The molecular mechanisms controlling “persistent” current through voltage-gated Na+ channels are poorly understood. Yan et al. show that apocalmodulin binding to the intracellular C-terminal domain limits persistent Na+ flux and accelerates inactivation across the voltage-gated Na+ channel family.
Abstract
Increased “persistent” current, caused by delayed inactivation, through voltage-gated Na+ (NaV) channels leads to cardiac arrhythmias or epilepsy. The underlying molecular contributors to these inactivation defects are poorly understood. Here, we show that calmodulin (CaM) binding to multiple sites within NaV channel intracellular C-terminal domains (CTDs) limits persistent Na+ current and accelerates inactivation across the NaV family. Arrhythmia or epilepsy mutations located in NaV1.5 or NaV1.2 channel CTDs, respectively, reduce CaM binding either directly or by interfering with CTD–CTD interchannel interactions. Boosting the availability of CaM, thus shifting its binding equilibrium, restores wild-type (WT)–like inactivation in mutant NaV1.5 and NaV1.2 channels and likewise diminishes the comparatively large persistent Na+ current through WT NaV1.6, whose CTD displays relatively low CaM affinity. In cerebellar Purkinje neurons, in which NaV1.6 promotes a large physiological persistent Na+ current, increased CaM diminishes the persistent Na+ current, suggesting that the endogenous, comparatively weak affinity of NaV1.6 for apoCaM is important for physiological persistent current.
Introduction
Na+ influx through voltage-gated Na+ (NaV) channels drives the rapid rising phase of action potentials in neurons, cardiac myocytes, and skeletal muscle. There are nine NaV channel family members that share a common structure composed of four homologous transmembrane repeats, each containing six transmembrane segments, arranged around a central ion-permeating pore. The four transmembrane repeats are joined by intracellular loops, and the protein is capped on either end by intracellular N and C termini. These pore-forming subunits serve as the core of NaV channel macromolecular complexes containing components that allow targeting of NaV channels to specific subcellular domains such as the intercalated disks in cardiomyocytes or the axon initial segment in neurons and various regulatory proteins that fine-tune channel function. Among these critical regulatory proteins is calmodulin (CaM), which binds directly to the channel’s intracellular C-terminal domain (CTD). Although CaM is best appreciated for its ability to act as an intracellular Ca2+ transducer, for NaV channels growing evidence shows that Ca2+-free CaM also functions as both a structural component as well as a Ca2+ sensor, but the specific roles for CaM in NaV channel function are still not clearly understood (Pitt and Lee, 2016).
One strategy to define these NaV channel regulatory roles is to exploit disease mutations that affect CaM interaction. Mutations within the NaV channel CTDs in the region where CaM binds have been linked to several disorders. In the cardiac NaV1.5 channel, these CTD mutations are associated with inherited arrhythmia syndromes such as long QT type 3 (LQT3), and mutations in the central nervous system NaV1.2 and NaV1.6 channels are linked to various epilepsy syndromes. Common to these conditions is that the mutant NaV channels show a gain-of-function phenotype caused by a defect in channel inactivation. WT NaV channels activate rapidly in response to membrane depolarization and then quickly inactivate. For most of those mutant NaV channels, however, inactivation is defective, usually characterized as an increased “persistent” or “late” Na+ current (Antzelevitch et al., 2014). This additional inward Na+ current in cardiomyocytes, for example, drives the prolonged action potential that is the substrate for the life-threatening cardiac arrhythmias accompanying LQT3. The detailed molecular mechanisms responsible for persistent Na+ current are not known except for a small subset of mutants. The prototypical LQT3 mutation (ΔKPQ; Wang et al., 1995) in the NaV1.5 intracellular III-IV linker is a rare example. The III-IV linker is thought to serve as the inactivation particle, which is disrupted by the mutation. Although other LQT3 mutations are also believed to affect movement of the III-IV linker, how mutations outside of the III-IV linker affect inactivation is generally unknown.
Nevertheless, it is clear that the channel’s CTD and the associated CaM are important for persistent Na+ component. Truncation of the CTD to eliminate the CaM-binding site led to a marked enhancement of persistent current (Cormier et al., 2002). Focusing more directly on the role for CaM, we engineered point mutations in a NaV1.5 channel so that CaM binding to the CTD was diminished and observed a Ca2+-independent increase in the persistent Na+ current (Kim et al., 2004a). Ca2+ may also contribute to the CTD’s role as suggested by a recent study (Potet et al., 2015). However, the underlying molecular mechanisms for how Ca2+ or CaM control influence the role of the CTD in the regulation of persistent Na+ current are not known.
Recent structural information derived from x-ray crystallography provides new insights into CaM interaction with NaV channel CTDs (Wang et al., 2012, 2014; Gabelli et al., 2014) and offers novel opportunities to unravel the specific contributions of CaM to Na+ channel inactivation. Although some of these structures were obtained in the presence of fibroblast growth factor homologous factors, a set of NaV channel modulators that fine-tune inactivation (Wang et al., 2011; Venkatesan et al., 2014; Yan et al., 2014), here we isolated the effects of CaM by focusing on the structure obtained without a fibroblast growth factor homologous factor (PDB: 4OVN; Gabelli et al., 2014) and performed detailed functional analyses in a mammalian cell system devoid of fibroblast growth factor homologous factors. We then validated our findings in neurons to demonstrate the functional significance holds also in the presence of fibroblast growth factor homologous factors.
We started our analysis by focusing on the binary structure of the NaV1.5 CTD (residues 1,776–1,929) associated with CaM. In this structure, the CTD is comprised of a globular-shaped domain followed by an extended α-helix termed the “IQ domain” (Fig. S1, A and B). CaM has multiple points of contact. Most prominently, the NaV1.5 CTD IQ domain contains a canonical apoCaM “IQ” binding motif, represented by I1908 and Q1909 in the NaV1.5 CTD that the CaM C-lobe buries. A similar interaction has been observed between the CaM C-lobe and the IQ motif of NaV1.2 CTD (Feldkamp et al., 2011) and the NaV1.6 CTD (Reddy Chichili et al., 2013), which has a Leu for Ile substitution in the signature Ile. The potential significance of the IQ motif interaction with the apoCaM C-lobe is highlighted by several observations. First, mutation of the IQ residues in NaV1.5 to alanines, which disrupted CaM interaction, led to increased persistent Na+ current in a Ca2+-independent manner (Kim et al., 2004a). Second, a rare Q1909R mutation in NaV1.5 is associated with LQT3 (Kapplinger et al., 2015) and sudden infant death syndrome, and the mutation increased persistent Na+ current when examined in a heterologous expression system (Winkel et al., 2015). For NaV1.6, disruption of CaM interaction by mutation of the IQ residues reduced current density and slowed the rate of inactivation in a heterologous expression system (Herzog et al., 2003). Outside of the signature IQ residues, other residues in the IQ domain that contact the CaM C-lobe also appear important for channel function. For example, the E1901Q and R1913H mutations in NaV1.5 (Fig. S1 C) have been associated with LQT3 (Napolitano et al., 2005; Kapplinger et al., 2009). Whether these mutations affect CaM interaction and whether a perturbed interaction with CaM contributes more generally to the Na+ channel dysfunction are not known.
There are additional CaM interaction sites outside of the IQ domain, including the globular domain to which the CaM N-lobe binds (Fig. S1, B and D). The presence of LQT3 mutations within the globular domain (D1790G and Y1795C) raises the possibility that these mutations may also affect Na+ channel function through perturbation of CaM interaction. To the best of our knowledge, the contribution of CaM interaction in the context of these mutations has not been evaluated. Beyond the contacts between CaM and the NaV1.5 CTD mapped in the binary structure, another set of potential contacts include intercomplex interactions between the CaM C-lobe of one binary complex and the globular domain of a second binary complex. These interactions, observed in the x-ray crystal structure within the asymmetric unit but not confirmed biochemically or functionally (Gabelli et al., 2014), result from the IQ domain of one NaV1.5 CTD fitting into a groove on the globular domain of a second NaV1.5 CTD (Fig. S1, E and F). Whether these interprotein CTD to CTD interactions occur in vivo or have a consequence on channel function is not known. Nevertheless, previous studies of dominant-negative Brugada syndrome mutations hint that interprotein NaV1.5 α subunit interactions exist and influence channel function. Although those interactions have only been found between the cytoplasmic NaV1.5 N termini (Clatot et al., 2012; Hoshi et al., 2014), interactions between CTDs of the homologous voltage-gated L-type Ca2+ channel family are proposed to affect channel function by coupling the gating properties of the interacting channels (Dixon et al., 2015; Moreno et al., 2016). Intriguingly, these inter-CTD interactions and the consequent coupled gating in L-type Ca2+ channels are dependent on CaM, but the molecular details by which CaM affects coupling of CaV channels are not known.
Materials and methods
Molecular biology
Human NaV1.5 CTD (amino acids 1773–1940), human NaV1.2 (amino acids 1777–1937), and human NaV1.6 CTD (amino acids 1769–1926) were cloned into pET28. The NaV1.5 III-IV linker (amino acids 1471–1522) was cloned into pSMT. CaM was cloned into pSGC02 as previously described (Kim et al., 2004b). NaV1.2 and NaV1.5 were cloned into pcDNA3.1 and subjected to site-directed mutagenesis (C373Y in NaV1.5 to generate the tetrodotoxin [TTX]-sensitive NaV1.5TTX-S; Satin et al., 1992). Human NaV1.6 (containing the substitution Y371C to make it TTX resistant) was generated by gene synthesis using optimized codons for mammalian expression (Genewiz) and cloned into pcDNA3.1. All site-directed mutagenesis for the various mutations tested was performed with QuikChange (Agilent Technologies), and the mutations were verified by sequencing. CaM for mammalian expression was cloned into pCI.
Recombinant protein expression and purification
Plasmids were transformed into BL-21 (DE3) Escherichia coli. Bacteria were grown in Luria broth medium to OD600 = 0.6–0.8 before induction with 1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) at 16°C for 48 h or 20°C for 16 h. Cells were harvested, and the initial purification was performed as previously described (Wang et al., 2012). Additional purification was performed by gel filtration on a Superdex 200 10/300L column on an AKTA FPLC (GE Healthcare) in a buffer containing 300 mM NaCl, 20 mM Tris-HCl, 5 mM imidazole, and 5 mM EGTA, pH 7.5. Protein concentrations were determined by UV absorbance with a NanoDrop (Thermo Fisher Scientific). CaM protein was purified as previously described (Kim et al., 2004b) and further purified by FPLC with the above buffer. For the NaV1.5 III-IV linker interaction studies, the proteins were expressed and purified, mixed together in equimolar ratios, and incubated at 4°C for 1 h before chromatography on a Superdex S75 column.
Isothermal titration calorimetry (ITC)
Experiments were performed with an VP-ITC (MicroCal) at 20°C, as previously described (Wang et al., 2012). In brief, the NaV CTDs (20–45 µM) were titrated with 1 injection of 5 µl and 27 injections of 10 µl of solutions containing CaM (200–350 µM).
