Abstract
ADIPOQ gene polymorphisms have been indicated to be associated with hypertension; however, published studies have reported inconsistent results. Eligible studies were retrieved by searching the PubMed, Embase and China National Knowledge Infrastructure databases. The case group consisted of patients with hypertension, and the control group consisted of subjects with normal blood pressure. Based on eleven published articles, involving 4837 cases and 5618 controls, the pooled results from rs2241766 polymorphism showed increased risk in the allelic model (G VS T: OR = 1.16, 95%CI = 1.06–1.27), recessive model (GG VS GT + TT: OR = 1.34, 95%CI = 1.10–1.63), dominant model (GG + GT VS TT: OR = 1.15, 95%CI = 1.02–1.30) and homozygote model (GG VS TT: OR = 1.38, 95%CI = 1.21–1.69). In addition, rs266729 polymorphism showed increased risk for hypertension in the recessive model (GG VS GC + CC: OR = 1.43, 95%CI = 1.02–2.01). In the Caucasian subgroup, rs1501299 polymorphism showed decreased risk of hypertension in the allelic model (T VS G: OR = 0.75, 95%CI = 0.58–0.97), dominant model (TT + TG VS GG: OR = 0.83, 95%CI = 0.71–0.98) and heterozygote model (TG VS GG: OR = 0.82, 95%CI = 0.68–0.99). The rs2241766 polymorphism was associated with a significant increase in hypertension risk based on our analysis. Moreover, an increased risk of rs266729 in hypertension patients was also detected. Our meta-analysis suggests that the rs1501299 polymorphism may play a protective role in hypertension in Caucasian subgroup; however, this finding requires further study.
Hypertension is a major risk factor for vascular disease and the leading source of global health burden1. Although the aetiology of hypertension has not been fully elucidated, interactions between genes and environmental factors are suggested to play an important role in the pathological process of hypertension2. In recent years, genes and their polymorphisms have been found to be associated with blood pressure and susceptibility of hypertension3,4,5,6.
Adiponectin is an adipocyte-derived hormone and plays an important role in energy homeostasis by regulating glucose and lipid metabolism7. Decreases in plasma adiponectin levels are observed in cardiovascular disease, essential hypertension, obesity and type II diabetes8,9,10, which may suggest a special role of adiponectin in the pathogenesis of hypertension. The ADIPOQ gene is located on chromosome 3q27 and encodes the protein adiponectin. Several single-nucleotide polymorphisms (SNPs) in the ADIPOQ gene have been described11. These polymorphisms in the ADIPOQ gene can modulate circulating concentration of adiponectin12,13,14.
Most of the published studies have investigated the associations of rs2241766, rs1501299 and rs266729 polymorphisms in the ADIPOQ gene with risk factors of hypertension15,16,17,18. However, the results are inconsistent. Wei et al.17 reported that the polymorphisms of 45 T/G (rs2241766) and 276 G/T (rs1501299) in the ADIPOQ gene were not associated with hypertension, but Mousavinasab et al.19 reported opposite findings. Moreover, polymorphism of 11377 C/G (rs266729) in ADIPOQ gene was indicated to have independent effects on diastolic blood pressure reported by Avery et al.16. The small numbers and varying populations of the published studies may partially account for the controversial results. Therefore, we performed this meta-analysis to investigate the associations between ADIPOQ rs2241766, rs1501299 and rs266729 polymorphisms and hypertension.
Results
The Characteristics of the Include Studies
A total of 233 articles were obtained by online and manual search. After removing duplicates and screening the title and abstract, eighteen articles were selected. Seven articles were excluded due to lack of detailed genotype distribution. Finally, a total of eleven published articles10,17,18,20,21,22,23,24,25,26,27, involving 4837 cases and 5618 controls were included in this meta-analysis (Seen in the Table S2 PRISMA Flow Diagram28).
The characteristics of all the included articles are summarized in Table 1. For rs2241766, 12 studies were included with 2147 cases and 2601 controls; 9 studies with 1859 cases and 2109 controls are included for rs1501299; only 5 studies with 831 cases and 908 controls concerning the rs266729 and hypertension risk are included.
Table 1. Characteristics of included studies selected for meta-analysis.
| Study | Year | Country | Ethnicity | Age, mean ± SD, year |
Body mass index (kg/m2) |
Type of study | Sample size |
Control source | Genotype distributions in cases/controls |
Genotyping method | Quality score | HWE* | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | Case | Control | 11 | 12 | 22 | |||||||||
| rs2241766 | |||||||||||||||||
| Machado GH | 2014 | Brazil | Caucasian | 26 ± 4.5 | 24 ± 4 | 27.53 ± 5.15 | 22.80 ± 2.7 | Case-control | 113 | 169 | HB | 81/139 | 29/29 | 3/1 | PCR | 8 | 0.697 |
| Machado PE | 2014 | Brazil | Caucasian | 27 ± 4.5 | 24 ± 5 | 26.28 ± 4.8 | 22.80 ± 2.7 | Case-control | 127 | 169 | HB | 97/139 | 26/29 | 4/1 | PCR | 8 | 0.697 |
| Jiang HT | 2014 | China | Mongolian | 68.8 ± 6.7 | 67.1 ± 7.1 | 24.5 ± 4.3 | 23.1 ± 3.3 | Case-control | 202 | 166 | PB | 102/91 | 86/65 | 14/10 | PCR | 10 | 0.720 |
| Jiang HT + DM | 2014 | China | Mongolian | 68.2 ± 6.1 | 67.1 ± 7.2 | 26.0 ± 3.7 | 23.1 ± 3.3 | Case-control | 169 | 166 | PB | 90/91 | 67/65 | 12/10 | PCR | 10 | 0.720 |
| Kang | 2013 | China | Mongolian | NA | NA | 22.57 ± 2.27 | 21.53 ± 2.17 | Case-control | 153 | 126 | HB | 63/75 | 68/40 | 22/11 | PCR | 8 | 0.105 |
