Abstract
BACKGROUND
The Colorado potato beetle, Leptinotarsa decemlineata (Say), is a major agricultural pest of commercial potatoes. Pest managers use a combination of control tactics to limit populations, including multiple insecticides. Finding a window of insecticide susceptibility and understanding genetic responses to insecticide exposure during a growing season may provide novel management recommendations for L. decemlineata.
RESULTS
We examined temporal changes (during one growing season) in phenotypic response between a susceptible population and an imidacloprid‐resistant population. Beetles remained more susceptible to imidacloprid in the susceptible population throughout the growing season. Estimated mean LC50 values varied throughout the growing season in the resistant population, with increased susceptibility among overwintered and recently emerged adult beetles compared with a heightened level of resistance in the second generation. RNA transcript abundance was compared among multiple time points through the growing season, showing that cuticular proteins and cytochrome p450s were highly upregulated during peaks of measured resistance.
CONCLUSION
Temporal variation in imidacloprid susceptibility of L. decemlineata was observed, which included early time points of susceptibility and later peaks in resistance. Heightened resistance occurred during the second generation and correlated to increased transcript abundance of multiple mechanisms of resistance, including multiple cuticular protein and cytochrome p450 transcripts. © 2016 The Authors. Pest Management Science published by John Wiley & Sons Ltd on behalf of Society of Chemical Industry.
Keywords: Colorado potato beetle, imidacloprid, transcript abundance, cuticular protein, insecticide resistance
1. INTRODUCTION
The Colorado potato beetle (CPB), Leptinotarsa decemlineata (Say), is a key agricultural pest of potatoes (Solanum tuberosum), tomatoes (Solanum lypcopersicum), eggplants (Solanum melongena) and peppers (Solanum annuum),1 causing significant crop loss and direct damage that can lead to loss of revenue for commercial growers. Leptinotarsa decemlineata has become a global pest, occupying over 16 million km2,1, 2 and impacting potato production in North America and Eurasia. According to the United Nations Food and Agricultural Organization, the USA produced 19.8 million tons of potatoes in 2013, and it is one of the leading vegetable crops in the country.3 The impact of L. decemlineata on individual state agricultural markets is also significant, especially in Wisconsin, where potato production accounts for more than $310 million annually.4
The history of insecticidal inputs for control of L. decemlineata is a story retold in many potato‐producing areas of the country, where many classes of insecticides have been effective for short periods of time, before the beetles become resistant.2 Recent estimates suggest that populations of L. decemlineata have now become resistant to more than 56 insecticides.2, 5 More recently (1995), the registration and introduction of the neonicotinyl insecticide class (IRAC Classification, Group 4A, nicotinic acetylcholine receptor (nAChR) agonists) has resulted in the use of active ingredients, which include, but are not limited to, imidacloprid, thiamethoxam, clothianadin, and dinotefuran.2, 6 Since the initial introduction of this insecticide class in the mid‐1990s, populations of L. decemlineata have steadily developed resistance, but it remains the principal insecticidal tool used for crop protection in potato.2, 7, 8, 9, 10, 11
Temporal patterns of phenotypic variation in resistance to insecticides within populations of L. decemlineata has been previously suggested between generations in this pest species.2, 7 Specifically, these studies suggest that the second‐generation population is significantly more resistant when compared to its first‐generation counterpart.12 In the current investigation, we hypothesize that temporal patterns in phenotypic variation in imidacloprid resistance may not be limited to the differences between the first and second generation, but may also vary at additional time points throughout the growing season. Uncovering new information about the susceptibility at specific time points would provide insights into the design of pest management strategies and enhance the effectiveness of insecticide deployment.
Recent investigations have examined transcriptomic data to classify possible mechanisms of pesticide resistance in L. decemlineata.11, 13, 14 These studies examined transcript abundance in relation to insecticide‐resistant populations independent of collection time, with the goal of classifying over‐expressed transcripts and contigs in resistant populations. These upregulated transcripts provide an initial glimpse into enzymatic detoxification mechanisms, such as those mediated by cytochrome p450s and glutathione S‐transferases, taking place within a select set of adult L. decemlineata, but are limited to a discrete time point over the growing season. We hypothesize that at time points of increased imidacloprid resistance transcripts that encode for known mechanisms of resistance, such as cytochrome p450s and glutathione S‐transferases, will be expressed with increased abundance. Furthermore, these peaks in resistance and the associated upregulated transcripts are known to differ between resistant and susceptible populations.11 Uncovering up‐regulated transcripts provides important new information on how this species combats insecticide inputs and could lead to improvements in our ability to manage resistant populations of this damaging pest.
2. EXPERIMENTAL METHODS
2.1. Beetle collection
Two populations of L. decemlineata were collected from two field locations in the Central Sands region of Wisconsin in the spring and summer of 2015. The first population represents a documented imidacloprid‐susceptible population collected from the University of Wisconsin's Arlington Agricultural Research Station (AARS), Arlington, which is located approximately 5.5 km SE of the city of Arlington, Wisconsin. The second population was collected from a commercial agricultural field with a previously documented history of imidacloprid resistance and termed ‘Systemic‐3’,7 and this field is located approximately 4.8 km SW of the city of Hancock, Wisconsin. From the two populations, adult beetles were collected at four time points, representing: (i) early emergence from diapause (28 May to 1 June); (ii) late emergence from diapause (16–20 June); (iii) conclusion of first generation (26–30 June); and (iv) emergence of second generation (10 July to 10 September). Efforts were made to ensure that each collection represented the aforementioned distinct subgroups of adult beetles at each of the two experimental locations by staggering collections due to the natural differences in phenology of emergence. This staggered collection resulted from the fact that the two locations differed only very slightly in longitude (0.153°), but did depart further in latitude (0.796°). On the first collection dates, approximately 2000 adult beetles were collected from each field within the first 48 h of initial adult emergence and field colonization. On the second collection dates, an estimated 2000 adult beetles were again collected from the Systemic‐3 population and approximately 1900 adult beetles were collected from the AARS site. On the third range of dates of adult collection, approximately 500 beetles were obtained from both sites. Taken together, these first three collection dates effectively encompass the first generation (post‐diapause) of L. decemlineata present at each experimental field location. The fourth and final set of collections represented adult emergence and colonization by the second generation of L. decemlineata, where approximately 300 beetles were obtained from each location. Due to the low number of beetles in the second‐generation populations, we were unable to collect more than 300 beetles at this time. Furthermore, scouting of the beetles was performed regularly through each of the field locations in order to ensure that adult beetles were collected at appropriate times to represent the categories described previously. Due to the low number of beetles present in the second‐generation population at the AARS, we were unable to collect beyond the fourth collection date (10 July). However, due to the more abundant populations at the Systemic‐3 field site, we continued collections at three additional time points (20 July, 27 August, and 10 September) during the late summer and collectively refer to these additional collections as the fourth collection interval for this specific location. Following collection from the field, all adults representing unique phenological time points were held on pesticide‐free, field‐grown potato plants located in an agricultural field and secured within separate 1 m3 mesh cages (BioQuip Products, Inc., Rancho Dominguez, CA, USA).