HEK293T cell culture and transfection
HEK293T cells were maintained in DMEM containing 10% FBS at 37°C. The cells were plated on 60-mm tissue culture dishes, grown to 50–70% confluency, and transfected with Lipofectamine 2000 (Invitrogen) in serum-reduced medium (Opti-MEM; Invitrogen) according to the manufacturer’s instructions. The total amount of cDNA used per dish was 3 µg for NaV1.2, NaV1.5, or NaV1.6; 1.5 µg of NaV β1 (for NaV1.5) or 1.5 µg of NaV β2 (for NaV1.2 or NaV1.6); 2 µg of CaM or empty vector in 5 ml of transfection medium. After 24 h, the cells were replated on coverslips coated with 50 µg/ml poly-d-lysine (Sigma-Aldrich) at a low density for electrophysiology.
Quantification of CaM overexpression in HEK293
CaM, NaV1.5, and NaV β1 were transfected into HEK293T cells. Cells were lysed in buffer containing 150 mM NaCl, 5 mM EGTA, and 50 mM Tris-HCl, pH 7.5, supplemented with protease inhibitor (Roche) and 1% Triton X-100. Lysates were centrifuged at 4°C at 16,000 g for 10 min, and the protein concentration was determined by the bicinchoninic acid (BCA) assay (Thermo Fisher Scientific). A total of 20 µg of protein was loaded onto an 8–16% SDS-PAGE gel, and the proteins were subsequently transferred to a nitrocellulose membrane. CaM was identified by immunoblot with an anti-CaM antibody (EMD Millipore). A CaM standard was run on the same gel using purified recombinant CaM.
Protein cross-linking with disuccinimidyl glutarate (DSG) and liquid chromatography electrospray ionization tandem mass spectrometry (LC-MS/MS) analysis
The 6×His-tagged NaV1.5 CTD and CaM or NaV1.5 only were coexpressed and copurified by metal affinity chromatography as described in Recombinant protein expression and purification. The purified protein was exchanged into PBS buffer supplemented with 5 mM EGTA and concentrated ∼1,000-fold using 10K MWCO centrifugal filters (Amicon Ultra concentrator; EMD Millipore). DSG (Thermo Fisher Scientific) was added to the proteins (concentration ∼10 µM) at 0.1, 0.3, or 3 mM at 25°C for 45 min. Cross-linking was terminated by the addition of 1 M Tris-Cl, pH 7.5. The cross-linked proteins were separated by SDS-PAGE, and the bands were excised and processed for mass spectrometry by the Duke Proteomics Core facility. In brief, the excised bands were subjected to an in-gel reduction, iodoacetamide alkylation, and trypsin digestion as previously described (Wilm et al., 1996). Extracted peptides were lyophilized to dryness and then resuspended in 20 µl of 2% acetonitrile, 0.1% formic acid before LC-MS/MS analysis. Chromatographic separation was performed on a Waters NanoAcquity UPLC equipped with a 1.7 µm BEH130 C18 75 µm I.D. X 250 mm reversed-phase column. The mobile phase consisted of (A) 0.1% formic acid in water and (B) 0.1% formic acid in acetonitrile. After a 4-µl injection, peptides were trapped for 5 min on a 5 µm Symmetry C18 180 µm I.D. X 20 mm column at 5 µl/min in 99.9% A. The analytical column was then switched in-line, and a linear elution gradient of 5% B to 40% B was performed over 90 min at 400 nl/min. The analytical column was connected to a fused silica PicoTip emitter (New Objective) with a 10-µm tip orifice and coupled to a Thermo QExactive Plus high-resolution accurate mass tandem mass spectrometer (Thermo Fisher Scientific) through an electrospray interface. The instrument was operated in data-dependent mode of acquisition with precursor MS scans from m/z 375–1,600 and the top 10 most abundant precursor ions being subjected to MS/MS fragmentation. For all experiments, charge-dependent CID energy settings were used, and a 20-s dynamic exclusion was used for previously fragmented precursor ions.
Qualitative identifications and cross-linked peptide identification from raw LC-MS/MS data
Raw LC-MS/MS data files were processed in Mascot distiller (Matrix Science) and then submitted to independent Mascot database searches (Matrix Science) against a Swiss-Prot human database appended with the reverse sequence of all of the forward entries. Search tolerances were 5 ppm for precursor ions and 0.02 D for product ions using trypsin specificity with up to two missed cleavages. Carbamidomethylation (+57.0214 D on C) was set as a fixed modification, whereas oxidation (+15.9949 D on M) and hydrolyzed DSG (+114.031694 D on K) were considered as variable modifications. All searched spectra were imported into Scaffold (Proteome Software), and protein confidence thresholds were set using a Bayesian statistical algorithm based on the PeptideProphet and ProteinProphet algorithms, which yielded a peptide and protein false discovery rate <1% (Keller et al., 2002; Nesvizhskii et al., 2003). To identify cross-linked species, Mascot distiller–generated MGF files were submitted to MassMatrix (v 2.4.2, Feb 2012) searches against a forward/reverse database containing only the proteins of interest (Xu and Freitas, 2007). Search mass tolerances and modifications were as described for Mascot searches, with the “advanced search” option enabled to allow for inter- or intrapeptide cross-linking of DSG (+96.0211 D). Specificity of the cross-linker was confined to lysine–lysine residues. Trypsin rules were set to not allow cleavage at a cross-linked modified residue, and only one cross-link per peptide pair was allowed. A peptide match within MassMatrix was only considered if peptide scoring thresholds were above that required for a matching probability less than p-value <0.05.
Primary cerebellar culture and transfection
Primary dissociated cerebellar cultures were prepared using minor modifications of our previously described procedure (Yan et al., 2013, 2014). In brief, the cerebellum cortex was dissected on ice from P6–P8 male or female WT C57BL/6J (The Jackson Laboratory), digested with 0.25% trypsin for 10–15 min at 37°C with Dulbecco’s modified Eagle’s medium (DMEM; Sigma-Aldrich), and dissociated into single cells by gentle trituration. The cells were seeded onto coverslips coated with 50 µg/ml poly-d-lysine (Sigma-Aldrich) and 20 µg/ml laminin (Sigma-Aldrich) at a density of 2.5–3.0 × 105 cells/per coverslip (12-mm diameter) in DMEM supplemented with 10% heat-inactivated FBS. The cells were maintained in a humidified incubator in 5% CO2 at 37°C. After 15–16 h, the medium was replaced with basal medium Eagle (BME; Sigma-Aldrich) supplemented with 2% B27 (Invitrogen), 5% FBS, 25 mM uridine, 70 mM 5-fluorodeoxyuridine, and 20 mM KCl. After 5–7 d in vitro (DIV), the neurons were transiently transfected with 1 µg plasmid DNA (CaM or empty vector; NaV1.6TTX-R or pcDNA3; and 0.1 µg pEGFP) per coverslip with calcium phosphate. Recordings were performed 7–12 d after transfection.
Electrophysiological recordings
Whole-cell currents and membrane voltage from HEK293T cells and cerebellar neurons were obtained at room temperature (∼25°C) using an EPC 10 USB patch amplifier (HEKA). The signal was filtered at 2.9 Hz and digitized at 20 Hz. Transient and persistent Na+ currents were recorded from transfected HEK293T cells with an extracellular solution containing (mM) 124 NaCl, 5 KCl, 2 CaCl2, 1 MgCl2, 20 TEA-Cl, 5 HEPES, and 10 glucose. Transient and persistent Na+ currents were recorded from cerebellar Purkinje neurons with the same solution supplemented with (mM) 0.3 CdCl2, 2 BaCl2, and 5 4-aminopyridine (4-AP), whereas the KCl and CaCl2 were removed. NaOH was added to achieve pH 7.3 (300–310 mOsm). Borosilicate glass patch pipettes (resistance 2–3 MΩ for HEK293T cells and 3–4 MΩ for cultured cerebellar Purkinje neurons) were filled with the following internal solution (mM): 125 CsF, 10 NaCl, 10 HEPES, 15 TEA-Cl, 1.1 EGTA, and 0.5 Na-GTP, adjusted to pH 7.3 with CsOH (290–300 mOsm). All drugs were from Sigma-Aldrich, except for TTX (Abcam). The persistent Na+ current was isolated by subtraction after addition of 1 µM TTX. Series resistance was compensated >80% for HEK293T cells and >70% for Purkinje neurons. The liquid junction potentials were not corrected.
For peak current amplitude measurements, HEK293T cells and neurons were held at −120 mV and −90 mV, respectively. Persistent Na+ current was measured at 150 ms during a 200-ms depolarizing pulse to −20 or −10 mV (NaV1.6) and expressed as a normalized value to the peak Na+ current amplitude (percentage of peak Na+ current). The rate of decay (τ) of the NaV1.2 transient Na+ current (at −20 mV) was obtained by fitting with a single exponential function, I(t) = INa exp(−t/τ) + ISS, where I(t) is the amplitude of the current at time t and ISS is the steady-state current during a single voltage step. The transient Na+ current was elicited by depolarizing pulses of 40 ms from −120 (HEK293T) or −90 mV (cerebellar Purkinje neurons) to 55 mV in 5-mV increments. Current density was obtained by normalizing peak Na+ current to membrane capacitance. Na+ activation curves were obtained by transforming current data to conductance (G), with the equation G = INa/(Em − Erev), where INa is the peak current, Em is the membrane potential, and Erev is the reversal potential of INa, and fitted with a Boltzmann equation of the form: G = Gmax/[1 + exp(V1/2 − V)/k], where Gmax is the extrapolated maximum Na+ conductance, V is the test voltage, V1/2 is the half-activation voltage, and k is the slope factor. For Na+ steady-state inactivation, a voltage step to −20 mV for 20 ms was applied from a holding potential of −120 mV (for HEK293T cells) or −90 mV (for cerebellar Purkinje neurons) to preferentially activate INa after prepulse conditioning voltage steps of 500 ms in duration (ranging from −110 or 90 mV to 20 mV) in 5-mV increments. Steady-state inactivation curves were constructed by plotting the normalized peak current amplitude elicited during the test pulse as a function of the conditioning prepulse. A Boltzmann relationship, I/Imax = (1 + exp((V − V1/2)/k))−1 was used to fit the data, where Imax is current elicited by the test pulse after a −90-mV prepulse, V1/2 is half-inactivation voltage, and k is the slope.
Statistical analyses
Data analysis was performed using FitMaster (HEKA), Excel (Microsoft), and Origin 8 software. All averaged data are presented as mean ± SEM. Statistical significance was determined using Student’s t test or one-way ANOVA. Calculated p-values of ≤0.05 were accepted as evidence of statistically significant differences.