| Leu HT | 2011 | China | Mongolian | 41.4 ± 0.7 | 46.7 ± 0.8 | 25.4 ± 0.3 | 23.5 ± 0.1 | Family-based | 159 | 446 | PB | 74/233 | 70/181 | 15/32 | PCR | 10 | 0.696 |
| Leu HT + Mets | 2011 | China | Mongolian | 41.7 ± 0.6 | 57.5 ± 1.0 | 28.3 ± 0.3 | 27.1 ± 0.3 | Family-based | 192 | 165 | PB | 92/78 | 81/75 | 19/12 | PCR | 9 | 0.696 |
| Youpeng | 2010 | China | Mongolian | 30.9 ± 6.0 | 30.0 ± 4.5 | 22.05 ± 2.65 | 20.1 ± 1.8 | Case-control | 107 | 81 | HB | 74/53 | 26/27 | 7/1 | PCR | 7 | 0.228 |
| Wang | 2008 | China | Mongolian | 46.4 ± 14.0 | 46.8 ± 15.4 | 26.82 ± 2.44 | 22.43 ± 3.99 | Case-control | 80 | 160 | HB | 41/87 | 34/60 | 5/13 | PCR | 8 | 0.561 |
| Tang | 2008 | China | Mongolian | 63.5 ± 6.5 | 60.5 ± 7.1 | NA | NA | Case-control | 151 | 100 | HB | 99/63 | 32/28 | 20/9 | PCR | 8 | 0.036 |
| Jeng | 2007 | China | Mongolian | 51.6 ± 14.7 | 50.7 ± 12.1 | 25.7 ± 3.6 | 23.8 ± 3.4 | Case-control | 212 | 356 | HB | 82/126 | 99/173 | 31/57 | PCR | 9 | 0.853 |
| Yan | 2006 | China | Mongolian | 49 ± 9 | 48 ± 10 | 30.8 ± 2.5 | 22.4 ± 1.8 | Case-control | 482 | 497 | HB | 201/222 | 186/203 | 95/72 | PCR | 9 | 0.024 |
| rs1501299 | |||||||||||||||||
| Machado GH | 2014 | Brazil | Caucasian | 26 ± 4.5 | 24 ± 4 | 27.53 ± 5.15 | 22.80 ± 2.7 | Case-control | 113 | 161 | HB | 59/68 | 46/74 | 8/19 | PCR | 8 | 0.868 |
| Machado PE | 2014 | Brazil | Caucasian | 27 ± 4.5 | 24 ± 5 | 26.28 ± 4.8 | 22.80 ± 2.7 | Case-control | 127 | 161 | HB | 66/68 | 47/74 | 14/19 | PCR | 8 | 0.868 |
| Kang | 2013 | China | Mongolian | NA | NA | 22.57 ± 2.27 | 21.53 ± 2.17 | Case-control | 153 | 126 | HB | 100/82 | 46/38 | 7/6 | PCR | 8 | 0.560 |
| Leu HT | 2011 | China | Mongolian | 41.4 ± 0.7 | 46.7 ± 0.8 | 25.4 ± 0.3 | 23.5 ± 0.1 | Family-based | 159 | 446 | PB | 85/228 | 60/178 | 14/40 | PCR | 9 | 0.536 |
| Leu HT + Mets | 2011 | China | Mongolian | 41.7 ± 0.6 | 57.5 ± 1.0 | 28.3 ± 0.3 | 27.1 ± 0.3 | Family-based | 192 | 165 | PB | 117/93 | 68/56 | 7/16 | PCR | 9 | 0.089 |
| Youpeng | 2010 | China | Mongolian | 30.9 ± 6.0 | 30.0 ± 4.5 | 22.05 ± 2.65 | 20.1 ± 1.8 | Case-control | 107 | 81 | HB | 19/19 | 60/49 | 28/13 | PCR | 7 | 0.051 |
| Wang | 2008 | China | Mongolian | 46.4 ± 14.0 | 46.8 ± 15.4 | 26.82 ± 2.44 | 22.43 ± 3.99 | Case-control | 80 | 160 | HB | 41/74 | 28/73 | 11/13 | PCR | 8 | 0.392 |
| Yan | 2006 | China | Mongolian | 49 ± 9 | 48 ± 10 | 30.8 ± 2.5 | 22.4 ± 1.8 | Case-control | 482 | 497 | HB | 259/274 | 184/187 | 39/36 | PCR | 9 | 0.599 |
| Iwashima | 2004 | Japan | Mongolian | 59.4 ± 0.5 | 57.1 ± 0.6 | 24.4 ± 0.1 | 23.1 ± 0.2 | Case-control | 446 | 312 | HB | 225/165 | 180/124 | 41/23 | PCR | 9 | 0.964 |
| rs266729 | |||||||||||||||||
| Machado GH | 2014 | Brazil | Caucasian | 26 ± 4.5 | 24 ± 4 | 27.53 ± 5.15 | 22.80 ± 2.7 | Case-control | 113 | 161 | HB | 61/93 | 36/57 | 16/11 | PCR | 8 | 0.577 |
| Machado PE | 2014 | Brazil | Caucasian | 27 ± 4.5 | 24 ± 5 | 26.28 ± 4.8 | 22.80 ± 2.7 | Case-control | 127 | 161 | HB | 81/93 | 27/57 | 19/11 | PCR | 8 | 0.577 |
| Leu HT | 2011 | China | Mongolian | 41.4 ± 0.7 | 46.7 ± 0.8 | 25.4 ± 0.3 | 23.5 ± 0.1 | Family-based | 198 | 159 | PB | 85/84 | 90/64 | 23/11 | PCR | 9 | 0.800 |
| Leu HT + Mets | 2011 | China | Mongolian | 41.7 ± 0.6 | 57.5 ± 1.0 | 28.3 ± 0.3 | 27.1 ± 0.3 | Family-based | 173 | 159 | PB | 93/84 | 68/64 | 12/11 | PCR | 9 | 0.800 |
| Zhang | 2010 | China | Mongolian | 52.39 ± 9.60 | 49.25 ± 10.84 | NA | NA | Case-control | 220 | 268 | HB | 138/163 | 70/84 | 12/21 | PCR | 8 | 0.035 |
For rs2241766 variant, 11, 12 and 22 represent TT, GT, GG, respectively; for rs1501299 variant, 11, 12 and 22 represent GG, TG and TT, respectively; for rs266729 variant, 11, 12 and 22 represent CC, CG, and GG.
GH = gestational hypertension; PE = pre-eclampsia; HT = hypertension; DM = type 2 diabetes; Mets = metabolic syndrome; HB = hospital based; PB = population based; NA = not available.
*P value for Hardy–Weinberg equilibrium test in controls.
Meta-Analysis Results and Heterogeneity Analysis
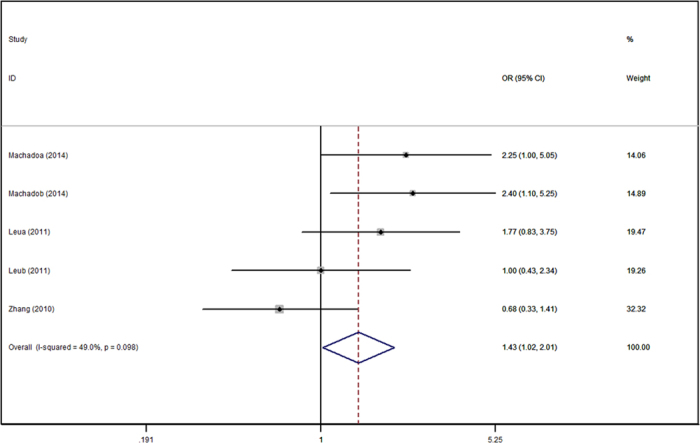
Table 2 shows the main results of this meta-analysis and the heterogeneity of the ADIPOQ gene polymorphisms and hypertension risk. The fixed effect model was adopted to calculate the pooled ORs for each individual polymorphism lacking heterogeneity. The rs2241766 polymorphism was associated with increased risk of hypertension in the allelic model (G VS T: OR = 1.16, 95%CI = 1.06–1.27), recessive model (GG VS GT + TT: OR = 1.34, 95%CI = 1.10–1.63) (Fig. 1), dominant model (GG + GT VS TT: OR = 1.15, 95%CI = 1.02–1.30), and homozygote model (GG VS TT: OR = 1.38, 95%CI = 1.21–1.69). For rs266729, increased risk was only found in the recessive model (GG VS GC + CC: OR = 1.43, 95%CI = 1.02–2.01) (Fig. 2). Association was not detected in rs1501299 polymorphism, and subsequently, subgroup analysis was introduced to uncover some potential details concerning associations between rs1501299 and hypertension risk. Table 3 summarizes the results of the stratified analyses of ADIPOQ polymorphisms and hypertension risk. As stratified by ethnicity for rs1501299, significant associations were found in the Caucasian group. A decreased risk of hypertension was observed in the allelic model (T VS G: OR = 0.75, 95%CI = 0.58–0.97) (Fig. 3), dominant model (TT + TG VS GG: OR = 0.83, 95%CI = 0.71–0.98), and heterozygote model (TG VS GG: OR = 0.82, 95%CI = 0.68–0.99). However, when stratified by age, body mass index, source of controls and sample size, no significant association was found. Significant results were also observed in both Caucasian and Mongolian subgroups in rs2241766 and rs266729 polymorphisms (Table 3).
