Abstract
In the South China Sea, coastal eutrophication in the Beibu Gulf has seriously threatened reef habitats by subjecting corals to chronic physiological stress. To determine how coral holobionts may tolerate such conditions, we examined the transcriptomes of healthy colonies of the galaxy coral Galaxea fascicularis and its endosymbiont Symbiodinium from two reef sites experiencing pristine or eutrophied nutrient regimes. We identified 236 and 205 genes that were differentially expressed in eutrophied hosts and symbionts, respectively. Both gene sets included pathways related to stress responses and metabolic interactions. An analysis of genes originating from each partner revealed striking metabolic integration with respect to vitamins, cofactors, amino acids, fatty acids, and secondary metabolite biosynthesis. The expression levels of these genes supported the existence of a continuum of mutualism in this coral-algal symbiosis. Additionally, large sets of transcription factors, cell signal transduction molecules, biomineralization components, and galaxin-related proteins were expanded in G. fascicularis relative to other coral species.
Due to the high risk of land-based sources of pollution, the most biodiverse coral reefs in Southeast Asia have so far been neglected1. Beibu Gulf is a semi-enclosed gulf located in the northwest of the South China Sea, which is surrounded by Vietnam, Guangxi, Leizhou Peninsula and Hainan Island. The deteriorating eutrophication in Beibu Gulf due to the increasing nutrient and organic matter fluxes over the past 30 years is one of the major threats to coral reefs2. Eutrophication and turbidity enrichment directly affect coral physiology, including photosynthesis, respiration, calcification and reproduction, resulting in coral diseases and the microbial community shift of corals3. Most coral species appear to be sensitive to eutrophication4, while some corals, such as Galaxea retiformis and Turbinaria mesenterina, are considered to be more resistant due to a higher trophic plasticity5. To comprehensively understand the mechanism of eutrophication tolerance, fundamental studies are needed to shed light on the perception of the eutrophic disturbance and the corals’ holobiont metabolic and molecular resilient regimens.
Galaxea fascicularis is a type of massy reef-building coral that is widely distributed in the tropic Indo-Pacific area6. G. fascicularis is classified into soft and hard types based on nematocyst morphology7. Because of genetic differentiation8, their morphological characteristics are correlated with the length of the noncoding region between the mitochondrial genes cytb and nad29. A more recent study reported several cryptic types found in G. fascicularis based on genetic information from microsatellite loci8,10. G. fascicularis shows intraspecific genetic diversity and connectivity and appears to have the potential for the recovery of populations after disturbance8,10. Additionally, G. fascicularis is relatively resilient to bleaching and sedimentation11 and is therefore widely used for comparative analyses of coral responses to thermal, acidulated and light stresses12,13. Despite the unique and interesting biological features of G. fascicularis, molecular information about its responses to stressors is still limited.
Coral bleaching often results from increasing environmental perturbation, which causes corals to lose their vital photosynthetic dinoflagellate symbionts (Symbiodinium)14. Despite the importance of the mutualism between coral and Symbiodinium, it is surprising that very limited information is available about the molecular events underlying the establishment, interaction and collapse of the mutualism under stress. Here, we used a transcriptomic approach to elucidate the transcriptional profiles of native G. fascicularis and its endosymbionts in Beibu Gulf of China, and furthermore, we identified specific genes and possible metabolic pathways involved in tolerance to mid to long-term eutrophic stress. The availability of transcriptome sequences and gene annotation enabled us to study the molecular interactions between the host coral and its endosymbiont Symbiodinium. Our study demonstrates that the usefulness of the G. fascicularis transcriptome will accelerate the understanding of the mechanisms of reef coral resistance on the impacts of anthropogenic global climate changes.
Results
Transcriptome assembly and annotation
To globally characterize the transcriptome of G. fascicularis and its Symbiodinium endosymbionts with enhanced sequence coverage, 12 cDNA libraries were constructed using G. fascicularis polys obtained from eastern and western pools of Hainan Island, China. In total, 289,711,812 Illumina paired-end raw reads were generated. After discarding adaptor and low-quality sequences, we obtained 281,021,672 clean reads (accounting for 42.15 Gb). The percentages of clean reads among the raw tags (Q20) ranged from 96.74 to 97.02% (Table S1). The de novo assembly of clean reads resulted in 347,084 unigenes in the range of 201–50,485 bp, with an N50 length of 1,259 bp (Fig. S1). In total, 150,771 (43.44%) unigenes were successfully annotated in at least one of the Nr, Nt, Swiss-Prot, KEGG, GO, COG and Pfam public databases (Table S2).
Dissecting G. fascicularis and its Symbiodinium endosymbiont transcriptomes
BLASTN analysis of the G. fascicularis symbiosis transcriptome against the custom genome database sets of Nematostella vectensis, Acropora digitifera and Symbiodinium (e-value < 1E-10) indicated that 13,293 (3.83%), 11,098 (3.20%) and 25,030 (7.21%) unigenes had significant matches to the genomes of these species, respectively. Among the remaining 297,664 (85.76%) unigenes, approximately 88,415 (25.47%) of them were annotated in the NCBI nr database. Figure S2 depicts the species distribution of the top BLAST results. Of these 88,415 protein-encoding unigenes, 31,844 (36.02%) matched metazoan genes (i.e., invertebrate and vertebrate) and thus were presumably of coral (G. fascicularis) origin15. Likewise, 24,189 (27.36%) unigenes were significantly aligned to genes from alveolates, other algae, or green plants. These unigenes were assumed to be of Symbiodinium origin (Fig. S3A)16. Additionally, approximately 32,382 (36.63%) of these nr database-matching unigenes aligned to bacteria or undetermined taxonomic assignment and were not used in the subsequent analyses.
A previous study has suggested that variations in the GC content are correlated with the origin and evolution of species’ life-history features17. The mean GC content corresponding to typical exons for anthozoan cnidarians (the mean GC contents of A. digitifera and Nematostella are similar) was approximately 40%. In comparison, the mean GC content of the Symbiodinium genome is over 50%18,19,20. We further investigated the GC content distribution of all the sequences in the assembly. Two clear peaks at approximately 43 and 55% were detected (Fig. S3B), suggesting that the two peaks possibly resulted from mixed transcriptomes of coral (G. fascicularis) and Symbiodinium. The peak GC content and N50 in the components of the G. fascicularis and Symbiodinium origin assembly were approximately 42.96% (N50 length 2,735 bp) and 54.91% (N50 length 1,893 bp), respectively (Table S3). These results were largely congruent with the fact that the two peaks were detected in the whole transcriptome, indicating that we effectively separated G. fascicularis and Symbiodinium-originated unigenes.
Transcriptome-wide differential expression in response to eutrophication
Eutrophic environment in the west coast of Hainan Island
As a prerequisite for this study, we investigated the eutrophication status of the two reef sampling sites (western and eastern pools of Hainan Island) (Fig. 1A,B). There were significant differences in the concentrations of the total alkalinity (TA), dissolved inorganic nitrogen (DIN), NO3−, NO2−, PO4−, suspended particulate matter (SPM) and chlorophyll a between eastern and western pools during the period of our sampling (p < 0.05) (Fig. 1C). As expected, high concentrations of dissolved inorganic nitrogen (DIN, NO3− and NO2−), phosphate (PO4−) and chlorophyll a at western sampling sites reflected eutrophic conditions in western coastal waters21. Consistently, many studies have shown that in recent decades, strong human impacts on Beibu gulf areas have resulted in a high abundance of nitrogen and phosphorus signals, SPM and phytoplankton, which may contribute to coastal eutrophication2. This process motivated us to explore the molecular basis of coral tolerance to chronic seawater eutrophication.