Online supplemental material
Fig. S1 shows CaM interaction with the NaV CTD. Fig. S2 shows the measurement of persistent Na+ current from NaV1.5TTX-S. Fig. S3 shows structural parallels between the NaV1.5 and CaV1.1 CTDs.
Results
LQT3 mutations within the IQ motif increase the persistent Na+ current and affect apoCaM interaction
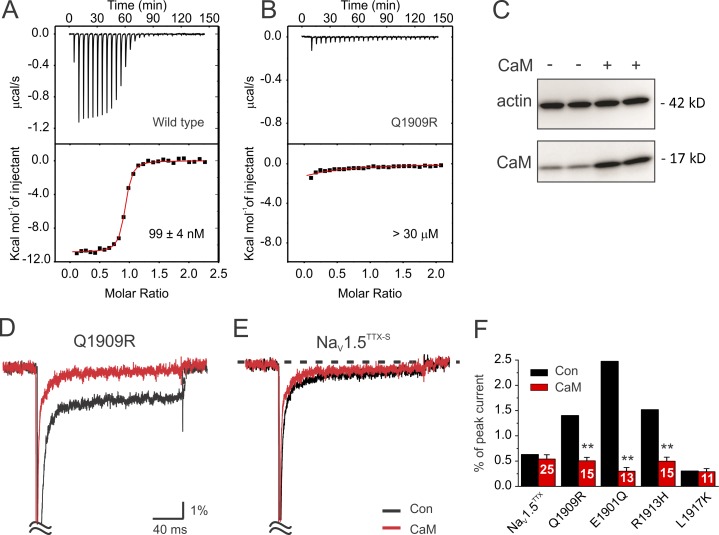
We first tested the consequences of LQT3 mutations within the IQ motif, where the CaM C-lobe binds. We used a heterologous expression system in which we expressed a NaV1.5 mutant channel with a Y373C substitution that confers an ∼1,000-fold increase in TTX sensitivity. The Y373C mutant NaV1.5 channel (NaV1.5TTX-S) otherwise displays WT NaV1.5 channel properties (Satin et al., 1992), so the addition of TTX allows background subtraction and consequent accurate quantification of the relatively small amplitude persistent Na+ current that is characteristic of LQT3 mutations (Fig. S2). All recordings were performed with CsF and EGTA in the patch pipette to chelate intracellular Ca2+ and thereby remove any contribution of Ca2+ action through endogenous CaM. Under these conditions (and with coexpression of an empty plasmid “Con” as a control for experiments in which CaM is overexpressed; e.g., Fig. 2), the pseudo-WT NaV1.5TTX-S channel displayed a normalized (to the peak current) persistent Na+ current amplitude of 0.63 ± 0.08% (n = 35), as shown in Fig. 1 A and Table 1. In contrast, the persistent Na+ current amplitude for the LQT3-associated mutant of the signature Gln residue within the IQ motif to Arg (Q1909R) was more than double (1.40 ± 0.12%, n = 15, P < 0.01 compared with NaV1.5TTX-S; Fig. 1 A and Table 1), consistent with previous studies (Kapplinger et al., 2015; Winkel et al., 2015). Because the Q1909R mutation did not significantly alter peak current density (Fig. 1 A, inset; and Table 1), the mutation-dependent increase in the normalized persistent Na+ current amplitude thus reflects a specific change in the persistent Na+ current properties. Two other LQT3 mutations within the IQ motif (E1901Q and R1913H) also increased the persistent Na+ current amplitude (Fig. 1, B and C; and Table 1), again without any effect on the peak current density. As a control, we mutated L1917, which is just distal to the apoCaM C-lobe–binding site. Compared with NaV1.5TTX-S, a L1917K mutant channel did not show an increased persistent Na+ current (Fig. 1, B and C; and Table 1). Peak current density was also unaffected.
Figure 2.
LQT3 mutations in the IQ domain decrease apoCaM binding affinity and show reduced persistent Na+ current after CaM overexpression. (A and B) Exemplar ITC traces for apoCaM and a WT NaV1.5 CTD (A) or a Q1909R NaV1.5 CTD (B). (C) Western blot of CaM from lysates of HEK293T cells expressing NaV1.5TTX-S, NaV β1, and empty vector (−) or CaM (+). (D) Exemplar traces of NaV1.5TTX-S with an additional Q1909R mutation coexpressed with CaM (red) or empty vector (black), showing rescue of the persistent Na+ current. (E) Exemplar traces recorded from cells expressing the pseudo-WT NaV1.5TTX-S coexpressed with CaM (red) or empty vector (black), showing no effect on the persistent Na+ current. (F) Quantification of persistent Na+ current amplitude as a percentage of peak current after overexpression of CaM (red) compared with Con (black; data from Fig. 1 C). Data are presented as mean ± SEM. **, P < 0.01.
Figure 1.
LQT3 mutations in the IQ domain increase the persistent Na+ current amplitude. (A) Exemplar traces of the pseudo-WT NaV1.5TTX-S and Q1909R (magnified region on the right). Y axis of scale bars represents the percentage of persistent current amplitude normalized to the peak Na+ current. The inset shows I-V plots of peak current amplitude for NaV1.5TTX-S and Q1909R, demonstrating no significant difference in peak current amplitude between NaV1.5TTX-S and Q1909R. (B) Magnified region of exemplar traces showing persistent Na+ current for the LQT3 mutations in the CaM C-lobe–binding site and the L1917K. (C) Quantification of persistent Na+ current (as % peak of current). The white number in each bar indicates the number of cells, n. Data are presented as mean ± SEM. **, P < 0.01.
Table 1. Peak Na+ current density and persistent Na+ current.
| Na+ current tested | Con | CaM | |||||
|---|---|---|---|---|---|---|---|
| Peak current density | Persistent Na+ current | n | Peak current density | Persistent Na+ current | n | ||
| pApF | % of peak | pApF | % of peak | ||||
| NaV1.5TTX-S | −210 ± 13 | 0.63 ± 0.08 | 35 | −211 ± 16 | 0.54 ± 0.09 | 25 | |
| D1790G | −170 ± 22 | 0.76 ± 0.11 | 22 | −180 ± 22 | 0.57 ± 0.09 | 17 | |
| Y1795C | −205 ± 21 | 1.74 ± 0.19a | 18 | −192 ± 12 | 0.48 ± 0.07 | 18 | |
| E1901Q | −175 ± 18 | 2.47 ± 0.53a | 13 | −176 ± 24 | 0.30 ± 0.07 | 13 | |
| Q1909R | −188 ± 21 | 1.40 ± 0.12a | 15 | −200 ± 28 | 0.50 ± 0.07 | 15 | |
| R1913H | −184 ± 16 | 1.51 ± 0.21a | 15 | −188 ± 20 | 0.50 ± 0.08 | 15 | |
| L1917K | −185 ± 21 | 0.27 ± 0.06 | 11 | −175 ± 25 | 0.29 ± 0.06 | 11 | |
| ΔKPQ | −171 ± 19 | 1.90 ± 0.19a | 14 | −170 ± 27 | 1.95 ± 0.21a | 14 | |
| NaV1.2 WT | −228 ± 37 | 0.66 ± 0.17 | 16 | −193 ± 16 | 0.50 ± 0.13 | 16 | |
| H1853R | −204 ± 17 | 1.41 ± 0.20b | 15 | −181 ± 27 | 0.61 ± 0.13b | 15 | |
| R1918H | −158 ± 27 | 3.17 ± 0.53b | 20 | −162 ± 27 | 0.80 ± 0.12b | 20 | |
| NaV1.6 WT | −70 ± 4.7 | 13.20 ± 2.34 | 24 | −79 ± 13 | 5.01 ± 0.92c | 24 | |
| NaV1.6TTX-R in Purkinje neurons | −119 ± 16 | 6.96 ± 0.91 | 12 | −130 ± 10 | 3.36 ± 0.45c | 10 | |
| Purkinje neurons | −137 ± 13 | 9.03 ± 0.74 | 22 | −132 ± 11 | 3.75 ± 0.54c | 18 | |
P < 0.01 compared with NaV1.5TTX-S.
P < 0.01 compared with NaV1.2.
P < 0.01 compared with Con.
Based on the previous observation that the result of mutation of I1908 and Q1909 to Ala (IQ/AA) was increased persistent Na+ current and markedly reduced CaM binding to the NaV1.5 CTD (Kim et al., 2004a), we hypothesized that the LQT3 mutations within the IQ motif, associated with increases in persistent Na+ current, adversely affected the affinity of CaM for the mutant NaV1.5 CTD. In that previous study (Kim et al., 2004a), we had tested whether CaM interacted with the NaV1.5 CTD by expressing both CaM and a 6×His-tagged NaV1.5 CTD in E. coli and assaying for copurification of the untagged CaM with the 6×His-tagged NaV1.5 CTD isolated by metal affinity chromatography. Although we observed stoichiometric purification of CaM with the WT NaV1.5 CTD, for the IQ/AA mutant, we detected no copurification of CaM. Here, we used ITC to obtain a quantitative assessment of binding. Experiments were performed in EGTA to mimic the functional studies and to eliminate any of the previously observed Ca2+-dependent conformational changes between CaM and CTD (Wang et al., 2014). We measured a Kd of 99 ± 4 nM for the WT NaV1.5 CTD and apoCaM (Fig. 2 A and Table 2), in excellent agreement with previous measurements (Gabelli et al., 2014; Wang et al., 2014). Consistent with our previous result showing no apoCaM interaction with the IQ/AA mutant in the copurification assay (Kim et al., 2004a), we detected no apoCaM binding with an IQ/AA NaV1.5 CTD mutant (Table 2) by ITC. Similarly, for the LQT3 mutant Q1909R, we detected only minimal apoCaM binding to the mutant CTD (Fig. 2 B and Table 2). We remain cautious about assigning a specific Kd to this mutant, however, because the Q1909R mutant CTD showed decreased solubility, as indicated by a relatively poor recovery of the recombinant CTD protein from the high-speed supernatant compared with its abundance in the cell lysate (not depicted), analogous to our previous observation with the IQ/AA CTD (Kim et al., 2004a). Thus, folding of the Q1909R mutant CTD may be compromised, and the effect on apoCaM binding may be exaggerated. Supporting our overall hypothesis, the apoCaM affinity was also reduced by the E1901Q and R1913H LQT3 mutations, albeit to levels that were measurable by ITC (Table 2). However, the effect of the R1913H mutation on the Kd value for apoCaM may be exaggerated, as the reduced n value (0.73 ± 0.03) compared with the WT (0.99 ± 0.02) suggests that folding of the R1913H mutant NaV1.5 CTD may be compromised even though recovery of both R1913H and E1901Q mutant CTD proteins in the high-speed supernatant was similar to the WT protein (not depicted). For the L1917K control, however, the affinity was similar to the WT NaV1.5 CTD (Table 2). Thus, LQT3 mutations within the apoCaM C-lobe–binding site on the NaV1.5 CTD reduce apoCaM affinity.