Table 2. Pooled ORs and 95%CIs of the associations between three polymorphisms in ADIPOQ gene and hypertension risk.
| Genetic model | NO. of studies | OR | 95%CI | Pmeta-analysis | Bon | FDR | Statistical method | I2 (%) | Paheterogeneity |
|---|---|---|---|---|---|---|---|---|---|
| rs2241766 | |||||||||
| G VS T | 12 | 1.16 | 1.06–1.27 | 0.002 | 0.010 | 0.007 | Fixed | 22.90 | 0.218 |
| GG VS GT + TT | 12 | 1.34 | 1.10–1.63 | 0.004 | 0.020 | 0.007 | Fixed | 0.00 | 0.597 |
| GG + GT VS TT | 12 | 1.15 | 1.02–1.30 | 0.024 | 0.120 | 0.030 | Fixed | 24.20 | 0.206 |
| GT VS TT | 12 | 1.09 | 0.96–1.23 | 0.204 | 1.000 | 0.204 | Fixed | 25.00 | 0.196 |
| GG VS TT | 12 | 1.38 | 1.12–1.69 | 0.003 | 0.015 | 0.007 | Fixed | 0.00 | 0.457 |
| rs1501299 | |||||||||
| T VS G | 9 | 0.98 | 0.88–1.08 | 0.646 | 1.000 | 0.838 | Fixed | 28.90 | 0.188 |
| TT VS TG + GG | 9 | 1.05 | 0.84–1.32 | 0.670 | 1.000 | 0.838 | Fixed | 33.50 | 0.15 |
| TT + TG VS GG | 9 | 0.95 | 0.83–1.08 | 0.404 | 1.000 | 0.838 | Fixed | 0.00 | 0.458 |
| TG VS GG | 9 | 0.94 | 0.82–1.08 | 0.363 | 1.000 | 0.838 | Fixed | 0.00 | 0.647 |
| TT VS GG | 9 | 1.00 | 0.79–1.26 | 0.988 | 1.000 | 0.988 | Fixed | 37.50 | 0.119 |
| rs266729 | |||||||||
| G VS C | 5 | 1.10 | 0.94–1.28 | 0.219 | 1.000 | 0.365 | Fixed | 29.70 | 0.224 |
| GG VS GC + CC | 5 | 1.43 | 1.02–2.01 | 0.040 | 0.200 | 0.140 | Fixed | 49.00 | 0.098 |
| GG + GC VS CC | 5 | 1.04 | 0.86–1.26 | 0.704 | 1.000 | 0.732 | Fixed | 19.70 | 0.289 |
| GC VS CC | 5 | 0.96 | 0.79–1.18 | 0.732 | 1.000 | 0.732 | Fixed | 41.90 | 0.142 |
| GG VS CC | 5 | 1.41 | 0.99–2.00 | 0.056 | 0.280 | 0.140 | Fixed | 44.20 | 0.127 |
OR = odd ratio; CI = confidence interval.
aP value for between-study heterogeneity based on Q test; Bon = p value in Bonferroni test; FDR = false discovery rate.
Significant results are marked in bold.
Figure 1. Forest plot of hypertension risk associated with the GG genotype in ADIPOQ rs2241766 polymorphism.
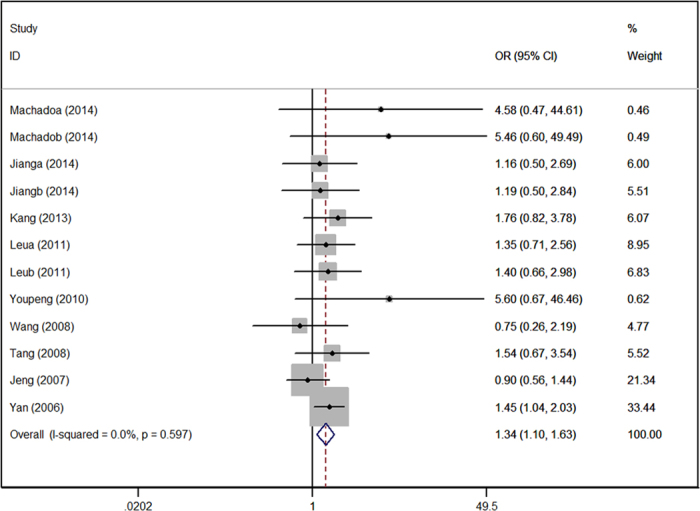
OR = odd ration, CI = confidence interval.
Figure 2. Forest plot of hypertension risk associated with the T allele compared with the G allele in ADIPOQ rs1501299 polymorphism.
OR = odd ration, CI = confidence interval.
Table 3. Stratified analysis of the associations of ADIPOQ polymorphsims with hypertension.