2.2. LC50 assays
After beetles were placed in their respective cages, median lethal concentration (LC50) assays were performed as previously described by Zhao et al.9 and results were used to characterize variation in resistant phenotypes throughout the growing season. Every 4 days, 90–270 adult beetles were randomly selected from each field cage, representing a different field collection date and location, to conduct LC50 assays. Initially we aimed to assay approximately 225–270 adult beetles for each LC50 assay, but as the season progressed and mortality increased only assays with smaller sample sizes were possible on later assay dates. To establish an initial dose range for the study, a pre‐screening assay was conducted on 1 June from among a randomly selected set of adults from each collection site and were dosed with a concentration gradient of imidacloprid (Technical grade 98.80%) carried in acetone (imidacloprid contents of 0.0034–1.74 µg µL−1). Specifically, 1 µL of solution was placed on the first abdominal sternite of a subset of adult beetles and the material was absorbed within 3–5 s following topical application. From the pre‐screening assays, we determined that the adult beetles from the AARS would be serially dosed with concentrations of 0, 0.00034, 0.0034, 0.034, and 0.17 µg µL−1 of imidacloprid in acetone in order to accurately estimate LC50 values, whereas adult beetles from the imidacloprid‐resistant, Systemic‐3 population were dosed with concentrations of 0, 0.034, 0.17, 0.69, and 1.74 µg µL−1. The pre‐screening assay process was intermittently performed throughout the growing season as beetles became more or less responsive to the initially predetermined dose ranges, and in multiple cases the dose range was adjusted for increased imidacloprid resistance. Prior to the full screening assay with the full range of serial imidacloprid doses, adult beetles were first placed into Petri dishes (five beetles per dish) and equally divided for the assay containing five serial concentrations. One microliter of the imidacloprid solution was topically applied to replicate sets of five adult beetles/dose, and adults were held dorsal side down until the solution had been completely absorbed (e.g., 3–5 s) and were then placed back into their respective Petri dishes. Following topical application, all Petri dishes containing adults, plus fresh, untreated potato foliage, were held in an incubator at 26 °C, 72% relative humidity, and a photoperiod of 16/8 h (light/dark). Adult beetles were maintained under these conditions for 7 days before any response (e.g., mortality) was assessed (Proc Probit, SAS).15 In total, 15 time points were assessed throughout the growing season for the imidacloprid‐susceptible, AARS population, whereas a total of 17 time points were assessed for the imidacloprid‐resistant Systemic‐3 population.
2.3. RNA extraction and RNA sequencing
At similar time points for which LC50 assays were performed, randomly selected subgroups of (N = 3 each site) untreated beetles were similarly collected and later used for RNA extraction. Total RNA was extracted using Trizol (Life Technology, Grand Island, NY, USA) and stored at −80 °C for later analysis. The University of Wisconsin–Madison, Biotechnology Center was contracted to isolate and generate mRNA libraries and run Illumina HiSeq 2500 1X100bp sequencing. We conducted RNA sequencing (RNA‐seq) to examine transcript abundance throughout the growing season in the imidacloprid‐resistant population, and available funding only allowed us to examine 11 of the 17 time points that corresponded to times when LC50 measurements were performed in the imidacloprid‐resistant population. Prior to submitting samples to the Biotechnology Center, RNA was initially pre‐treated with TurboDNase (Life Technology, Grand Island, NY, USA), and the DNA‐free RNA was cleaned from protein with a phenol–chloroform extraction, and an EtOH precipitation was conducted to remove any other contaminates. Approximately 1500 ng RNA was submitted to the Biotechnology Center, and the RNA was analyzed with a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) before RNA sequencing was conducted.
2.4. Differential transcript comparison and enrichment analysis
After high‐quality reads were generated, the University of Wisconsin–Madison, Biotechnology Center further cleaned and aligned the raw reads to unannotated L. decemlineata genomic scaffolds available from Baylor College.16 As a research participant, the Biotechnology Center also examined transcript abundance and the difference in transcript abundance between different collections through the use of RSEM,17 EdgeR,18 and EBSeq.19 This was quantified using FPKM (fragments per kilobase of exon per million fragments mapped), TPM (transcripts per million), and read counts to generate a ‘gene count’ to examine differentially expressed transcripts. Finally, the Biotechnology Center compared transcript abundance between first and second generations of collected adult L. decemlineata in three contrasts: early emergence from diapause versus second generation, late emergence versus second generation, conclusion of first generation versus second generation. These comparisons were conducted by examining all the ‘gene counts’ of the first‐generation collection versus all the ‘gene counts’ of the second generation. Fold change and an FDR (false discovery rate) were also calculated for each transcript. Transcripts that had a fold change greater than 2 and an FDR less than ($P ≤ 0.049$) were considered differentially expressed and upregulated. Using standalone blast with BLASTx, all transcripts including the upregulated transcripts were compared to reference proteins (E value < 10−3). Transcripts were classified based on the NCBI nomenclature returned by BLASTx. The database of Reference Protein Sequences (Refseq) from Tribolium castaneum, Acyrthosiphon pisum, Anopheles gambiae, Drosophila melanogaster, and Pediculus humanus was downloaded from NCBI for a total of 80,49 8 sequences and used to classify transcripts. BLASTx results were examined for upregulated transcripts known to play a role in insecticide resistance including cuticular proteins, cytochrome p450s, glutathione S‐transferases, ABC transporters, and carboxylesterases. BLASTx results were uploaded into Blast2Go20 for further data analysis. Upregulated transcripts were first analyzed and the components were mapped to the corresponding GO terms. The annotation step was run with a cut‐off of E value < 1E‐3, annotation cut‐off > 45, and GO weight > 5. An enrichment analysis was performed between all the upregulated transcripts in the second‐generation collection and all aligned transcripts were run using a two‐tailed FDR test with a 0.005 cut‐off in order to determine whether any group of GO terms were differentially expressed in the upregulated components.
