Abstract
The interleukin 1 family plays an important role in the immune and inflammatory responses. Coronary artery disease (CAD) is a chronic inflammatory disease. However, the genetic association between IL-37, the seventh member of the IL-1 family, and CAD is unknown. Here we show that a single nucleotide polymorphism in the IL-37 gene (rs3811047) confers a significant risk of CAD. We have performed an association analysis between rs3811047 and CAD in two independent populations with 2,501 patients and 3,116 controls from China. Quantitative RT-PCR analysis has been performed to determine if the IL-37 expression level is influenced by rs3811047. We show that the minor allele A of rs3811047 is significantly associated with CAD in two independent populations under a recessive model (Padj = 5.51 × 10−3/OR = 1.56 in the GeneID Northernern population and Padj = 1.23 × 10−3/OR = 1.45 in the GeneID Central population). The association became more significant in the combined population (Padj = 9.70 × 10−6/OR = 1.47). Moreover, the association remains significant in a CAD case control population matched for age and sex. Allele A of rs3811047 shows significant association with a decreased mRNA expression level of IL-37 (n = 168, P = 3.78 × 10−4). These data suggest that IL37 is a new susceptibility gene for CAD, which provides a potential target for the prevention and treatment of CAD.
Coronary artery disease (CAD) is the leading cause of morbidity and mortality in the world1,2. Epidemiological and family studies revealed that CAD has a strong genetic component3. The heritability of CAD was shown to be from 40% to 60%4. Understanding the genetic risk factors of CAD or atherosclerosis (the cause of CAD)5 may provide important information on the biological pathways of CAD pathogenesis.
Recently, large-scale genome-wide association studies (GWAS) have identified more than 50 risk loci for CAD and provided new information to the biological pathways that were not associated with traditional risk factors of CAD before6,7,8,9,10,11,12. Although genome-wide approaches have provided novel insights into the genetic basis of CAD etiology, the GWAS risk loci have the modest effects and explain approximately 10% of CAD heritability9. Furthermore, GWAS may miss some specific candidate genetic variants13. Therefore, the genetic architecture of CAD remains to be further defined.
Cytokines and chronic inflammation play an important role in the pathogenesis of atherosclerosis and CAD14. Recent studies have found that many variants of genes encoding cytokines, such as IL-6, IL-10, IL-16, IL-17A, IL-18 and IL-3315,16,17,18,19,20, are genetically associated with atherosclerosis and CAD in humans. Evidence from animal studies has demonstrated that many cytokines regulated by interleukins participate in the pathological inflammatory processes involved in atherosclerosis21.
Interleukin-37 (IL-37/IL-1F7) is a member of the IL-1 cytokine family22, which broadly modulates inflammatory and immune responses23. IL-37 has a caspase-1 site and can be processed by caspase-124. After caspase-1 processing, IL-37 is translocated to the nucleus and reduces the production of pro-inflammatory cytokines in murine RAW cells following LPS stimulation25. In 2010, Nold et al. observed that IL-37 was an anti-inflammatory cytokine, affected a broad spectrum of pro-inflammatory cytokines, and suppressed immune responses26. Subsequently, McNamee et al. observed that IL-37 played a protective and anti-inflammatory role by decreasing recruitment of leukocytes into the inflamed tissue. Furthermore, Ge et al. reported that SNP rs3811047, a functional tagSNP of IL-37, was associated with ankylosing spondylitis, which is an idiopathic inflammatory disease affecting the axial and/or peripheral skeleton with an increased risk of atherosclerosis and cardiovascular mortality and morbidity27,28. Based on these data, we hypothesized that IL-37 may play a role in the pathogenesis of CAD and the functional tagSNP of IL-37, rs3811047, may be genetically associated with CAD. In the present study, we genotyped SNP rs3811047 in IL-37 in two independent case control populations of CAD, and performed an association study to test whether the genetic variant in IL37 confers a risk to CAD.
Results
Characteristics of study populations and power analysis
The demographic and clinical characteristics of two independent CAD populations used for case control analysis, including the GeneID Northern population and the GeneID Central population, are shown in Table 1. The GeneID Northern population included 1,038 CAD patients and 1,076 controls. The mean age was 62.01 ± 12.70 in cases and 50.52 ± 16.71 in controls. The proportion of males was 65.51% in cases and 69.33% in controls. The GeneID Central population enrolled 1,463 cases and 2,040 controls. The mean age was 64.21 ± 12.44 in cases and 49.02 ± 13.88 in controls, respectively. The proportion of males was 65.21% and 61.76% in cases and controls, respectively. The percentage of males in cases was higher than in controls because the male gender is a well-known risk factor for the development of CAD. The average age of the control populations is younger than that in the case populations because most controls are study subjects who had free physical examinations offered to active working individuals by their respective working institutions. The differences of age and sex between cases and controls were adjusted in later statistical analysis. To further reduce the confounding of age and sex, we generated a case control population by randomly matching each individual case to a control based on age and sex. The data on demographic and clinical characteristics of the matched case control population are shown in Table 1.