Table 2. Summary of ITC data.
| Cell | Kd (nM) | ΔH | ΔS | n value | n |
|---|---|---|---|---|---|
| Kcal/mol | cal/mol/deg | ||||
| NaV1.5 | |||||
| WT | 99 ± 4 | −10.5 ± 0.1 | −3.6 ± 0.2 | 0.99 ± 0.02 | 4 |
| D1790G | 128 ± 36 | −10.5 ± 0.6 | −2.4 ± 1.1 | 0.25 ± 0.04 | 3 |
| Y1795C | 283 ± 8a | −7.43 ± 0.2 | 4.6 ± 0.5 | 0.92 ± 0.02 | 3 |
| Y1795H | 103 ± 4 | −10.2 ± 0.2 | −3.0 ± 0.6 | 0.76 ± 0.02 | 3 |
| E1901Q | 243 ± 9a | −9.0 ± 0.2 | −0.5 ± 0.6 | 0.85 ± 0.00 | 3 |
| Q1909R | 32,600 ± 1,300a | −14.9 ± 3.9 | −37 ± 10 | 0.04 ± 0.02 | 3 |
| R1913H | 2,980 ± 570a | −9.0 ± 0.3 | −5.3 ± 1.2 | 0.73 ± 0.03 | 3 |
| L1917K | 75 ± 29 | −9.8 ± 1.0 | −3.0 ± 0.6 | 0.98 ± 0.08 | 3 |
| IQ/AA | Not detected | - | - | - | 3 |
| NaV1.2 | |||||
| WT | 36 ± 6 | −4.54 ± 0.1 | 18.6 ± 0.6 | 1.02 ± 0.02 | 3 |
| H1853R | 172 ± 20a | −8.3 ± 0.2 | 2.6 ± 1.0 | 0.30 ± 0.05 | 3 |
| R1918H | 112 ± 20a | −7.5 ± 0.2 | 6.2 ± 0.9 | 0.90 ± 0.40 | 3 |
| NaV1.6 | |||||
| WT | 276 ± 63b | −17.5 ± 0.3 | −29.5 ± 1.1 | 0.24 ± 0.01 | 3 |
IQ/AA, IQ1908-1909AA.
P < 0.01 compared with WT.
P < 0.01 compared with NaV1.5 WT.
We next investigated whether the decrease in apoCaM binding affinity was causal for the LQT3 mutants’ increased persistent Na+ current by testing whether overexpressed CaM could, by mass action, surmount the decreased apoCaM affinity and thereby reduce the persistent Na+ current. We achieved a 107 ± 26% (n = 3) relative increase in cellular CaM concentration by overexpression under the conditions used for electrophysiology, as indicated by immunoblotting for CaM (Fig. 2 C). With this background, we then examined the normalized persistent Na+ current for the Q1909R mutant and found that CaM overexpression reduced the persistent Na+ current to 0.50 ± 0.07% (Fig. 2 D and Table 1), a level similar to that observed for the pseudo-WT NaV1.5TTX-S in the absence of CaM overexpression. In contrast, we observed no effect on persistent Na+ current amplitude when CaM was overexpressed with NaV1.5TTX-S (0.54 ± 0.09% of peak current, n = 25; Fig. 2 E and Table 1). The two other mutants showing increased persistent Na+ current and decreased CaM binding affinity (E1901Q and R1913H) were also rescued by CaM overexpression, but persistent Na+ current for the control L1917K mutation that did not affect CaM binding affinity was not rescued by CaM overexpression (Fig. 2 F and Table 1). Thus, the mutation-induced decrease in CaM affinity is causal for increased persistent Na+ current for these CTD mutants.
A LQT3 mutation reveals CTD–CTD interactions that affect apoCaM and channel function
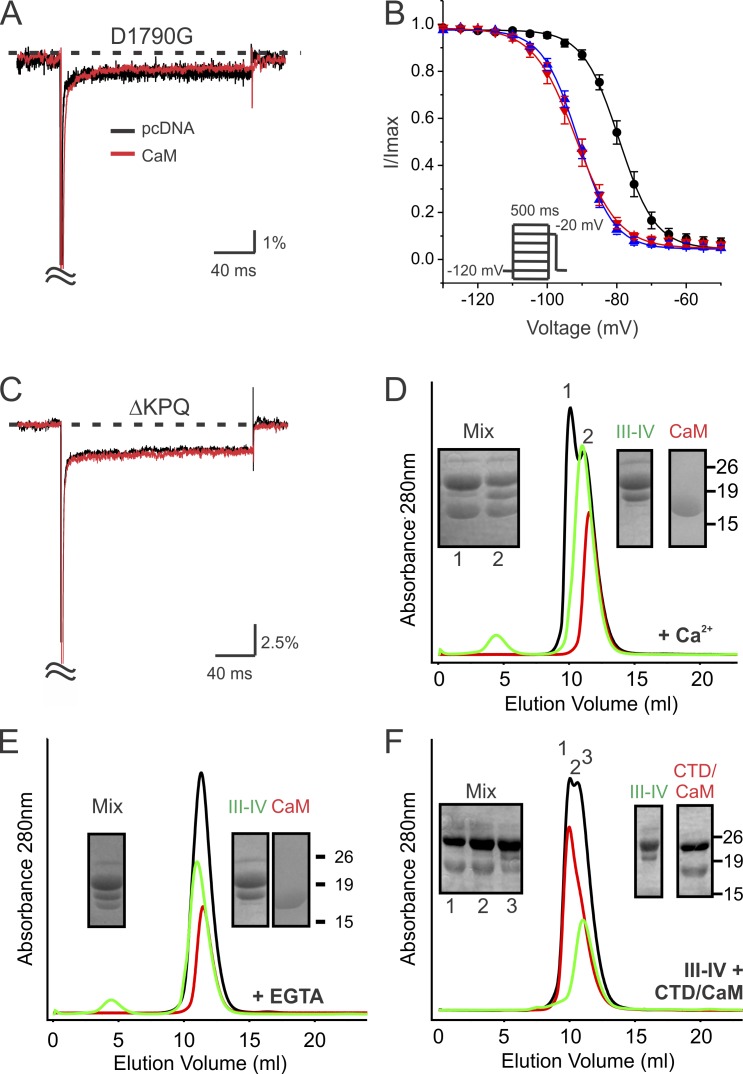
Because the CaM N-lobe contacts the NaV1.5 CTD globular domain (Fig. S1), we next tested whether an LQT3 mutation in the globular domain also affected apoCaM binding and persistent Na+ current. We focused on the consequences of the well-characterized LQT3 mutation Y1795C in the globular domain. A previous investigation showed that Y1795C significantly increased the persistent Na+ current amplitude (Rivolta et al., 2001), so we hypothesized that Y1795C likewise perturbed the interaction between the NaV1.5 CTD and apoCaM. Indeed, ITC showed that the mutant NaV1.5 CTD had a reduced affinity for apoCaM (Table 2). Y1795 sits atop of the globular domain and is remote from apoCaM in the NaV1.5 CTD–CaM binary complex (Fig. S1 D), yielding no ready explanation for the mutant’s reduced apoCaM affinity except for an allosteric effect. However, when we focused on the location of Y1795 in the context of the multiple interacting binary complexes observed in the crystal’s asymmetric unit, in which Y1795C sits within a groove that envelopes the IQ domain of a second binary complex (Fig. S1 E; Gabelli et al., 2014), another possible rationale emerged. This arrangement, which nestles the apoCaM of the second binary complex against the globular domain of the first (Fig. 3, A and B), suggested to us that Y1795C interfered with this intercomplex interaction and thus affected the interaction between the globular domain of the CTD in one binary complex and the apoCaM of a second complex. Although size exclusion chromatography failed to detect multimerization of binary complexes (Gabelli et al., 2014), we reasoned that the interaction might be dynamic or that detection required conditions closer to those that supported the multimerization observed in the crystals than those used for chromatography. We therefore looked for any evidence of inter-CTD interaction by using the lysine cross-linker DSG on concentrated recombinant material. Specifically, we attempted to exploit the proximity of Lys1878 on the surface of the globular domain of one NaV1.5 CTD and Lys1922 on the IQ domain of a second NaV1.5 CTD (Fig. 3 B), two lysines that are otherwise remote within one CTD, but the only two lysines within an appropriate distance for intersubunit cross-linking. We coexpressed a 6×His tagged NaV1.5 CTD (∼23 kD) with an untagged CaM (∼17 kD), purified the complex by metal affinity chromatography as we previously reported (Kim et al., 2004a), and concentrated the material to ∼10 µM. After adding DSG to the purified complex, we observed that some material migrated as a higher molecular mass species of ∼45 kD (Fig. 3 C) on SDS-PAGE, consistent with a CTD–CTD interaction. Importantly, we did not observe a band near 40 kD or 34 kD, the expected sizes for a CaM–CTD cross-linking or a CaM–CaM cross-linking, respectively. The material was excised from the gel, subject to trypsin proteolysis, and analyzed by mass spectrometry. The detected peptides were scored for intensity in a pairwise analysis of all possible Lys–Lys interactions (Fig. 3 D). Analyses of these pairs revealed a low abundance of cross-linking of adjacent Lys residues in the CaM N-lobe (Lys22 to Lys31). We also observed cross-linking of adjacent Lys residues in the NaV1.5 CTD (Lys1872 to Lys1878, or Lys1878 to Lys1886, which are in flexible loops within the CTD globular domain). Additionally, there was a low abundance of cross-linking of peptides in the CTD to CaM, such as Lys1922 in the IQ domain to Lys149 in the CaM C terminus were detected. Notably, however, mass spectrometry showed a relatively high abundance of cross-linking between peptides containing Lys1878 and Lys1922 of the NaV1.5 CTD (Fig. 3 D, circle/arrow). Thus, these data suggest that binary CTD complexes can multimerize in solution, albeit at low efficiency.
Figure 3.
NaV1.5 CTD–CaM heterodimer interaction is disrupted by the Y1795C LQT3 mutation. (A and B) Proposed interaction between two NaV1.5 CTD–CaM heterodimers and position of Y1795C in one of the NaV1.5 CTDs. The positions of Lys1878 in one NaV1.5 CTD globular domain (sky blue) and Lys1922 in the IQ domain of a second NaV1.5 CTD (green), which are available for cross-linking by DSG, are indicated. (C) Coomassie-blue–stained gel of NaV1.5 CTD and CaM after cross-linking with DSG or buffer control (Con). Molecular weight markers are indicated. (D and F) XMapper display of LC/MS data showing DSG cross-linked peptide 1 for the WT NaV1.5 CTD and CaM (WT) and the Y1795C mutant NaV1.5 CTD and CaM. The intensity score (color code) indicates the number of peptides identified for each pairwise interaction. The position of cross-linking between Lys1878 and Lys1922 is circled and indicated by an arrow. (E) LC/MS data showing the cross-linked peptide. (G) Exemplar traces showing increased persistent Na+ current for the Y1795C and rescue by CaM overexpression. (H) Quantification of persistent Na+ current for the pseudo-WT NaV1.5TTX-S and the Y1795C NaV1.5 mutant with and without CaM overexpression. Data are presented as mean ± SEM. **, P < 0.01.