| Subgroup analysis | N | T VS G |
TT VS TG + GG |
TT + TG VS GG |
TG VS GG |
TT VS GG |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | Ph (I2) | OR (95%CI) | Ph (I2) | OR (95%CI) | Ph (I2) | OR (95%CI) | Ph (I2) | OR (95%CI) | Ph (I2) | ||
| rs1501299 | |||||||||||
| Ethnicity | |||||||||||
| Caucasians | 2 | 0.75 (0.58,0.97) | 0.694 (0.0%) | 0.77 (0.47,1.27) | 0.396 (0.0%) | 0.83 (0.71,0.98) | 0.975 (0.0%) | 0.82 (0.68,0.99) | 0.789 (0.0%) | 0.68 (0.42,1.10) | 0.445 (0.0%) |
| Mongolian | 7 | 1.03 (0.92,1.15) | 0.405 (2.6%) | 1.05 (0.85,1.28) | 0.157 (35.5%) | 1.00 (0.93,1.08) | 0.807 (0.0%) | 1.00 (0.92,1.08) | 0.891 (0.0%) | 1.08 (0.87,1.34) | 0.207 (29.0%) |
| Age | |||||||||||
| ≥40 | 5 | 1.00 (0.89,1.13) | 0.398 (1.5%) | 1.06 (0.80,1.39) | 0.105 (47.7%) | 0.99 (0.85,1.15) | 0.705 (0.0%) | 0.98 (0.84,1.15) | 0.729 (0.0%) | 1.04 (0.79,1.38) | 0.137 (42.6%) |
| <40 | 3 | 0.89 (0.71,1.10) | 0.044 (67.9%) | 1.05 (0.68,1.61) | 0.114 (54.0%) | 0.77 (0.57,1.04) | 0.179 (41.8%) | 0.76 (0.55,1.05) | 0.369 (0.0%) | 0.89 (0.55,1.44) | 0.064 (63.7%) |
| BMI of cases (kg/m2) | |||||||||||
| ≥26 | 5 | 0.91 (0.79,1.04) | 0.190 (34.8%) | 0.91 (0.67,1.23) | 0.074 (53.2%) | 0.88 (0.74,1.04) | 0.324 (14.2%) | 0.88 (0.74,1.06) | 0.360 (8.1%) | 0.84 (0.62,1.16) | 0.082 (51.7%) |
| <26 | 4 | 1.07 (0.92,1.25) | 0.482 (0.0%) | 1.25 (0.90,1.75) | 0.595 (0.0%) | 1.04 (0.86,1.27) | 0.696 (0.0%) | 1.01 (0.82,1.25) | 0.874 (0.0%) | 1.24 (0.87,1.77) | 0.508 (0.0%) |
| Source of controls | |||||||||||
| Hospital based | 7 | 1.01 (0.90,1.13) | 0.205 (29.3%) | 1.17 (0.91,1.50) | 0.445 (0.0%) | 0.97 (0.84,1.12) | 0.299 (17.1%) | 0.94 (0.81,1.10) | 0.429 (0.0%) | 1.11 (0.85,1.45) | 0.308 (16.0%) |
| Population based | 2 | 0.86 (0.69,1.07) | 0.313 (1.8%) | 0.69 (0.41,1.16) | 0.072 (69.1%) | 0.87 (0.66,1.15) | 0.738 (0.0%) | 0.93 (0.69,1.24) | 0.828 (0.0%) | 0.66 (0.39,1.14) | 0.087 (65.8%) |
| Sample size | |||||||||||
| Large (≥300) | 4 | 1.00 (0.89,1.14) | 0.255 (26.1%) | 1.00 (0.75,1.33) | 0.110 (50.4%) | 1.01 (0.86,1.18) | 0.650 (0.0%) | 1.01 (0.86,1.19) | 0.914 (0.0%) | 1.00 (0.67,1.48) | 0.160 (39.1%) |
| Small (<300) | 5 | 0.93 (0.78,1.10) | 0.156 (39.8%) | 1.14 (0.79,1.64) | 0.224 (29.6%) | 0.82 (0.65,1.04) | 0.387 (3.5%) | 0.80 (0.62,1.02) | 0.561 (0.0%) | 1.00 (0.74,1.34) | 0.101 (51.8%) |
| rs2241766 | G VS T | GG VS GT + TT | GG + GT VS TT | GT VS TT | GG VS TT | ||||||
| Ethnicity | OR (95%CI) | Ph (I2) | OR (95%CI) | Ph (I2) | OR (95%CI) | Ph (I2) | OR (95%CI) | Ph (I2) | OR (95%CI) | Ph (I2) | |
| Caucasians | 2 | 1.67 (1.16,2.40) | 0.648 (0.0%) | 4.92 (1.03,23.54) | 0.915 (0.0%) | 1.46 (1.07,2.00) | 0.571 (0.0%) | 1.37 (0.99,1.90) | 0.511 (0.0%) | 5.29 (1.11,25.28) | 0.948 (0.0%) |
| Mongolian | 10 | 1.13 (1.03,1.24) | 0.353 (9.7%) | 1.26 (1.06,1.50) | 0.697 (0.0%) | 1.05 (0.99,1.12) | 0.281 (17.6%) | 1.03 (0.96,1.10) | 0.244 (21.7%) | 1.25 (1.06,1.48) | 0.527 (0.0%) |
| rs266729 | G VS C | GG VS GC + CC | GG + GC VS CC | GC VS CC | GG VS CC | ||||||
| Ethnicity | OR (95%CI) | Ph (I2) | OR (95%CI) | Ph (I2) | OR (95%CI) | Ph (I2) | OR (95%CI) | Ph (I2) | OR (95%CI) | Ph (I2) | |
| Caucasians | 2 | 1.13 (0.93,1.38) | 0.423 (0.0%) | 2.13 (1.28,3.54) | 0.915 (0.0%) | 0.97 (0.80,1.18) | 0.238 (28.3%) | 0.81 (0.63,1.04) | 0.125 (57.4%) | 1.87 (1.14,3.07) | 0.859 (0.0%) |
| Mongolian | 3 | 1.05 (0.91,1.20) | 0.109 (54.8%) | 1.06 (0.71,1.59) | 0.204 (37.1%) | 1.04 (0.92,1.19) | 0.230 (31.9%) | 1.05 (0.91,1.21) | 0.451 (0.0%) | 1.10 (0.74,1.63) | 0.129 (51.1%) |
OR = odd ratio; CI = confidence interval; BMI = Body mass index; Ph = P value for heterogeneity test; N = the number of studies.
Figure 3. Forest plot of hypertension risk associated with the GG genotype in ADIPOQ rs266729 polymorphism in subgroup analysis of ethnicity.
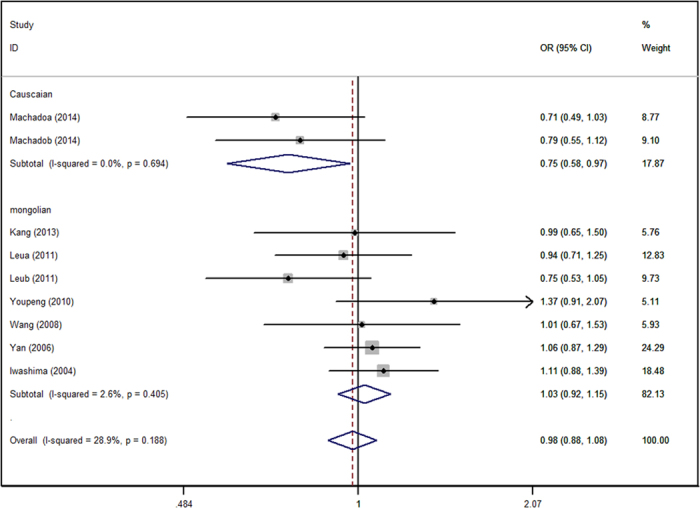
OR = odd ration, CI = confidence interval.
