. We can also expect fruitful marriages of higher resolution structures from crystallography with cryo-EM structures in multiple conformational states and also with computer simulations of conformational transitions. All of these advances should serve to advance fundamental understanding of membrane biology.
Acknowledgments
I thank Youzhong Guo for producing Figure 2 and Yigong Shi for providing a high resolution copy for Figure 3b; and I thank Filippo Mancia, Qun Liu, Lawrence Shapiro, Brian Kloss, Renato Bruni, Ravi Kalathur and others from the NYCOMPS team for numerous contributions to the experience reported here. This work was supported in part by NIH grants GM095315 and GM107462.
Footnotes
The author declares no competing financial interests.
References
- 1.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 2.Henderson R, Unwin PN. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- 3.Deisenhofer J, Epp O, Miki K, Huber R, Michel H. Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridis at 3Å resolution. Nature. 1985;318:618–624. doi: 10.1038/318618a0. [DOI] [PubMed] [Google Scholar]
- 4.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 5.Rasmussen SG, et al. Crystal structure of the β2 adrenergic receptor – Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White SH. Biophysical dissection of membrane proteins. Nature. 2009;459:344–346. doi: 10.1038/nature08142. [DOI] [PubMed] [Google Scholar]
- 7.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levitt M. Growth of novel protein structural data. Proc Natl Acad Sci USA. 2007;104:3183–3188. doi: 10.1073/pnas.0611678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruni R, Kloss B. High-throughput cloning and expression of integral membrane proteins in Escherichia coli. Curr Protoc Protein Sci. 2013;74 doi: 10.1002/0471140864.ps2906s74. Unit 29.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goehring A, et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc. 2014;9:2574–2585. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Chae PS, et al. Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat Methods. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popot JL. Amphipols from A to Z. Annu Rev Biophys. 2011;40:379–408. doi: 10.1146/annurev-biophys-042910-155219. [DOI] [PubMed] [Google Scholar]
- 14.Love J, et al. The New York Consortium on Membrane Protein Structure (NYCOMPS): a high-throughput platform for structural genomics of integral membrane proteins. J Struct Funct Genomics. 2010;11:191–199. doi: 10.1007/s10969-010-9094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevens RC, et al. The GPCR Network: a large-scale collaboration to determine human GPCR structure and function. Nat Rev Drug Discov. 2013;12:25–34. doi: 10.1038/nrd3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorsen TS, Matt R, Weis WI, Kobilka BK. Modified T4 Lysozyme Fusion Proteins Facilitate G Protein-Coupled Receptor Crystallogenesis. Structure. 2014;22:1657–1664. doi: 10.1016/j.str.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caffrey M. A comprehensive review of the lipid cubic phase or in meso method for crystallizing membrane and soluble proteins and complexes. Acta Crystallogr F. 2015;71:3–18. doi: 10.1107/S2053230X14026843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith JL, Fischetti RF, Yamamoto M. Micro-crystallography comes of age. Curr Opin Struct Biol. 2012;22:602–612. doi: 10.1016/j.sbi.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weierstall U, et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat Commun. 2014;5:3309. doi: 10.1038/ncomms4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Q, et al. Structures from anomalous diffraction of native biological macromolecules. Science. 2012;336:1033–1037. doi: 10.1126/science.1218753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinert T, et al. Fast native-SAD phasing for routine macromolecular structure determination. Nat Methods. 2015;12:131–133. doi: 10.1038/nmeth.3211. [DOI] [PubMed] [Google Scholar]
- 22.Liu Q, Hendrickson WA. Crystallographic phasing from weak anomalous signals. Curr Opin Struct Biol. 2015;34:99–107. doi: 10.1016/j.sbi.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng Y, Grigorieff N, Penczek PA, Walz T. A primer to single-particle cryo-electron microscopy. Cell. 2015;161:438–449. doi: 10.1016/j.cell.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bai XC, McMullan G, Scheres SH. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci. 2015;40:49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zalk R, et al. Structure of a mammalian ryanodine receptor. Nature. 2015;517:44–49. doi: 10.1038/nature13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan Z, et al. Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution. Nature. 2015;517:50–55. doi: 10.1038/nature14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bai XC, et al. An atomic structure of human γ-secretase. Nature. 2015;525:212–217. doi: 10.1038/nature14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hite RK, et al. Cryo-electron microscopy structure of the Slo2.2 Na(+)-activated K(+) channel. Nature. 2015;527:198–203. doi: 10.1038/nature14958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Y, et al. Structure and Activity of Tryptophan-rich TSPO Proteins. Science. 2015;347:551–555. doi: 10.1126/science.aaa1534. [DOI] [PMC free article] [PubMed] [Google Scholar]