Table 1. Demographic and clinical characteristics of study populations.
| Characteristics | GeneID Northern Population |
GeneID Central Population |
GeneID Combined Population |
Sex- and Age-Matched Population |
||||
|---|---|---|---|---|---|---|---|---|
| CAD | Control | CAD | Control | CAD | Control | CAD | Control | |
| N | 1,038 | 1,076 | 1,463 | 2,040 | 2,501 | 3,116 | 1,596 | 1,596 |
| Age (years) | 62.01 ± 12.70*** | 50.52 ± 16.71 | 64.21 ± 12.44*** | 49.02 ± 13.88 | 63.30 ± 13.25*** | 49.54 ± 15.58 | 60.05 ± 11.35 | 60.05 ± 11.35 |
| Male N (%) | 680 (65.51) | 746 (69.33) | 954 (65.21)** | 1260 (61.76) | 1,634 (65.33) | 2,006 (64.38) | 1,049 (65.73) | 1,049 (65.73) |
| Hypertension N (%) | 594 (57.23)*** | 202 (18.77) | 1007(68.83)*** | 187 (9.17) | 1601 (64.01)*** | 389 (12.48) | 853 (53.45) | 208 (13.03) |
| Diabetes N (%) | 145 (13.99)*** | 84 (7.80) | 362 (24.74)*** | 74 (3.60) | 507 (20.27)*** | 158 (5.07) | 316 (19.80) | 108 (6.77) |
| Tch (mmol/L) | 4.40 ± 1.08 | 4.32 ± 1.16 | 4.68 ± 0.99** | 4.45 ± 1.11 | 4.56 ± 1.08*** | 4.40 ± 1.07 | 4.66 ± 1.12*** | 4.46 ± 1.20 |
| TG (mmol/L) | 1.62 ± 1.11*** | 1.44 ± 1.07 | 1.59 ± 1.14*** | 1.46 ± 1.08 | 1.60 ± 1.12*** | 1.45 ± 1.10 | 1.57 ± 1.18*** | 1.44 ± 1.11 |
| HDL-c (mmol/L) | 1.10 ± 0.38** | 1.17 ± 0.50 | 1.14 ± 0.52** | 1.20 ± 0.55 | 1.12 ± 0.40*** | 1.19 ± 0.41 | 1.14 ± 0.45** | 1.19 ± 0.40 |
| LDL-C (mmol/L) | 2.85 ± 0.81*** | 2.62 ± 0.91 | 2.88 ± 0.84*** | 2.62 ± 0.84 | 2.87 ± 0.82*** | 2.62 ± 0.88 | 2.79 ± 0.76*** | 2.52 ± 0.81 |
Data are shown as means ± standard deviation (SD) for quantitative variables and n (%) for binary traits.
**P < 0.01 between cases and controls for quantitative variables and percent (%) for qualitative variables.
***P < 0.001 between cases and controls for quantitative variables and percent (%) for qualitative variables.
Under the population parameter setting of the effect size or odds ratio (OR) of 1.2 for CAD29, and the minor allele frequency of 0.209 for rs3811047 (HapMap CHB data sets), our samples provide a statistical power of 70% in the Northern population, and 88% in the Central population to detect an association between rs3811047 and CAD with a type I error of 0.05. The combined population has 2,501 cases and 3,116 controls and can provide a statistical power of 98%. The matched case control population has a power of 83%. Therefore, our GeneID samples are sufficiently large to test the association between SNP rs3811047 and CAD.
Significant association between SNP rs3811047 in IL37 and CAD in two independent Chinese populations
The genotyping data for SNP rs3811047 showed no deviation from the Hardy-Weinberg equilibrium in the control populations (P > 0.05). In the GeneID Northern population, the minor allele A of rs381047 confers a significant risk of CAD (Pobs = 6.72 × 10−4, OR = 2.45) under a recessive model (Table 2). After adjustment for covariates of age, sex, hypertension, diabetes, and lipid concentrations (TG, Tch, LDL-c and HDL-c), the significant association remained (Padj = 5.51 × 10−3, OR = 1.56).
Table 2. Genotypic analysis of IL-37 SNP rs3811047 with CAD in the GeneID Chinese Han population under three genetic models.
| Cohort (N, Case/Control) | Genotype |
Model | Pobs | OR (95% CI) | Padj | OR (95% CI) | ||
|---|---|---|---|---|---|---|---|---|
| Cases (N) | Controls (N) | |||||||
| GeneID Central (1,503/2,040) | AA | 77 | 60 | Additive | 1.31 × 10−3 | n.a | 0.46 | 1.06 (0.91–1.24) |
| AG | 402 | 607 | Dominant | 0.98 | 1.00 (0.87–1.16) | 0.64 | 0.98 (0.89–1.07) | |
| GG | 984 | 1373 | Recessive | 4.72 × 10–4 | 1.83 (1.30–2.59) | 1.23 × 10−3 | 1.45 (1.16–1.83) | |
| GeneID Northern (998/1,076) | AA | 46 | 20 | Additive | 5.80 × 10−5 | n.a | 0.02 | 1.27 (1.04–1.55) |
| AG | 349 | 310 | Dominant | 3.49 × 10−4 | 1.39 (1.16–1.66) | 0.11 | 1.10 (0.98–1.24) | |
| GG | 643 | 746 | Recessive | 6.72 × 10−4 | 2.45 (1.44–4.17) | 5.51 × 10−3 | 1.56 (1.14–2.13) | |
| GeneID Combined (2,501/3,116) | AA | 123 | 80 | Additive | 9.21 × 10−6 | n.a | 1.28 × 10−2 | 1.16 (1.03–1.30) |
| AG | 751 | 917 | Dominant | 1.97 × 10−2 | 1.14 (1.02–1.28) | 0.24 | 1.04 (0.97–1.11) | |
| GG | 1627 | 2119 | Recessive | 2.72 × 10−6 | 1.96 (1. 47–2.61) | 9.70 × 10−6 | 1.47 (1.24–1.74) | |
Pobs, P value from Chi square tests with 2 × 3 contingency tables without adjustment for covariates;
Padj, P value adjusted by covariates of sex, age, hypertension, T2D and lipid concentrations by multiple logistic regression analysis;
OR, odds ratio;
95% CI, 95% confidence interval;
Additive model = AA/AG/GG; Dominant model = AA + AG/GG; Recessive model = AA/AG + GG.
