Abstract
Mammary gland development, various stages of mammary tumorigenesis and breast cancer progression have the peptidyl-prolyl cis/trans isomerase PIN1 at their centerpiece, in virtue of the ability of this unique enzyme to fine-tune the dynamic crosstalk between multiple molecular pathways. PIN1 exerts its action by inducing conformational and functional changes on key cellular proteins, following proline-directed phosphorylation. Through this post-phosphorylation signal transduction mechanism, PIN1 controls the extent and direction of the cellular response to a variety of inputs, in physiology and disease. This review discusses PIN1's roles in normal mammary development and cancerous progression, as well as the clinical impact of targeting this enzyme in breast cancer patients.
Facts
PIN1 mediates pathway crosstalk following oncogenic phosphorylation of several key cellular proteins involved in different aspects of breast cancer malignancy.
PIN1 is overexpressed in the majority of human breast cancers and is essential for breast cancer development and progression, as well as for Cancer Stem Cell maintenance.
Genetic or pharmacological inactivation of PIN1 in preclinical models is sufficient to block breast cancer growth and dissemination as well as to recover chemosensitivity.
Open questions
A detailed scenario of how PIN1-catalyzed prolyl-isomerization of key proteins might function as a timing mechanism to differentially turn on or off protein functions during dynamic cellular processes, such as mammary gland morphogenesis, is still missing.
PIN1 function in the normal mammary stem cell compartment is only poorly understood; in particular there is a limited knowledge of the pathways that are subjected to PIN1 activity and that have a role in the correct maintenance of mammary stem cell and progenitor compartments.
Despite the relevance of PIN1 for human breast carcinogenesis, the regulation of its expression is scarcely defined and the biological outcome of combined post-translational modifications is still an unanswered question.
The impact of prolyl-isomerization on different classes of PIN1 substrates in cancer is still scarcely addressed.
Breast cancer (BC) is the most common cancer among women worldwide.1 Despite significant improvements, still a large number of patients relapse after treatment, thus indicating pitfalls in diagnosis and therapy. Major obstacles reside in genetic and phenotypic heterogeneity that characterize BCs. BC-related morbidity and mortality after therapeutic failure is attributable to tumor recurrence and dissemination of metastases. Thus, to tackle BC malignancy, comprehension of the underpinning molecular mechanisms is paramount.2
The prolyl-isomerase PIN1 is a major player in sensing and coordinating the cellular responses to phosphorylation-dependent signals, both in physiologic and pathologic contexts.3 Given its enzymatic nature and depending on the cellular context, PIN1 exerts opposite functions by simultaneously modulating both growth-promoting and growth-suppressive pathways, integrating cellular responses to different stimuli. In cancer cells, PIN1 is required for boosting oncogenic signals, while it blocks proteins with tumor suppressor functions.3 In addition, PIN1 function serves the tumor suppressing activities of p53 family members, such as p53 itself and p73, to induce growth arrest and apoptosis following oncogenic or genotoxic stress signals.4, 5, 6, 7, 8 However, in established cancers, where key tumor suppressors like p53 become inactivated, while oncoproteins are upregulated, PIN1 activity may tip the balance toward pro-oncogenic signaling. Accordingly, PIN1 expression has been found associated with high-grade BCs.9
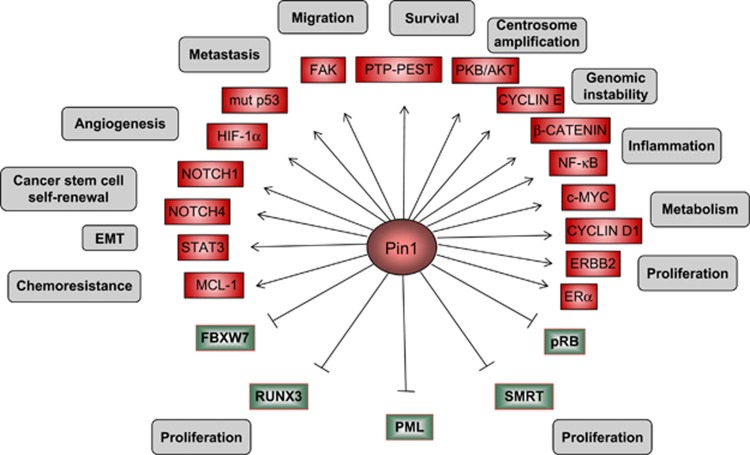
We and others have described PIN1 as an essential factor for mammary tumor development and progression.9, 10, 11, 12, 13, 14, 15 In this context, PIN1 is upregulated by activated oncogenes12, 14, 16 and mediates crucial pathway crosstalk following oncogenic phosphorylation of several proteins involved in different aspects of malignancy, such as Cyclin D1, c-MYC, β-catenin, NF-κB, STAT3, MCL-1, ERBB2/HER2/NEU, ERα, PKB/AKT, mutant p53 proteins, Notch1 and Notch4, etc.3, 14, 17 (Figure 1 and Table 1). The relevance of PIN1-mediated protein conformational changes in the development and progression of BCs is underscored by the observation that PIN1 inactivation is sufficient to block BC growth and dissemination, and to recover chemosensitivity in defined in vitro and in vivo models.9, 10, 11, 12, 13, 14, 15, 18
Figure 1.
PIN1 regulates several cellular processes in breast cancer depending on the substrates. Schematic representation of breast cancer-specific PIN1 substrates and the involved biological processes. Products of oncogenes and tumor suppressors are indicated in red and green boxes, respectively. Arrows and blunted lines indicate a positive or negative action of PIN1, respectively, eliciting a particular cellular outcome, as indicated in the gray boxes
Table 1. List of PIN1 actions on cellular substrates identified in breast cancer.
| Protein | PIN1-binding motifs | Kinase | PIN1 function | Cellular effect/phenotype in BC cells | Synergy between PIN1 inhibition and known drugs | Refs. |
|---|---|---|---|---|---|---|
| AKT/PKB | Thr92, Thr450 | ND | Stabilization | Cell survival | 65 | |
| β-catenin | Ser246 | ND | Stabilization and nuclear localization | Enhanced transcriptional activity | 3, 17 | |
| Cyclin D1 | Thr286 | GSK-3β | Increased expression, stabilization, and nuclear localization | Cell cycle progression, enhanced proliferation | 17, 18 | |
| Cyclin E | Ser384 | CDK2 | Regulation of protein stability | Cell cycle progression | 3, 17, 53 | |
| c-JUN | Ser63, Ser73 | JNK | Enhancement of transcriptional activity | Enhanced proliferation | 9, 17 | |
| c-MYC | Thr58, Ser62 | ERK, GSK-3β | Regulation of protein stability and transcriptional activity | Enhanced growth and metabolism | 15, 53 | |
| ERα | Ser118, Ser294 | MAPK, ERK, CDK2 | Stabilization, enhanced DNA binding and transcriptional activity | Enhanced proliferation of ER+ BC cells, resistance to hormone therapy | Tamoxifen | 17, 26 |
| ERBB2/HER2 | ND | ND | Stabilization | Enhanced proliferation of HER2+ BC | Rapamycin | 10 |
| FAK | Ser910 | ERK | Promotes interaction with PTP-PEST | Enhanced HRAS-dependent migration, invasion and metastatic spread | 62 | |
| FBXW7 | Thr205 | ND | Degradation | Enhanced cell proliferation | 55 | |
| HIF-1α | Ser641, Ser643 | ERK MAPK | Stabilization and promotion of transcriptional activity | Induction of angiogenesis and endocrine therapy resistance | 34 | |
| MCL-1 | Thr92, Thr163 | ERK | Stabilization | Chemoresistance | Sorafenib | 11 |
| Mutant p53 | Ser46, Ser315 | AURKA/B | Enhances transcriptional activity and sequestration of p63 | Enhanced mutant p53 pro-metastatic activities | 13 | |
| STAT3 | Ser727 | ND | Promotes transcriptional activity | Promotion of EMT | 60 | |
| NF-κB | Thr254 | ND | Stabilization and nuclear localization | Enhanced transcriptional activity | 3, 17 | |
| Notch1 | Ser2122, Thr2133, Ser2137, Thr2512 | ND | Processing of the membrane bound receptor, stabilization of the intracellular domain | Enhanced transcription, proliferation, CSCs self-renewal, tumorigenicity, chemoresistance | GSI (gamma secretase inhibitors) | 12, 14 |
| Notch4 | ND | ND | Stabilization of the intracellular domain | Enhanced CSCs self-renewal and tumorigenicity | 14 | |
| PML | Ser403, Ser505, Ser518, Ser527 | ERK | Degradation | Enhanced cell proliferation | 3, 17 | |
| pRB | Ser608, Ser612 | CDK2/4/6 | Hyper-phosphorylation and inactivation | Cell cycle progression, growth of Tamoxifen- resistant cells | PI3 Kinase Inhibitors | 3, 17, 52 |
| PTP-PEST | Ser571 | ERK | Promotes interaction with FAK | HRAS-dependent migration, invasion and metastatic spread | 62 | |
| RUNX3 | Thr209, Thr212, Thr231, Ser214 | ND | Degradation | Repression of RUNX3 transcriptional activity | 3, 17 | |
| SMRT | Ser1241, Ser1469 | CDK2 | Degradation | Increased ER gene expression and cell proliferation | 29, 31 |
Abbreviation: ND, not determined
Along with the key findings in BC, here we will review recent advancements in the understanding of the physiologic roles of PIN1 in mammary gland development. The clinical relevance of PIN1 activities and the potential impact of its inhibition in selected BC patients will be finally addressed.