Sensitivity analysis
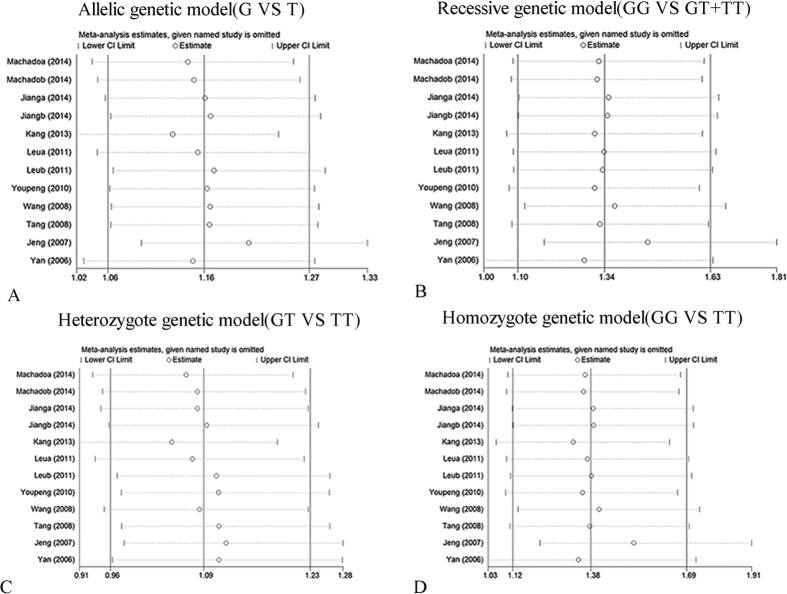
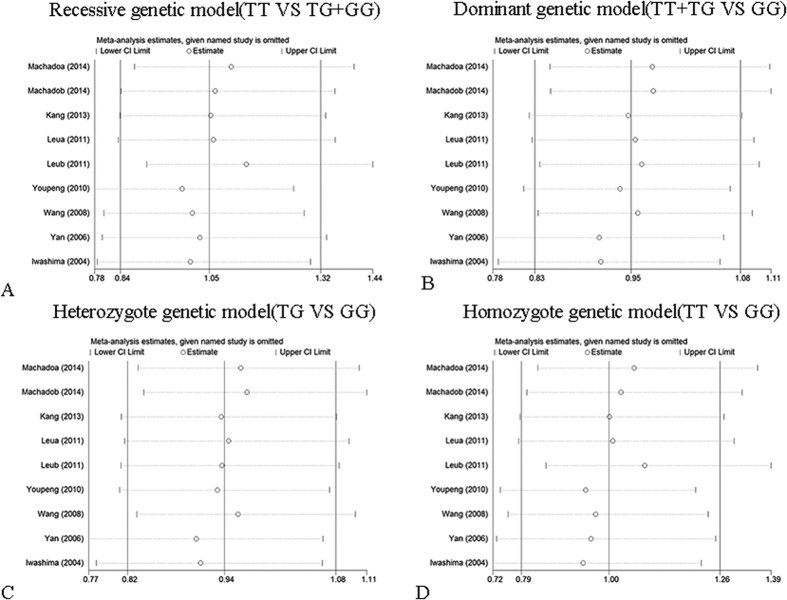
Sensitivity analysis was performed by sequentially omitting 1 individual study; in order to detect the influence of each study on the overall meta-analysis. No heterogeneity was observed in the polymorphisms (Figs 4, 5 and 6), thus the results of our meta-analysis were stable.
Figure 4. Influence analysis of rs2241766 polymorphism and hypertension risk.
(A) Allelic genetic model analysis. (B) Recessive genetic model analysis. (C) Heterozygote genetic model analysis. (D) Homozygote genetic model analysis.
Figure 5. Influence analysis of rs1501299 polymorphism and hypertension risk.
(A) Recessive genetic model analysis. (B) Dominant genetic model analysis. (C) Heterozygote genetic model analysis. (D) Homozygote genetic model analysis.
Figure 6. Influence analysis of rs266729 polymorphism and hypertension risk.
(A) Recessive genetic model analysis. (B) Dominant genetic model analysis. (C) Heterozygote genetic model analysis. (D) Homozygote genetic model analysis.
Publication bias
No publication bias was detected among studies regarding the association between the rs2241766 polymorphism and hypertension risk (Fig. 7). However, for rs1501299 and rs266729, publication bias was not evaluated because the number of studies included was less than 1029.
Figure 7. Begg’s funnel plot for contrast in overall analysis in recessive model.
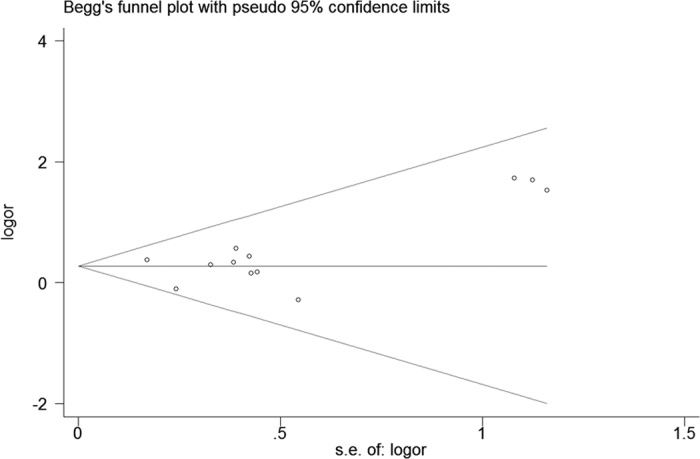
Each point represents a separate study for the indicated association. Size graph symbol by weighs. Log [OR] natural logarithm of OR. Horizontal line means effect size.
Discussion
Endothelial dysfunction plays an important role in the pathogenesis of hypertension30,31,32,33. Adiponectin modulates the endothelial inflammatory response in vitro, and its concentration is decreased in patients with coronary artery disease and hypertension34,35. The low circulating level of adiponectin was found to be related to hypertension36,37, and polymorphisms of the ADPIOQ gene was reported to have a strong association with adiponectin concentration38. Whether the polymorphisms of the ADIPOQ gene are associated with hypertension have attracted growing increased attention. Although there are many researches about the SNPs in ADIPOQ with hypertension, three common gene variants of ADIPOQ, which are rs2241766 (+45 T > G in exon2), rs1501299 (+276 G > T in intron2) and rs266729 (−11377 C > G in proximal promoter region), are widely deeply studied and full of inconsistent results, besides, the data about the three SNPs is sufficient to conduct meta-analysis or subgroup-analysis. Therefore, we choose these three polymorphisms to explore the associations between ADIPOQ gene polymorphisms with hypertension risk in our meta-analysis.
Our results show that the rs2241766 polymorphism in the ADIPOQ gene is significantly associated with hypertension. Increased risk is observed in the allele, recessive, dominant and homozygote genetic model. The minor G allele of ADIPOQ rs2241766 may increase the risk of hypertension by 16% (P = 0.002, OR = 1.16, 95%CI = 1.06–1.27), and GG genotype may increase the risk of hypertension by 34% compared to the GT and TT genotype (P = 0.004, OR = 1.34, 95%CI = 1.10–1.63). When compared with the TT genotype alone, a 38% increase in risk was detected in GG genotype group (P = 0.003, OR = 1.38, 95%CI = 1.21–1.69). In the ethnicity subgroup analysis, increased risk is not only observed in the allele, recessive, dominant and homozygote genetic models in the Caucasian group but also in allele, recessive and homozygote genetic model in Mongolian population. In the eleven included studies, four studies from the Chinese population are consistent with our results18,21,23,26. An increase in hypertension risk attributed to rs2241766 polymorphism was found in the Chinese Bai population and a significant association with serum total triglyceride and low density lipoprotein was also detected in patients with high blood pressure reported by Kang et al.26. In addition, the GG + GT genotype was related to high hypertension susceptibility and low circulating adiponectin level when compared to the TT genotype reported by Tang et al.23. However, the association between rs2241766 polymorphism and plasma adiponectin level was found to be only significant in normal blood pressure group reported by Jeng et al.18. In his opinion, no association between hypertension and rs2241766 polymorphism may be attribute to environmental factors such as diet-induced obesity. The rs2241766 polymorphism at exon2 in ADIPOQ gene may have an influence on the mRNA level in adipose tissue39. It is postulated that the rs2241766 polymorphism may affect the expression of ADIPOQ gene and contribute to the low level of adiponectin in plasma.