The association between SNP rs3811047 and CAD is the first time finding, therefore, we need to replicate the finding in another independent population. The replication study using the GeneID Central population showed that SNP rs3811047 was a significant risk factor for CAD (Pobs = 4.72 × 10−4, OR = 1.83; Padj = 1.23 × 10−3, OR = 1.45) (Table 2) in the GeneID Central populaion, also under the recessive model (Table 2). Therefore, the association between rs3811047 and CAD was independently confirmed in the replication population.
Combination of the two independent populations together resulted in a larger population with 2,501 cases and 3,116 controls. The association between SNP rs3811047 and CAD became more significant in the combined population. Minor allele A of SNP rs3811047 showed a much more significant risk of CAD (Padj = 9.70 × 10−6 with an OR of 1.47) under the recessive model (Table 2).
We also analyzed allelic association between SNP rs3811047 and CAD in the combined population (Table 3). A significant allelic association was found between rs3811047 and CAD (Pobs = 3.20 × 10−4, OR = 1.19). After adjustment for covariats of age, sex, hypertension, diabetes, and lipid concentrations (TG, Tch, LDL-c and HDL-c), the significant association remained (Padj = 0.01, OR = 1.16) (Table 3).
Table 3. Significant allelic association of IL-37 SNP rs3811047 with CAD in the GeneID Chinese Han population.
| Population | N Case/Control | Frequency of Minor Allele A (Case/Control) | Phwe | Pobs | OR (95% CI) | Padj | OR (95% CI) |
|---|---|---|---|---|---|---|---|
| GeneID Combined | 2,501/3,116 | 0.20/0.17 | 0.12 | 3.20 × 10−4 | 1.19 (1.08–1.31) | 0.01 | 1.16 (1.03–1.30) |
Phwe, P value from Hardy-Weinberg equilibrium tests;
Pobs, P value from Chi square tests with 2 × 2 contingency tables without adjustment for covariates;
Padj, P value adjusted by covariates of sex, age, hypertension, T2D and lipid concentrations by multiple logistic regression analysis;
OR, odds ratio;
95% CI, 95% confidence interval.
The significant association between SNP rs3811047 and CAD remained significant after adjustment for age and sex, suggesting that the differences of age and the percentage of males between cases and controls did not affect the conclusion of our case control analysis. To further minimize the confounding of age and sex, we generated a case control population with each case randomly matched to a control by exact matching or one-by-one matching. Statistical analysis was then carried out to further test whether IL-37 SNP rs3811047 is still significantly associated with CAD. As shown in Table 4, the minor allele A of rs3811047 conferred a significant risk of CAD in the matched case control population under a recessive model (Pobs = 4.57 × 10−9, OR = 2.82) and under an additive model (Pobs = 3.66 × 10−8). The association remained significant after adjustment of other covariates, including hypertension, diabetes, and lipid concentrations (TG, Tch, LDL-c and HDL-c) (Padj = 2.23 × 10−4, OR = 1.99 under a recessive model; Padj = 1.02 × 10−3, OR = 1.45 under an additive model) (Table 4). Allelic association analysis also showed that the minor allele A of SNP rs3811047 conferred a significant risk of CAD in the matched case control population before (Pobs = 1.60 × 10−5, OR = 1.31) and after adjustment of hypertension, diabetes, and lipid concentrations (TG, Tch, LDL-c and HDL-c) (Padj = 6.43 × 10−3, OR = 1.26) (Table 5). We also analyzed the association between rs3811047 and CAD in a male only population and in a female only population, and significant association was identified in both populations (Table 5).
Table 4. Genotypic analysis of IL-37 SNP rs3811047 with CAD in age- and sex-matched case control populations under three genetic models.
| Population (N, Case/Control) | Genotype |
Model | Pobs | OR (95% CI) | Padj | OR (95% CI) | ||
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | |||||||
| Sex- and age-matched population (1,596/1,596) | AA | 113 | 42 | Additive | 3.66 × 10−8 | n.a | 1.02 × 10−3 | 1.45 (1.15–1.81) |
| AG | 471 | 487 | Dominant | 0.06 | 1.16 (0.99–1.34) | 0.17 | 1.10 (0.96–1.29) | |
| GG | 1012 | 1067 | Recessive | 4.57 × 10−9 | 2.82 (1.96–4.05) | 2.23 × 10−4 | 1.99 (1.45–3.02) | |
Pobs, P value from Chi square tests with 2 × 3 contingency tables without adjustment for covariates;
Padj, P value adjusted by covariates of sex, age, hypertension, T2D and lipid concentrations by multiple logistic regression analysis;
OR, odds ratio;
95% CI, 95% confidence interval;
Additive model = AA/AG/GG; Dominant model = AA + AG/GG; Recessive model = AA/AG + GG.