PIN1: A Critical Hub of Phosphorylation-Dependent Pathway Crosstalk
Protein phosphorylation is a reversible post-translational modification (PTM) implicated in a variety of cellular processes, that impacts on protein activity, dictates subcellular localization and induces the establishment of recruitment platforms for interacting proteins. Frequently altered as a downstream consequence of oncogenic driver mutations, protein phosphorylation is central to cancer biology and treatment.19
Among the possible phosphorylation sites, Serines or Threonines preceding a Proline (Ser/Thr-Pro) that are targeted by Proline-directed kinases (CDKs, MAPKs, and GSK-3β) deserve particular attention. Prolines can adopt either a cis or trans conformation with consequences on protein folding and function. The spontaneous conversion of one isomer into the other is a very slow process and phosphorylation of Ser/Thr-Pro moieties per se further decreases the isomerization rate of Prolines.20 The intervention of the phosphorylation-dependent prolyl-isomerase (PPIase) PIN1 enables the occurrence of the cis–trans conversion in a biologically relevant timescale, adding a further layer of post-translational control over client proteins.3 Among all PPIases, PIN1 is the sole that specifically recognizes phosphorylated Ser/Thr-Pro moieties (phospho-Ser/Thr-Pro). Such unique substrate specificity is conferred by its highly conserved two-domain structure consisting of an N-terminal WW domain binding specific phospho-Ser/Thr-Pro modules and a C-terminal PPIase domain catalyzing their cis-trans isomerization.3, 17
PIN1-induced conformational changes on certain substrates are required for the correct sequence of PTMs, where other enzymes, such as phosphatases, are specific for the cis or trans conformation of the prolyl peptide bond.3, 17 Since the discovery of PIN1 20 years ago, major findings have now clearly demonstrated that phosphorylation-dependent prolyl-isomerization is a crucial signal transduction mechanism that, depending on the context, functions like a cellular rheostat for fine-tuning or amplification of phosphorylation signaling.3
PIN1 is Required for Normal Breast Development
Mammary gland development occurs through well-defined stages throughout embryonic and pubertal development as well as during reproductive life. In the course of the adult female life cycle, the mammary gland undergoes dynamic changes that range from ductal elongation and branching during pre-puberty through pregnancy, to lactation and involution.21
Major insights into the role of PIN1 in organismal homeostasis in mammals stem from constitutive Pin1 knockout (Pin1−/−) mice.22 These mice are viable and reach the adulthood like their wild-type littermates, but display a range of developmental and age-related abnormalities.3, 8 In particular, the mammary glands of Pin1−/− female mice display an impaired pregnancy-induced expansion of the mammary epithelium.23 In vivo and ex vivo assays allowed to demonstrate that Pin1−/− mice have an impaired mammary stem cell compartment. Indeed, they display less adult mammary stem cells with reduced self-renewal capacity and regenerative potential, without altering the luminal or myoepithelial fate commitment.14, 24
Actually, it is not known which are the pathways responsible for the correct maintenance of the mammary stem cell and progenitor compartments that are subjected to PIN1 activity (Figure 2). One possible determinant of PIN1 function in these compartments is Cyclin D1. Indeed, Pin1−/− mammary glands have decreased Cyclin D1 levels, and Cyclin D1 knockout mice display a similar mammary gland developmental defect.23 In addition to this genetic evidence, PIN1 was shown to enhance Cyclin D1 function, promoting both its transcription, through β-catenin and c-JUN, and its nuclear localization following growth factor pathway stimulation.9 Moreover, also Cyclin D1 was shown to control the mammary stem cell compartment,25 further supporting a PIN1-Cyclin D1 interplay in mammary gland development. Other likely candidates are the Notch1 and Notch4 receptors, key determinants of mammary stem cell maintenance and luminal/myoepithelial commitment, respectively. We found that their levels were decreased in Pin1−/− mouse mammary epithelial cells, suggesting that PIN1 may regulate stage-specific Notch functions in this compartment.14
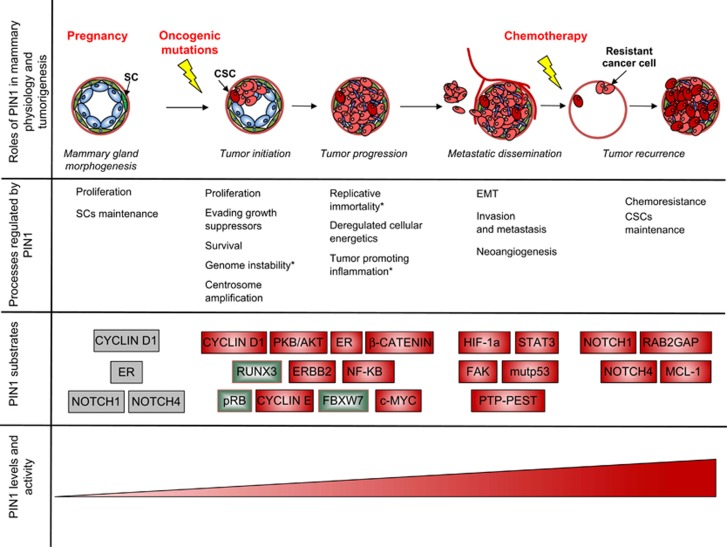
Figure 2.