With this demonstration, we then tested the consequence of the Y1795C mutation on cross-linking. Similar to the WT, we observed that some material migrated around ∼45 kD. The pairwise interaction data in Fig. 3 E show cross-linking between CaM N-lobe lysines and NaV1.5 CTD adjacent lysines, similar to the patterns observed for the WT NaV1.5 CTD. However, no detectable cross-linking was observed between Lys1878 in the NaV1.5 CTD globular domain and Lys1922 in the NaV1.5 CTD IQ domain. Thus, the Y1795C-induced disruption of interaction between heterodimers and the consequent loss of contact between a CTD of one heterodimer and CaM in the other provide one possible explanation for the decreased affinity between apoCaM and the Y1795C mutant NaV1.5 CTD observed by ITC.
With this biochemical support, we then assessed whether the resultant decreased association between the NaV1.5 CTD and apoCaM was causative for the pathological persistent Na+ current by examining the consequences on the persistent Na+ current after CaM overexpression. As shown in Fig. 3 (E and F), CaM overexpression reduced the persistent Na+ current for the Y1795C mutant to WT levels. Thus, these data suggest that the Y1795C may increase persistent Na+ current by reducing CaM interaction in an interchannel manner. Interestingly, mutation of the same amino acid to histidine (Y1795H) is associated with Brugada syndrome rather than LQT3, and the Y1795H mutant channel showed a minimal effect on the persistent Na+ current (Rivolta et al., 2001). Our model then predicts that the Y1795H mutation would have a smaller effect on apoCaM affinity compared with Y1795C. Indeed, we observed minimal consequences upon apoCaM affinity by ITC (Table 2) for the Y1795H mutant. Together, our results suggest that an allosteric effect of the Y1795C mutation on apoCaM interaction increases persistent Na+ current and provide an alternative mechanistic hypothesis to the proposal that Y1795C increases persistent Na+ current by forming an intrachannel disulfide bond with C1850 (Tateyama et al., 2004).
ApoCaM does not affect LQT3 mutations outside of the known apoCaM interaction sites
The remarkable ability of CaM to rescue functional defects associated with various LQT3 mutations in the NaV1.5 CTD led us to consider whether CaM overexpression was a nonspecific stabilizer of NaV1.5 channel function rather than a rescuer of a reduced CaM affinity. We therefore tested two additional LQT3 mutants to address this question. First, we examined D1790G, a mutation originally described in a large multigenerational cohort (Benhorin et al., 1993, 1998). D1790 is located within the interior of the globular domain and distant from any CaM contact, and ITC measurement of apoCaM affinity did not show any defect for the D1790G mutant NaV1.5 CTD (Table 2). The biophysical defect associated with D1790G is atypical for a reported LQT3 mutation, in that the D1790G channels display a β subunit–dependent hyperpolarizing shift (compared with WT) in the V1/2 of steady-state inactivation and no effect upon persistent Na+ current (An et al., 1998). We observed the same effect (Fig. 4 A and Tables 1 and 3). After CaM overexpression, the shifted V1/2 of steady-state inactivation was unaffected (Fig. 4 B and Table 3). Thus, for a CTD mutant that does not perturb CaM binding, CaM overexpression does not affect NaV1.5 function.
Figure 4.
CaM overexpression does not affect channel function for mutants that do not display abnormal interactions with CaM. (A) Exemplar traces showing no increased persistent Na+ current for the D1790G and no change after CaM overexpression. (B) Hyperpolarizing shift in the V1/2 of steady-state inactivation for the D1790G mutant NaV1.5 and absence of an effect by CaM overexpression. Data are presented as mean ± SEM. (C) Exemplar traces showing increased persistent Na+ current for the ΔKPQ mutant and no change after CaM overexpression. (D) Gel filtration profiles of the III-IV linker fusion protein (green), Ca2+/CaM (red), and the mixture of the III-IV linker protein and Ca2+/CaM (black). (E) Gel filtration profiles of the III-IV linker fusion protein (green), apoCaM (red, treated with EGTA), and the mixture of the III-IV linker protein and apoCaM (black). (F) Gel filtration profiles of the III-IV linker fusion protein (green), the NaV1.5 CTD (amino acids 1773–1940) complexed with apoCaM (red), and a mixture of the III-IV linker protein and the NaV1.5 CTD–CaM after the addition of 5 mM Ca2+ (black). Insets show Coomassie blue–stained polyacrylamide gels; lane numbers correspond to the fractions labeled in the chromatograms. Molecular weight markers are indicated.
Table 3. Activation and inactivation.
| NaV1.5 channel (with control plasmid or CaM overexpression plasmid tested) | Activation | Inactivation | |||||
|---|---|---|---|---|---|---|---|
| V1/2 | k | n | V1/2 | k | n | ||
| mV | mV | ||||||
| NaV1.5TTX-S/Con | −48.9 ± 1.3 | 2.1 ± 3.1 | 15 | −79.3 ± 1.2 | 4.7 ± 0.4 | 14 | |
| /CaM | −50.8 ± 1.2 | 1.8 ± 3.6 | 15 | −81.2 ± 1.6 | 4.5 ± 0.2 | 10 | |
| D1790G/pcNDA | −43.8 ± 1.1 | 5.1 ± 0.3 | 15 | −94.1 ± 1.6a | 5.1 ± 0.2 | 15 | |
| /CaM | −44.1 ± 1.5a | 5.3 ± 0.5 | 13 | −94.4 ± 1.5a | 5.1 ± 0.2 | 14 | |
| Y1795C/Con | −54.2 ± 1.6 | 1.6 ± 2.9 | 16 | −82.4 ± 1.4 | 4.9 ± 0.3 | 15 | |
| /CaM | −50.1 ± 1.7 | 2.5 ± 4.0 | 15 | −81.5 ± 1.1 | 4.3 ± 0.1 | 15 | |
| E1901Q/Con | −51.6 ± 1.9 | 2.2 ± 0.3 | 16 | −84.0 ± 2.2 | 4.7 ± 0.1 | 13 | |
| /CaM | −51.0 ± 2.2 | 2.4 ± 0.4 | 16 | −83.0 ± 1.8 | 4.6 ± 0.2 | 15 | |
| Q1909R/Con | −52.2 ± 1.5 | 1.8 ± 0.4 | 14 | −84.9 ± 1.9 | 4.9 ± 0.3 | 13 | |
| /CaM | −49.6 ± 1.6 | 2.5 ± 0.3 | 16 | −84.3 ± 1.2 | 4.6 ± 0.2 | 13 | |
| R1913H/Con | −51.4 ± 1.4 | 1.7 ± 0.3 | 17 | −79.0 ± 0.9 | 4.7 ± 0.1 | 15 | |
| /CaM | −49.4 ± 1.6 | 2.0 ± 0.4 | 15 | −82.5 ± 0.7 | 5.0 ± 0.2 | 15 | |
| L1917K/Con | −46.2 ± 1.7 | 2.5 ± 0.3 | 13 | −74.2 ± 1.0 | 4.3 ± 0.2 | 13 | |
| /CaM | −46.3 ± 1.6 | 2.2 ± 0.3 | 16 | −77.2 ± 1.2 | 4.0 ± 0.1 | 16 | |
| ΔKPQ/Con | −42.5 ± 1.3a | 5.3 ± 0.4 | 15 | −88.1 ± 1.6b | 4.6 ± 0.1 | 15 | |
| /CaM | −42.3 ± 1.9a | 4.8 ± 0.4 | 15 | −84.8 ± 1.3 | 4.7 ± 0.2 | 14 | |
P < 0.01 compared with NaV1.5TTX-S.
P < 0.05 compared with NaV1.5TTX-S.
We next evaluated whether CaM affected the prototypical LQT3 mutation (ΔKPQ) in the NaV1.5 intracellular III-IV linker (Wang et al., 1995). Motivation for analyzing a mutation in the III-IV linker derived in part from the striking parallels revealed by the recent structural characterization of the homologous CaV1.1 Ca2+ channel (Wu et al., 2016). The proximal portion of that channel’s CTD shares an identical fold with the NaV1.5 CTD globular domain (Fig. S3 A), and the CaV1.1 channel’s III-IV linker lies within a groove on CaV1.1 CTD, analogous to the manner in which the IQ domain of one NaV1.5 CTD fits into the globular domain of a second NaV1.5 CTD (Fig. S3 B). In the context of this parallel is a previous report that the NaV1.5 III-IV linker can bind CaM in a Ca2+-dependent manner and a proposed model in which CaM acts as a bridge between the NaV1.5 III-IV linker and CTD to influence NaV1.5 channel inactivation properties (Sarhan et al., 2012). We therefore tested whether CaM overexpression affected the persistent Na+ current associated with the ΔKPQ mutation. Although we recorded the expected large amplitude persistent Na+ current for ΔKPQ (1.90 ± 0.19%), overexpression of CaM did not rescue it (1.95 ± 0.21%), as shown in Fig. 4 C. Thus, the structural parallels with the CaV1.1 channel may not include an interaction between the CTD and the III-IV linker. Nevertheless, the ΔKPQ recordings were performed in Ca2+-free conditions, yet central to the bridging model was a proposed Ca2+-dependent ternary complex of Ca2+/CaM, the III-IV linker, and the NaV1.5 CTD. We therefore queried directly whether a ternary complex containing the NaV1.5 CTD, CaM, and the III-IV linker formed in the presence of Ca2+. Consistent with the previous study (Sarhan et al., 2012), we were able to generate a binary complex between CaM and the III-IV linker in the presence of Ca2+, but not in its absence: a fraction of a mixture of CaM and a SUMO-tagged III-IV peptide (amino acids 1471–1522) eluted from a size exclusion column earlier than either of the individual components in Ca2+ (Fig. 4 D), but we observed no shift in elution volume when the experiment was repeated in EGTA (Fig. 4 E). We then examined whether Ca2+ drove CaM binding to the III-IV linker when CaM is prebound to the CTD. We first generated a binary complex containing CaM and the NaV1.5 CTD without Ca2+, to which we then added the SUMO-tagged III-IV peptide in the presence of 5 mM Ca2+. Fig. 4 F shows no evidence for interaction, as no material eluted earlier than the NaV1.5 CTD–CaM binary complex. Thus, our biochemical investigations did not support a model in which CaM bridges the III-IV linker to the CTD and therefore provide a rationale for the inability of CaM overexpression to rescue the ΔKPQ mutant.