Table 5. Significant allelic association of IL-37 SNP rs3811047 with CAD in an age- and sex-matched case control population.
| Population | N Case/Control | Frequency of Minor Allele A (Case/Control) | Phwe | Pobs | OR (95% CI) | Padj | OR (95% CI) |
|---|---|---|---|---|---|---|---|
| Age- and sex-matched population | 1,596/1,596 | 0.22/0.18 | 0.86 | 1.60 × 10−5 | 1.31 (1.16–1.48) | 6.43 × 10−3 | 1.26 (1.08–1.40) |
| Males | 1,049/1,049 | 0.22/0.18 | 0.81 | 1.31 × 10−3 | 1.30 (1.12–1.52) | 0.01 | 1.24 (1.05–1.43) |
| Females | 547/547 | 0.22/0.18 | 0.80 | 8.01 × 10−3 | 1.33 (1.08–1.64) | 0.02 | 1.28 (1.06–1.45) |
Phwe, P value from Hardy-Weinberg equilibrium tests;
Pobs, P value from Chi square tests with 2 × 2 contingency tables without adjustment for covariates;
Padj, P value adjusted by covariates of sex, age, hypertension, T2D and lipid concentrations by multiple logistic regression analysis;
OR, odds ratio;
95% CI, 95% confidence interval.
Real time RT-PCR analysis idenfified significant association between SNP rs3811047 and the expression level of IL-37 mRNA
We carried out real time RT-PCR to analyze whether the expression level of IL-37 is associated with the genotype of SNP rs3811047 using blood samples from 168 study subjects. The results showed that the expression level of the IL-37 mRNA was significantly different among different genotypes under a recessive genetic model (P = 3.78 × 10−4) (Fig. 1). The expression level of the IL-37 mRNA was significantly lower in carriers with the AA genotype than carriers with GG and GA genotypes (Fig. 1). Together, these results suggest that the minor allele A of SNP rs3811047 is significantly associated with a decreased expression of IL-37.
Figure 1. Assessment of the relationship between IL-37 SNP rs3811047 and the expression level of IL-37 mRNA by real time RT-PCR analysis.
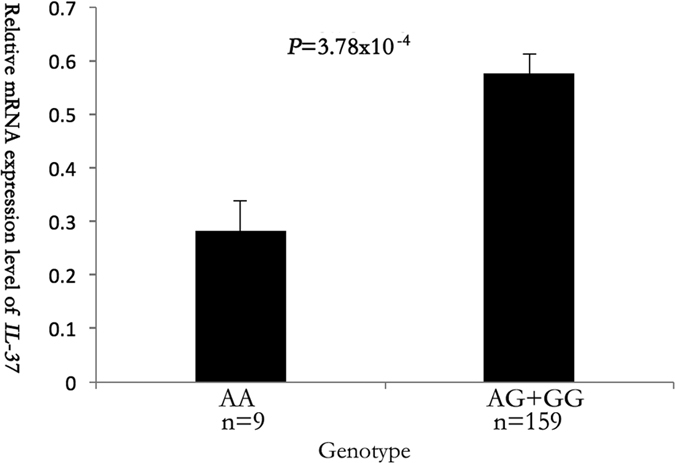
Total RNA samples were isolated from 168 blood samples (lymphocytes), converted into cDNA, and used for real time PCR analysis. Genomic DNA samples were isolated from the 168 study subjects and genotyped for SNP rs3811047 by HRM analysis. A linear regression was used to compare the differences for the mean RQ values between different genotypes (AA and AG + GG) of SNP rs3811047.
Discussion
In the present study, we genotyped SNP rs3811047 in the IL-37 gene in two independent case control populations of CAD, and performed an association study to test whether the genetic variant in IL37 confers risk of CAD. Here we provide genetic evidence that minor allele A of rs3811047 in the IL-37 gene was significantly associated with the risk of CAD in two independent case control populations (Tables 2 and 3). The association between SNP rs3811047 and CAD became even more significant in the combined population (Tables 2 and 3). The association between SNP rs3811047 and CAD remained significant after adjustment of covariates of age, sex, hypertension, diabetes, triglyceride, total cholesterol, LDL-cholesterol and HDL-cholesterol levels (Tables 2 and 3). Significant allelic and genotypic association between SNP rs3811047 and CAD was also identified in a case control population matched by age and sex (Tables 4 and 5). The significant allelic association remained in the separated male population and the female population (Table 5). These data suggest that SNP rs3811047 in IL-37 is a risk factor of CAD independent from age, gender, hypertension, diabetes, and lipid levels. Moreover, we found that the minor allele A of SNP rs3811047 was associated with a decreased expression level of the IL-37 mRNA. These results suggest that IL-37 is a susceptibility gene for CAD. This is the first study that establishes the significant association between an IL-37 variant and CAD. Our studies used cases and controls from the GeneID Chinese Han population. We hope that the significant association between an IL-37 variant and CAD can be replicated in other Chinese populations and even in other ethnic populations. In addition, future studies can also analyze whether the IL-37 variant is also associated with the severity of CAD and other diseases such as ischemic stroke associated with inflammation.
SNP rs3811047 was first associated with human leukocyte antigen-B27 positive ankylosing spondylitis in the Chinese population30. The previous study observed that there was an interaction between IL-37 gene and alcohol drinking in ankylosing spondylitis patients in a case-only study31. Ankylosing spondylitis is one of the most common chronic inflammatory autoimmune diseases affecting the axial and/or peripheral skeleton with an estimated prevalence of 0.1–0.9%32. Chronic inflammation and cytokines play important roles in the pathogenesis of atherosclerosis and CAD. There is evidence that ankylosing spondylitis patients have a higher risk of mortality and morbidity compared to the general population and also a higher rate of cardiovascular death28. Several studies also showed that cardiovascular diseases are more common in patients with ankylosing spondylitis33,34. These findings suggested that a common mechanism, such as chronic inflammation, may be shared between CAD and ankylosing spondylitis. We hypothesized that the genetic risk factors of ankylosing spondylitis may also play a role in the pathogenesis of atherosclerotic CAD. In the present study, for the first time, we show that SNP rs3811047 in IL-37 is indeed a risk factor for CAD.