PIN1 is required for both normal mammary gland morphogenesis and breast cancer progression through regulation of multiple cellular processes. Top: the phases of normal breast development and tumor progression in which a role of PIN1 has been demonstrated are schematically represented from left to right. Specific conditions, where coordination of different pathways requires PIN1 activity, are indicated in red. Below, specific cellular processes occurring during the abovementioned phases and the involved PIN1 substrates are indicated. Oncogenes and tumor suppressors are indicated in red and green boxes, respectively. In gray boxes putative PIN1 substrates involved in human breast morphogenesis. Bottom: The levels/activity of PIN1 that increase from normal mammary gland to tumors progressing to high-grade cancers are represented by a red wedge. Asterisks indicate cellular processes regulated by PIN1 in contexts different fom breast tissue. SC, stem cells
PIN1 may coordinate growth factor/hormone stimulation of the mammary gland
From puberty on, mammary gland morphogenesis depends on the responsiveness of mammary epithelial cells to hormones and growth factors and on the activation of various intracellular signaling pathways in a timely concerted action. Under the influence of estrogens and progesterone, the ducts begin to expand, invade the surrounding stroma and develop the lobulo-alveolar end buds.21
A growing body of evidence supports the idea that PIN1 might coordinate hormone receptor with growth factor signaling. For instance, downstream of stimulated growth factor receptor pathways, activation of Proline-directed kinases leads to ERα phosphorylation in its N-terminal Ser/Thr-Pro motifs.26 These modifications render ERα, a putative PIN1 substrate. In accordance, PIN1 was shown to bind and catalyze a conformational change of the ERα N-terminal domain upregulating both protein levels and transcriptional activity.26, 27 Furthermore, PIN1 synergistically acts with CDK2 to regulate ERα basal transcriptional activity,28 and, downstream of HER2 activation, PIN1 activates ERα transcriptional activity by downregulating NCOR2/SMRT and enhancing SRC-3 and p300 co-activators' recruitment on cognate promoters.29, 30, 31 As many of the studies supporting this evidence were made in BC cells, the relevance of PIN1 for ERα expression and function in normal mammary epithelial cells remains to be demonstrated. In addition, PIN1 has multiple roles in the function of other steroid receptors, such as PR, by influencing the activity of one or more of their co-activators.30 On these bases, it can be argued that the mammary phenotype of Pin1−/− mice may also derive in part from the lack of a correct response to mammary-specific hormones.
Stage- and tissue-specific conditional Pin1−/− mice need to be generated in order to confirm these hypotheses and outline a more detailed scenario of how PIN1-catalyzed prolyl-isomerization after phosphorylation of key proteins might function as a timing mechanism during dynamic cellular processes such as mammary gland morphogenesis.
PIN1 has Critical Roles in BC Development and Progression
BC is a heterogeneous disease consisting of different subtypes that are characterized by different histo-pathological features, specific genetic and epigenetic alterations, and diverse aggressive traits acquired during malignant progression.32 PIN1 is involved in all main cellular processes of BC development and progression9, 11, 12, 13, 14, 15, 18, 24, 33, 34, 35 (Figure 2). PIN1 is overexpressed in the majority of BCs and correlates with worse clinical outcome,9, 10, 11, 12, 13, 14, 24 pointing to its essential role in oncogenic phosphorylation-dependent events, from the very early steps of breast carcinogenesis.
Regulation of PIN1 gene expression in human BC
PIN1 is not annotated as a cancer gene, as it is mutated only at very low frequency in few tumor types, according to cancer databases like CONAN, COSMIC and TGCA (Figure 3). PIN1 alterations may derive from gene amplification, as found in cancers of the female urogenital system. In BCs, instead, the frequently observed mRNA upregulation is most likely due to transcriptional mechanisms (Figure 4). In fact, PIN1 expression is promoted by the transcription factor E2F,16 possibly via activated ERBB2/HER2/NEU, RAS, and IGF-1R pathways.18, 35 Moreover, PIN1 was unveiled as a direct target of two members of the Notch receptor family in BC.12, 14 Of note, the human PIN1 promoter polymorphism -842G>C;rs2233678 is associated to decreased gene expression and to a lower risk of BC incidence.36 In BC cells, PIN1 mRNA levels were found downmodulated by the miR-200c small non-coding RNA, a well-known keeper of epithelial fate and repressor of metastasis.24
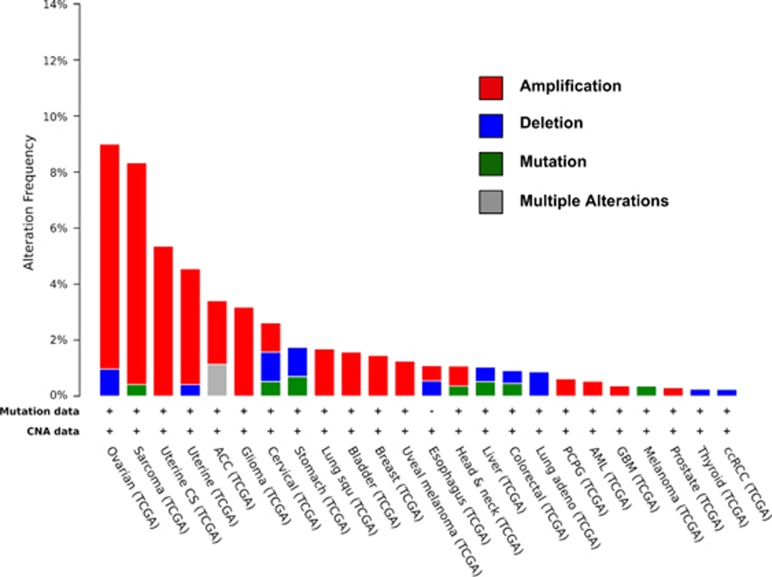
Figure 3.
PIN1 is not a frequently altered gene in human cancers. Analysis of mutations (Mutation data) and copy number (CNA data) of human PIN1 was performed across all human tumor types in the TCGA cancer panel (obtained from cbioportal cross-cancer mutations). In the histogram only those tumors are indicated, where alterations of human PIN1 were found: alteration frequencies along with copy number variation data are provided. Breast cancers display amplifications of PIN1 in only 1.5% of the analyzed cases
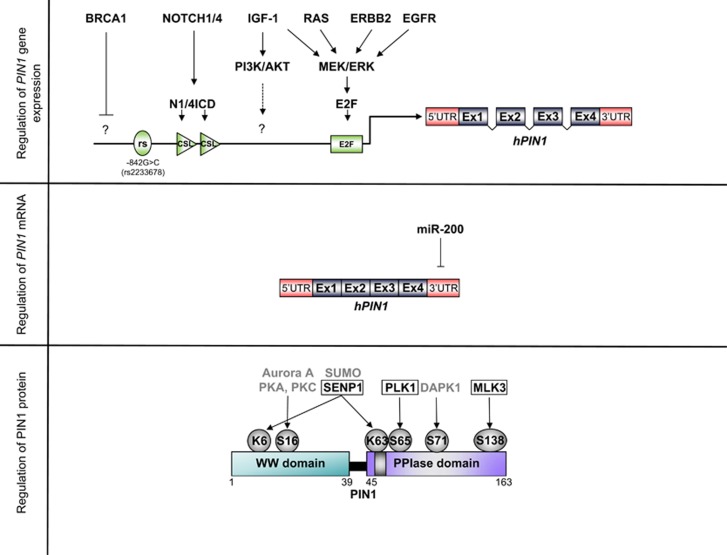
Figure 4.