Figure 1.
(A) Overview of the location of the study areas at Beibu Gulf of the South China Sea. The map was created using ArcGIS 10.3.1 (http://www.esri.com/software/arcgis). (B) Location of the coral sampling sites of the western and eastern pool of Hainan island. The map was drawn using DIVA-GIS (Version 7.5.0, http://www.diva-gis.org/) software based on coordinates recorded for each locality with a GPS device. ●: point of G. fascicularis samples. (C) Mean environmental conditions in eastern and western points of water sampling (sample sizes are n = 13 at the eastern pool and n = 15 at the western pool). Values represent the mean ± SD data for analysing the statistical significance by Student’s t-test. Asterisks indicate significant differences (*p < 0.05, **p < 0.01). Abbreviations are as follows: Deg: Degrees Celsius; DIC: Dissolved inorganic carbon; TA: Total alkalinity; DO: Dissolved oxygen; DIN: Dissolved inorganic nitrogen; SPM: Suspended particulate matter; Ω calcite: calcite saturation state; Ω arag: Aragonite saturation state.
Differential gene expression patterns of coral
Our transcriptome dataset included the expression levels of unigenes from G. fascicularis and its endosymbiont Symbiodinium. We performed a principal component analysis (PCA) on the normalized counts for G. fascicularis holobiont unigenes to visualize the extent of variation between the two pools and found that the samples were distinctly separated by geographical location between the eastern and western pools (Fig. 2A). Substantial differences in the gene expression patterns of G. fascicularis holobionts perhaps represented signs of genetically based or mid- to long-term adaptive acquisition of tolerance to eutrophication in western sites. Coral gene expression data revealed that the expression of 236 unigenes (1.06%) differed significantly between the two sample sets with pristine versus eutrophied conditions (origin: east vs west; |log2 fold change| > 1.0 expression difference, FDR–corrected P value < 0.05). Of these unigenes, 56 had higher expression levels at the pristine (east) pool, while the remaining 180 unigenes had higher expression levels at the eutrophied (west) pool (Fig. 2B,C). Out of 236 unigenes, 117 were annotated with the Swiss-Prot database (Table S4). Among the annotated genes, we found that many of them encode transcription factors (11 genes), cell signalling proteins (20 genes), innate immunity related factors (e.g., NF-κB and TNF receptor–associated TRAF–type proteins, 4 genes), solute carriers (6 genes), cytochrome P450 (2 genes), components of the TCA cycle (3 genes), lipid or fatty acid synthases (5 genes), and regulators of cell cycle and apoptosis (3 genes). In particular, a large number of genes (43 genes, 24%) with increased expression in the eutrophied pool were involved in innate immunity, cell apoptosis and energy metabolism.
Figure 2.
(A) Principal component analysis based on the expression level data of the G. fascicularis symbioses transcriptome from six sites. Gene expression was computed as FPKM with a pseudocount of 0.01. Principal component analysis was performed in Origin Pro 9.1. The first two principal components explained 95.45% of the total variance. (B) Overview of the number of differentially expressed genes. The differentially expressed genes of the G. fascicularis host and its endosymbiont Symbiodinium were identified by comparing the east and west transcriptome with |log2 fold change| > 1.0 and FDR adjusted p-values ≤ 0.05. Heatmap analysis of differentially expressed genes identified in the G. fascicularis host (C) and its endosymbiont Symbiodinium (D). The heatmap was created using heatmap2 in the R statistical computing environment, and the gene expression was characterized as the log2 of the ratio between expression levels in the 6 sites. Higher expression levels are shown in pink, while lower expression levels are shown in green. (E1, E2 and E3: sampling sites in the east pool. W1, W2 and W3: sampling sites in the west pool).
Expression variation of different Symbiont types
The Symbiodinium types in each coral sample were determined by a molecular approach22. The alignment results of the Symbiodinium internal transcribed spacer 2 (ITS2) sequences reflected that symbiont types C1, D1a, and D1 were detected, and type C1 was dominant in all colonies examined (Table S5). It is likely that the eutrophied environment did not change the dominant type of Symbiodinium.
Our transcriptome data showed that 139 and 66 unigenes were up-regulated and down-regulated in the pristine pool compared to the eutrophied pool, respectively (|log2 FC| > 1.0, FDR–corrected P value < 0.05) (Fig. 2B and D). Only 39 of these unigenes were annotated with KEGG terms. The affected genes could be assigned to the biosynthesis of secondary metabolites (6 genes), transportation (3 genes), protein processing in endoplasmic reticulum (2 genes), ribosome (2 genes), photosynthesis (LHCA1 gene), circadian clock regulation (CSNK1E gene) and were associated with physiological plasticity under environmental stressors (4 genes) (Table S6), suggesting that the metabolic and cellular processes of symbionts were also affected under long-term eutrophic stress.
Functional analysis of the unigene sets originating from G. fascicularis and Symbiodinium
To investigate the comprehensive transcriptome of the holobiont consisting of G. fascicularis and its symbionts from the perspective of systems biology, we first categorized the unigene sets originated from G. fascicularis and Symbiodinium separately by gene ontology (GO) analysis. We assigned 8,460 and 17,177 unigenes to one or more GO terms for G. fascicularis and Symbiodinium, respectively (Fig. S4). In comparison, we observed that for most functional categories that are essential for normal cell activities, Symbiodinium have lower unigene percentages than the coral host, except for the three categories related to the extracellular matrix, catalytic activity and transporter activity.
The unigenes originated from each component were subjected to a search against the KOG database for functional classification. We subdivided 6,752 and 9,307 non-redundant unigenes of G. fascicularis and Symbiodinium, respectively, into 26 classifications (Fig. 3). Interestingly, G. fascicularis-host and Symbiodinium had different percentages of unigenes involved in metabolic pathways, suggesting that the metabolic components of G. fascicularis-host and Symbiodinium had distinct enrichment patterns. Of these pathways, 7 Symbiodinium showed much higher percentages than that in G. fascicularis-host, including (A) RNA processing and modification, (C) energy production and conversion, (G) carbohydrate transport and metabolism, (I) lipid transport and metabolism, (M) cell wall/membrane/envelope biogenesis, (O) posttranslational modification, protein turnover, chaperones and (P) inorganic ion transport and metabolism. In contrast, several other pathways were particularly abundant in G. fascicularis-host, whereas they were represented in much lower percentages in Symbiodinium. These pathways included (B) chromatin structure and dynamics, (D) cell cycle control, cell division, chromosome partitioning, (K) transcription, (R) general function prediction and (U) intracellular trafficking, secretion, and vesicular transport.
Figure 3. Large-scale comparison of KOG categories assigned to G. fascicularis and its Symbiodinium symbiont transcriptomes.
The enrichment analysis (y-axis) is expressed as the percentage of sequences in the G. fascicularis and its Symbiodinium symbiont transcriptome sets, respectively.
In addition, KEGG enrichment analysis for G. fascicularis and its Symbiodinium symbiont transcriptomes was performed. Symbiodinium retained very abundant genes involved in nutrient provision to their hosts, which was associated with carbohydrate metabolism, energy metabolism, glycan biosynthesis, lipid metabolism and nucleotide metabolism (Fig. S5). These findings are consistent with a previous report showing that the coral host acquired and utilized sugars and carbon sources for energy metabolism from its Symbiodinium23. In addition to the above-mentioned metabolic pathways, we were also surprised by the observation that the Symbiodinium transcriptome had an ample amount of genes associated with other metabolic pathways, including amino acid metabolism, vitamin and cofactor metabolism, biosynthesis of secondary metabolites and metabolism of terpenoids and polyketides. Many of these genes were not found in the G. fascicularis-host transcriptome, which pointed to the genetic complementarity between G. fascicularis and Symbiodinium.