IL-37 is the seventh member of the IL-1 family, and considered as an anti-inflammatory cytokine which mainly inhibits the expression, production and function of other pro-inflammatory cytokines35. IL-37 is normally expressed at a low level in peripheral blood monocytes, but its expression is rapidly up-regulated in monocytes as well as dendritic cells under an inflammatory context26. This leads to suppression of the production of other IL-1 family of pro-inflammatory cytokines36. Transgenic mice expressing human IL-37 exhibited an anti-inflammatory function by directly inhibiting the production of pro-inflammatory cytokines37. IL-37 was also shown as a key modulator of intestinal inflammation by decreasing IL-1β and TNFα38. In addition, IL-37 effectively inhibits the activation of dendritic cells35. As IL-37 is involved in anti-inflammation, reduced expression of IL-37 associated with SNP rs3811047 as found in this study (Fig. 1) may cause inflammation, increasing risk of atherosclerosis and CAD.
In conclusion, through a case control association study in two independent populations with a total of 2,501 cases and 3,116 controls, we found significant association between SNP rs3811047 in the IL37 gene and CAD. We also demonstrated that the minor allele of SNP rs3811047 was associated with a decreased expression level of IL-37. Our results suggest that IL37 is a new susceptibility gene for CAD and that SNP rs3811047 in IL37 is a new genetic risk factor of CAD.
Methods
Study populations
The study subjects were selected from the GeneID database, which is a large ongoing database with clinical data and DNA samples from more than 80,000 Chinese patients and controls. The major goal of GeneID database is to identify susceptibility genes for various cardiovascular diseases in the Chinese Han population7,29,39,40.
The diagnosis of CAD was based on coronary angiography, and followed the standard guidelines by the ACC/AHA41. The diagnosis was made by more than two independent expert cardiologists. We classified patients with >70% of luminal stenosis in at least one main vessel by coronary angiography, coronary artery bypass graft, percutaneous coronary intervention, and/or a myocardial infarction (MI) as CAD cases41. The diagnosis of MI was based on typical chest pain of ≧30 min duration, characteristic electrocardiographic patterns of acute MI, and significant elevation of cardiac enzymes (creatine kinase-MB, lactate dehydrogenase) and troponin I or T41. We excluded patients with myocardial spasma and myocardial bridge identified by angiography and those subjects with congenital heart disease, childhood hypertension, and type I diabetes mellitus.
The study included two independent populations. To avoid geographical confounding, we selected one population with study subjects recruited from Northern China and another population with study subjects recruited from Central China. The GeneID Northern population was enrolled from the Northern area of China and had 1,038 CAD cases and 1,076 controls. The GeneID Central population was enrolled from Wuhan in central China and had 1,463 CAD cases and 2,040 controls. In total, our study population included 2,501 CAD cases and 3,116 controls. All subjects were reported to be of Chinese Han origin by self-description or self-report.
The basic demographic and clinical charateristics of the subjects, including the age, gender, hypertension, type 2 diabetes (T2D) and lipid profiles, were obtained from medical records. Hypertension was defined as a systolic blood pressure of ≥140 mm Hg or a diastolic blood pressure ≥90 mm Hg. T2D was diagnosed as a fasting plasma glucose concentration ≥126 mg/dL after at least 8 hours of fasting or a 2-hour plasma glucose level of ≥200 mg/dL during an oral glucose tolerance test (OGTT).
This study was approved by the Ethics Committees on human subject research of Huazhong University of Science and Technology and local institutions and conformed to guidelines set forth by the Declaration of Helsinki. Written informed consent was obtained from subjects following instructions approved by the Ethics Committees.
SNP selection and genotyping
We selected a non-synonymous taqSNP (rs3811047 in IL-37) for this study. SNP rs3811047 is located in the second exon and caused a transition of threonine to alanine at the 42th amino acid residue of IL-37.
The human genomic DNA of each study subject was extracted from the peripheral whole blood samples using the Wizard Genomic DNA Purification Kit (Promega Corporation).
We used the Syto 9 fluorescent dye-based high resolution melt (HRM) method on a Rotor-gene 6200 System (Corbett Life Science) to genotype SNP rs3811047 as described by us16,40,42,43,44,45,46,47. Primers were designed by software Genetool. The fragment flanking rs3811047 was amplified with the forward primer 5′-AGCCCCCTGGAACCAGGC-3′ and the reverse primer 5′-TCAGCCACCCCCATCACC-3′, together with a final concentration of 5 μmol/L Syto 9 fluorescent dye. The polymerase chain reaction (PCR) was performed with a reaction of a total volume of 25 μL containing 2.5 μL of 10 × PCR buffer, 1.5 mmol/L MgCl2, 5 mmol/L dNTPs, 5 pmol of each primer, 25 ng of genomic DNA, 1 μL of Syto 9 fluorescent dye and 1 U of Taq DNA polymerase. The PCR profile was 5 min at 94 °C, 39 cycles of 94 °C for 10 s, 63 °C for 10 s and at 72 °C for 10 s, and a final elongation step at 72 °C for 10 min. Four positive controls were included in each run. The HRM genotyping was verified by direct Sanger sequence analysis of 52 randomly selected samples.