Regulation of PIN1 expression in breast cancer. Upper part: Schematic representation of the human PIN1 gene, encompassing the promoter sequence, the 5' and 3' untranslated regions (UTRs), and exons 1–4 (Ex). Known cis-elements and the upstream signals and trans-acting factors are indicated; CSL Notch-responsive element. Middle part: Scheme of the mature PIN1 mRNA targeted by mir-200. Lower part: Schematic of the PIN1 protein structure with the sites of regulatory post-translational modifications and the responsible enzymes. The type IV WW domain, the linker region in between, and the PPIase domain are indicated. The nuclear localization signal is depicted as a small gray box within the PPIase domain. Modified aminoacid residues of PIN1 are indicated with a circle, enzymes acting on the specific residue with activating or repressing function on PIN1 are indicated in black boxed or in gray letters, respectively
PIN1 protein expression, PTMs and subcellular localization in human BC
In human BC deregulation of PIN1 protein levels and activity are most relevant. In fact, while normal breast epithelial cells express low levels of nuclear PIN1, BC cells instead stain strongly both in the nucleus and cytoplasm and display highly phosphorylated PIN1.9
Different phosphorylation and other PTM sites of PIN1 have been identified so far and shown to impact on its stability, subcellular localization, substrate binding and catalytic activity (Figure 4,Table 2). Interestingly, whereas the levels of PIN1 do not vary significantly during cell cycle progression, its phosphorylation status is highly modulated. Indeed, PLK1, an early trigger for G2/M transition involved in centrosome maturation and mitotic spindle establishment, has been found to phosphorylate Ser65 in the catalytic domain of PIN1.37 This phosphorylation increases PIN1 stability, a relevant event during mitosis.37 On the other hand, Ser16 phosphorylation by PKA and AURKA abolishes PIN1 ability to bind its substrates and is required for exit from mitosis.38 Phosphorylation by PKA has been shown to impact also on PIN1 localization in nuclear speckles, by facilitating its nuclear export.39 Of note, Gonadotropin signaling regulates the activity of PIN1 through a nuclear-cytoplasmic shuttling mechanism based on Ser16 phosphorylation by PKA or PKC and its subsequent dephosphorylation by calcineurin.40 The tumor suppressor DAPK1 phosphorylates PIN1 at Ser71, fully inhibiting its isomerase activity, nuclear localization and cellular function.41 DAPK1 expression correlates with phospho-Ser71-PIN1 levels in human normal breast tissues and tumors. Accordingly, reduction of PIN1 expression or restoration of DAPK1 in cancer cells effectively suppress tumorigenic phenotypes.41 MLK3, a MAP3K family member, was shown to phosphorylate PIN1 on Ser138, fostering PIN1 catalytic activity and nuclear localization. A significant difference in the levels of phospho-Ser138-PIN1 between normal and cancer tissue has been observed, although total protein levels were comparable, suggesting that MLK3-induced phosphorylation of PIN1 could be an early event in oncogenesis and a reliable marker for PIN1 activation in BC.42 Finally, sumoylation of Lys6 or Lys63 is another important PTM that inhibits PIN1 functions, and is reverted by de-sumoylation through SENP1.43
Table 2. Post-translational regulation of PIN1.
| Modification | Enzyme | Modified aminoacid | Consequence | Refs. |
|---|---|---|---|---|
| Phosphorylation | PKA | Ser16 | Abolished substrate recognition. Nuclear-cytoplasmic shuttling | 39, 40 |
| PKC | Ser16 | Abolished substrate recognition. Nuclear-cytoplasmic shuttling | 40 | |
| AURKA | Ser16 | Suppresses PIN1 function in G2/M | 38 | |
| PLK1 | Ser65 | Halts PIN1 protein turnover | 37 | |
| DAPK1 | Ser71 | Inhibition of PIN1 nuclear localization and activity | 41 | |
| MLK3 | Ser138 | Induces PIN1 nuclear localization and catalytic activity | 42 | |
| Dephosphorylation | Calcineurin | Ser16 | Reactivates PIN1 nuclear localization and activity | 40 |
| Sumoylation | SUMO1 | Lys6, Lys63 | Inhibits PIN1 protein activity and oncogenic function | 43 |
| De-sumoylation | SENP1 | Lys6, Lys63 | Increases PIN1 protein stability and activity | 43 |
The above-mentioned PTMs might influence the interaction of the nuclear localization signal sequence, contained in the PPIase domain of PIN1, with proteins of the nuclear import machinery.44 Thus, PIN1 nuclear-cytoplasmic shuttling may represent a further level of control. In the nucleus, PIN1 activity affects both specific transcription factors (e.g., p53, p63, p73, ERα, Notch1, NF-κB, c-MYC, MEF2C, etc.)3, 8, 17, 45 and global transcriptional as well as post-transcriptional regulators (e.g., Histones, RNApolII, RNA binding proteins, ADAR1, etc.).3, 17, 46, 47, 48, 49 PIN1 functions in the cytoplasm range from cytoskeletal remodeling (e.g., PTP-PEST, TAU, etc.)3, 17 to mitochondrial functions (p66SHC, p53, Bax, Bcl-2, BIMEL, etc.),3, 8, 7, 50 plasma-membrane associated protein regulation (e.g., Notch1, Notch4, ERBB2, etc.)10, 12, 14 and vesicular trafficking (RAB2A).33 Although nuclear PIN1 functions are clearly associated to BC, the biological significance of its accumulation in the cytoplasm for BC cells remains substantially elusive.
Genomic instability in BC initiation and progression: PIN1 is a fundamental player of cell proliferation and centrosome amplification
A fundamental trait of cancer cells involves their ability to sustain uncontrolled proliferation.51 PIN1 regulates a number of proteins important for cell cycle entry, progression through and exit from mitosis and is presumed to operate as a molecular timer of this important process.3 The best-described scenario involves the induction of PIN1 expression by activated ERBB2/HER2/RAS via E2F that, in turn, leads to Cyclin D1 upregulation.3, 16, 17 In addition, the Cyclin D1/CDK pathway is responsible for the interaction of PIN1 and pRB, which boosts the switch from hypo- to hyper-phosphorylated pRB in tumor cells, triggering cell proliferation.52 PIN1 is also required for FBXW7-mediated degradation of c-MYC and Cyclin E to coordinate the timing of entry and progression through the G1/S phase, respectively.53 In cancer cells, control over c-MYC and Cyclin E degradation is upset15, 54 and for c-MYC it was clearly demonstrated that PIN1 facilitates the activation of c-MYC target genes leading to enhanced proliferative activity in BC.15 Intriguingly, PIN1 overexpression in BC causes enhanced degradation of the FBXW7 ub-ligase,55 further supporting a positive connection between PIN1 and c-MYC/Cyclin E in this context.
Another candidate 'hallmark' of cancer cells is centrosome amplification (CA), frequently detected in human tumors.56 CA constitutes a major mechanism leading to chromosomal instability and aneuploidy and is associated to poor patient prognosis.56 The centrosome duplication cycle is regulated in part by the cell cycle machinery and their uncoupling may result in abnormal centrosome numbers and aberrant mitosis. PIN1 has a key role in the coordination between DNA and centrosome synthesis, but its overexpression induces CA, abnormal mitotic spindle formation, and chromosome missegregation, leading to aneuploidy and oncogenic transformation, as shown in MMTV-PIN1 mice57 and in human BC cells.41, 42, 43, 56
A major event controlling the correct duplication of centrosomes is the activation of E2F and of G1 and G1/S CDK/Cyclin complexes during G1/S transition.56 Cyclin E is a well-known PIN1 substrate involved in centrosome duplication after CDK2 phosphorylation.3, 53 In BC, overexpression of both PIN1 and Cyclin E is associated to CA, and oncogenic HRAS activity, known to boost PIN1 levels and activity, was shown to potentiate Cyclin E-induced genetic instability, indicating a possible PIN1-Cyclin E cooperation in CA.54
The tight relationship between PIN1 and AURKA or PLKs in the regulation of cell cycle progression might also have an impact on CA, considering that overexpression of all three proteins is causally involved in the aberrant centrosome hyper-replication in BC.3, 56 Other intriguing connections may be directed to the tumor suppressor BRCA1, a negative regulator of PIN1 gene transcription58 (Figure 4). Inhibition of BRCA1 in breast cell lines causes rapid centrosome over-replication, suggesting that lack of PIN1 suppression might also contribute to CA in BRCA1-mutated BCs.