2.5. Statistical analysis
Statistical analyses were conducted to determine whether the three biological comparisons were dissimilar using a Bray–Curtis dissimilarity index to examine transcripts upregulated by a fold change of 2 and a fold change of 100. In order to incorporate a ‘gene count’ dissimilarity comparison between the three biological conditions, we initially subtracted the second‐generation ‘gene count’ from that of the first generation to obtain an absolute value. The absolute value of the resulting difference in gene count was then divided by the average of both gene counts, resulting in a value that ranged between 0 and 2, with 0 representing identical transcript expression values and 2 representing completely different values. All of the upregulated transcripts were then assigned as similar or dissimilar, using a cut‐off of <0.5 to represent similar values. Finally, estimates were input into a Bray–Curtis analysis to examine dissimilarities between time points, which provides an output between 0 and 1, with 0 being completely similar and 1 being completely dissimilar (UW Statistical Consulting, Bray–Curtis analysis).21
2.6. Quantitative polymerase chain reaction (qPCR)
To confirm transcript abundance from RNA‐seq data, qPCR was conducted. Three technical replicates of pooled RNA from each unique collection time point in 2015 were used in cDNA synthesis for qPCR. Total RNA from each population was quantified using a Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA), and DNA contamination was removed using TurboDNase (Life Technology, Grand Island, NY, USA).Total RNA was purified through EtOH precipitation, air dried until no visible liquid was observed, and then suspended in 100 µL DNase/RNase‐free H2O. All RNA concentrations were equalized before input into the cDNA synthesis kit, and the subsequent cDNA was generated with a Super Script III kit (ThermoFisher Scientific). The cDNA was diluted to a final concentration of 5 ng µL−1 RNA equivalent input for qPCR. β‐Actin was used as a reference gene in these investigations, and the β‐actin primers were shortened versions of those previously described by Zhu et al.22 The qPCR reaction was run on a CFX‐96 platform (Bio‐Rad Laboratories, Hercules, CA, USA) with a master mix of Bullseye EverGreen (MIDSCI, Valley Park, MO, USA). Genes of interest (GOI) were selected based on their relevance to this study and primers were designed to contigs found in the generated transcripts. The qPCR reactions were conducted using the Pfaffl efficiency calibrated methodology; primer and primer efficiency (amplification efficiency of reactions as described by Pfaffl23) are found in Supplementary Table 1 (supporting information). Primer specificity was checked against the transcriptome using BLAST. Triplicate reactions were run at 95 °C for 10 min, followed by 95 °C for 15 s, and 62 °C for 60 s for a total of 40 cycles. Data were collected for each biological replicate, and the relative expression of resistant time points to susceptible time points was calculated using the Pfaffl methodology.23 The Pfaffl methodology takes into consideration the efficiency of the primer sets and provides the ratio of the target gene to the reference population.
Table 1.
Regression estimates for median lethal concentration assays (LC50) resulting from topical bioassays of adult L. decemlineata for the Arlington Agricultural Research Station (AARS), imidacloprid‐susceptible and Systemic‐3, imidacloprid‐resistant populations during summer 2015
| Population | Assay date | N | LC50 (µg µL−1) | 95% CIa | χ 2 b | PR > χ 2 |
|---|---|---|---|---|---|---|
| Early emergence | ||||||
| Arlington | 9 June | 225 | 0.0052 | (0.00051–0.021) | 19.43 | <0.0001 |
| Arlington | 13 June | 225 | 0.058 | (0.0015–0.61) | 8.35 | 0.0038 |
| Arlington | 21 June | 270 | 0.15 | (0.0100.73) | 11.47 | 0.0007 |
| Arlington | 25 June | 270 | 0.099 | (NA*) | 3.80 | 0.051 |
| Arlington | 29 June | 180 | 0.021 | (NA*–0.14) | 11.62 | 0.0006 |
| Systemic‐3 | 9 June | 225 | 1.5 | (0.18–19) | 7.28 | 0.007 |
| Systemic‐3 | 13 June | 225 | 3.5 | (1.1–53) | 13.54 | 0.0002 |
| Systemic‐3 | 21 June | 270 | 7.5 | (3.0–49) | 19.53 | <0.0001 |
| Systemic‐3 | 25 June | 270 | 7.5 | (4.5–19) | 26.29 | <0.0001 |
| Systemic‐3 | 29 June | 180 | 2.4 | (1.6–3.8) | 32.28 | <0.0001 |
| Late emergence | ||||||
| Arlington | 25 June | 270 | 0.18 | (0.13–0.24) | 23.79 | <0.0001 |
| Arlington | 29 June | 270 | 0.23 | (0.067–0.68) | 11.56 | 0.0007 |
| Arlington | 3 July | 270 | 0.073 | (NA*–0.24) | 4.96 | 0.030 |
| Arlington | 6 July | 270 | 0.099 | (0.0059–0.40) | 10.50 | 0.0012 |
| Arlington | 10 July | 270 | 0.11 | (0.015–0.55) | 14.31 | 0.0002 |
| Arlington | 14 July | 90 | 0.18 | (0.020–13) | 8.73 | 0.0031 |
| Systemic‐3 | 25 June | 270 | 6.8 | (5.5–10) | 11.11 | 0.0009 |
| Systemic‐3 | 29 June | 270 | 8.7 | (6.3–30) | 8.29 | 0.004 |
| Systemic‐3 | 3 July | 270 | 8.7 | (6.0–13.9) | 9.84 | 0.0017 |
| Systemic‐3 | 6 July | 270 | 11 | (4.7–40) | 4.79 | 0.028 |
| Systemic‐3 | 10 July | 270 | 9.0 | (2.4–15) | 5.62 | 0.017 |
| Systemic‐3 | 14 July | 180 | 4.1 | (3.1–5.5) | 46.31 | <0.0001 |
| Conclusion of first generation | ||||||
| Arlington | 10 July | 180 | 0.18 | (0.12–0.27) | 20.73 | <0.0001 |
| Arlington | 14 July | 180 | 0.14 | (0.063–0.36) | 28.32 | <0.0001 |
| Arlington | 18 July | 90 | 0.030 | (NA*) | 2.68 | 0.10 |
| Sytemic‐3 | 10 July | 180 | 5.7 | (4.2–8.9) | 29.14 | <0.0001 |
| Sytemic‐3 | 14 July | 180 | 8.7 | (0.050–23) | 4,7 | 0.03 |
| Sytemic‐3 | 18 July | 90 | 5.3 | (3.7–8.3) | 19.81 | <0.0001 |
| Second generation | ||||||
| Arlington | 21 July | 270 | 0.28 | (0.016–1.3) | 5.65 | 0.017 |
| Systemic‐3 | 21 July | 270 | 8.4 | (4.1–20) | 19.18 | <0.0001 |
| Systemic‐3 | 27 August | 270 | 15 | (7.0–28) | 16.58 | <0.0001 |
| Systemic‐3 | 10 September | 270 | 14 | (4.9–28) | 13.77 | 0.0002 |
95% confidence interval (CI) estimates around mean LC50 estimates.