CaM regulates persistent Na+ current in vivo and affects neuronal function
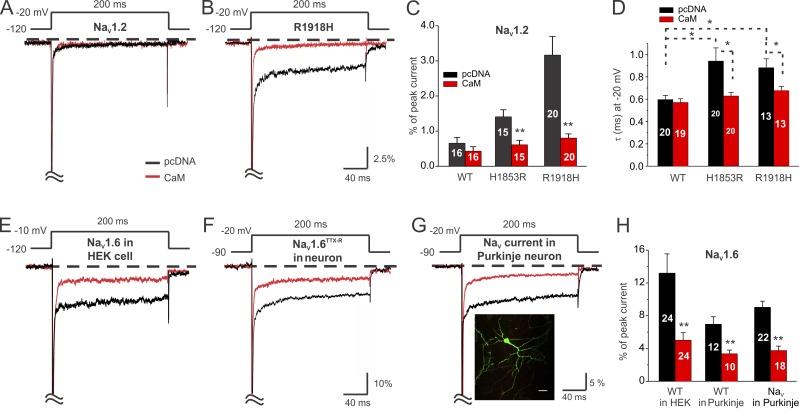
Having established a strong correlation among LQT3 mutations within the NaV1.5 CTD, abnormal inactivation, and decreased apoCaM affinity, we wondered whether CaM similarly modulated the persistent Na+ current in other NaV channels and whether a change in CaM affinity was likewise associated with disease mutations. We turned first to NaV1.2, for which epilepsy mutations such as an Ohtahara syndrome mutation at H1853R in the globular domain (Martin et al., 2014) and an idiopathic generalized epilepsy mutation at R1918H (Haug et al., 2001) have been associated with an increase in the persistent Na+ current. Consistent with the overall pattern observed with LQT3 mutations in NaV1.5, we found that both of the NaV1.2 mutant channels showed increased persistent Na+ current and that this increased persistent Na+ current was rescued by CaM overexpression (Fig. 5, A–C). Moreover, the decay time of inactivation for both NaV1.2 mutants was prolonged, and CaM overexpression restored the decay time to the WT rate (Fig. 5 D). Additionally, both mutant NaV1.2 CTDs displayed reduced CaM affinity compared with the WT NaV1.2 CTD (Table 2), thereby extending the overall association of decreased CaM affinity and increased persistent Na+ current to NaV1.2.
Figure 5.
ApoCaM regulates persistent Na+ current in neuronal NaV channels. (A and B) Exemplar traces for NaV1.2TTX-R (WT) and the NaV1.2 mutant R1918H showing increased persistent Na+ current for the R1918H mutant (but not for WT) and rescue by CaM overexpression for the R1918H mutant. (C) Summary data showing rescue by CaM for the NaV1.2 mutants H1853R and R1918H. (D) Summary data showing increased τ of inactivation and rescue by CaM for the NaV1.2 mutants H1853R and R1918H. (E) Exemplar traces for NaV1.6TTX-R (WT) expressed in HEK293T cells showing reduced persistent Na+ current by CaM overexpression. (F) Exemplar traces for NaV1.6TTX-R expressed in cultured cerebellar Purkinje neurons and reduced persistent Na+ current by CaM overexpression. (G) Exemplar traces of total NaV Na+ current in cultured cerebellar Purkinje neuron showing reduction in persistent Na+ current after CaM overexpression. The inset shows GFP-expressing cultured cerebellar Purkinje neuron. Bar, 20 µm. (H) Summary data for NaV1.6 expressed in HEK293T cells or in cultured cerebellar Purkinje neurons or total NaV Na+ current in cultured cerebellar Purkinje neurons. Data are presented as mean ± SEM. *, P < 0.05; **, P < 0.01.
We also tested whether CaM regulates the persistent Na+ current for NaV1.6. As shown by a study in NaV1.6 knockout mice, NaV1.6 is the major contributor to the large overall NaV channel persistent Na+ current observed in cerebellar Purkinje neurons (Raman et al., 1997). The large contribution of NaV1.6 to the persistent Na+ current becomes immediately obvious when analyzing the Na+ current of isolated NaV1.6 expressed in HEK293T cells (Fig. 5 E). Given the correlation between decreased CaM affinity and an increased magnitude of persistent Na+ current established by our data with NaV1.5 and NaV1.2 mutants, we therefore expected that the WT NaV1.6 CTD had a lower affinity for CaM than NaV1.5 or NaV1.2 CTDs. Indeed, ITC measurement showed a reduced affinity of CaM for the NaV1.6 CTD compared with either the NaV1.2 or NaV1.5 CTDs (Table 2). To test whether this reduced CaM affinity contributed to the persistent Na+ current, we overexpressed CaM with NaV1.6 in HEK293T cells and measured the resulting effect on the late Na+ current. As with NaV1.5 and NaV1.2 disease mutants that had increased persistent Na+ currents and decreased apoCaM affinity, CaM overexpression reduced the persistent Na+ current amplitude for NaV1.6 (Fig. 5, E and H; and Table 1).
The large amplitude persistent Na+ current from NaV1.6 offered an opportunity to test whether CaM modulated persistent Na+ current in native cells. First, we expressed a TTX-resistant NaV1.6 (NaV1.6TTX-R) in the cultured cerebellar Purkinje neurons and isolated this exogenous NaV1.6 current by applying TTX to silence the endogenous NaV channels. The persistent Na+ current was then identified by background subtraction after application of 5 mM lidocaine to the remaining current. As in HEK293T cells, exogenous NaV1.6 (NaV1.6TTX-R) expressed in cerebellar Purkinje neurons displayed a large amplitude persistent Na+ current that was reduced by CaM overexpression (Fig. 5, F and H). With this foundation, we then measured the endogenous persistent Na+ current in cultured cerebellar Purkinje neurons in the presence and absence of CaM overexpression. The large amplitude endogenous persistent Na+ current observed at baseline in cultured cerebellar Purkinje neurons was markedly reduced after CaM overexpression (Fig. 5, G and H).
Discussion
Our data demonstrate that the interaction between apoCaM and the NaV CTD regulates persistent Na+ current and the rate of inactivation across multiple NaV family members. Specifically, we found that disease mutations in NaV CTDs that are associated with perturbed inactivation—increased persistent Na+ current or slowed rate of inactivation—correlated with decreased CaM binding affinity and that CaM overexpression rescued the mutations’ effects. Thus, a decreased affinity for apoCaM binding appears to be a common feature for LQT3 mutations within the NaV1.5 CTD and epilepsy mutations in the NaV1.2 CTD.
In this regard, our data showing the lack of an apoCaM influence on the D1790G mutation in NaV1.5 are informative. Although originally identified in a large LQT3 cohort, this family was considered remarkable for unusual phenotypic heterogeneity (Benhorin et al., 1993). Moreover, a recent study found that D1790G is relatively benign compared with other LQT3 mutations (Wilde et al., 2016). With this context, previous analyses of D1790G (and recapitulated in our studies here; Fig. 4 and Table 3) showed that the most prominent biophysical defect, a β subunit–dependent hyperpolarizing shift in the V1/2 of steady-state inactivation (An et al., 1998), was atypical for LQT3. In fact, such a biophysical defect would cause decreased Na+ current at physiological membrane potentials—a loss-of-function effect that is more consistent with a Brugada syndrome phenotype than LQT3. Recently, the D1790G mutation was indeed reported in a Brugada syndrome patient (Blich et al., 2015). Thus, the association with a relatively benign LQT3 phenotype and separately with Brugada syndrome, and the absence of increased persistent Na+ current for the NaV1.5 D1790G mutant channel, suggest that this mutation more likely causes a mixed “overlap syndrome” (Remme, 2014) rather than a pure LQT3 phenotype and perhaps should be reclassified.
Our analyses also verified, functionally and biochemically, the multiple apoCaM contacts observed in the crystal structure of the NaV1.5 CTD and apoCaM (Gabelli et al., 2014). Although the functional consequences of the interaction between the apoCaM C-lobe and the IQ motif had been previously tested—and it was shown that disrupting CaM interaction through mutation of the signature IQ residues increased persistent Na+ current (Kim et al., 2004a)—the consequences of perturbing neither the CaM N-lobe interaction with the globular domain nor the interaction between CaM from one binary complex to the CTD in a different binary complex had been examined. Our ability to detect the CTD–CTD interaction, not previously observed, likely derives from the use of DSG to trap what may be a dynamic and/or low-affinity interaction. Although inefficient, these interactions may be functionally relevant. For channels with limited diffusion in the context of the plasma membrane, especially in certain subcellular domains with highly concentrated Na+ channels like the intercalated disks in cardiac myocytes or the axon initial segment in neurons, the effective concentration may be closer to that in the crystal than in solution. Furthermore, previous studies investigating dominant-negative mutations in NaV1.5 reported interactions between intact NaV1.5 channels through coimmunoprecipitation (Clatot et al., 2012; Ziyadeh-Isleem et al., 2014). Whether these interchannel interactions are mediated at least in part by the CTDs, as observed in the crystal structure, has not yet been determined, as previous studies have only reported such interactions between the cytoplasmic N termini of two intact channels (Clatot et al., 2012; Hoshi et al., 2014). Our findings provide the basis for testing additional domains and specifically whether mutations in the NaV1.5 CTD associated with Brugada syndrome likewise exert a dominant-negative effect.
Nevertheless, such interchannel interactions are of particular interest because their possible existence echoes recent findings of similar CTD to CTD interactions between voltage-gated L-type Ca2+ channels. Remarkably, although the molecular and structural details are not understood for the interchannel interactions between Ca2+ channels, those interactions are also dependent on CaM (Navedo et al., 2010; Dixon et al., 2015; Moreno et al., 2016). It is interesting to speculate that interactions between L-type Ca2+ channel CTDs use similar structural features to those observed for NaV1.5 to NaV1.5 CTDs, a hypothesis supported by the structural homology between the globular domains of CaV1.1 and NaV1.5 (Fig. S3). Although the CaV1.1 structure (Wu et al., 2016) revealed an interaction with the III-IV linker rather than a second CTD, the CaV1.1 structure likely differs from the CaV1.2 and CaV1.3 (the L-type channels in which interchannel interactions have been studied). Specifically, CaV1.1 CTD does not bind CaM despite the homology in the CaM-binding IQ domains (Ohrtman et al., 2008), and the structural homology between NaV1.5 and CaV1.1 diverges just before the IQ domains (Fig. S3). Perhaps for CaV1.2 and CaV1.3, which do bind CaM, the structural homology extends more distally and thereby provides an interaction for a second CTD analogous to NaV1.5.
Our data also highlight how the consequences of CaM regulation on persistent Na+ current extend beyond NaV1.5, namely to NaV1.2 and NaV1.6 that were studied here. Similar to our findings with LQT3 mutations in NaV1.5, we observed a correlation between epilepsy mutations in NaV1.2 that decreased apoCaM affinity and an increase in persistent Na+ current. Furthermore, our data show that neuronal NaV channels are likewise sensitive to apoCaM (Fig. 5) and reveal a similar correlation between apoCaM affinity and persistent current amplitude. Notably, NaV1.6 shows a relatively large persistent current and a relatively low affinity for apoCaM.