Integration of metabolic pathways between coral and Symbiodinium
The analysis of a metabolic pathway map based on G. fascicularis and its Symbiodinium symbiont transcriptomes revealed that high integration and interdependence at the metabolic level occurred in the coral-symbiotic system (Fig. S6). Of note, G. fascicularis holobiont showed a striking metabolic complementarity in vitamin and cofactor biosynthesis. A large number of the Symbiodinium unigene-encoded proteins were predicted to play important roles in the synthesis pathways of a diverse set of vitamins, cofactors, prosthetic groups and other related compounds. However, the paucity of vitamin and cofactor synthesis pathways in the G. fascicularis host suggested the possibility that unigenes in G. fascicularis and Symbiodinium may play distinct and collaborative roles in this process24 (Fig. 4A). Furthermore, G. fascicularis showed an incomplete fatty acid biosynthetic pathway due to the lack of the β-ketoacyl-ACP synthase I and II genes (fabF and fabB). These two genes can be found in the Symbiodinium based on our transcriptome data. Our data also provided molecular evidence of a mutualistic continuum between coral hosts and Symbiodinium in amino acid synthesis. The amino acid synthesis pathways showed a Symbiodinium-dominated tendency as the majority of genes participating in these pathways were only found in Symbiodinium (Fig. 4B). The coral holobiont had complete pathways for the biosynthesis of most standard amino acids but lacked several functional genes that are important for the biosynthesis of histidine (hisB gene is missing), tryptophan (trp1 gene is missing), tyrosine (tyrA, tyr B and tyrC genes are missing), and phenylalanine (tyrA, tyrB and tyrC genes are missing). To test these hypotheses, we investigated whether any critical genes necessary for the biosynthesis of vitamins and cofactors and amino acids were also lost in the A. digitifera genome. Indeed, the biosynthesis pathways of vitamins and cofactors and amino acids were also incomplete in A. digitifera according to the KEGG annotation, which is consistent with our observation. Additionally, G. fascicularis holobiont also showed remarkable integration in the biosynthesis of secondary metabolites (Fig. S7) to produce the diverse and complex compounds required by members of the consortium. However, it is still unclear how the coral host and Symbiodinium coordinate their respective systems in the biosynthesis of vitamins and cofactors, fatty acids, amino acids and secondary metabolites to establish mutualistic nutritional benefits.
Figure 4.
(A) Vitamin-related gene expression and retention patterns for the G. fascicularis and Symbiodinium symbiont transcriptomes. (B) The predicted amino acid biosynthesis pathways of G. fascicularis and Symbiodinium. Blue lines indicate genes present in both the Symbiodinium and G. fascicularis transcriptome. Red lines indicate genes only present in the G. fascicularis transcriptome. Green lines indicate genes only present in the Symbiodinium transcriptome. Grey lines indicate genes absent in the Symbiodinium and coral (G. fascicularis) transcriptome.
Comparative analysis of prominent gene families revealed significant differences among related species
A transcriptome-scale comparison of major gene families in related species suggested that some families were expanded in G. fascicularis. PCA allowed the estimation of the depth of the divergence among corals and other Cnidaria species. The domain similarities composition of the G. fascicularis was closely related to that of Porites australiensis and N. vectensis among the species examined (Fig. 5). The numbers of predicted transcription factor genes and cell signalling molecules in G. fascicularis were significantly higher in comparison with those in P. australiensis and A. digitifera but lower than that in N. vectensis (Tables S7 and S8). We applied a comparative analysis of the predicted biomineralization-related proteins involved in the formation of inorganic skeletons and organic matrices in G. fascicularis and other species. The results showed that the number of biomineralization-related proteins in the G. fascicularis transcriptome was expanded to 68, compared to 48 in P. australiensis and 43 in A. digitifera (Table S9). Additionally, there were only 289 unigenes with conserved innate immunity and the apoptosis domain found in the G. fascicularis transcriptome (Table S10). The number was significantly less than that in P. australiensis (379 unigenes) and A. digitifera (833 unigenes).
Figure 5. Principal component analysis depicting the correlations between predictor variables and the relative abundance of domain similarities of G. fascicularis and other related corals.

Principal component analysis performed in Origin Pro 9.1. The first two principal components explain 95.58% of the total variance. Abbreviations are as follows: Acropora: Acropora digitifera, Porites: Porites australiensis, Galaxea: Galaxea fasciculari, Nematostella: Nematostella vectensis, Hydra: Hydra magnipapillata.
We further searched for the unigenes and pathways potentially involved in the G. fascicularis symbiosis. The symbiosis-associated genes fell into several broad functional categories as listed in Table S11 25. These genes were identified by the presence of characteristic domains in the predicted proteins by BLASTx analysis. These genes were then compared to homologous sequences in publicly available resources (genomes and transcriptomes) for A. digitifera, A. hyacinthus, A. millepora, A. palmata, M. faveolata, P. astreoides, P. damicornis, and the more distantly related species N. vectensis, A. pallida and A. viridis. The symbiosis-associated genes of G. fascicularis were more similar to those of the other corals (A. hyacinthus, A. millepora, P. astreoides and A. digitifera) than to the highly different sets found in anemones such as N. vectensis, A. pallida and A. viridis (Fig. S8). This exceptionally large and diverse inventory of cnidarian genes associated with symbiosis will help to elucidate molecular models of host-symbiont interactions.
Large expansion of galaxin genes for coral calcification
Six unigenes encoding galaxin proteins were identified in the G. fascicularis transcriptome based on similarity searching, including one galaxin protein and five galaxin-like proteins. A phylogenetic tree was constructed based on the predicted galaxin and galaxin-like protein sequences. Our results revealed a species-specific pattern in galaxin and galaxin-like proteins between G. fascicularis and Acropora (A. digitifera and A. millepora). No galaxin 2 homolog was found in G. fascicularis (Fig. 6). The location of the signal peptides and N-glycosylation sites of galaxin and galaxin-like proteins were predicted using the SignalP 3.0 Serve and NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services), respectively. The signal peptides of both the galaxin and galaxin-like proteins contained Cys repeat-like regions26. Different from galaxin, the galaxin-like precursor proteins of G. fascicularis possessed acidic amino acid regions (Asp- and Glu-rich motifs were present in the galaxin-like proteins) that varied widely in sizes ranging from 11 to 135 aa (Fig. S9).
Figure 6. Phylogeny of Galaxin and Galaxin-like proteins.

A phylogenetic tree of Galaxin proteins in the corals was created based on the maximum likelihood analysis with the Kimura 2 parameter method. Full–length amino acid sequences were used in this analysis. Percentages of 1,000 bootstrap replicates are indicated next to the tree nodes. Sequences from the present study are marked with solid triangles.