Quantitative RT-PCR analysis
We performed quantitative RT-PCR analysis to evaluate whether SNP rs3811047 was associated with the mRNA expression level of IL-37. The ΔΔCq method was used to determine the difference of the mean expression levels of IL-37 among study subjects with different genotypes for rs381104716,29,46. Quantitative real-time PCR analysis was carried out according to the MIQE guidelines48. Total RNA samples were extracted from human peripheral blood leukocytes using Trizol reagent (Life Technologies, Gaithersburg, MD). Quantification of RNA samples was performed using a spectrophotometer (NanoDrop, Thermo Scientific, Hudson, NH). One μg of total RNA was used for reverse transcription with Superscript II reverse transcriptase (Life Technologies, Gaithersburg, MD) and oligo (dT)18. A standard two step real-time PCR assay was performed using an ABI 7900-HT Genetic Analyzer (Applied Biosystems, Gaithersburg, MD). Each PCR reaction was performed in a final volume of 10 μL reaction mixture containing 5 μL of 2X PCR master mixture with ROX (Faststart Universal SYBR Green Master Kit, Roche Applied Science, Indianapolis, IN), 2 μL of cDNA, 0.4 μL of 10 pM primers, and 2.6 μL of ddH2O. Each reaction was performed in triplicate. The cycling conditions were 95 °C for 10 minutes and 40 cycles of 95 °C for 15 seconds and 60 °C elongation for 45 seconds. After the PCR reaction, Cq values (threshold cycle) of a target gene (IL-37) (Cq T) or reference gene GAPDH (Cq E) were computed using the RQ Manager program (version 1.3) and SDS (version 2.3). Reaction with a Cq of ≥40 or with the difference between Cq and mean Cq greater than 0.5 were excluded for further analysis. For each individual, the relative expression level △Cq (Cq T-Cq E) of a target gene was normalized with the reference gene and then transformed into relative quantity using RQ formula (RQ = 2−△△Cq, ΔΔCq = individual’s ΔCq-calibrator’s ΔCq)46. The calibrator was a mixed cDNA sample pooled from 10 randomly selected individuals. The RQ value for the calibrator was normalized to 1. After outliers were excluded, linear regression was used to compare the differences for mean RQ values of IL-37 between different genotypes of SNP rs3811047.
The sequences for qRT-PCR primers are 5′-AGCTGAAGAAGGAGAAACT-3′ (forward primer) and 5′-CGCCGACTCCAGCATGTTC-3′ (reverse primer) for IL-37 and 5′-AAGGTGAAGGTCGGAGTCAAC-3′ (forward primer) and 5′- GGGGTCATTGATGGCAACAATA -3′ (reverse primer) for GAPDH.
Statistical analysis
We used PS software 3.0.12 to calculate the statistical power and sample sizes for the case-control design (http://biostat.mc.vanderbilt.edu/wiki/Main/PowerSampleSize). The statistical power of a case control study can be calculated with special parameters, including the minor allele frequency (0.209 for rs3811047 in our study), OR, the numbers of cases and controls, and the Type I error of 0.05. The null hypothesis can be rejected if the odds ratio equals 1 with probability (power). The program uses an uncorrected chi-squared statistic method to evaluate the null hypothesis (http://biostat.mc.vanderbilt.edu/wiki/Main/PowerSampleSize).
The Hardy-Weinberg linkage disequilibrium test among control groups was performed using PLINK 1.06.
For association analysis, χ2 tests were performed using Pearson’s 2 × 2 and 2 × 3 contingency tables to calculate the P values and corresponding odds ratios (OR) with 95% confidential intervals (CI) by PLINK 1.06 as described by us7,49,50,51. Multivariate logistic regression analysis was performed to adjust for some risk factors (age, gender, hypertension, diabetes mellitus and lipid concentrations) using SPSS version 17.0. We used a student’s t-test to compare the continuous variables between cases and controls. Linear regression was used to assess the association between gene expression levels and SNP genotypes. A P value of 0.05 or less was considered to be statistically significant.
Additional Information
How to cite this article: Yin, D. et al. Genomic Variant in IL-37 Confers A Significant Risk of Coronary Artery Disease. Sci. Rep. 7, 42175; doi: 10.1038/srep42175 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
We thank the study subjects for their participation and support of this study and all members of the GeneID team for help and assistance. This study was supported by the China National Natural Science Foundation Key Program (31430047), Chinese National Basic Research Programs (973 Programs 2013CB531101 and 2012CB517801), Hubei Province’s Outstanding Medical Academic Leader Program, Hubei Province Natural Science Key Program (2014CFA074), the China National Natural Science Foundation grant (91439129, NSFC-J1103514), NIH/NHLBI grants R01 HL121358 and R01 HL126729, Specialized Research Fund for the Doctoral Program of Higher Education from the Ministry of Education, and the “Innovative Development of New Drugs” Key Scientific Project (2011ZX09307-001-09).
Footnotes
The authors declare no competing financial interests.
Author Contributions Design of the study: T.X., C.X. and Q.K.W. Experiments and data analysis: D.Y., D.H.N., Y.X., S.L., Y.B., G.J., Y.Z., X.W., Y.H., S.C., J.F., C.T., M.Z., Y.Z., L.W., Y.L., J.L., Q.C., X.T., C.X., and Q.K.W. Drafting of the manuscript: D.Y. and C.X. Critical revision of the manuscript: C.X., Q.C. and Q.K.W. Study supervision: Q.K.W.