Chi‐square analysis of effects of the proc probit regression.
NA represents a probit mortality regression estimate without a 95% confidence interval (CI).
3. RESULTS
3.1. LC50 comparison
Median lethal concentration (LC50) assays were conducted on both an imidacloprid‐susceptible and a resistant population of L. decemlineata to determine temporal patterns of phenotypic variation in imidacloprid susceptibility throughout a growing season. During the growing season, four collection times were imposed on both susceptible and imidacloprid‐resistant field populations. The collections represent a first‐generation population of beetles sampled at time points corresponding to the earliest emerging beetles from diapause (first quartile), late‐emerging beetles from diapause (second and third quartile) and finally beetles emerging from the soil at the conclusion of the first generation (fourth quartile). A second generation of adult beetles, which were offspring of the first generation population, were also sampled. The beetles collected were held in mesh cages in an agricultural potato field. Subsets of individuals were then removed to determine LC50 values throughout the life cycle for each collection time. LC50 assays were conducted at 15 time points for the imidacloprid‐susceptible (AARS) population and 17 time points for the imidacloprid‐resistant (Systemic‐3) population (Table 1). The mean LC50 estimates uncovered considerable phenotypic variation in imidacloprid susceptibility over the four collection intervals, specifically representing the three time points of first‐generation emergence (e.g., early, late, and conclusion emergence periods) compared to the second‐generation emergence (Table 1 and Fig. 1). With few exceptions, the LC50 estimates were statistically higher during the growing season in the imidacloprid‐resistant population when compared to the susceptible population over the entire sampling season (Table 1 ). Further examination of the mean LC50 values observed in the imidacloprid‐resistant population demonstrate that adult beetles appear to be more susceptible immediately after emergence and colonization, and again later in the development of their adult life cycle before they reach the end of their adult lives.
Figure 1.
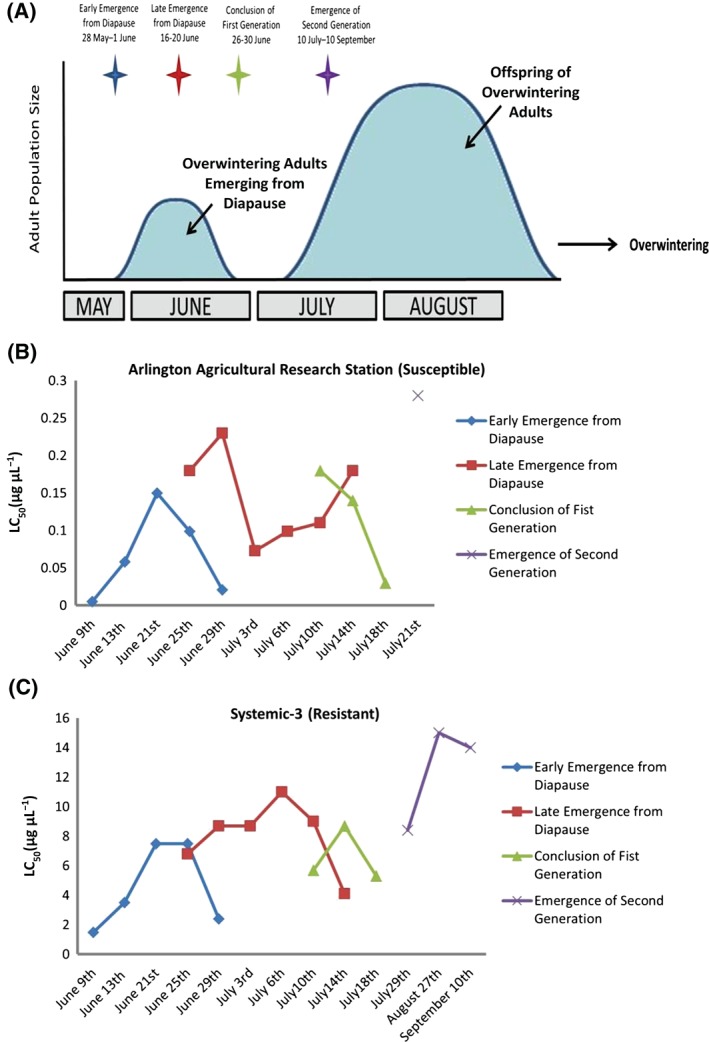
(A) Dynamics of adult population size throughout the growing season, as suggested by Huseth et al.,24 along with collection dates. (B) Median lethal concentration (LC50) estimates representing the four different sampling intervals plotted over the season for the imidacloprid‐susceptible and (C) imidacloprid‐ resistant populations of L. decemlineata. Note significant differences in the scale of median LC50 (µg µL−1) estimates for each population.
3.2. Differential transcript comparison
From the imidacloprid‐resistant, Systemic‐3 population, 11 unique time points (nine from the first generation and two from the second generation) were sequenced to examine transcript abundance (Fig. 2). Time points were selected to cover all four collection intervals including time points with high and low imidacloprid susceptibility. We conducted three transcript abundance comparisons between the specific emergence periods in the first generation versus the second generation (Supplementary Fig. 1, supporting information) to determine upregulated transcripts in the second‐generation population that could partially explain the phenotypic increases in levels of measured resistance. These comparisons examined all ‘gene counts’ across generational comparisons. From the three comparisons, candidate molecular mechanisms of resistance were classified (Table 2). These mechanisms of resistance were classified based on significant levels of fold change and FDR. Here again, a transcript was considered upregulated if there was a fold change greater than 2 and an FDR < 0.049. A candidate list of possible mechanisms of resistance can be seen in Supplementary Table 2 (supporting information). Similarly, highly upregulated transcripts were classified in the second‐generation population to uncover trends in transcript abundance. This was done by examining transcripts encoding possible mechanisms of resistance which were upregulated greater than 100‐fold and FDR < 0.049 (Table 3). The results from the three comparisons demonstrate a set of 13 cuticular proteins and a cytochrome p450 which were highly upregulated in the second generation when compared with the discrete emergence intervals (early, middle, and late) of the first generation. We further conducted an enrichment analysis between the transcripts that were upregulated in the second‐generation population and all the transcripts assembled from RNA sequencing to determine whether there were any apparent trends in the upregulated transcripts (Supplementary Table 3, supporting information). Both over‐ and under‐expression of 51 GO terms was observed in this analysis, including over‐expression in the levels of oxidoreductase activity, monoooxygenase activity and structural constituents of the cuticle.