Discussion
Molecular basis for coral tolerance to chronic eutrophication
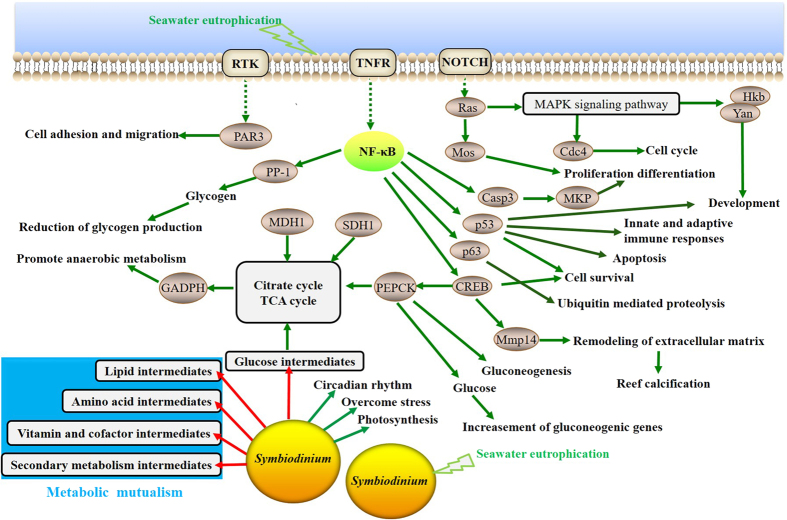
The water qualities in the eastern and western pools of Hainan were comprehensively analysed in this study. Nitrogen, phosphorus, and chlorophyll-a, which are the most significant factors causing eutrophication, were much more abundant in the western pools. The transcriptome data revealed that 236 genes (FDR corrected P < 0.05) were potentially associated with the chronic eutrophic condition, which demonstrated that this approach provided new insights into the study of the molecular basis of coral eutrophication tolerance. The expression levels of transcription factors and cell signalling molecules associated with tolerance processes changed significantly in coral. The classic transcription factor NF-κB showed remarkable variations in expression levels, which could be used as the ‘smoke-detector’ due to the strong link between NF-κB and different stress signals27. Once activated, NF-κB regulates the expression of a large number of downstream processes involved in immunity, apoptosis, cell structuring, adhesion, cell cycle and development, maintenance of energy homeostasis, and response to stress (Fig. 7)28. The gene expression pattern of signal transduction in the coral immune response was largely congruent with previous observations in Acropora palmata29.
Figure 7. An integrative model summarizing the roles of genes involved in G. fascicularis tolerance to coastal chronic eutrophication.
The known functional pathways are represented as green lines. The possible functional pathways are represented by red lines.
The overloading of nutrients favours the growth of algae, aquatic macrophytes and other microbes. Prodigious dissolved oxygen is consumed by the algae and microbes in the respiration and decomposition of organic matter, triggering aquatic hypoxia and cyanotoxin stress in the eutrophication area30. We hypothesized that such an environment disfavoured the survival of corals and may result in the stress/immune response of corals. Several receptors showed increased expression in the eutrophic condition, such as DRD1, HTR1, NPFFR2, NPYNR, TNFR1, RTK and Notch. It is very likely that under eutrophication, TNF signalling activated the NF-κB pathway (increased 1.17- and 1.97-fold in eutrophied sites, respectively), which subsequently transduced the signal to its primary target proteins, such as p53, p63 and Casp3 elements. These proteins contributed to governing innate and ‘adaptive-like’ immunity responses, the control of cell survival, differentiation and proliferation. Lesser et al. previously detected similar patterns invoked by heat and light stresses in Montastraea faveolata31. The ubiquitin-mediated proteolysis pathway targets many short-lived cell cycle regulatory proteins, such as cdc14 (1.59-fold), cdk5R1 (1.46-fold) and TOB (1.65-fold)32, allowing for quick transitions between cell cycle stages33. The expression levels of three calcification genes (Mmp14, SNX10 and SOX9) were also highly variable, changing 2–4 fold in our analysis26,29.
Maintaining the metabolic balance and energy homeostasis between coral and Symbiodinium as the basis for symbiotic systems ensures the survival of individuals. Herein, the KEGG term enrichment analysis (p < 0.05) for the coral samples collected from the western pool versus the eastern pool was performed. The eutrophied pool showed increased expression of genes involved in several pathways, including the TCA cycle, glucose metabolism and gluconeogenesis, and decreased expression of the lipid biosynthesis pathway. These genes could also be categorized according to their roles in energy metabolism. Additionally, NF-κB directly or indirectly participates in the regulation of several insulin-related proteins (such as PEPCK, CREB, PP-1, GADPH, KLF3, MDH1 and SDH1) and signalling pathways that are in control of energy demand34. Meanwhile, corals have coordinated the expression of genes involved in intermediate transport and exchange between the coral host and Symbiodinium during adaptation to eutrophic conditions, enabling them to efficiently reconstruct mutualistic ecology and share metabolism balance35. Solute carriers (SLCs) embrace transporters for inorganic substrates or other small organic molecules, i.e., nitrogen, Na+, Ca+ and amino acid carriers. CYP1C and CYP3A, which belong to the cytochrome P450 family genes, typically catalyse mono-oxygenase reactions involving the metabolism of intermediate substrates36. We observed that six SLC genes and these two cytochrome P450 family genes exhibited strikingly different expression levels (fold change > 1.5) in transcriptomic analysis. Our study also indicated that eutrophication induced the expression of several proteins responsible for activating cell self-protection, such as HSP27 (1.03-fold) and opuD (4.01-fold). HSP27 is essential for the response to a wide variety of unfavourable environmental conditions to finely regulate the physiological balance37. OpuD is a major osmoprotectant glycine betaine transporter required for early osmotic adjustment38. Overall, these results implied the potential for transcriptomic plasticity in the coral and indicated the underlying molecular mechanisms for chronic eutrophication tolerance.
Major Symbiodinium shifts of corals occur in severe bleaching events or extreme high temperature episodes28. Our study showed that the chronic eutrophied coral colonies did not shift the tendency of Symbiodinium C to be the dominant symbiont, but approximately 0.82% (205 genes) of symbiotic Symbiodinium genes in the eutrophic state showed differential expression compared to that in the relative pristine state. It is possible that both the coral and Symbiodinium have adapted to the eutrophic environment under long-term exposure. Consistently, Daniel et al. suggested that Symbiodinium is able to adjust gene expression and transcriptional acclimation over long time scales in response to shifting environmental conditions39. Our results indicated that eutrophication had significant influences on the photosynthesis (LHCA1, 1.01-fold), circadian rhythm (CSNK1E, 2.07-fold) and stress response (STIP1, 1.83-fold) of symbiotic Symbiodinium, in line with the symptoms of eutrophic stress, which include decreased photosynthetic efficiency and increased consumption4. Another group was found to be associated with the biosynthesis of secondary metabolites (E1.1.1.122, E3.2.1.21, dxs, PLSC, GPI and MetH) and sucrose metabolism (beta-glucosidase) adjustment for concordance with the coral host gene expression, which may relate to metabolic modulation for the mutualistic continuum of host nutrients/intermediates (Fig. 7). Our results suggested that chronic eutrophication influenced multiple specific biological processes by altering transcription accumulation40 and coordinating the expression of two aspects of the coral host and Symbiodinium symbionts. However, the mechanisms of global transcriptional changes and their physiological consequences are not yet fully understood.