References
- He J. et al. Major causes of death among men and women in China. The New England journal of medicine 353, 1124–1134, doi: 10.1056/NEJMsa050467 (2005). [DOI] [PubMed] [Google Scholar]
- Lopez A. D., Mathers C. D., Ezzati M., Jamison D. T. & Murray C. J. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet 367, 1747–1757, doi: 10.1016/S0140-6736(06)68770-9 (2006). [DOI] [PubMed] [Google Scholar]
- Lloyd-Jones D. M. et al. Parental cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults: a prospective study of parents and offspring. JAMA: the journal of the American Medical Association 291, 2204–2211, doi: 10.1001/jama.291.18.2204 (2004). [DOI] [PubMed] [Google Scholar]
- Zdravkovic S. et al. Heritability of death from coronary heart disease: a 36-year follow-up of 20 966 Swedish twins. Journal of internal medicine 252, 247–254 (2002). [DOI] [PubMed] [Google Scholar]
- Duvall W. L. & Vorchheimer D. A. Multi-bed vascular disease and atherothrombosis: scope of the problem. Journal of thrombosis and thrombolysis 17, 51–61, doi: 10.1023/B:THRO.0000036029.56317.d1 (2004). [DOI] [PubMed] [Google Scholar]
- Samani N. J. et al. Genomewide association analysis of coronary artery disease. The New England journal of medicine 357, 443–453, doi: 10.1056/NEJMoa072366 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F. et al. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nature genetics 43, 345–349, doi: 10.1038/ng.783 (2011). [DOI] [PubMed] [Google Scholar]
- Lu X. et al. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nature genetics 44, 890–894, doi: 10.1038/ng.2337 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium C. A. D. et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nature genetics 45, 25–33, doi: 10.1038/ng.2480 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke R. et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. The New England journal of medicine 361, 2518–2528, doi: 10.1056/NEJMoa0902604 (2009). [DOI] [PubMed] [Google Scholar]
- Erdmann J. et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nature genetics 41, 280–282, doi: 10.1038/ng.307 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schunkert H. et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nature genetics 43, 333–338, doi: 10.1038/ng.784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden J. F. & Farrall M. Thirty-five common variants for coronary artery disease: the fruits of much collaborative labour. Human molecular genetics 20, R198–205, doi: 10.1093/hmg/ddr384 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson G. K. Inflammation, atherosclerosis, and coronary artery disease. The New England journal of medicine 352, 1685–1695, doi: 10.1056/NEJMra043430 (2005). [DOI] [PubMed] [Google Scholar]
- Guo L., Zhou X., Guo X., Zhang X. & Sun Y. Association of interleukin-33 gene single nucleotide polymorphisms with ischemic stroke in north Chinese population. BMC medical genetics 14, 109, doi: 10.1186/1471-2350-14-109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X. et al. The IL-33-ST2L pathway is associated with coronary artery disease in a Chinese Han population. American journal of human genetics 93, 652–660, doi: 10.1016/j.ajhg.2013.08.009 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Minkelen R. et al. Haplotypes of the interleukin-1 receptor antagonist gene, interleukin-1 receptor antagonist mRNA levels and the risk of myocardial infarction. Atherosclerosis 203, 201–205, doi: 10.1016/j.atherosclerosis.2008.06.029 (2009). [DOI] [PubMed] [Google Scholar]
- Chen Y. et al. IL-16 rs11556218 gene polymorphism is associated with coronary artery disease in the Chinese Han population. Clinical biochemistry 44, 1041–1044, doi: 10.1016/j.clinbiochem.2011.06.010 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Interleukin-17A gene variants and risk of coronary artery disease: a large angiography-based study. Clinica chimica acta; international journal of clinical chemistry 412, 327–331, doi: 10.1016/j.cca.2010.10.027 (2011). [DOI] [PubMed] [Google Scholar]
- Tiret L. et al. Genetic analysis of the interleukin-18 system highlights the role of the interleukin-18 gene in cardiovascular disease. Circulation 112, 643–650, doi: 10.1161/CIRCULATIONAHA.104.519702 (2005). [DOI] [PubMed] [Google Scholar]
- Kleemann R., Zadelaar S. & Kooistra T. Cytokines and atherosclerosis: a comprehensive review of studies in mice. Cardiovascular research 79, 360–376, doi: 10.1093/cvr/cvn120 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. et al. Identification and initial characterization of four novel members of the interleukin-1 family. The Journal of biological chemistry 275, 10308–10314 (2000). [DOI] [PubMed] [Google Scholar]
- van de Veerdonk F. L. & Netea M. G. New Insights in the Immunobiology of IL-1 Family Members. Frontiers in immunology 4, 167, doi: 10.3389/fimmu.2013.00167 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. et al. Interleukin-1F7B (IL-1H4/IL-1F7) is processed by caspase-1 and mature IL-1F7B binds to the IL-18 receptor but does not induce IFN-gamma production. Cytokine 18, 61–71 (2002). [DOI] [PubMed] [Google Scholar]
- Sharma S. et al. The IL-1 family member 7b translocates to the nucleus and down-regulates proinflammatory cytokines. Journal of immunology 180, 5477–5482 (2008). [DOI] [PubMed] [Google Scholar]
- Nold M. F. et al. IL-37 is a fundamental inhibitor of innate immunity. Nat Immunol 11, 1014–1022 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters M. J., van der Horst-Bruinsma I. E., Dijkmans B. A. & Nurmohamed M. T. Cardiovascular risk profile of patients with spondylarthropathies, particularly ankylosing spondylitis and psoriatic arthritis. Seminars in arthritis and rheumatism 34, 585–592 (2004). [DOI] [PubMed] [Google Scholar]