Figure 2.
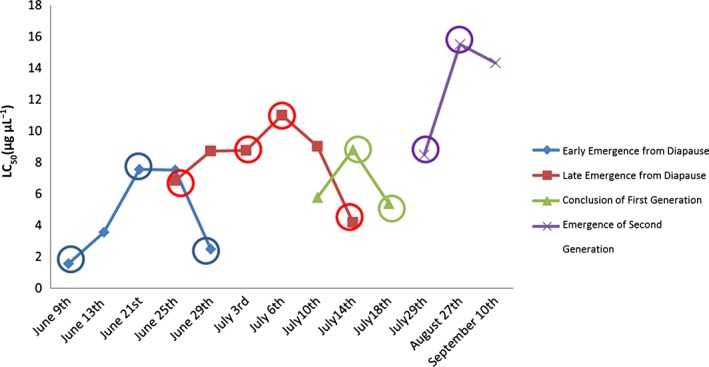
Time points of adult L. decemlineata collection chosen from the Systemic‐3 field location for RNA sequencing. Colored circles correspond to the specific time points chosen for RNA sequencing.
Table 2.
Number of differentially expressed transcripts between the three discrete emergence time points of first‐generation compared to second‐generation collection time points in the imidacloprid‐resistant field population. Transcripts were considered upregulated if there was a fold change of 2 and FDR value ≤ 0.049
| Total upregulated Transcripts in the second‐generation population compared to the first generation | Transcripts encoding for cuticular proteins | Transcripts encoding for cytochrome P450 | Transcripts encoding for glutathione S‐transferase | Transcripts encoding for ABC transporters | Transcripts encoding for carboxylesterase | |
|---|---|---|---|---|---|---|
| Early emergence vs. second generation | 469 | 38 | 10 | 2 | 0 | 0 |
| Late emergence vs. second generation | 624 | 44 | 11 | 2 | 2 | 1 |
| Conclusion of first generation vs. second generation | 423 | 40 | 13 | 2 | 2 | 0 |
| Total unique genes | 728 |
Table 3.
Upregulated transcripts with a log fold change > 10 and FDR ≤ 0.049 that could encode for increased imidacloprid resistance in the second generation of L. decemlineata at the Systemic‐3 location. NCBI accession numbers represent the BLAST hit
| Transcript blast × Result | Fold change | NCBI accession number | Transcript ID |
|---|---|---|---|
| Early emergence from diapause vs. second generation | |||
| Cuticular protein | 993.839 | XP_966639.1 | LDEC003961 |
| Cuticular protein ld‐cp1v1 | 473.248 | XP_970573.1 | LDEC005679 |
| Cuticular protein isoform a | 437.191 | NP_647668.1 | LDEC024510 |
| Cuticular protein 62bc cg1919‐pa | 226.513 | XP_968445.1 | LDEC003211 |
| Cuticular protein 92f cg5494‐pa | 202.820 | XP_969801.1 | LDEC014400 |
| Cytochrome p450 | 167.761 | XP_973153.1 | LDEC016769 |
| Cuticle protein cp5 | 166.128 | XP_973942.1 | LDEC006896 |
| Cuticle protein 1 | 110.520 | XP_970381.1 | LDEC003423 |
| Cuticular protein rr‐1 family (agap000344‐pa) | 108.965 | XP_971011.1 | LDEC013734 |
| Late emergence from diapause vs. second generation | |||
| Cuticular protein ld‐cp1v1 | 1804.609 | XP_970573.1 | LDEC005679 |
| Cuticular protein | 1411.250 | XP_966639.1 | LDEC003961 |
| Cuticular protein ld‐cp3 | 1212.622 | XP_973909.1 | LDEC006898 |
| Cuticular protein precursor | 835.095 | NP_001161316.1 | LDEC013733 |
| Cuticular protein 92f cg5494‐pa | 655.360 | XP_969801.1 | LDEC014400 |
| Cuticle protein 1 | 631.133 | XP_970381.1 | LDEC003423 |
| Cuticular protein rr‐1 family (agap000344‐pa) | 549.102 | XP_971011.1 | LDEC013734 |
| Cytochrome p450 | 199.748 | XP_973153.1 | LDEC016769 |
| Cuticular protein precursor | 133.291 | NP_001161313.1 | LDEC014399 |
| Conclusion of first‐generation emergence from diapause vs. second generation | |||
| Cuticular protein 92f cg5494‐pa | 1399.634 | XP_969801.1 | LDEC014400 |
| Cuticular protein | 1166.062 | XP_966639.1 | LDEC003961 |
| Cuticular protein 62bc cg1919‐pa | 624.162 | XP_968445.1 | LDEC003211 |
| Cuticular protein ld‐cp3 | 536.339 | XP_973909.1 | LDEC006898 |
| Cuticular protein ld‐cp1v1 | 399.977 | XP_970573.1 | LDEC005679 |
| Cuticular protein rr‐1 family (agap000344‐pa) | 371.136 | XP_971011.1 | LDEC013734 |
| Cuticular protein isoform a | 355.026 | NP_647668.1 | LDEC024510 |
| Cytochrome p450 | 271.828 | XP_973153.1 | LDEC016769 |
| Cuticle protein 1 | 260.013 | XP_970381.1 | LDEC003423 |
| Cuticular protein precursor | 202.230 | NP_001161316.1 | LDEC013733 |
| Cuticle protein cp5 | 141.040 | XP_973942.1 | LDEC006896 |
| Cuticle protein 20 | 105.933 | XP_968593.1 | LDEC017994 |
3.3. Statistical analysis
To determine whether there were dissimilarities in transcript abundance from among the three time point comparisons to the second‐generation collection, a Bray–Curtis dissimilarity analysis was performed. The analysis was conducted at a fold change greater than 2 and a fold greater than 100 (Supplementary Table 4, supporting information). At a fold change of greater than 2, we concluded that the comparison results were similar, with values between 0.25 and 0.32 for the three comparisons, suggesting that the upregulated transcripts were rather similar at a fold change greater than 2. The Bray–Curtis analysis of transcripts with a fold change greater than 100 demonstrated substantially more dissimilarities, with values between 0.30 to 0.73, suggesting that the upregulated transcripts with a fold change greater than 100 were much more dissimilar, and that as the fold change increases the transcripts become more dissimilar.