Metabolic mutualism between coral and Symbiodinium partners
Metabolic mutualism, where each partner produces gene products necessary for mutual survival, reflects the shared metabolites of the partners41. Metabolic mutualism is maintained under a plot of reciprocal exploitative interactions simultaneously keeping the contribution of each partner to a minimum while selfishly maximizing their own fitness42. Recently, Shinzato and Lin found support for a potentially genetically complementary supply between the coral and Symbiodinium genomes in terms of essential amino acids and major photoprotector mycosporine-like amino acids (MMA), forming an interdependent metabolic patchwork for essential nutrient provisioning and photo-protection19,20,43. Many pathways of amino acid synthesis require gene products produced by coral and Symbiodinium, while G. fascicularis and symbiont seem to lack the expression of genes necessary for histidine, tryptophan, tyrosine and phenylalanine biosynthesis, suggesting yet another source or some yet elusive metabolic patchwork of these amino acids (i.e., bacterial endosymbionts)44. In our transcriptomics study, we firstly proposed that such a coral host-Symbiodinium metabolic mutualism pattern involved vitamin and cofactor, fatty acid and secondary metabolites biosynthesis. Previously, we considered that Symbiodinium was a complement machine and provided different resources (i.e., energy source, amino acids, vitamins and cofactors) for the host coral. However, based on the current evidence that little overlap occurs in the biosynthetic genes, the coral and Symbiodinium appear to depend on each other for necessary compounds and intermediates in the metabolic processes. These mutualism associations suggest that gene products or intermediates for vitamin and cofactor biosynthesis are shared between coral and Symbiodinium. In this regard, interaction intermediates could occur in many aspects of metabolism, and required intermediates or enzymes are somehow available in both systems23,45, which contributed to create a strong metabolic plasticity and continuity in the face of environmental change. The mutualistic characteristics may extend to the mutual compounds that were redundant in the symbionts or the hosts46. Typically, the Symbiodinium transcriptome has an ample amount of genes associated with pathways involved in DNA replication, protein processing, transcription, translation, and nutrient provisioning to host47,48. To our knowledge, the coral host acquires carbon source for energy and nutrition from its symbionts. However, of particular interest to us are the roles of Symbiodinium in translocating photosynthetic products as well as the recycling of host metabolic products35.
It is worth mentioning that in this study, the coral samples were collected with limited time points and environmental conditions, and it is possible that the completeness of the gene repertoire of the assembled transcriptome is low and that some metabolic genes in the coral or Symbiodinium are missing in the dataset. Nevertheless, we discuss the evidence supporting the existence of a mutualistic continuum in Symbiodinium-coral interactions and propose a consideration of the evolutionary ecology of these associations42. The oligotrophic habitat makes the association between coral host and symbionts driven by metabolic integration desirable49. Thus, a metabolic mutualism continuum reflects long-term cooperation and coevolution of the two lineages represented by the coral host and symbiont Symbiodinium50. There is a more recent hypothesis that vertical transmission of symbionts generally represented a greater number of mutualistic associations, which favours increased metabolic integration that (i) leads to symbiont genome reduction and obligate dependency for a specific host and (ii) genetic uniformity of symbionts and reduction of the number of competitive phenotypes42. Most horizontally transmitted symbionts (i.e., free-living Symbiodinium) are facultative or shift to parasitism, which leads to a reduction in host fitness and steadiness51. There is evidence that some species of Pocillopora and Porites are vertical transmitters and are more resistant to environmental stress52. If true, then this could provide novel insights into the potential fitness consequences of the coral-symbiotic mutualistic system, specifically when these associations are exposed to a range of environmental stresses.
G. fascicularis transcriptome characteristics
Here, we reported the first transcriptome generated for a galaxy coral, G. fascicularis. In total, we obtained 42.15 Gb of raw sequence data that was assembled into 347,084 unigenes ( ≥ 200 bp). We further characterized 150,771 unigenes based on functional annotation. The BLAST search against the A. digitifera and N. vectensis datasets identified 24,391 G. fascicularis unigenes with hits, a similar number of shared unigenes with other Anthozoa species. However, a large amount of sequences could not be matched to the reference genomes. There could be several possibilities for the unmatched reads, such as sequencing and assembling errors, the presence of contaminated sequences and the incompleteness of the reference genomes. In particular, because the coral we used in the transcriptome sequencing was G. fascicularis, which taxonomically differs from the reference coral A. digitifera19, the unblasted unigenes may be derived from the presence of a considerable number of G. fascicularis-specific genes or divergently transcribed genes53. Because total RNA was isolated from the adult G. fascicularis polyps, the transcriptome assembly may contain unigenes from both coral and its zooxanthellae symbionts. Indeed, 25,030 sequences were successfully matched to Symbiodinium (clade C and D, C as the dominant symbiont type). In addition, approximately 31,844 and 24,189 unigenes were assumed to be of coral and Symbiodinium origin, respectively. All of these data will lay the foundation for further studies on the ecology of interactions between G. fascicularis and Symbiodinium.
Surveys for major gene families showed that the G. fascicularis genome encoded a larger number of transcription factors and signal transduction molecules than did P. australiensis and A. digitifera. It is argued that as organism complexity increases during evolution, more regulators are required for the regulatory network manifold54. The comparisons revealed that biomineralization-related proteins are particularly expanded in G. fascicularis. Galaxin, which encodes for a matrix protein suspected to be involved in calcification, is originally identified from the coral G. fascicularis55. It is hypothesized that the coral galaxin homologs are recruited as biomineralization proteins when Scleractinia diverged from non-biomineralizing taxa during the Triassic56 because galaxin-like proteins are also found in non-calcifying taxa outside Cnidaria50. Six galaxin coding sequences is a surprisingly high number in G. fascicularis, exhibiting a far greater calcific potential26. Such enhanced biomineralization and calcification machinery indicates the enhanced reef-building capacity of G. fascicularis. Herein, we also identified and comparatively analysed a number of immune-related and symbiosis-associated genes, which revealed significant differences between related coral species. These genes will be of great importance to study the basis of the coral innate immunity network and symbiont recognition and maintenance.
Conclusions
In this study, the main findings are as follows: (i) Transcriptome-wide gene differential expression analysis of G. fascicularis and its endosymbiont Symbiodinium was used to determine multiple specific biology processes that play important roles in coral holobiont chronic tolerance to coastal eutrophication. (ii) Metabolic integration was observed in vitamin and cofactor, amino acid, fatty acid, and secondary metabolite biosynthesis between the G. fascicularis host and its symbionts at the expression level, providing evidence to support the existence of a metabolic continuum in coral-Symbiodinium systems. (iii) Surveys of the G. fascicularis transcriptome showed that large sets of transcription factors, cell signal transduction molecules, biomineralization machinery, and galaxin-related proteins were expanded to a comparative scale with other associated coral species. We envision that analysis of the molecular ecology bases of G. fascicularis may provide new insights that are critical for the future protection of coral reefs from global marine environmental pollution.
Materials and Methods
Ecological setting and experimental design
The western pool of Hainan lies in the Beibu Gulf, which is a semi-enclosed shallow bay bounded by Vietnam, Guangxi, Leizhou Peninsula and Hainan island (Fig. 1A). The Beibu Gulf is subjected to land-based sources of pollutants from industrial, agricultural and aquaculture activities on the northeastern side, which is one of the most industrialized areas in China. Our focus is on three main points of the seawater eutrophication area at the western pool of Hainan, including the Haitou/Yangpu port and an adjacent pool zone, which is fed by two lowland rivers (Zhubi River and Beimen River) that drain agriculture and industry areas of the coastal plain. It is noted that the rivers bring nutrients (especially nitrates and phosphates) from the land to the Gulf. To decipher the transcriptomic landscape of coral tolerance to eutrophication in the western pool, we used the corals originating from the eastern pool of Hainan at the same latitude as the reference to estimate different expression patterns of unigenes. The east coast of Hainan is open to the west-Pacific. Sewage discharge and other forms of pollution have not yet been reported in this area. The two pools we have focused on represent two distinct coral reef water qualities that are similar in depth (1.5–2 m), wave exposure and water delivery57. As a prerequisite for this study, we further confirmed by examining the water samples from the eastern and western pools the nutrient concentrations. The results showed that strongly higher concentrations of nutrients were present in western sites due to anthropogenic eutrophication and poor flow in the semi-enclosed pool in the Beibu Gulf. No significant difference in the sea-surface temperature, salinity or water pH was found between the two pools in our investigation. As the environmental condition of the coral reef is a dynamic system, we did not rule out the possibility of other environmental differences, such as minor variability in oxygen or water flow. We used G. fascicularis as the coral model in this study because this species lives from shallow water backreefs to deeper fore reef habitats and is highly abundant across the South Sea of China.