PIN1 acts as an amplifier of oncogenic signaling to elicit migratory and invasive phenotypes of BC cells
In spite of their enormous clinical relevance, the events that govern metastatic tumor progression still remain poorly understood. The metastatic cascade includes cancer cell detachment from the tumor, migration, invasion, survival in the vascular and lymphatic circulation, and colonization of distant organ sites.2 PIN1 deregulation acts on crucial steps of this cascade, likely through modulation of several key transcription factors that control cell plasticity, shape and migratory capability. In particular, PIN1 overexpression promotes EMT through downregulation of E-cadherin in normal and cancerous breast epithelial cells.9, 14, 24 At the same time, overexpression of the mesenchymal markers N-cadherin and Vimentin as well as increased SNAIL transcriptional activity and protein levels were shown to depend on PIN1 via different pathways, such as PI3K-AKT, NF-κB59 or Notch1.14 In BC cells, PIN1 also enhances STAT3-mediated EMT induced by Oncostatin M60 indicating that PIN1 promotes EMT via different mechanisms.
Cytoskeletal responses contribute to the development of a migratory phenotype.61 Exciting insights in the regulation of cytoskeletal remodeling and cell migration have been provided by studies on FAK, a cytoplasmic protein kinase that has an essential role during embryonic development and pathogenesis of cancer.62 In response to oncogenic HRAS, both FAK and PTP-PEST phosphatase are phosphorylated by ERK1/2 and isomerized by PIN1, which favors both their interaction and FAK dephosphorylation by PTP-PEST, promoting migration, invasion, and metastasis.62
Other crucial promoters of the metastatic process fostered by PIN1 are missense mutant forms of the p53 protein. In cancer cells, a majority of point mutations hitting the TP53 tumor suppressor gene generate proteins that loose wild-type functions and acquire new oncogenic properties. This gain of new functions is mainly exerted through aberrant protein–protein interactions, resulting in altered protein partner functions and cellular transcriptional outputs.63 p53 point mutants were demonstrated to be essential promoters of transformation and metastasis in BC,63 and we have shown that PIN1 acts as a central transducer of oncogenic signaling converging on them, both in mice and humans.13 In particular, PIN1 interacts with mutant p53 and boosts a mutant p53 dependent transcriptional program that correlates with worse clinical outcome in BC. Moreover, mutant p53 was shown to sequester the metastasis suppressor p63, thereby neutralizing its function, and high PIN1 levels favor their association. Thus, PIN1 silencing was sufficient to impair migration and invasion of mutant p53 harboring BC cells, as well as metastasis growth in immune-compromised mice.13
Clinical Implications of PIN1 Overexpression in BC
Histologic and molecular pathology-driven classification of BC has been refined by classification through integrated genomic and transcriptomic analyses, revealing the presence of at least 10 different BC subtypes with distinct clinical outcome.64 Nevertheless, current therapeutic approaches pertain mainly to three principal and oversimplified BC subtypes, namely ER+ receiving endocrine therapy, ERBB2/HER2+ treated with therapeutic targeting of HER2, and triple negative BCs (TNBC) relying only on chemotherapeutics.64
Overexpression of PIN1 has been observed in all these BC subtypes9, 10, 13, 14, 15 and correlates with the expression of subtype-specific molecular markers, such as ERα,27 ERBB2,10 PKB/AKT,65 mutant p53,13 Notch1,14 c-MYC15 and FOXC1/NF-κB.66 Moreover, quantification of PIN1 expression improves the prognostic value of substrates with strong oncogenic activity, such as MCL-1,11 mutant p53,13 PKB/AKT,65 c-MYC,15 and Notch1.14
PIN1 mediates drug resistance of BCs
Intrinsic or acquired drug resistance limits the effectiveness of chemo- and molecularly targeted cancer therapies. Mechanisms of resistance rely on drug efflux, modification of the drug target, activation of pro-survival pathways, and ineffective induction of cell death. In addition, cancer cell plasticity and tumor heterogeneity contribute to elude anti-cancer therapies.67 In particular, cancer stem cells (CSCs), that are already a source of heterogeneity, are also intrinsically more resistant to chemotherapy and some targeted therapies. These cells need to be eradicated in order to provide long-term disease-free survival.68
We have demonstrated that, like in the normal mammary gland, PIN1 acts as a fundamental regulator of stem cell features in the breast CSCs compartment. In fact, PIN1 critically controls CSC self-renewal, replicative potential, frequency and drug resistance, while its ablation causes tumor shrinkage, decreased metastatic spread and synergizes with conventional therapy in eradicating CSCs.14
From a molecular viewpoint, PIN1 elicits its functions in the breast CSCs compartment primarily by sustaining the levels and transcriptional activities of Notch1 and Notch414 and through increased expression of the small GTPase RAB2A.33 Moreover, ectopic overexpression of PIN1 in normal and transformed BC cells evokes an increase of the stem cell population, suggesting that PIN1 could impact also on molecular and phenotypic plasticity of the bulk cells giving rise to new tumor-initiating cells for instance through induction of EMT.14, 24 PIN1 might directly mediate chemoresistance sustaining PKB/AKT,65 MCL-111 or Notch1, that promotes the expression of cell survival (e.g., SURVIVIN and BCL-2) and drug efflux pump genes (e.g., ABCG2).14
The majority of BCs in post-menopausal women are ER+. Hence, anti-estrogens are employed as first-line treatment.69 After surgery these patients can be treated with hormonal therapy alone, resulting in a relatively favorable prognosis. Unfortunately, a substantial proportion of the hormone-sensitive BC patients undergo disease progression ascribable to endocrine treatment resistance.69 PIN1 was found overexpressed in ER+ tumors and cell lines9 and it is thought to confer treatment resistance to these tumors through induction of EMT and angiogenesis,34, 59 as well as degradation of CDK10,70 an important determinant of sensitization to endocrine therapy. Moreover, PIN1 sustains ERα levels and activity, by preventing ERα degradation26, 27 even under hormone- and growth factor-free conditions.28, 71 As a consequence, by keeping high both levels and activity of ERα, PIN1 might be responsible for reduced efficacy of hormone therapy. Despite PIN1-mediated chemoresistance acts also through ERα-independent mechanisms, altogether these data indicate that PIN1 might serve as a surrogate marker for predicting outcome of ER+ BC patients.27
Both preclinical and clinical data highlight the involvement of ERBB2/HER2 overexpression in the development of endocrine resistance, especially to tamoxifen.69 PIN1 functions as an essential catalyst of the ERα-HER2 crosstalk,31 supporting the idea that PIN1 inhibitors may re-sensitize tumors to endocrine therapies. Likewise, in ERBB2/HER2+ BC cells, where a strong correlation has been observed between ERBB2 and PIN1 levels, PIN1 inhibition greatly increased sensitivity to the mTOR inhibitor Rapamycin10 (Table 1).
TNBC patients, characterized by ER-, PR-, and HER2- tumors, have a higher risk of recurrence and their management still represents a clinical challenge. These tumors frequently bear p53 mutations63 or activated Notch receptors,14 that we found critically dependent on PIN1 for their oncogenic functions.13, 14 Inhibition of PIN1 carries therefore therapeutic implications also for this BC subtype.
Development of Clinical PIN1 Inhibitors
PIN1 still represents a challenge for drug discovery, since the mechanism of the isomerization reaction catalyzed by this enzyme has not yet been fully understood. Several modes of action of both the WW and PPIase domains have been proposed,72 and it appears that inhibition of either domain will have a therapeutic effect. Until recently many PIN1 inhibitors have been isolated (Table 3). However, due to unsatisfactory pharmacological characteristics, so far none of these has reached clinical trials as PIN1 inhibitors.73 In fact, difficulties have been encountered mainly due to low substrate affinity or specificity (e.g., EGCG, Juglone), poor solubility (e.g., PiB) or cell-permeability (e.g., peptide inhibitors).73 Also, literature data report the existence of relevant cellular off-targets, as for example for Juglone, whose anti-tumor effect was ascribed to production of ROS.74 Moreover, most of the compounds were tested in in vitro assays, hence still await validation in cell-based assays and in preclinical models.