3.4. Confirmation with qPCR
To confirm transcript abundance generated through the use of RNA sequencing, qPCR assays were performed between the late emergence time point of the first generation and the second generation collections in the Systemic‐3, imidacloprid‐resistant, population to confirm upregulated transcripts. We specifically focused on four transcripts that we classified as upregulated and that could play a role in imidacloprid resistance; they included three highly upregulated cuticular proteins and a single cytochrome p450. β‐Actin expression was used as a reference control in our investigations. We confirmed the transcripts were upregulated by calculating the transcript expression ratio with the Pfaffl methodology and the fold change of the ‘gene count’ (Table 4 ).
Table 4.
Transcript expression determined by quantitative PCR
| Late emergence | Second generation | |||
|---|---|---|---|---|
| Mean CT ± SD | Mean CT ± SD | Fold change ‘gene count’ | qPCR expression ratio | |
| β‐Actin (reference) | 21.32 ± 0.14 | 19.96 ± 0.01 | N/A | N/A |
|
LDEC003961 (cuticular protein) |
32.28 ± 0.25 | 21.12 ± 0.01 | 1411.250 | 752.12 |
|
LDEC014400 (cuticular protein) |
30.79 ± 0.32 | 20.86 ± 0.06 | 655.360 | 422.13 |
|
LDEC003423 (cuticular protein) |
31.42 ± 1.5 | 21.24 ± 0.03 | 631.133 | 388.94 |
|
LDEC016769 (cytochrome p450) |
37.48 ± 1.0 | 27.22 ± 0.17 | 199.748 | 325.65 |
4. DISCUSSION
The life cycle of L. decemlineata has been previously described in detail, including investigations into which specific developmental stages (e.g., eggs, larvae, adults) are the best targets for insecticide treatments.24, 25 However, the phenotypic variation in imidacloprid resistance over the growing season in adult L. decemlineata has yet to be examined. In this study, we examined phenotypic changes in imidacloprid resistance throughout a growing season in both an imidacloprid‐susceptible and resistant population through the use of imidacloprid LC50 assays. The imidacloprid‐susceptible population represents a field population that had not been previously exposed to neonicotinoids and, as a result, is still very susceptible to imidacloprid. The imidacloprid‐resistant population used in the current study was obtained from a working, agricultural operation and has had a lengthy exposure to common insecticidal inputs for over 50 years, including, but not limited to, synthetic pyrethroids, organophosphates, carbamates, chlorinated hydrocarbons, together with other insecticides, including at‐plant treatments of neonicotinoids. Previous results by Huseth et al. provide an example of the most common insecticide inputs that are typically applied to potato in this production region, which include both at‐plant, systemic treatments, as well as foliar applications of neonicotinoids throughout the growing season.24 Huseth et al. observed staggered, post‐diapause emergence of L. decemlineata in agricultural settings, which they hypothesized could partially explain the beetles' capacity to cope with systemic insecticides.7 Here, we have examined the effects of temporal patterns of phenotypic variation in insecticide resistance throughout a growing season using distinct collections of adult L. decemlineata with staggered emergence dates. Furthermore, we utilized RNA sequencing to classify candidate transcripts that could explain these temporal patterns in phenotypic variation in an imidacloprid‐resistant population.
Our study demonstrates that there is phenotypic variation in imidacloprid resistance during the growing season in adult L. decemlineata. In describing the temporal patterns in mean LC50 estimates over the growing season in the first generation set of collection time points (early, late, and conclusion), an imidacloprid‐resistant population generally followed a bell‐shaped distribution in susceptibility as the growing season progressed, with newly emerged and aging adults being the most susceptible. Dramatic levels of overall resistance were obvious in LC50 estimates that were two orders of magnitude (100×) higher in the resistant population when compared to the susceptible population. With the use of LC50 assays we were able to describe trends in resistances that give valuable insight to pest managers, including optimal windows of susceptibility to insecticides. Huseth et al. further hypothesized that trends in insecticide resistance are tied to the dynamic life history of L. decemlineata.7 In this study we observed that individuals in the late‐emergence time period of the first generation were considerably more resistant than the early emergence period, suggesting that staggered and later‐emerging beetles are potentially more resistant. The differential effects of pesticides applied as either systemic (targeting early emerging, overwintering adults beetles) or foliar protectants (targeting second generation and later‐emerging beetles) throughout the growing season may have an impact on these trends and need to be examined further. For example, as resistance increases during the growing season, further steps might need to be taken to control problematic populations, including adding new and unique insecticidal chemistries and monitoring field populations closely for insecticide resistance development.
Mean LC50 values of the Systemic‐3, imidacloprid‐resistant population demonstrate that first‐generation adults are consistently more susceptible than second‐generation adults, indicating that overwintering diapause might influence the relative fitness of first generation adults. However, there is significant variation in the estimated confidence intervals (CIs), potentially due to the differing sample sizes of adult beetles included in these assays. We also noted that field populations inherently contain a heterogeneous mixture of both resistant and susceptible individuals and variance is frequently high.7, 11 Despite significant variation in estimated CIs, several time points in the first generation remain statistically more susceptible than their second‐generation counterparts. Multiple factors could explain these findings, including the increased expression of enzymatic detoxification mechanisms in the second, and more resistant generation.
Recent investigations have classified potential mechanisms by which insecticide resistance develops in L. decemlineata.11, 13, 14 The general processes by which imidacloprid can be metabolized and broken down rely on multiple detoxification enzymes, including cytochrome p450 and glutathione S‐transferases.26 We therefore chose to conduct comparisons between multiple groups of the first‐generation collection to the second‐generation collection of the imidacloprid‐resistant population (early emergence from diapause vs. second generation, late emergence vs. second generation, conclusion of first generation vs. second generation) to determine if difference in resistance could be partially explained by transcript abundance of enzymatic detoxification mechanisms.