On the one hand, the homologous function of apoCaM across multiple NaV channels is not surprising given the sequence similarity in the CTDs among the different NaV channels, particularly in the IQ domain to which the CaM C-lobe binds. This is echoed by the nearly identical structures of apoCaM interacting with the respective IQ domains of NaV1.2 and NaV1.5 (Chagot and Chazin, 2011; Feldkamp et al., 2011; Wang et al., 2012). On the other hand, previous studies have noted channel-specific differences. For example, Ca2+ confers a CaM-dependent fast regulation on the muscle-specific NaV1.4, but not on the WT NaV1.5 (Ben-Johny et al., 2014). Whether mutant NaV1.5 channels are also insensitive to Ca2+-CaM has not been tested, but it is possible that some NaV1.5 mutants may acquire Ca2+-dependent regulation. We previously observed that Ca2+ induced a CaM-mediated shift in steady-state activation and inactivation in an autism-associated NaV1.2 mutant, whereas the WT NaV1.2 was insensitive (Wang et al., 2014). Thus, some regulatory features, while appearing lost in certain NaV isoforms, may be latent in the WT and only revealed by a mutation. Nevertheless, further evidence for some isoform-specific regulation comes from mutation of the CaM-binding IQ motif for NaV1.4 or NaV1.6, which reduced current density (Herzog et al., 2003), whereas here, we observed no decrease in current density for NaV1.2 or NaV1.5 mutations studied. Thus, determination that all of the features of apoCaM regulation are consistent across the family of NaV channels will require further structural and functional interrogation, but initial hints provided by comparing the structure of CaM bound to the NaV1.6 IQ motif with CaM bound to NaV1.2 or NaV1.5 suggest some basis for the channel-specific differences (Reddy Chichili et al., 2013).
CaM, bound to the intracellular CTD and thus close to the membrane, is well positioned to regulate NaV channel inactivation. Although the specific orientation of the CTD in relation to the bulk of the channel in the membrane is not known, CTD–CTD interactions (Fig. S1, E and F) place limits that orient the long IQ domain helix closer to parallel with the plane of the membrane than perpendicular. Indeed, such an arrangement is compatible with the identified structural homology between the proximal portion of the NaV CTDs with the CaV1.1 CTD (Wu et al., 2016), especially in the context of the postulated flexibility between the NaV CTD globular domain and the long IQ domain (Wang et al., 2014). In turn, this would locate CaM near the linkers between transmembrane repeats and the loops between individual transmembrane segments. In this regard, an interaction between the CTD-bound Ca2+/CaM and the III-IV intracellular linker, the putative inactivation particle, was an attractive hypothesis to explain how the CTD influenced inactivation and was supported by the demonstration that CaM can bind directly to the isolated III-IV linker (Sarhan et al., 2009, 2012). We were unable to demonstrate a tripartite interaction for NaV1.5, however. Moreover, the affinity of CaM for the NaV1.5 CTD (∼100 nM whether in the absence or presence of Ca2+ for a longer, more complete CTD) suggests that CaM is unlikely to release from the CTD to form a micromolar Ca2+-dependent interaction with the III-IV linker as proposed for the tripartite interaction (Sarhan et al., 2012). Furthermore, if the structure of CaV1.1 (Wu et al., 2016) predicts salient features for NaV channels—extending the observed structural homology between the proximal CaV1.1 CTD and the NaV CTDs—a tripartite interaction including CaM appears unlikely because in CaV1.1 the CTD binds the III-IV linker precisely where Ca2+/CaM binds the homologous region of the isolated III-IV linker peptide in NaV1.5 (Sarhan et al., 2012). An important caveat, however, is that our analyses eliminated Ca2+; the presence of elevated Ca2+ could lead to structural rearrangements in the NaV1.5 channel that cannot be gleaned from the CaV1.1 structure. Nevertheless, the apoCaM–CTD complex, in close proximity to intracellular loops and at the end of the fourth transmembrane repeat (IV), is ideally situated to influence the actions of the S4 voltage sensor in the fourth transmembrane repeat (IVS4) that regulates NaV channel inactivation (Chahine et al., 1994; Sheets et al., 1999).
Although we observed a tight correlation between increased persistent Na+ current and decreased apoCaM binding affinity among the mutants tested, our data suggest that additional factors beyond apoCaM interaction with the CTD contribute to the regulation of NaV channel inactivation. For example, the relationship between the magnitude of apoCaM affinity and the amplitude of persistent Na+ current is not monotonic, as illustrated by a comparison of the E1901Q and Q1909R mutations. The E1901Q has a larger relative persistent Na+ current amplitude, yet the reduction in affinity caused by the E1901Q mutation is modest compared with Q1909R. The underlying reasons for the lack of a monotonic relationship is not clear, and multiple factors may contribute on a mutation-specific basis. One possibility is that some of the mutations may adversely affect protein folding for the recombinant CTDs, thereby exaggerating the measured effect upon CaM binding. Another possibility is that specific locations may exert particularly potent effects on the persistent Na+ current. For example, E1901Q sits at a flexible hinge point between the IQ domain and the globular domain (Wang et al., 2014), so an effect on the inherent flexibility may impart the disproportionally large persistent Na+ current for this mutant.
In conclusion, our data demonstrated an intimate relationship between inactivation and apoCaM binding to the CTD. Disease mutation in the CTDs of NaV1.2 or NaV1.5 that reduce CaM affinity result in increased persistent Na+ current or slower inactivation, and these effects are restored to WT levels by CaM overexpression. The functional significance of the interaction with CaM is underscored by the reduction in persistent Na+ current and decreased action potential firing in cerebellar Purkinje neurons after CaM overexpression. Thus, our data show that CaM is a major regulator of inactivation properties across multiple members of the NaV channel family.
Supplementary Material
Acknowledgments
This work was supported by National Heart, Lung, and Blood Institute (NHLBI) grants R01 HL112918 (to G.S. Pitt) and R01 HL122967 (to G.S. Pitt and S.O. Marx).
Author contributions: G.S. Pitt and S.O. Marx conceived of the project. H. Yan performed the electrophysiology experiments and Purkinje neuron cell cultures. C. Wang generated the mutant constructs and performed the biochemistry experiments. H. Yan, C. Wang, S.O. Marx, and G.S. Pitt jointly analyzed the data and wrote the manuscript.
The authors declare no competing financial interests.
Kenton J. Swartz served as editor.
Footnotes
Abbreviations used:
- CaM
- calmodulin
- CTD
- C-terminal domain
- DSG
- disuccinimidyl glutarate
- ITC
- isothermal titration calorimetry
- LC-MS/MS
- liquid chromatography electrospray ionization tandem mass spectrometry
- NaV
- voltage-gated Na+
- TTX
- tetrodotoxin
References
- An R.H., Wang X.L., Kerem B., Benhorin J., Medina A., Goldmit M., and Kass R.S.. 1998. Novel LQT-3 mutation affects Na+ channel activity through interactions between α- and β1-subunits. Circ. Res. 83:141–146. 10.1161/01.RES.83.2.141 [DOI] [PubMed] [Google Scholar]
- Antzelevitch C., Nesterenko V., Shryock J.C., Rajamani S., Song Y., and Belardinelli L.. 2014. The role of late I Na in development of cardiac arrhythmias. Handbook Exp. Pharmacol. 221:137–168. 10.1007/978-3-642-41588-3_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhorin J., Kalman Y.M., Medina A., Towbin J., Rave-Harel N., Dyer T.D., Blangero J., MacCluer J.W., and Kerem B.S.. 1993. Evidence of genetic heterogeneity in the long QT syndrome. Science. 260:1960–1962. 10.1126/science.8316839 [DOI] [PubMed] [Google Scholar]
- Benhorin J., Goldmit M., MacCluer J.W., Blangero J., Goffen R., Leibovitch A., Rahat A., Wang Q., Medina A., Towbin J., and Kerem B.. 1998. Identification of a new SCN5A mutation, D1840G, associated with the long QT syndrome. Hum. Mutat. 12:72 [DOI] [PubMed] [Google Scholar]
- Ben-Johny M., Yang P.S., Niu J., Yang W., Joshi-Mukherjee R., and Yue D.T.. 2014. Conservation of Ca2+/calmodulin regulation across Na and Ca2+ channels. Cell. 157:1657–1670. 10.1016/j.cell.2014.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blich M., Efrati E., Marai I., Suleiman M., Gepstein L., and Boulous M.. 2015. Novel clinical manifestation of the known SCN5A D1790G mutation. Cardiology. 132:228–232. 10.1159/000437089 [DOI] [PubMed] [Google Scholar]
- Chagot B., and Chazin W.J.. 2011. Solution NMR structure of Apo-calmodulin in complex with the IQ motif of human cardiac sodium channel NaV1.5. J. Mol. Biol. 406:106–119. 10.1016/j.jmb.2010.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahine M., George A.L. Jr., Zhou M., Ji S., Sun W., Barchi R.L., and Horn R.. 1994. Sodium channel mutations in paramyotonia congenita uncouple inactivation from activation. Neuron. 12:281–294. 10.1016/0896-6273(94)90271-2 [DOI] [PubMed] [Google Scholar]
- Clatot J., Ziyadeh-Isleem A., Maugenre S., Denjoy I., Liu H., Dilanian G., Hatem S.N., Deschênes I., Coulombe A., Guicheney P., and Neyroud N.. 2012. Dominant-negative effect of SCN5A N-terminal mutations through the interaction of Nav1.5 α-subunits. Cardiovasc. Res. 96:53–63. 10.1093/cvr/cvs211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier J.W., Rivolta I., Tateyama M., Yang A.S., and Kass R.S.. 2002. Secondary structure of the human cardiac Na+ channel C terminus: evidence for a role of helical structures in modulation of channel inactivation. J. Biol. Chem. 277:9233–9241. 10.1074/jbc.M110204200 [DOI] [PubMed] [Google Scholar]
- Dixon R.E., Moreno C.M., Yuan C., Opitz-Araya X., Binder M.D., Navedo M.F., and Santana L.F.. 2015. Graded Ca2+/calmodulin-dependent coupling of voltage-gated CaV1.2 channels. eLife. 4:e05608 10.7554/eLife.05608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldkamp M.D., Yu L., and Shea M.A.. 2011. Structural and energetic determinants of apo calmodulin binding to the IQ motif of the NaV1.2 voltage-dependent sodium channel. Structure. 19:733–747. 10.1016/j.str.2011.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabelli S.B., Boto A., Kuhns V.H., Bianchet M.A., Farinelli F., Aripirala S., Yoder J., Jakoncic J., Tomaselli G.F., and Amzel L.M.. 2014. Regulation of the NaV1.5 cytoplasmic domain by calmodulin. Nat. Commun. 5:5126 10.1038/ncomms6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug K., Hallmann K., Rebstock J., Dullinger J., Muth S., Haverkamp F., Pfeiffer H., Rau B., Elger C.E., Propping P., and Heils A.. 2001. The voltage-gated sodium channel gene SCN2A and idiopathic generalized epilepsy. Epilepsy Res. 47:243–246. 10.1016/S0920-1211(01)00312-6 [DOI] [PubMed] [Google Scholar]