Table 3. PIN1 inhibitors*.
| Inhibitor | Mechanism of action Potency | Known targets | Origin | Refs. |
|---|---|---|---|---|
| Juglone | Competitive irreversible inhibitor Low μM range | PIN1 BUB1R Tubulin | Natural compound from Walnut tree | 73 |
| PiB | Competitive reversible inhibitor Low μM range | PIN1 PAR14 | Synthetic library | 12, 13, 14, 15, 73 |
| cis-locked Alkene peptidomimetics | Competitive reversible inhibitors Low μM range | PIN1 | Structure-based design | 73 |
| D-peptide inhibitors | Competitive reversible inhibitors Low nM range | PIN1 | Combinatorial peptide library with non-natural amino acids | 73 |
| Aryl 1-indanyl Ketones | Reversible inhibitors Low μM range | PIN1 | Structure-based transition state analogs | 73 |
| DTM | Competitive inhibitor Low μM range | PIN1 | PPIase activity screening of a random chemical library | 73 |
| Cyclic D-pThr-Pip-Nal peptides | Competitive reversible inhibitors nM range | PIN1 | Cyclic phosphorylated peptide synthetic library | 73 |
| Naphthyl-substituted amino acids | Competitive reversible inhibitors Low μM-nM range | PIN1 | NMR-based fragment screening of structure-guided designed competitor ligands | 73 |
| TME-001 | Competitive Inhibitor Low μM range | PIN1 CYPA | High throughput screening of chemical libraries with real time-fluorescence detection. | 73 |
| EGCG | Competitive and reversible inhibitor Low μM range | PIN1 DDC (Dopa decarboxylase) FAS | Natural compound from Green Tea | 73 |
| ATRA | Competitive and reversible inhibitor Low μM range | PIN1 RAR | Mechanism-based screening | 75 |
*Only the compounds that were characterized for their mechanism of inhibition and were tested in cells of mammalian origin or in vivo with a potency in the low micromolar (μM) or nanomolar (nM) range are listed
ATRA is a known therapeutic for APL by targeting RARα and causing degradation of the oncoprotein PML-RARα. Very recently the team of K.P. Lu uncovered that ATRA is also an inhibitor of active PIN1, that promotes downregulation of several of its oncogenic substrates, among them PML-RARα, giving an additional mechanistic explanation of its efficacy in APL. Thus, ATRA was tested for its effect on BC growth in vitro and in mouse xenografts raising the possibility to use this FDA approved molecule for treatment of BC patients.75
Conclusions
This review article covers a range of insights acquired in the past two decades into the molecular and biological functions played by PIN1 in breast development and cancer. The role of PIN1 in organismal development and tumorigenesis was firstly highlighted in Pin1 knockout mice, which are characterized by proliferative defects of the mammary gland and by a strong reduction of mammary carcinoma formation. Since then, many research groups have demonstrated how PIN1 impacts on mammary gland development and tumorigenesis, as well as on critical and clinically challenging phases of tumor progression. Despite these efforts a number of highly relevant questions remain unanswered.
First of all, despite increased levels of PIN1 are a critical requirement for its effect on oncogenic pathways and impact on breast cancer progression, we only have a scattered knowledge of the regulation of PIN1 expression at the gene and mRNA level. Likewise, the impact of the growing repertoire of PTMs on PIN1 and the mechanisms governing its protein stability and activity or subcellular localization is still scarcely addressed. In addition to its much known impact on nuclear functions, PIN1 may also contribute to the tumorigenic process by affecting cytoplasmic/mitochondrial functions. In cancer cells, it is conceivable that a variety of upstream regulatory inputs boost PIN1 expression or activity and determine its role in different subcellular compartments. However, the molecular scenario in which PIN1 exerts its functions still awaits a more exhaustive characterization.
Second, little is known on how the post-phosphorylation mechanism centered on PIN1 actually regulates protein function, producing strikingly opposite biochemical consequences on PIN1 client proteins. A paradigmatic example is provided by the unleashing of the oncogene potential of a multitude of proteins and concomitantly by the inactivation of a large number of tumor suppressors in cancer. An invaluable tool to understand the contribution of phosphorylation-dependent prolyl-isomerization on specific substrates and cell types in mammary gland development and tumors could be conditional Pin1 knockout mice, to the establishment of which part of the future research efforts should be dedicated. The identification of key PIN1 substrates in a specific breast cancer subtype, along with the characterization of the consequence of cis/trans isomerization on protein function for each specific substrate, will provide key novel and substrate-specific mechanistic insights.
Third, almost nothing is known about the role of PIN1 in the control of gene expression through epigenetic or editing mechanisms, and about its effect on the non-coding genome. All these aspects need to be elucidated for a better understanding of PIN1 and of the unique signaling mechanism centred on it.
PIN1 capacity to regulate oncogenic signaling at various levels, its requirement for both non-CSCs and CSCs, and its enzymatic nature render it an attractive pharmacological target in breast cancer. Indeed, ablation of PIN1 synergizes with chemotherapy in reducing tumor growth and eradicating tumor-initiating cells in vivo, as well as in reducing metastatic dissemination of breast cancer cells in xenograft experiments.14 PIN1 inhibition promises to simultaneously impact on multiple and cooperating oncogenic pathways, producing multi-target effects. However, potent and highly specific PIN1 inhibitors that effectively inhibit PIN1 functions in vivo still have to be identified.
Hence, a main goal of future work in this field should be the design and implementation of effective strategies to stably inhibit PIN1 catalytic activity in cancer or target particular post-translationally modified forms of PIN1 that might be present only in tumors or, alternatively, to selectively inhibit PIN1 activators.
Moreover, a better understanding of the substrate-specific cis/trans isomerization mechanisms and of the consequence of structural modifications on proteins may also provide relevant knowledge and possibly help developing substrate-specific inhibitors or conformation-specific antibodies against key PIN1 client oncoproteins, as done before for the cis or trans isomers of phosphorylated TAU.76 These and other approaches should be actively pursued in the future in order to fully leverage PIN1 potential as a molecular target for new anti-cancer drugs.
Acknowledgments
We thank A Testa for discussing and editing the manuscript. We apologize to the colleagues whose contributions have not been cited due to space constraints. Work in our lab is supported by the Italian Health Ministry (RF-2011-02346976), the Italian Association for Cancer Research (AIRC) Special Program Molecular Clinical Oncology '5 per mille' (Grant No. 10016), AIRC IG (Grant No. 17659), by the Cariplo Foundation (Grant No. 2014-0812), and European Fund for Regional Development—Cross-Border Cooperation Programme Italy-Slovenia 2007–2013, (Project PROTEO, Code No. CB166) to GDS. YC is recipient of an AIRC postdoctoral fellowship.
Author contributions
Conception and design: A Rustighi, A Zannini, G Del Sal. Writing of the manuscript: A Rustighi, A Zannini, E Campaner, G Del Sal. Acquisition and analysis of bioinformatics data: Y Ciani, S Piazza.