Examining upregulated transcripts that corresponded to imidacloprid resistance in the second, more resistant, generation uncovered multiple mechanisms of resistance that were upregulated in the second‐generation population compared to the first‐generation counterpart. Moreover, we classified highly upregulated transcripts (fold change greater than 100) in the second‐generation population. This revealed interesting trends in highly upregulated cuticular proteins and a single cytochrome p450. The cytochrome p450 had the highest BLAST match to L. decemlineata cytochrome P450 412a2 (NCBI accession KF044265.1). Both cuticular and cytochrome p450 have been previously suggested to play a large role in insecticide resistance in multiple insect taxa. In Anopheles funestus, increases in cuticular thickness were associated with pyrethroid resistance,27 and in Cimex lectularius the over‐expression of multiple cuticular proteins was observed in association with pyrethroid resistance.28 In L. decemlineata, elevated expression of mRNA transcripts encoding for cuticular proteins have been observed in adult beetles and have previously been associated with environmental stressors such as a dry environments and insecticidal exposure.11, 29 Mota‐Sanchez et al. demonstrated a phenotypic change back to susceptibility in a neonicotinoid‐resistant population with the use of piperonyl butoxide (PBO), an inhibitor of cytochrome p450, suggesting the importance of cytochrome p450 in neonicotinoid resistance.8
The relative changes in transcript abundance were compared between the first (overwintering), more imidacloprid‐susceptible generation, and the second, more imidacloprid‐resistant generation. It is possible that the beetles are upregulating these transcripts as a response to increased insecticidal pressure throughout the growing season. The elevated transcript abundance data indicates that both cuticular proteins and cytochrome p450s may play a role in the observed increase in resistance of the second generation. Moreover, it is possible that some of the trends in the upregulated cuticular proteins and cytochrome p450s could be due to other, non‐insecticidal exposures. Further investigation is needed to assess the true role of these upregulated transcripts in insecticide resistance using supplementary knock‐down assays. Previous studies have classified similar mechanisms of resistance in imidacloprid‐resistant populations of L. decemlineata using RNA sequencing. Zhu et al. classified upregulated cytochrome p450 in an imidacloprid‐resistant population; and many of the cytochrome p450s classified in our study belong to the same clans that Zhu observed to be important in imidacloprid resistance.14 Clements et al. also used RNA sequencing to classify upregulated transcripts in an imidacloprid‐resistant L. decemlineata population in Wisconsin. The upregulation of similar resistance mechanisms was observed in both investigations, including the upregulation of transcripts encoding for the specific cytochrome p450, 9z4.11
Examining the data further, we ran an enrichment analysis on the gene ontology terms for which the transcripts encoded. The enrichment analysis suggested that members of the second‐generation population over‐expressed multiple metabolic processes, including terms that correspond to increased oxidoreductase activity, monooxygenase activity, and structural constituents of the cuticle. This suggests that individuals representing the second generation upregulated molecular mechanisms of resistance that, in turn, gave rise to increased imidacloprid resistance. Although the stresses that beetles face in an agricultural setting are vast, including different xenobiotics and environmental stressors, the transcript abundance data clearly demonstrated that there are many differences in the gene regulation between the first‐ and second‐generation population, many of which can be tied to insecticide resistance.
Our results provide pest managers with valuable insight describing mechanisms by which beetles cope with insecticide insults, including the suggestion that to effectively control problematic populations of L. decemlineata the genetic mechanisms of resistance must be considered. Understanding the genetic response to specific insecticidal chemistries will allow growers to better determine a pest management strategy that may include a combination of other foliar and systemic insecticides that do not upregulate the same detoxification mechanisms. Further, the use of enzymatic detoxification inhibitors, such as PBO, may also be used as an insecticide synergist by inhibiting detoxification mechanisms, such as cytochrome p450.2, 8
5. CONCLUSIONS
Leptinotarsa decemlineata is a major agricultural pest of potatoes. It is of upmost importance to understand differences in levels of observed insecticide resistance that correspond to the phenology of L. decemlineata over a growing season. This study demonstrates that there is phenotypic variation in imidacloprid susceptibility within a resistant population over a growing season. The results of this study further demonstrate that researchers, producers, and pest management practitioners may benefit from an improved understanding of when, during the growing season, this insect may be better prepared to cope with insecticide inputs. This study also demonstrates the specific upregulation of a unique set of transcripts, a portion of which may encode the dominant mechanisms of insecticide resistance. The differential expression, and overall abundance, of these transcripts provide us a glimpse into how these economically important pests deal with insecticide insults and aid in our ability to determine the specific mechanisms of resistance, which may lead to more precision delivery of pest management options that slow the pace of resistance development.
Supporting information
Table S1. Quantitative PCR primers and primer efficiency
Table S2. List of up‐regulated transcripts in the second generation population that could encode for increased imidacloprid resistance and transcript ID's. NCBI accession numbers represent the BLAST hit.
Table S3. Enrichment analysis between GO terms from the up‐regulated transcripts of the second generation population compared to all transcripts.
Table S4. Bray Curtis analysis conducted to observe the dissimilarities between the three comparisons for investigating fold changes greater than 2 or fold changes greater than 100. Bray Curtis analysis output is expressed as a dissimilarity constant between 0 and 1, with 0 being completely similar and 1 completely different.
Figure S1. (A) Early, (B) Late and (C) Conclusion‐emergence time periods of the first generation of L. decemlineata used for discrete comparisons against second generation beetles using RNA sequencing to examine transcript abundance.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the Wisconsin Potato and Vegetable Growers Association and the associated producers in central Wisconsin, who provided access to collect L. decemlineata from their fields. This work could not have been completed without the help of numerous people, including those that helped collect L. decemlineata from around Wisconsin in 2015: Linda Crubaugh and Michael Crossley. The authors would also like to acknowledge individuals who contributed valuable criticisms to earlier versions of this paper, including Anna Clements. This research was partially supported by a combination of funding from the: (1) Wisconsin Potato Industry Board (133AAA9362), (2) National Potato Council's State Cooperative Research Program FY11‐13, and (3) University of Wisconsin, Hatch Formula 142 Funds (142PRJ72SV).
REFERENCES
- 1. Hare JD, Ecology and management of the Colorado potato beetle. Annu Rev Entomol 35:81–100 (1990). [Google Scholar]
- 2. Alyokhin A, Baker M, Mota‐Sanchez D, Dively G and Grafius E, Colorado potato beetle resistance to insecticides. Am J Potato Res 85:395–413 (2008). [Google Scholar]
- 3. Food and Agriculture Orginization of the United Nations (FAOSTAT) . [Online]. Available: http://faostat3.fao.org/home/E [23 July 2016].