- Herzog R.I., Liu C., Waxman S.G., and Cummins T.R.. 2003. Calmodulin binds to the C terminus of sodium channels Nav1.4 and Nav1.6 and differentially modulates their functional properties. J. Neurosci. 23:8261–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi M., Du X.X., Shinlapawittayatorn K., Liu H., Chai S., Wan X., Ficker E., and Deschênes I.. 2014. Brugada syndrome disease phenotype explained in apparently benign sodium channel mutations. Circ Cardiovasc Genet. 7:123–131. 10.1161/CIRCGENETICS.113.000292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapplinger J.D., Tester D.J., Salisbury B.A., Carr J.L., Harris-Kerr C., Pollevick G.D., Wilde A.A., and Ackerman M.J.. 2009. Spectrum and prevalence of mutations from the first 2,500 consecutive unrelated patients referred for the FAMILION long QT syndrome genetic test. Heart Rhythm. 6:1297–1303. 10.1016/j.hrthm.2009.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapplinger J.D., Giudicessi J.R., Ye D., Tester D.J., Callis T.E., Valdivia C.R., Makielski J.C., Wilde A.A., and Ackerman M.J.. 2015. Enhanced classification of Brugada syndrome–associated and long-QT syndrome–associated genetic variants in the SCN5A-encoded Nav1.5 cardiac sodium channel. Circ Cardiovasc Genet. 8:582–595. 10.1161/CIRCGENETICS.114.000831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A., Nesvizhskii A.I., Kolker E., and Aebersold R.. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383–5392. 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- Kim J., Ghosh S., Liu H., Tateyama M., Kass R.S., and Pitt G.S.. 2004a Calmodulin mediates Ca2+ sensitivity of sodium channels. J. Biol. Chem. 279:45004–45012. 10.1074/jbc.M407286200 [DOI] [PubMed] [Google Scholar]
- Kim J., Ghosh S., Nunziato D.A., and Pitt G.S.. 2004b Identification of the components controlling inactivation of voltage-gated Ca2+ channels. Neuron. 41:745–754. 10.1016/S0896-6273(04)00081-9 [DOI] [PubMed] [Google Scholar]
- Martin H.C., Kim G.E., Pagnamenta A.T., Murakami Y., Carvill G.L., Meyer E., Copley R.R., Rimmer A., Barcia G., Fleming M.R., et al. WGS500 Consortium . 2014. Clinical whole-genome sequencing in severe early-onset epilepsy reveals new genes and improves molecular diagnosis. Hum. Mol. Genet. 23:3200–3211. 10.1093/hmg/ddu030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno C.M., Dixon R.E., Tajada S., Yuan C., Opitz-Araya X., Binder M.D., and Santana L.F.. 2016. Ca2+ entry into neurons is facilitated by cooperative gating of clustered CaV1.3 channels. eLife. 5:e15744 10.7554/eLife.15744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano C., Priori S.G., Schwartz P.J., Bloise R., Ronchetti E., Nastoli J., Bottelli G., Cerrone M., and Leonardi S.. 2005. Genetic testing in the long QT syndrome: development and validation of an efficient approach to genotyping in clinical practice. JAMA. 294:2975–2980. 10.1001/jama.294.23.2975 [DOI] [PubMed] [Google Scholar]
- Navedo M.F., Cheng E.P., Yuan C., Votaw S., Molkentin J.D., Scott J.D., and Santana L.F.. 2010. Increased coupled gating of L-type Ca2+ channels during hypertension and Timothy syndrome. Circ. Res. 106:748–756. 10.1161/CIRCRESAHA.109.213363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii A.I., Keller A., Kolker E., and Aebersold R.. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646–4658. 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- Ohrtman J., Ritter B., Polster A., Beam K.G., and Papadopoulos S.. 2008. Sequence differences in the IQ motifs of CaV1.1 and CaV1.2 strongly impact calmodulin binding and calcium-dependent inactivation. J. Biol. Chem. 283:29301–29311. 10.1074/jbc.M805152200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt G.S., and Lee S.Y.. 2016. Current view on regulation of voltage-gated sodium channels by calcium and auxiliary proteins. Protein Sci. 25:1573–1584. 10.1002/pro.2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potet F., Beckermann T.M., Kunic J.D., and George A.L. Jr. 2015. Intracellular calcium attenuates late current conducted by mutant human cardiac sodium channels. Circ Arrhythm Electrophysiol. 8:933–941. 10.1161/CIRCEP.115.002760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman I.M., Sprunger L.K., Meisler M.H., and Bean B.P.. 1997. Altered subthreshold sodium currents and disrupted firing patterns in Purkinje neurons of Scn8a mutant mice. Neuron. 19:881–891. 10.1016/S0896-6273(00)80969-1 [DOI] [PubMed] [Google Scholar]
- Reddy Chichili V.P., Xiao Y., Seetharaman J., Cummins T.R., and Sivaraman J.. 2013. Structural basis for the modulation of the neuronal voltage-gated sodium channel NaV1.6 by calmodulin. Sci. Rep. 3:2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remme C.A. 2014. Cardiac sodium channel overlap syndrome. Card. Electrophysiol. Clin. 6:761–776. 10.1016/j.ccep.2014.08.005 [DOI] [Google Scholar]
- Rivolta I., Abriel H., Tateyama M., Liu H., Memmi M., Vardas P., Napolitano C., Priori S.G., and Kass R.S.. 2001. Inherited Brugada and long QT-3 syndrome mutations of a single residue of the cardiac sodium channel confer distinct channel and clinical phenotypes. J. Biol. Chem. 276:30623–30630. 10.1074/jbc.M104471200 [DOI] [PubMed] [Google Scholar]
- Sarhan M.F., Van Petegem F., and Ahern C.A.. 2009. A double tyrosine motif in the cardiac sodium channel domain III-IV linker couples calcium-dependent calmodulin binding to inactivation gating. J. Biol. Chem. 284:33265–33274. 10.1074/jbc.M109.052910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarhan M.F., Tung C.C., Van Petegem F., and Ahern C.A.. 2012. Crystallographic basis for calcium regulation of sodium channels. Proc. Natl. Acad. Sci. USA. 109:3558–3563. 10.1073/pnas.1114748109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satin J., Kyle J.W., Chen M., Bell P., Cribbs L.L., Fozzard H.A., and Rogart R.B.. 1992. A mutant of TTX-resistant cardiac sodium channels with TTX-sensitive properties. Science. 256:1202–1205. 10.1126/science.256.5060.1202 [DOI] [PubMed] [Google Scholar]
- Sheets M.F., Kyle J.W., Kallen R.G., and Hanck D.A.. 1999. The Na channel voltage sensor associated with inactivation is localized to the external charged residues of domain IV, S4. Biophys. J. 77:747–757. 10.1016/S0006-3495(99)76929-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateyama M., Liu H., Yang A.-S., Cormier J.W., and Kass R.S.. 2004. Structural effects of an LQT-3 mutation on heart Na+ channel gating. Biophys. J. 86:1843–1851. 10.1016/S0006-3495(04)74251-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan K., Liu Y., and Goldfarb M.. 2014. Fast-onset long-term open-state block of sodium channels by A-type FHFs mediates classical spike accommodation in hippocampal pyramidal neurons. J. Neurosci. 34:16126–16139. 10.1523/JNEUROSCI.1271-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Hennessey J.A., Kirkton R.D., Wang C., Graham V., Puranam R.S., Rosenberg P.B., Bursac N., and Pitt G.S.. 2011. Fibroblast growth factor homologous factor 13 regulates Na+ channels and conduction velocity in murine hearts. Circ. Res. 109:775–782. 10.1161/CIRCRESAHA.111.247957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Chung B.C., Yan H., Lee S.Y., and Pitt G.S.. 2012. Crystal structure of the ternary complex of a NaV C-terminal domain, a fibroblast growth factor homologous factor, and calmodulin. Structure. 20:1167–1176. 10.1016/j.str.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Chung B.C., Yan H., Wang H.G., Lee S.Y., and Pitt G.S.. 2014. Structural analyses of Ca2+/CaM interaction with NaV channel C-termini reveal mechanisms of calcium-dependent regulation. Nat. Commun. 5:4896 10.1038/ncomms5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Shen J., Splawski I., Atkinson D., Li Z., Robinson J.L., Moss A.J., Towbin J.A., and Keating M.T.. 1995. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 80:805–811. 10.1016/0092-8674(95)90359-3 [DOI] [PubMed] [Google Scholar]
- Wilde A.A., Moss A.J., Kaufman E.S., Shimizu W., Peterson D.R., Benhorin J., Lopes C., Towbin J.A., Spazzolini C., Crotti L., et al. . 2016. Clinical aspects of type 3 long-QT syndrome: An International multicenter study. Circulation. 134:872–882. 10.1161/CIRCULATIONAHA.116.021823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm M., Shevchenko A., Houthaeve T., Breit S., Schweigerer L., Fotsis T., and Mann M.. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 379:466–469. 10.1038/379466a0 [DOI] [PubMed] [Google Scholar]
- Winkel B.G., Yuan L., Olesen M.S., Sadjadieh G., Wang Y., Risgaard B., Jabbari R., Haunsø S., Holst A.G., Hollegaard M.V., et al. . 2015. The role of the sodium current complex in a nonreferred nationwide cohort of sudden infant death syndrome. Heart Rhythm. 12:1241–1249. 10.1016/j.hrthm.2015.03.013 [DOI] [PubMed] [Google Scholar]
- Wu J., Yan Z., Li Z., Qian X., Lu S., Dong M., Zhou Q., and Yan N.. 2016. Structure of the voltage-gated calcium channel Cav1.1 at 3.6 Å resolution. Nature. 537:191–196. 10.1038/nature19321 [DOI] [PubMed] [Google Scholar]
- Xu H., and Freitas M.A.. 2007. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics. 8:133 10.1186/1471-2105-8-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Pablo J.L., and Pitt G.S.. 2013. FGF14 regulates presynaptic Ca2+ channels and synaptic transmission. Cell Reports. 4:66–75. 10.1016/j.celrep.2013.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Pablo J.L., Wang C., and Pitt G.S.. 2014. FGF14 modulates resurgent sodium current in mouse cerebellar Purkinje neurons. eLife. 3:e04193 10.7554/eLife.04193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziyadeh-Isleem A., Clatot J., Duchatelet S., Gandjbakhch E., Denjoy I., Hidden-Lucet F., Hatem S., Deschênes I., Coulombe A., Neyroud N., and Guicheney P.. 2014. A truncating SCN5A mutation combined with genetic variability causes sick sinus syndrome and early atrial fibrillation. Heart Rhythm. 11:1015–1023. 10.1016/j.hrthm.2014.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.