Glossary
- PIN1
peptidyl-prolyl cis/trans isomerase, NIMA-interacting 1
- BC
breast cancer
- p53
tumor protein p53
- p73
tumor protein p73
- c-MYC
cellular myelocytomatosis proto-oncogene
- NF-κB
cytokine-nuclear factor NF-kappaB
- STAT3
signal transducer and activator of transcription 3
- MCL-1
myeloid cell leukemia-1
- ERBB2/HER2/NEU
erb-B2 receptor tyrosine kinase 2
- ER alpha
estrogen receptor α
- PKB/AKT
protein kinase B
- PTM
post-translational modification
- CDK
Cyclin-dependent kinase
- MAPK
mitogen-activated protein kinase
- GSK-3β
glycogen synthase kinase 3 beta
- PPIase
peptidyl-prolyl cis/trans isomerase
- NCOR2/SMRT
nuclear receptor co-repressor-2
- SRC-3
steroid receptor coactivator 3
- PR
progesterone receptor
- EMT
epithelial-to-mesenchymal transition
- miR
micro-RNA
- PLK1
Polo-like kinase 1
- PKA
protein kinase A
- PKC
protein kinase C
- AURKA
Aurora kinase A
- DAPK1
death associated protein kinase 1
- MLK3
mixed-lineage kinase 3
- MAP3K
mitogen-activated protein kinase kinase kinase
- SENP1
SUMO1/sentrin specific peptidase 1
- p63
tumor protein p63
- ADAR1
RNA-specific adenosine deaminase 1
- pRB
Retinoblastoma protein
- FBXW7
F-Box and WD40 repeat domain containing 7
- CA
centrosome amplification
- HRAS
Harvey rat sarcoma viral oncogene homolog
- BRCA1
breast cancer 1
- PI3K
phosphatidylinositol-4,5-biphosphate 3-kinase
- ERK1/2
extracellular signal-regulated kinase 1 and 2
- FAK
focal adhesion kinase
- PTP-PEST
protein tyrosine phosphatase, non-receptor type 12
- FOXC1
Forkhead box C1
- mTOR
mammalian target of rapamycin
- FAS
fatty acid synthase
- CSC
cancer stem cells
- ATRA
all-trans retinoic acid
- APL
acute promyelocytic leukemia
- RAR
retinoic acid receptor
- PML
promyelocytic leukemia protein
Footnotes
Edited by J-C Marine
The authors declare no conflict of interest.
References
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-tieulent J, Jemal A. Global Cancer Statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- Steeg P. Targeting metastasis. Nat Rev Cancer 2016; 16: 201–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZZ. The isomerase PIN1 controls numerous cancer-driving pathways and is a unique drug target. Nat Rev Cancer 2016; 16: 463–478. [DOI] [PubMed] [Google Scholar]
- Zacchi P, Gostissa M, Uchida T, Salvagno C, Avolio F, Volinia S et al. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature 2002; 419: 853–857. [DOI] [PubMed] [Google Scholar]
- Mantovani F, Piazza S, Gostissa M, Strano S, Zacchi P, Mantovani R et al. Pin1 links the activities of c-Abl and p300 in regulating p73 function. Mol Cell 2004; 14: 625–636. [DOI] [PubMed] [Google Scholar]
- Mantovani F, Tocco F, Girardini J, Smith P, Gasco M, Lu X et al. The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. Nat Struct Mol Biol 2007; 14: 912–920. [DOI] [PubMed] [Google Scholar]
- Sorrentino G, Mioni M, Giorgi C, Ruggeri N, Pinton P, Moll U et al. The prolyl-isomerase Pin1 activates the mitochondrial death program of p53. Cell Death Differ 2013; 20: 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani F, Zannini A, Rustighi A, Del Sal G. Interaction of p53 with prolyl isomerases: Healthy and unhealthy relationships. Biochim Biophys Acta 2015; 1850: 2048–2060. [DOI] [PubMed] [Google Scholar]
- Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V et al. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J 2001; 20: 3459–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam PB, Burga LN, Wu BP, Hofstatter EW, Lu KP, Wulf GM. Prolyl isomerase Pin1 is highly expressed in Her2-positive breast cancer and regulates erbB2 protein stability. Mol Cancer 2008; 7: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Huo L, Yang JY, Xia W, Wei Y, Liao Y et al. Down-regulation of myeloid cell leukemia-1 through inhibiting Erk/Pin 1 pathway by sorafenib facilitates chemosensitization in breast cancer. Cancer Res 2008; 68: 6109–6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustighi A, Tiberi L, Soldano A, Napoli M, Nuciforo P, Rosato A et al. The prolyl-isomerase Pin1 is a Notch1 target that enhances Notch1 activation in cancer. Nat Cell Biol 2009; 11: 133–142. [DOI] [PubMed] [Google Scholar]
- Girardini JE, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E et al. A Pin1/Mutant p53 Axis Promotes Aggressiveness in Breast Cancer. Cancer Cell 2011; 20: 79–91. [DOI] [PubMed] [Google Scholar]
- Rustighi A, Zannini A, Tiberi L, Sommaggio R, Piazza S, Sorrentino G et al. Prolyl-isomerase Pin1 controls normal and cancer stem cells of the breast. EMBO Mol Med 2014; 6: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell AS, Pelz C, Wang X, Daniel CJ, Wang Z, Su Y et al. Pin1 regulates the dynamics of c-Myc DNA binding to facilitate target gene regulation and oncogenesis. Mol Cell Biol 2013; 33: 2930–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryo A, Liou Y-C, Wulf G, Nakamura M, Lee SW, Lu KP. PIN1 is an E2F target gene essential for Neu/Ras-induced transformation of mammary epithelial cells. Mol Cell Biol 2002; 22: 5281–5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Hunter T. Prolyl isomerase Pin1 in cancer. Cell Res 2014; 24: 1033–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf G, Garg P, Liou Y-C, Iglehart D, Lu KP. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. EMBO J 2004; 23: 3397–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Wagih O, Bader GD. The mutational landscape of phosphorylation signaling in cancer. Sci Rep 2013; 3: 2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutkowski M, Bernhardt A, Zhou XZ, Shen M, Reimer U, Rahfeld JU et al. Role of phosphorylation in determining the backbone dynamics of the serine/threonine-proline motif and Pin1 substrate recognition. Biochemistry 1998; 37: 5566–5575. [DOI] [PubMed] [Google Scholar]
- Gjorevski N, Nelson CM. Integrated morphodynamic signalling of the mammary gland. Nat Rev Mol Cell Biol 2011; 12: 581–593. [DOI] [PubMed] [Google Scholar]
- Fujimori F, Takahashi K, Uchida C, Uchida T. Mice lacking Pin1 develop normally, but are defective in entering cell cycle from G(0) arrest. Biochem Biophys Res Commun 1999; 265: 658–663. [DOI] [PubMed] [Google Scholar]
- Liou Y-C, Ryo A, Huang H-K, Lu P-J, Bronson R, Fujimori F et al. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc Natl Acad Sci USA 2002; 99: 1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ML, Gong C, Chen CH, Lee DY, Hu H, Huang P et al. Prolyl isomerase pin1 acts downstream of mir200c to promote cancer stem-like cell traits in breast cancer. Cancer Res 2014; 74: 3603–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeselsohn R, Brown NE, Arendt L, Klebba I, Hu MG, Kuperwasser C et al. Cyclin D1 kinase activity is required for the self-renewal of mammary stem and progenitor cells that are targets of MMTV-ErbB2 tumorigenesis. Cancer Cell 2010; 17: 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajbhandari P, Finn G, Solodin NM, Singarapu KK, Sahu SC, Markley JL et al. Regulation of Estrogen Receptor N-Terminus Conformation and Function by Peptidyl Prolyl Isomerase Pin1. Mol Cell Biol 2012; 32: 445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajbhandari P, Schalper K a, Solodin NM, Ellison-Zelski SJ, Ping Lu K, Rimm DL et al. Pin1 modulates ERα levels in breast cancer through inhibition of phosphorylation-dependent ubiquitination and degradation. Oncogene 2014; 33: 1438–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetti C, Caligiuri I, Toffoli G, Giordano A, Rizzolio F. The prolyl isomerase Pin1 acts synergistically with CDK2 to regulate the basal activity of estrogen receptor? in breast cancer. PLoS One 2013; 8: e55355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanya KJ, Liu Y, Means AR, Kao HY. Cdk2 and Pin1 negatively regulate the transcriptional corepressor SMRT. J Cell Biol 2008; 183: 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P, Wu R-C, Sandquist J, Wong J, Tsai SY, Tsai M-J et al. Peptidyl-prolyl isomerase 1 (Pin1) serves as a coactivator of steroid receptor by regulating the activity of phosphorylated steroid receptor coactivator 3 (SRC-3/AIB1). Mol Cell Biol 2005; 25: 9687–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryo A, Wulf G, Lee TH, Lu KP. Pinning down HER2-ER crosstalk in SMRT regulation. Trends Biochem Sci 2009; 34: 162–165. [DOI] [PubMed] [Google Scholar]