Significant association between rs266729 and hypertension is found in the recessive genetic model in our study. The GG genotype may increase the risk of hypertension by 43% compared to GC and CC genotype (P = 0.040, OR = 1.43, 95%CI = 1.02–2.01). In the ethnicity subgroup analysis, increased risk was only detected in the recessive and homozygote genetic model. Four of five included studies addressing the association between rs266729 and hypertension risk were consistent with our result. Haplotype is also an important risk factor in hypertension and the −11426G −11377C haplotype was found to be associated with low plasma adiponectin level in the hypertension group, as reported by Zhang et al.20. The CG genotype of rs266729 was associated with high blood pressure in female group reported by Machado et al.24. In addition, a tendency of increased in DBP towards the CG and GG genotypes of the rs266729 polymorphism in healthy pregnancy women was also detected in his study. The G allele of rs266729 was significantly associated with the increased risk of hypertension by Jiang et al.27. Moreover, not only in Chinese group, an increase in SBP16 and DBP15 caused by rs266729 polymorphism was detected in a British population. Replacing a C with a G in the rs266729 polymorphism occurs in the proximal promoter region and genetic polymorphisms in the promoter region of ADIPOQ are associated with low serum adiponectin level and increased risk of hypertension in a Hong Kong Chinese population15. Based on the present evidence, rs266729 polymorphism is significantly associated with hypertension, however, the mechanism of whether the rs266729 polymorphism has an influence on hypertension susceptibility through the low levels of plasma adiponectin warrants further study.
In the Caucasian subgroup, the rs1501299 polymorphism in the ADIPOQ gene was significantly associated with hypertension. The minor T allele of ADIPOQ rs266729 may decrease the risk of hypertension by 25% (P = 0.026, OR = 0.75, 95%CI = 0.58–0.97), and the TG genotype may decrease the risk of hypertension by 18% compared to GG genotype (P = 0.112, OR = 0.82, 95%CI = 0.68–0.99). The same decreased risk of hypertension was also observed in the dominant genetic model. For the rs1501299 polymorphism, a protective role of rs1501299 polymorphism in hypertension group is found in the Caucasian subgroup, which is contrary to the results of nine included studies. An association between the rs1501299 polymorphism and adiponectin concentration was found in obese subjects (BMI ≥ 26.7 Kg/m2)40. High serum adiponectin in T-allele carriers of rs1501299 polymorphism in the severe preeclampic group was observed by Youpeng et al.21. However, in our study, no significant association was found in the BMI subgroup and gender could not be stratified by subgroup analysis due to lack of details regarding the gender distribution. Although the plasma adiponectin level is low in hypertension patients, we hypothesized that the ability of rs1501299 polymorphism to reduce adiponectin level is weaker than the other polymorphisms, thereby allowing for a relatively higher level of serum adiponectin, which suggests to a protective role for this polymorphism hypertension. More studies are required to explore the precise protective mechanism of rs1501299 polymorphism in hypertension.
There were several limitations in this meta-analysis. First, only English and Chinese articles were included, which may bias the results. Second, patient heterogeneity and confounding factors might have distorted the analysis. Third, the number of included studies was relatively small in some subgroups, and, the results should be interpreted with caution. Fourth, the deviation of genotype distributions from HWE in the control populations in two studies addressing the association between rs2241766 and hypertension may imply genotyping errors or control selection bias in those studies. Fifth, only three common SNPs were evaluated in our study and other relevant SNPs in ADIPOQ which are unknown or understudied or not studies at all may also have potential associations with hypertension. Sixth, most included studies were researches about the ADIPOQ polymorphisms with systolic blood pressure. Whether there are potential relationship between the ADIPOQ polymorphisms with specific hypertension and related outcomes (diastolic blood pressure and pulse pressure) requires additional study. In addition, the potential influence on genotype-hypertension associations by environment factors is worthy of consideration.
In conclusion, the rs2241766 polymorphism was found to be associated with a significant increase in hypertension risk based on our analysis. Moreover, an increased risk in rs266729 in hypertension patients was also detected. Our meta-analysis suggests that rs1501299 polymorphism may play a protective role in hypertension in the Caucasian subgroup; however, additional studies are required to support this finding.
Methods
The systematic review was written in adherence to the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-analyses) checklist41. Ethical approval was not necessary according to local legislation because of the type of study (meta-analysis)42.
Identification of the related Studies
PubMed, Embase, VIP, Wanfang and China National Knowledge Infrastructure databases were thoroughly searched in March 2016 by the first two investigators to identify potential studies addressing the associations between the ADPIOQ gene polymorphisms and hypertension. The terms “hypertension,” “high blood pressure,” “adiponectin,” “ADIPOQ,” “APM1,” “polymorphism,” and “polymorphisms” were used. The missing data (the data that we failed to identify during the electronic search) were obtained by reviewing the citations of review articles and all eligible studies.
Inclusion and Exclusion criteria
To be included in this meta-analysis, studies met the following inclusion criteria: (1) evaluation of the association between the ADIPOQ gene polymorphisms and hypertension; (2) case-control study or cohort design; (3) detailed genotype data could be acquired to calculate odds ratios (ORs) and 95% confidence intervals (CIs); Exclusion criteria: (1) duplication of previous publications; (2) comment, review and editorial; (3) study without detailed genotype data. The selection of the studies was achieved by two investigators independently, according to the inclusion and exclusion criteria by screening the title, abstract and full-text. Any dispute was solved by discussion.
Data Extraction
From each study, the following data were independently extracted by the first two investigators using a standardized form: first author’s last name, year of publication, study country, ethnicity, genotyping methods, age, body mass index, sample size, source of controls, Hardy-Weinberg equilibrium, number of cases and controls, and genotype frequency in cases and controls for ADIPOQ gene. Different ethnicity descents were classified as Caucasian and Mongolian. Disagreements were resolved through discussion.
Quality assessment
Two reviewers independently assessed the quality of the included studies, according to a set of criteria (Table S1) modified on the basis of the Newcastle-Ottawa quality assessment scale. Scores ranged from 0 to 10, with 0 as the lowest and 10 as the highest quality.