- 4. Kashian R, Depas J, Fogarity P and Peterson J, Potato production in Wisconsin: analyzing the economic impact (2014). [Online]. Available: http://www.uww.edu/Documents/colleges/cobe/ferc/PotatoesInWI.pdf [23 July 2016].
- 5. Whalon M, Mota‐Sanchez D and Hollingworth R, Arthropod Pesticide Resistance Database. [Online]. Available: http://www.pesticideresistance.org/display.php?page=species&arId=141 [1 May 2016].
- 6. IRAC Mode of Action Classification Scheme . [Online]. Available: http://www.irac‐online.org/documents/moa‐classification/ [23 July 2016].
- 7. Huseth AS and Groves RL, Effect of insecticide management history on emergence phenology and neonicotinoid resistance in Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). J Econ Entomol 106:2491–2505 (2013). [DOI] [PubMed] [Google Scholar]
- 8. Mota‐Sanchez D, Hollingworth RM, Grafius EJ and Moyer DD, Resistance and cross‐resistance to neonicotinoid insecticides and spinosad in the Colorado potato beetle, Leptinotarsa decemlineata (Say) (Coleoptera: Chrysomelidae). Pest Manag Sci 62:30–37 (2006). [DOI] [PubMed] [Google Scholar]
- 9. Zhao JZ, Bishop BA and Grafius EJ, Inheritance and synergism of resistance to imidacloprid in the Colorado potato beetle (Coleoptera: Chrysomelidae). J Econ Entomol 93:1508–1514 (2000). [DOI] [PubMed] [Google Scholar]
- 10. Alyokhin A, Dively G, Patterson M, Mahoney M, Rodgers D and Wollam J, Susceptibility of imidacloprid‐resistant Colorado potato beetles to non‐neonicotinoid insecticides in the laboratory and field trials. Am J Potato Res 83:485–494 (2006). [Google Scholar]
- 11. Clements J, Schoville S, Peterson N, Lan Q and Groves RL, Characterizing molecular mechanisms of imidacloprid resistance in select populations of Leptinotarsa decemlineata in the Central Sands region of Wisconsin. PLoS One 11:e0147844 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zehnder GW, Timing of insecticides for control of Colorado potato beetle (Coleoptera: Chrysomelidae) in eastern Virginia based on differential susceptibility of life stages. J Econ Entomol 79:851–856 (1986). [Google Scholar]
- 13. Kumar A, Congiu L, Lindström L, Piiroinen S, Vidotto M and Grapputo A, Sequencing, de novo assembly and annotation of the Colorado potato beetle, Leptinotarsa decemlineata, transcriptome. PLoS One 9:e86012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu F, Moural TW, Nelson DR and Palli SR, A specialist herbivore pest adaptation to xenobiotics through up‐regulation of multiple cytochrome P450s. Sci Rep 6:20421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Institute SAS, SAS/STAT 9.3 User's Guide. SAS Institute, Cary, NC: (2011). [Google Scholar]
- 16. Schoville S and Chen Y, Colorado Potato Beetle Genome Project. [Online]. https://www.hgsc.bcm.edu/arthropods/colorado‐potato‐beetle‐genome‐project [23 July 2016].
- 17. Li B and Dewey CN, RSEM: accurate transcript quantification from RNA‐Seq data with or without a reference genome. BMC Bioinf 12:323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robinson MD, McCarthy DJ and Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BMG et al, EBSeq: an empirical Bayes hierarchical model for inference in RNA‐seq experiments. Bioinformatics 29:1035–1043 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Conesa A, Götz S, Garcia‐Gómez JM, Terol J, Talón M and Robles M, Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676 (2005). [DOI] [PubMed] [Google Scholar]
- 21. Beals EW, Bray–Curtis ordination: an effective strategy for analysis of multivariate ecological data. Adv Ecol Res 14:1–55 (1984). [Google Scholar]
- 22. Zhu F, Xu J, Palli R, Ferguson J and Palli SR, Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag Sci 67:175–182 (2011). [DOI] [PubMed] [Google Scholar]
- 23. Pfaffl MW, A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Res 29:e45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huseth AS, Groves RL, Chapman SA, Alyokhin A, Kuhar TP, Macrae IV et al, Managing Colorado potato beetle insecticide resistance: new tools and strategies for the next decade of pest control in potato. J Integr Pest Manag 5(4):A1–A8 (2014). [Google Scholar]
- 25. Foster R and Flood B (eds), Vegetable Insect Management. Meister Media Worldwide, Willoughby, OH: (2005). [Google Scholar]
- 26. Casida JE, Neonicotinoid metabolism: compounds, substituents, pathways, enzymes, organisms, and relevance. J Agric Food Chem 59:2923–2931 (2011). [DOI] [PubMed] [Google Scholar]
- 27. Wood OR, Hanrahan S, Coetzee M, Koekemoer LL and Brooke BD, Cuticle thickening associated with pyrethroid resistance in the major malaria vector Anopheles funestus . Parasit Vectors 3:67 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu F, Gujar H, Gordon JR, Haynes KF, Potter MF and Palli SR, Bed bugs evolved unique adaptive strategy to resist pyrethroid insecticides. Sci Rep 3:1456. doi:10.1038/srep01456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang J, Goyer C and Pelletier Y, Environmental stresses induce the expression of putative glycine‐rich insect cuticular protein genes in adult Leptinotarsa decemlineata (Say). Insect Mol Biol 17:209–216 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Quantitative PCR primers and primer efficiency
Table S2. List of up‐regulated transcripts in the second generation population that could encode for increased imidacloprid resistance and transcript ID's. NCBI accession numbers represent the BLAST hit.
Table S3. Enrichment analysis between GO terms from the up‐regulated transcripts of the second generation population compared to all transcripts.
Table S4. Bray Curtis analysis conducted to observe the dissimilarities between the three comparisons for investigating fold changes greater than 2 or fold changes greater than 100. Bray Curtis analysis output is expressed as a dissimilarity constant between 0 and 1, with 0 being completely similar and 1 completely different.
Figure S1. (A) Early, (B) Late and (C) Conclusion‐emergence time periods of the first generation of L. decemlineata used for discrete comparisons against second generation beetles using RNA sequencing to examine transcript abundance.