- Papagoras C., Voulgari P. V. & Drosos A. A. Atherosclerosis and cardiovascular disease in the spondyloarthritides, particularly ankylosing spondylitis and psoriatic arthritis. Clinical and experimental rheumatology 31, 612–620 (2013). [PubMed] [Google Scholar]
- Cheng X. et al. The same chromosome 9p21.3 locus is associated with type 2 diabetes and coronary artery disease in a Chinese Han population. Diabetes 60, 680–684 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F. et al. Association of IL-1F7 gene with susceptibility to human leukocyte antigen-B27 positive ankylosing spondylitis in Han Chinese population. Clinica chimica acta; international journal of clinical chemistry 411, 124–126 (2010). [DOI] [PubMed] [Google Scholar]
- Ge R. et al. Analysis on the interaction between IL-1F7 gene and environmental factors on patients with ankylosing spondylitis: a case-only study. Molecular biology reports 38, 2281–2284 (2011). [DOI] [PubMed] [Google Scholar]
- Yang Z., Ren Y., Liu D., Lin F. & Liang Y. Prevalence of systemic autoimmune rheumatic diseases and clinical significance of ANA profile: data from a tertiary hospital in Shanghai, China. APMIS, doi: 10.1111/apm.12564 (2016). [DOI] [PubMed] [Google Scholar]
- Mathieu S., Gossec L., Dougados M. & Soubrier M. Cardiovascular profile in ankylosing spondylitis: a systematic review and meta-analysis. Arthritis care & research 63, 557–563 (2011). [DOI] [PubMed] [Google Scholar]
- Szabo S. M. et al. Increased risk of cardiovascular and cerebrovascular diseases in individuals with ankylosing spondylitis: a population-based study. Arthritis and rheumatism 63, 3294–3304 (2011). [DOI] [PubMed] [Google Scholar]
- Tete S. et al. IL-37 (IL-1F7) the newest anti-inflammatory cytokine which suppresses immune responses and inflammation. Int J Immunopathol Pharmacol 25, 31–38 (2012). [DOI] [PubMed] [Google Scholar]
- Boraschi D. et al. IL-37: a new anti-inflammatory cytokine of the IL-1 family. Eur Cytokine Netw 22, 127–147 (2011). [DOI] [PubMed] [Google Scholar]
- Teng X. et al. IL-37 ameliorates the inflammatory process in psoriasis by suppressing proinflammatory cytokine production. J Immunol 192, 1815–1823 (2014). [DOI] [PubMed] [Google Scholar]
- McNamee E. N. et al. Interleukin 37 expression protects mice from colitis. Proceedings of the National Academy of Sciences of the United States of America 108, 16711–16716 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L. et al. Assessment of association of rs2200733 on chromosome 4q25 with atrial fibrillation and ischemic stroke in a Chinese Han population. Human genetics 126, 843–849 (2009). [DOI] [PubMed] [Google Scholar]
- Xu C. et al. Minor allele C of chromosome 1p32 single nucleotide polymorphism rs11206510 confers risk of ischemic stroke in the Chinese Han population. Stroke 41, 1587–1592 (2010). [DOI] [PubMed] [Google Scholar]
- Krumholz H. M. et al. ACC/AHA clinical performance measures for adults with ST-elevation and non-ST-elevation myocardial infarction: a report of the American College of Cardiology/American Heart Association Task Force on Performance Measures (Writing Committee to Develop Performance Measures on ST-Elevation and Non-ST-Elevation Myocardial Infarction). Circulation 113, 732–761 (2006). [DOI] [PubMed] [Google Scholar]
- Wang P. et al. Association of SNP Rs9943582 in APLNR with Left Ventricle Systolic Dysfunction in Patients with Coronary Artery Disease in a Chinese Han GeneID Population. PLoS One 10, e0125926 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. et al. Molecular Basis of Gene-Gene Interaction: Cyclic Cross-Regulation of Gene Expression and Post-GWAS Gene-Gene Interaction Involved in Atrial Fibrillation. PLoS Genet 11, e1005393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. et al. Significant Association Between CAV1 Variant rs3807989 on 7p31 and Atrial Fibrillation in a Chinese Han Population. J Am Heart Assoc 4, e001980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X. et al. BRG1 variant rs1122608 on chromosome 19p13.2 confers protection against stroke and regulates expression of pre-mRNA-splicing factor SFRS3. Hum Genet 133, 499–508 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y. et al. Regulation of CARD8 expression by ANRIL and association of CARD8 single nucleotide polymorphism rs2043211 (p.C10X) with ischemic stroke. Stroke 45, 383–388 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C. et al. Candidate pathway-based genome-wide association studies identify novel associations of genomic variants in the complement system associated with coronary artery disease. Circ Cardiovasc Genet 7, 887–894 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson G. L., Bibby D. F., Wong S., Agrawal S. G. & Bustin S. A. A MIQE-compliant real-time PCR assay for Aspergillus detection. PLoS One 7, e40022 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. et al. Identification of rare variants in TNNI3 with atrial fibrillation in a Chinese GeneID population. Mol Genet Genomics 291, 79–92 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. et al. Significant association of SNP rs2106261 in the ZFHX3 gene with atrial fibrillation in a Chinese Han GeneID population. Hum Genet 129, 239–246 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X. et al. Identification of NPPA variants associated with atrial fibrillation in a Chinese GeneID population. Clin Chim Acta 411, 481–485 (2010). [DOI] [PubMed] [Google Scholar]