Statistics analysis
Hardy–Weinberg equilibrium (HWE) was evaluated for each study by Chi-square test in control groups, and P < 0.05 was considered as a significant departure from HWE. Odds ratio (OR) and 95% confidence intervals (CIs) were calculated to evaluate the strength of the association between ADIPOQ gene polymorphisms and hypertension risk. Pooled ORs were performed for the allelic model (rs2241766: G versus T; rs1501299: T versus G; rs266729: G versus C), recessive model (rs2241766: GG versus GT + TT; rs1501299: TT versus TG + GG; rs266729: GG versus GC + CC), dominant model (rs2241766: GG + GT versus TT; rs1501299: TT + TG versus GG; rs266729: GG + GC versus CC), heterozygote model (rs2241766: GT versus TT; rs1501299: TG versus GG; rs266729: GC versus CC), and homozygote model (rs2241766: GG versus TT; rs1501299: TT versus GG; rs266729: GG versus CC), respectively. Heterogeneity was evaluated by Q statistic (significance level of P < 0.1) and I2 statistic (greater than 50% as evidence of significant inconsistency). Heterogeneity between studies was evaluated with the I2 test, and a higher I2 values means higher levels of heterogeneity (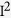 > 90%: extreme heterogeneity;
> 90%: extreme heterogeneity; 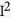 = 70% to 90%: large heterogeneity;
= 70% to 90%: large heterogeneity; 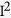 =50% to 70%: moderate heterogeneity;
=50% to 70%: moderate heterogeneity; 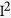 < 50%: no heterogeneity). In heterogeneity evaluation, when the
< 50%: no heterogeneity). In heterogeneity evaluation, when the 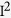 < 50%, the fixed-effects model would be used; if the
< 50%, the fixed-effects model would be used; if the  =50% to 90%, a random-effects model was used; if the
=50% to 90%, a random-effects model was used; if the 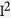 > 90%, the studies would not be pooled. Whenever heterogeneity was significant, sensitivity analysis was performed to detect the heterogeneity by omitting each study in each turn. Besides, subgroup analyses were stratified by ethnicity (Caucasian, Mongolian), age, body mass index, source of controls and sample size. The potential for publication bias was assessed with Begg’s funnel plot and Egg’s test. All the tests in this meta-analysis were conducted with the STATA software (version 12.0; State Corporation, College Station, Texas, USA). To adjust for multiple comparisons, we applied the Bonferroni method43, which control for the false discovery rate (FDR). The power of meta-analysis for each SNP to detect some effect size was estimated according to the method recommended by Hedges and Piggott, given a significant value of 0.0544.
> 90%, the studies would not be pooled. Whenever heterogeneity was significant, sensitivity analysis was performed to detect the heterogeneity by omitting each study in each turn. Besides, subgroup analyses were stratified by ethnicity (Caucasian, Mongolian), age, body mass index, source of controls and sample size. The potential for publication bias was assessed with Begg’s funnel plot and Egg’s test. All the tests in this meta-analysis were conducted with the STATA software (version 12.0; State Corporation, College Station, Texas, USA). To adjust for multiple comparisons, we applied the Bonferroni method43, which control for the false discovery rate (FDR). The power of meta-analysis for each SNP to detect some effect size was estimated according to the method recommended by Hedges and Piggott, given a significant value of 0.0544.
Additional Information
How to cite this article: Fan, W. et al. Associations between polymorphisms of the ADIPOQ gene and hypertension risk: a systematic and meta-analysis. Sci. Rep. 7, 41683; doi: 10.1038/srep41683 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Footnotes
The authors declare no competing financial interests.
Author Contributions Weina Fan, Xiaowei Qu and Liu Wei, Qiang Ma wrote the main manuscript text and Jing Li, Xingning Wang, Yanping Bai and Qingmei Cao prepared Figures 1–4. Figures 5–7 were prepared by Liqun Ma, Xiaoyao Zhou and Wei Zhu. All authors reviewed the manuscript.
References
- Lim S. S. et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet (London, England) 380, 2224–2260, doi: 10.1016/s0140-6736(12)61766-8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M., Maresso K. & Broeckel U. Genetic determinants of hypertension: an update. Current hypertension reports 10, 488–495 (2008). [DOI] [PubMed] [Google Scholar]
- Iwai N. et al. Genetic analysis of 22 candidate genes for hypertension in the Japanese population. Journal of hypertension 22, 1119–1126 (2004). [DOI] [PubMed] [Google Scholar]
- Zadjali F., Al-Yahyaee S., Hassan M. O., Albarwani S. & Bayoumi R. A. Association of adiponectin promoter variants with traits and clusters of metabolic syndrome in Arabs: family-based study. Gene 527, 663–669, doi: 10.1016/j.gene.2013.06.057 (2013). [DOI] [PubMed] [Google Scholar]
- Miyanaga K. et al. C allele of angiotensin II type 1 receptor gene A1166C polymorphism affects plasma adiponectin concentrations in healthy young Japanese women. Hypertension research: official journal of the Japanese Society of Hypertension 32, 901–905, doi: 10.1038/hr.2009.111 (2009). [DOI] [PubMed] [Google Scholar]
- Chung C. M. et al. A genome-wide association study reveals a quantitative trait locus of adiponectin on CDH13 that predicts cardiometabolic outcomes. Diabetes 60, 2417–2423, doi: 10.2337/db10-1321 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Cheng K. K., Vanhoutte P. M., Lam K. S. & Xu A. Vascular effects of adiponectin: molecular mechanisms and potential therapeutic intervention. Clinical science (London, England: 1979) 114, 361–374, doi: 10.1042/cs20070347 (2008). [DOI] [PubMed] [Google Scholar]
- Hotta K. et al. Plasma concentrations of a novel, adipose-specific protein, adiponectin, in type 2 diabetic patients. Arteriosclerosis, thrombosis, and vascular biology 20, 1595–1599 (2000). [DOI] [PubMed] [Google Scholar]
- Pischon T. et al. Plasma adiponectin levels and risk of myocardial infarction in men. Jama 291, 1730–1737, doi: 10.1001/jama.291.14.1730 (2004). [DOI] [PubMed] [Google Scholar]
- Iwashima Y. et al. Hypoadiponectinemia is an independent risk factor for hypertension. Hypertension 43, 1318–1323, doi: 10.1161/01.HYP.0000129281.03801.4b (2004). [DOI] [PubMed] [Google Scholar]
- Vasseur F. et al. Single-nucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Human molecular genetics 11, 2607–2614 (2002). [DOI] [PubMed] [Google Scholar]
- Menzaghi C. et al. Multigenic control of serum adiponectin levels: evidence for a role of the APM1 gene and a locus on 14q13. Physiological genomics 19, 170–174, doi: 10.1152/physiolgenomics.00122.2004 (2004). [DOI] [PubMed] [Google Scholar]
- Park J. W., Park J. & Jee S. H. ADIPOQ Gene Variants Associated with Susceptibility to Obesity and Low Serum Adiponectin Levels in Healthy Koreans. Epidemiology and health 33, e2011003, doi: 10.4178/epih/e2011003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H. K. et al. Influence of adiponectin gene polymorphisms on adiponectin level and insulin resistance index in response to dietary intervention in overweight-obese patients with impaired fasting glucose or newly diagnosed type 2 diabetes. Diabetes care 32, 552–558, doi: 10.2337/dc08-1605 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong K. L. et al. Association of genetic variants in the adiponectin gene with adiponectin level and hypertension in Hong Kong Chinese. European journal of endocrinology/European Federation of Endocrine Societies 163, 251–257, doi: 10.1530/eje-10-0251 (2010). [DOI] [PubMed] [Google Scholar]