Coral collection, identification and ethical statement
Coral polyps were collected from 12 healthy-looking colonies of G. fascicularis living in the eastern and western pools of Hainan island, China, at noon in July 2015 (E1, E2 and E3: samplings from the eastern pool. W1, W2 and W3: samplings from the western pool, see Table S1). The locations of the six sampling sites are shown in Fig. 1B. Two colonies were collected from each sampling site, which yielded 12 samples that were separately prepared for gene expression analysis via RNA-Seq. The polyps were rapidly immersed in 1 mL of TRIzol reagent and frozen in liquid nitrogen. The samples were then stored at −80 °C until processing. The field studies and sample collection activities were necessarily permitted by the Chinese Hainan Government Department of Coral Reefs Protection Provisions. Genomic DNA from the colonies of G. fascicularis, which were used in the transcriptome study, was extracted, and the noncoding regions of the mitochondrial genes cytb/nad2 were amplified using PCR with the primers (188-2: 5′-TCCTGTAGAATAGGGTATAC-3′) and (188-R2: 5′ TTTGCCTTTCCGTATCCACCAT-3′)9. The cytb/nad2 sequence data from G. fascicularis colonies as well as reference sequences were aligned using DNAMAN version 4.0 (Lynnon Biosoft, San Ramon, CA) (Fig. S10). The result presented here confirmed that the 12 corals used in the experiment were from the same lineage and were 100% identical to the sequences of mt-L1 type of Galaxea at GenBank (accession number: LC155810 and AB109376)8,10.
Water sampling and analysis
Water samples were collected from the western pool, eastern pool and coastal sites of Hainan. Front-reef and Back-reef waters were sampled randomly by boat along a land-sea gradient. Discrete water samples were collected, beginning per coral sampling within 1.0 m of the colony. For dissolved nutrients, near-bottom (1.00 m above bottom) water samples were filtered through 0.45-μm GF/F filters and collected in triplicate in clean 1-L Nalgene bottles. They were frozen in darkness until analysis in the Key Lab of Marine Biogenetic Resources at the Third Institute of Oceanography (TIO). Mercuric chloride (0.03% total volume) was added to the water samples to measure the total dissolved inorganic carbon (DIC) and total alkalinity (TA). For each discrete water sample, DIC was measured using an Apollo SciTech model AS-C3 DIC analyser (Apollo SciTech, DE, USA). TA was measured by open-cell potentiometric titration, and salinity was measured with a Guildline Autosal salinometer58. The initial seawater pCO2, pH, HCO3−, CO32−, Ω aragonite and Ω calcite were calculated by measurement of TA, DIC and salinity using the CO2 SYS program59. For chlorophyll a analysis, we collected 800-mL water samples in triplicate of near-bottom water that were filtered (after adding 1 mg of MgCO3) through GF/F glass fibre filters. The filters were extracted for 30 min with 10 mL of dimethyl sulfoxide, and we then added 15 mL of 90% acetone at 4 °C overnight. They were measured fluorometrically before and after acidification for the measurement of chlorophyll a and phaeopigment concentrations60. The samples were analysed for NH4+ –N, NO3−–N, NO2−–N, PO43− –P and SiO32−-Si on a model 7230 Spectrophotometer at the Marine Chemistry Laboratory in TIO. The concentration of dissolved inorganic nitrogen (DIN) is the sum of NO3−, NO2−, and NH4+. After a test for variances within location points, the significance of differences between the location points was assessed using 1-way ANOVA and a t-test via the Tukey method. All statistical tests were performed using IBM SPSS Statistics 23.
Transcriptome Sequencing and Assembly
Total RNA was extracted from each coral sample using the TRIzol® Reagent RNA Isolation Kit (Invitrogen, Grand Island, NY) following the manufacturer’s protocol. RNA degradation and contamination were detected on 1.2% agarose gels. A total amount of 1.5 μg RNA per sample was used as input material for the RNA sample preparations. mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Sequencing libraries were generated using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, all 12 libraries were sequenced on an Illumina Hiseq 2500 platform at Novogene Biological Information Technology Co., Ltd. (Beijing, China), and paired-end reads were generated.
Raw reads in the fastq format were first processed through in-house perl scripts. Clean reads were obtained by removing reads containing adapter, reads containing poly-N and low quality reads from raw data. At the same time, the Q20, Q30, GC-content and sequence duplication level of the clean data were calculated. Transcriptome de novo assembly was accomplished based on the left.fq and the right.fq files from all libraries using Trinity61 with min_kmer_cov set to 2 by default and all other parameters set to the default. The obtained sequences were defined as unigenes. Sequence redundancy was removed by searching similar sequences with a minimum similarity cut-off of 95% using CD-HIT-EST. CD-HIT was used for further clustering with a 90% similarity cut-off ref. 62. Each cluster represented by its longest sequence was then combined into one fasta file and used to compare the gene numbers with other organisms.
BLAST of the transcriptome with genomes from Anthozoa and Symbiodinium
To characterize the ‘coral host’ and ‘Symbiodinium spp.’ transcriptome assembly, unigenes of the metatranscriptome were first matched using BLASTN (E-value < 1E-10) to genome and transcriptome data from Anthozoa (A. digitifera and N. vectensis)19,63. Unigenes not present in Anthozoa were searched against the database containing Symbiodinium (S. minutum, clade B and S. kawagutii, clade F) genome sequences18,43. The remaining umatched unigenes were aligned with the NCBI NR protein database using BLASTX (with 90% identity and E-value < 1E-10). The resulting outputs were used to examine the taxonomic assignment15,22,53.
Gene annotation, gene ontology and metabolic pathway analysis
Gene function was annotated using BLAST with a cutoff E-value of 10−10 based on the following seven databases: Nr (NCBI non-redundant protein sequences), Nt (NCBI non-redundant nucleotide sequences), Pfam (Protein family), KOG/COG (Clusters of Orthologous Groups of proteins), Swiss-Prot (a manually annotated and reviewed protein sequence database), KO (KEGG Ortholog database) and GO (Gene Ontology). A. digitifera genomic metabolic pathways were predicted using the KEGG Species Server: Acropora digitifera (http://www.kegg.jp/kegg-bin/show_organism?menu_type=pathway_maps&org=adf).
Quantification and differential expression analysis
Gene expression levels were measured in terms of the fragments per kilobase of exon per million fragments mapped (FPKM) as described by Mortazavi et al.64. The FPKM for each unigene was calculated by Cufflinks and Cuffdiff65. For these analyses, genes with extremely low expression in all libraries (FPKM less than 0.1) were excluded. Differential expression analysis was performed using the DESeq R package (1.10.1)66. DESeq provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate67. A corrected P-value of 0.05 and log2 (fold change) of ±1 were set as the thresholds for the selection of unigenes with significantly differential expression. We used KOBAS software to test the statistical enrichment of differentially expressed genes in KEGG pathways68.