- Avery P. J., Patel S. K., Ibrahim I. M., Walker M. & Keavney B. D. Common variation in the adiponectin gene has an effect on systolic blood pressure. Journal of human hypertension 25, 719–724, doi: 10.1038/jhh.2010.122 (2011). [DOI] [PubMed] [Google Scholar]
- Yan W. L. et al. Common SNPs of APM1 gene are not associated with hypertension or obesity in Chinese population. Biomedical and environmental sciences: BES 19, 179–184 (2006). [PubMed] [Google Scholar]
- Jeng J. R. Plasma adiponectin, T94G gene polymorphism and PAI-1 in patients with and without hypertension. Cardiology 107, 30–37, doi: 10.1159/000093610 (2007). [DOI] [PubMed] [Google Scholar]
- Mousavinasab F. et al. Common polymorphisms (single-nucleotide polymorphisms SNP + 45 and SNP + 276) of the adiponectin gene regulate serum adiponectin concentrations and blood pressure in young Finnish men. Molecular genetics and metabolism 87, 147–151, doi: 10.1016/j.ymgme.2005.08.010 (2006). [DOI] [PubMed] [Google Scholar]
- Zhang Z. B. et al. [Association between single nucleotide polymorphisms (SNPs) at the promoter of adiponectin gene and essential hypertension in Chinese Korean and Han of Yanbian region]. Yi chuan = Hereditas/Zhongguo yi chuan xue hui bian ji 33, 54–59 (2011). [DOI] [PubMed] [Google Scholar]
- Youpeng B. et al. Relationships among adiponectin gene polymorphisms, proteinuria and increased blood pressure in the context of placental diseases. Hypertension research: official journal of the Japanese Society of Hypertension 33, 1066–1070, doi: 10.1038/hr.2010.134 (2010). [DOI] [PubMed] [Google Scholar]
- Wang Z. L., Xia B., Jiang L., Hu Z. G. & Cao P. Association between Adiponectin Gene Polymorphisms and Hypertension. Journal of New Medicine 25–26+29 (2008). [Google Scholar]
- Tang X. M., Li J. Q., Wu L. R., Zhuang M. & Lu Z. Relationship between Adiponectin + 45 Nucleotide T/G Polymorphism and Essential Hypertension. Tianjin Medical Journal 762–764 (2008). [Google Scholar]
- Machado J. S. et al. Polymorphisms of the adiponectin gene in gestational hypertension and pre-eclampsia. Journal of human hypertension 28, 128–132, doi: 10.1038/jhh.2013.53 (2014). [DOI] [PubMed] [Google Scholar]
- Leu H. B. et al. Adiponectin gene polymorphism is selectively associated with the concomitant presence of metabolic syndrome and essential hypertension. PLoS One 6, e19999, doi: 10.1371/journal.pone.0019999 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Z. et al. Genetic association analysis of variation in the adiponectin gene promoter with essential hypertension in Dali Bai populations. Guangdong Medical Journal 2957–2959 (2013). [Google Scholar]
- Jiang B. et al. Association of four insulin resistance genes with type 2 diabetes mellitus and hypertension in the Chinese Han population. Molecular biology reports 41, 925–933, doi: 10.1007/s11033-013-2937-0 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J. & Altman D. G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS medicine 6, e1000097, doi: 10.1371/journal.pmed.1000097 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F., Eastwood A. J., Gilbody S., Duley L. & Sutton A. J. Publication and related biases. Health technology assessment (Winchester, England) 4, 1–115 (2000). [PubMed] [Google Scholar]
- Perticone M. et al. Additive Effect of Non-Alcoholic Fatty Liver Disease on Metabolic Syndrome-Related Endothelial Dysfunction in Hypertensive Patients. International journal of molecular sciences 17, doi: 10.3390/ijms17040456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak K. L., Farmer H., Cadnapaphornchai M. A., Gitomer B. & Chonchol M. Vascular dysfunction in children and young adults with autosomal dominant polycystic kidney disease. Nephrology, dialysis, transplantation: official publication of the European Dialysis and Transplant Association - European Renal Association, doi: 10.1093/ndt/gfw013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumral Z. N. et al. Regular exercise alleviates renovascular hypertension-induced cardiac/endothelial dysfunction and oxidative injury in rats. Journal of physiology and pharmacology: an official journal of the Polish Physiological Society 67, 45–55 (2016). [PubMed] [Google Scholar]
- Inan B. et al. Are increased oxidative stress and asymmetric dimethylarginine levels associated with masked hypertension? Clinical and experimental hypertension, 1–5, doi: 10.3109/10641963.2015.1089883 (2016). [DOI] [PubMed] [Google Scholar]
- Ouchi N. et al. Novel modulator for endothelial adhesion molecules: adipocyte-derived plasma protein adiponectin. Circulation 100, 2473–2476 (1999). [DOI] [PubMed] [Google Scholar]
- Chu C. et al. Genetic variants in adiponectin and blood pressure responses to dietary sodium or potassium interventions: a family-based association study. Journal of human hypertension, doi: 10.1038/jhh.2016.5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi M. et al. Blockade of the renin-angiotensin system increases adiponectin concentrations in patients with essential hypertension. Hypertension 42, 76–81, doi: 10.1161/01.hyp.0000078490.59735.6e (2003). [DOI] [PubMed] [Google Scholar]
- Adamczak M. et al. Decreased plasma adiponectin concentration in patients with essential hypertension. American journal of hypertension 16, 72–75 (2003). [DOI] [PubMed] [Google Scholar]
- Heid I. M. et al. Genetic architecture of the APM1 gene and its influence on adiponectin plasma levels and parameters of the metabolic syndrome in 1,727 healthy Caucasians. Diabetes 55, 375–384 (2006). [DOI] [PubMed] [Google Scholar]
- Yang W. S. et al. Allele-specific differential expression of a common adiponectin gene polymorphism related to obesity. Journal of molecular medicine 81, 428–434, doi: 10.1007/s00109-002-0409-4 (2003). [DOI] [PubMed] [Google Scholar]
- Hara K. et al. Genetic variation in the gene encoding adiponectin is associated with an increased risk of type 2 diabetes in the Japanese population. Diabetes 51, 536–540 (2002). [DOI] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D. G. & Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. International journal of surgery (London, England) 8, 336–341, doi: 10.1016/j.ijsu.2010.02.007 (2010). [DOI] [PubMed] [Google Scholar]
- Sun Y., Chen J., Li H., Jiang J. & Chen S. Steroid Injection and Nonsteroidal Anti-inflammatory Agents for Shoulder Pain: A PRISMA Systematic Review and Meta-Analysis of Randomized Controlled Trials. Medicine 94, e2216, doi: 10.1097/MD.0000000000002216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X. Y., Ding J. Q. & Chen S. D. Apolipoprotein E promoter polymorphisms and risk of Alzheimer’s disease: evidence from meta-analysis. Journal of Alzheimer’s disease: JAD 19, 1283–1294, doi: 10.3233/jad-2010-1329 (2010). [DOI] [PubMed] [Google Scholar]
- Hedges L. V. & Pigott T. D. The power of statistical tests in meta-analysis. Psychological methods 6, 203–217 (2001). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.