Phylogenetic analysis
The amino acid sequences of the galaxin family from G. fascicularis, A. digitifera and A. millepora were used for phylogenetic analysis. Multiple sequence alignment was carried out using ClustalW, and the phylogenetic tree was generated by the Maximum likelihood method using MEGA version 6.0 69. Bootstrap values were calculated from 1,000-replicate analyses.
Additional Information
How to cite this article: Lin, Z. et al. Transcriptome profiling of Galaxea fascicularis and its endosymbiont Symbiodinium reveals chronic eutrophication tolerance pathways and metabolic mutualism between partners. Sci. Rep. 7, 42100; doi: 10.1038/srep42100 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the International Science & Technology Cooperation Program of China (2015DFA20500), the National Programme on Global Change and Air-Sea Interaction, Xiamen Southern Oceanographic Center (14CZY037HJ11), China NSF grants (31271567 and 31171366) and the Aquatic Sanxin Engineering Project of Jiangsu Province (D2015-11-5). We thank Dr. Chuya Shinzato and two anonymous reviewers for their constructive comments on the manuscript. This research is part of the requirements for the Ph.D. thesis for Zhenyue Lin at Xiamen University.
Footnotes
The authors declare no competing financial interests.
Author Contributions Z.Y.L., M.L.C. and J.M.C. conceived of the study and participated in its design and coordination. Z.Y.L. drafted the manuscript text. M.L.C. and J.M.C. reviewed and revised the manuscript. Z.Y.L. performed the transcriptomic analysis. Z.Y.L., M.L.C., X.D., H.N.H. and X.Q.Z. carried out the experiments. J.M.C. and X.X. contributed reagents/materials/analysis tools. All authors read and approved the final manuscript.
References
- Hughes T. P., Huang H. U. I. & Young M. A. L. The Wicked problem of China’s disappearing coral reefs. Conservation Biology 27, 261–269 (2013). [DOI] [PubMed] [Google Scholar]
- Wang Y. et al. Phosphorus, nitrogen and chlorophyll-a are significant factors controlling ciliate communities in summer in the Northern Beibu Gulf, South China Sea. PLoS One 9, e101121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen C. et al. In-situ effects of eutrophication and overfishing on physiology and bacterial diversity of the Red Sea coral Acropora hemprichii. PLoS One 8, e62091 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawall Y. et al. Nutritional status and metabolism of the coral Stylophora subseriata along a eutrophication gradient in Spermonde Archipelago (Indonesia). Coral Reefs 30, 841–853 (2011). [Google Scholar]
- Houlbrèque F. & Ferrier-Pagès C. Heterotrophy in Tropical Scleractinian Corals. Biological Reviews 84, 1–17 (2009). [DOI] [PubMed] [Google Scholar]
- Abe M., Watanabe T., Hayakawa H. & Hidaka M. Breeding experiments of hermatypic coral Galaxea fascicularis: partial reproductive isolation between colonies of different nematocyst types and enhancement of fertilization success by presence of parental colonies. Fisheries science 74, 1342–1344 (2008). [Google Scholar]
- Hidaka M. Use of nematocyst morphology for taxonomy of some related species of scleractinian corals. Galaxea, Journal of Coral Reef Studies 11, 21–28 (1992). [Google Scholar]
- Nakajima Y., Shinzato C., Satoh N. & Mitarai S. Novel polymorphic microsatellite markers reveal genetic differentiation between two sympatric types of Galaxea fascicularis. PLoS One 10, e0130176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Nishida M., Watanabe K., Wewengkang D. S. & Hidaka M. Polymorphism in nucleotide sequence of mitochondrial intergenic region in Scleractinian Coral (Galaxea fascicularis). Marine Biotechnology 7, 33–39 (2005). [DOI] [PubMed] [Google Scholar]
- Nakajima Y., Zayasu Y., Shinzato C., Satoh N. & Mitarai S. Genetic differentiation and connectivity of morphological types of the broadcast-spawning coral Galaxea fascicularis in the Nansei Islands, Japan. Ecology and Evolution 6, 1457–1469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe M. J. C. & Smith D. J. Modelling variations in corallite morphology of Galaxea fascicularis coral colonies with depth and light on coastal fringing reefs in the Wakatobi Marine National Park (SE Sulawesi, Indonesia). Computational Biology and Chemistry 30, 155–159 (2006). [DOI] [PubMed] [Google Scholar]
- Al-Horani F. A. Effects of changing seawater temperature on photosynthesis and calcification in the scleractinian coral Galaxea fascicularis, measured with O2, Ca2+ and pH microsensors. Scientia Marina 69, 347–354 (2005). [Google Scholar]
- Al-Horani F., Al-Moghrabi S. & De Beer D. The mechanism of calcification and its relation to photosynthesis and respiration in the scleractinian coral Galaxea fascicularis. Marine Biology 142, 419–426 (2003). [Google Scholar]
- Schwarz J. A. et al. Coral life history and symbiosis: functional genomic resources for two reef building Caribbean corals, Acropora palmata and Montastraea faveolata. BMC Genomics 9, 1 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield A. B., Wang Y. B., Chen C. S., Lin C. Y. & Chen S. H. Compartment‐specific transcriptomics in a reef‐building coral exposed to elevated temperatures. Molecular Ecology 23, 5816–5830 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield A., Fan T.-Y. & Chen C.-S. Physiological acclimation to elevated temperature in a reef-building coral from an upwelling environment. Coral Reefs 32, 909–921 (2013). [Google Scholar]
- Romiguier J., Ranwez V., Douzery E. J. & Galtier N. Contrasting GC-content dynamics across 33 mammalian genomes: relationship with life-history traits and chromosome sizes. Genome Research 20, 1001–1009 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoguchi E. et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Current Biology 23, 1399–1408 (2013). [DOI] [PubMed] [Google Scholar]
- Shinzato C. et al. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323 (2011). [DOI] [PubMed] [Google Scholar]
- Shinzato C., Inoue M. & Kusakabe M. A snapshot of a coral “holobiont”: a transcriptome assembly of the scleractinian coral, Porites, captures a wide variety of genes from both the host and symbiotic zooxanthellae. PLoS One 9, e85182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D., Unger D. & Qiu G. Particulate organic matter dynamics in coastal systems of the northern Beibu Gulf. Continental Shelf Research 82, 99–118 (2014). [Google Scholar]
- Pinzón J. H. et al. Whole transcriptome analysis reveals changes in expression of immune-related genes during and after bleaching in a reef-building coral. Royal Society Open Science 2, 140214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnik F. et al. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153, 1567–1578 (2013). [DOI] [PubMed] [Google Scholar]
- Wu D. et al. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biology 4, e188 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E. & Weis V. M. Study of cnidarian-algal symbiosis in the “omics” age. The Biological Bulletin 223, 44–65 (2012). [DOI] [PubMed] [Google Scholar]
- Reyes-Bermudez A., Lin Z., Hayward D. C., Miller D. J. & Ball E. E. Differential expression of three galaxin-related genes during settlement and metamorphosis in the scleractinian coral Acropora millepora. BMC Evolutionary Biology 9, 178 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn K. S. & Aggarwal B. B. Transcription Factor NF-κB: A Sensor for Smoke and Stress Signals. Annals of the New York Academy of Sciences 1056, 218–233, doi: 10.1196/annals.1352.026 (2005). [DOI] [PubMed] [Google Scholar]
- Palumbi S. R., Barshis D. J., Traylor-Knowles N. & Bay R. A. Mechanisms of reef coral resistance to future climate change. Science 344, 895–898 (2014). [DOI] [PubMed] [Google Scholar]
- DeSalvo M. K., Sunagawa S., Voolstra C. R. & Medina M. Transcriptomic responses to heat stress and bleaching in the elkhorn coral Acropora palmata. Marine Ecology Progress Series 402, 97–113 (2010). [Google Scholar]
- Rymuszka A. & Adaszek Ł. Pro- and anti-inflammatory cytokine expression in carp blood and head kidney leukocytes exposed to cyanotoxin stress – An in vitro study. Fish & Shellfish Immunology 33, 382–388, doi: 10.1016/j.fsi.2012.05.021 (2012). [DOI] [PubMed] [Google Scholar]
- Lesser M. P. & Farrell J. H. Exposure to solar radiation increases damage to both host tissues and algal symbionts of corals during thermal stress. Coral Reefs 23, 367–377 (2004). [Google Scholar]
- Tirone F. The gene PC3TIS21/BTG2, prototype member of the PC3/BTG/TOB family: regulator in control of cell growth, differentiation, and DNA repair ? Journal of Cellular Physiology 187, 155–165 (2001). [DOI] [PubMed] [Google Scholar]
- Rosic N. et al. Early transcriptional changes in the reef-building coral Acropora aspera in response to thermal and nutrient stress. BMC Genomics 15, 1052 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro C. et al. NF-[kappa] B controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nature Cell Biology 13, 1272–1279 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon B. R. & Leggat W. Symbiodinium—invertebrate symbioses and the role of metabolomics. Marine Drugs 8, 2546–2568 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno T., Ishizuka M. & Itakura T. Cytochrome P450 (CYP) in fish. Environmental Toxicology and Pharmacology 34, 1–13 (2012). [DOI] [PubMed] [Google Scholar]
- Acunzo J., Katsogiannou M. & Rocchi P. Small heat shock proteins HSP27 (HspB1), αB-crystallin (HspB5) and HSP22 (HspB8) as regulators of cell death. The International Journal of Biochemistry & Cell Biology 44, 1622–1631 (2012). [DOI] [PubMed] [Google Scholar]
- Boscari A., Mandon K., Dupont L., Poggi M.-C. & Le Rudulier D. BetS is a major glycine betaine/proline betaine transporter required for early osmotic adjustment in Sinorhizobium meliloti. Journal of Bacteriology 184, 2654–2663 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barshis D. J., Ladner J. T., Oliver T. A. & Palumbi S. R. Lineage-Specific transcriptional profiles of Symbiodinium spp. unaltered by heat stress in a coral host. Molecular Biology and Evolution 31, 1343–1352, doi: 10.1093/molbev/msu107 (2014). [DOI] [PubMed] [Google Scholar]
- Lin Z. et al. Deciphering the transcriptomic response of Fusarium verticillioides in relation to nitrogen availability and the development of sugarcane pokkah boeng disease. Scientific Reports 6, 29692, doi: 10.1038/srep29692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerardo N. M. The give and take of host-microbe symbioses. Cell host & microbe 14, 1–3 (2013). [DOI] [PubMed] [Google Scholar]
- Lesser M. P., Stat M. & Gates R. D. The endosymbiotic dinoflagellates (Symbiodinium sp.) of corals are parasites and mutualists. Coral Reefs 32, 603–611, doi: 10.1007/s00338-013-1051-z (2013). [DOI] [Google Scholar]
- Lin S. et al. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 350, 691–694 (2015). [DOI] [PubMed] [Google Scholar]
- Husnik F. & McCutcheon J. P. Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proceedings of the National Academy of Sciences 113, E5416–E5424, doi: 10.1073/pnas.1603910113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon J. P., McDonald B. R. & Moran N. A. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proceedings of the National Academy of Sciences 106, 15394–15399 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon J. P. & Moran N. A. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proceedings of the National Academy of Sciences 104, 19392–19397 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon J. P. & Moran N. A. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biology and Evolution 2, 708–718 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon J. P. & Moran N. A. Extreme genome reduction in symbiotic bacteria. Nature Reviews Microbiology 10, 13–26 (2012). [DOI] [PubMed] [Google Scholar]
- Thompson J. R., Rivera H. E., Closek C. J. & Medina M. Microbes in the coral holobiont: partners through evolution, development, and ecological interactions. Frontiers in Cellular and Infection Microbiology 4, 176 doi: 10.3389/fcimb.2014.00176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D. et al. Comparative genomics explains the evolutionary success of reef-forming corals. eLife 5, e13288, doi: 10.7554/eLife.13288 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs J. L. & Wilcox T. P. A shift to parasitism in the jellyfish symbiont Symbiodinium microadriaticum. Proceedings of the Royal Society B: Biological Sciences 273, 425 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oppen M. J. H., Souter P., Howells E. J., Heyward A. & Berkelmans R. Novel genetic diversity through somatic mutations: fuel for adaptation of reef corals? Diversity 3, doi: 10.3390/d3030405 (2011). [DOI] [Google Scholar]
- Kawahara Y. et al. Simultaneous RNA-seq analysis of a mixed transcriptome of rice and blast fungus interaction. PloS One 7, e49423 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoutzias G. et al. One billion years of bZIP transcription factor evolution: conservation and change in dimerization and DNA-binding site specificity. Molecular Biology and Evolution 24, 827–835 (2007). [DOI] [PubMed] [Google Scholar]
- Karako-Lampert S. et al. Transcriptome analysis of the scleractinian coral Stylophora pistillata. PloS One 9, e88615 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forêt S. et al. New tricks with old genes: the genetic bases of novel cnidarian traits. Trends in Genetics 26, 154–158, doi: 10.1016/j.tig.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Monismith S. G. Flow through a rough, shallow reef. Coral Reefs 33, 99–104, doi: 10.1007/s00338-013-1107-0 (2014). [DOI] [Google Scholar]
- Ruiz-Jones L. J. & Palumbi S. R. Transcriptome-wide changes in coral gene expression at noon and midnight under field conditions. The Biological Bulletin 228, 227–241 (2015). [DOI] [PubMed] [Google Scholar]
- Takahashi A. & Kurihara H. Ocean acidification does not affect the physiology of the tropical coral Acropora digitifera during a 5-week experiment. Coral Reefs 32, 305–314 (2013). [Google Scholar]
- Lapointe B. E., Barile P. J. & Matzie W. R. Anthropogenic nutrient enrichment of seagrass and coral reef communities in the Lower Florida Keys: discrimination of local versus regional nitrogen sources. Journal of Experimental Marine Biology and Ecology 308, 23–58 (2004). [Google Scholar]
- Grabherr M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. & Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659, doi: 10.1093/bioinformatics/btl158 (2006). [DOI] [PubMed] [Google Scholar]
- Putnam N. H. et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317, 86–94 (2007). [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L. & Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- Trapnell C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols 7, 562–578, doi: 10.1038/nprot.2012.016 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S. & Huber W. Differential expression analysis for sequence count data. Genome Biology 11, R106, doi: 10.1186/gb-2010-11-10-r106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 1–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Cai T., Olyarchuk J. G. & Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793, doi: 10.1093/bioinformatics/bti430 (2005). [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 12, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.