- Vargo-Gogola T, Rosen JM. Modelling breast cancer: one size does not fit all. Nat Rev Cancer 2007; 7: 659–672. [DOI] [PubMed] [Google Scholar]
- Luo ML, Gong C, Chen CH, Hu H, Huang P, Zheng M et al. The Rab2A GTPase promotes breast cancer stem cells and tumorigenesis via erk signaling activation. Cell Rep 2015; 11: 111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MR, Choi HS, Heo T-H, Hwang SW, Kang KW. Induction of vascular endothelial growth factor by peptidyl-prolyl isomerase Pin1 in breast cancer cells. Biochem Biophys Res Commun 2008; 369: 547–553. [DOI] [PubMed] [Google Scholar]
- You H, Zheng H, Murray SA, Yu Q, Uchida T, Fan D et al. IGF-1 induces Pin1 expression in promoting cell cycle S-phase entry. J Cell Biochem 2001; 84: 211–216. [DOI] [PubMed] [Google Scholar]
- Han CH, Lu J, Wei Q, Bondy ML, Brewster AM, Yu TK et al. The functional promoter polymorphism (-842G>C) in the PIN1 gene is associated with decreased risk of breast cancer in non-Hispanic white women 55 years and younger. Breast Cancer Res Treat 2010; 122: 243–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerdt F, Yuan J, Saxena K, Martin B, Kappel S, Lindenau C et al. Polo-like kinase 1-mediated phosphorylation stabilizes Pin1 by inhibiting its ubiquitination in human cells. J Biol Chem 2005; 280: 36575–36583. [DOI] [PubMed] [Google Scholar]
- Lee YC, Que J, Chen YC, Lin JT, Liou YC, Liao PC et al. Pin1 acts as a negative regulator of the G2/M transition by interacting with the Aurora-A-Bora complex. J Cell Sci 2013; 126: 4862–4872. [DOI] [PubMed] [Google Scholar]
- Lu PJ, Zhou XZ, Liou YC, Noel JP, Lu KP. Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. J Biol Chem 2002; 277: 2381–2384. [DOI] [PubMed] [Google Scholar]
- Luo Z, Wijeweera A, Oh Y, Liou Y-C, Melamed P. Pin1 facilitates the phosphorylation-dependent ubiquitination of SF-1 to regulate gonadotropin beta-subunit gene transcription. Mol Cell Biol 2010; 30: 745–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Chen CH, Suizu F, Huang P, Schiene-Fischer C, Daum S et al. Death-Associated Protein Kinase 1 Phosphorylates Pin1 and Inhibits Its Prolyl Isomerase Activity and Cellular Function. Mol Cell 2011; 42: 147–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangasamy V, Mishra R, Sondarva G, Das S, Lee TH, Bakowska JC et al. Mixed-lineage kinase 3 phosphorylates prolyl-isomerase Pin1 to regulate its nuclear translocation and cellular function. Proc Natl Acad Sci USA 2012; 109: 8149–8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Chang CC, Lee TH, Luo M, Huang P, Liao PH et al. SENP1 deSUMOylates and regulates pin1 protein activity and cellular function. Cancer Res 2013; 73: 3951–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lufei C, Cao X. Nuclear import of Pin1 is mediated by a novel sequence in the PPIase domain. FEBS Lett 2009; 583: 271–276. [DOI] [PubMed] [Google Scholar]
- Magli A, Angelelli C, Ganassi M, Baruffaldi F, Matafora V, Battini R et al. Proline isomerase pin1 represses terminal differentiation and myocyte enhancer factor 2C function in skeletal muscle cells. J Biol Chem 2010; 285: 34518–34527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanes SD. Prolyl isomerases in gene transcription. Biochim Biophys Acta 2015; 1850: 2017–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, Titus MA, Thapar R. The prolyl isomerase pin1 regulates mRNA levels of genes with short half-lives by targeting specific RNA binding proteins. PLoS One 2014; 9: e85427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z-J, Malter J. Regulation of AU-Rich Element RNA Binding Proteins by Phosphorylation and the Prolyl Isomerase Pin1. Biomolecules 2015; 5: 412–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci R, Brindle J, Paro S, Casadio A, Hempel S, Morrice N et al. Pin1 and WWP2 regulate GluR2 Q/R site RNA editing by ADAR2 with opposing effects. EMBO J 2011; 30: 4211–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinton P, Rimessi A, Marchi S, Orsini F, Migliaccio E, Giorgio M et al. Protein kinase C beta and prolyl isomerase 1 regulate mitochondrial effects of the life-span determinant p66Shc. Science 2007; 315: 659–663. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Rizzolio F, Lucchetti C, Caligiuri I, Marchesi I, Caputo M, Klein-Szanto a J et al. Retinoblastoma tumor-suppressor protein phosphorylation and inactivation depend on direct interaction with Pin1. Cell Death Differ 2012; 19: 1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh ES, Means AR. PIN1, the cell cycle and cancer. Nat Rev Cancer 2007; 7: 381–388. [DOI] [PubMed] [Google Scholar]
- Minella AC, Welcker M, Clurman BE. Ras activity regulates cyclin E degradation by the Fbw7 pathway. Proc Natl Acad Sci USA 2005; 102: 9649–9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min SH, Lau AW, Lee TH, Inuzuka H, Wei S, Huang P et al. Negative Regulation of the Stability and Tumor Suppressor Function of Fbw7 by the Pin1 Prolyl Isomerase. Mol Cell 2012; 46: 771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JY. A clinical overview of centrosome amplification in human cancers. Int J Biol Sci 2011; 7: 1122–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suizu F, Ryo A, Wulf G, Lim J, Lu KP. Pin1 regulates centrosome duplication, and its overexpression induces centrosome amplification, chromosome instability, and oncogenesis. Mol Cell Biol 2006; 26: 1463–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLachlan TK, Somasundaram K, Sgagias M, Shifman Y, Muschel RJ, Cowan KH et al. BRCA1 effects on the cell cycle and the DNA damage response are linked to altered gene expression. J Biol Chem 2000; 275: 2777–2785. [DOI] [PubMed] [Google Scholar]
- Kim MR, Choi HK, Cho K, Bin, Kim HS, Kang KW. Involvement of Pin1 induction in epithelial-mesenchymal transition of tamoxifen-resistant breast cancer cells. Cancer Sci 2009; 100: 1834–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lufei C, Koh TH, Uchida T, Cao X. Pin1 is required for the Ser727 phosphorylation-dependent Stat3 activity. Oncogene 2007; 26: 7656–7664. [DOI] [PubMed] [Google Scholar]
- Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev 2009; 28: 15–33. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Yang W, Xia Y, Hawke D, Liu DX, Lu Z. Ras-induced and extracellular signal-regulated kinase 1 and 2 phosphorylation-dependent isomerization of protein tyrosine phosphatase (PTP)-PEST by PIN1 promotes FAK dephosphorylation by PTP-PEST. Mol Cell Biol 2011; 31: 4258–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walerych D, Napoli M, Collavin L, Del Sal G. The rebel angel: Mutant p53 as the driving oncogene in breast cancer. Carcinogenesis 2012; 33: 2007–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012; 486: 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Wei Y, Zhou X, Yang J-Y, Dai C, Chen Y-J et al. Peptidyl-prolyl cis/trans isomerase Pin1 is critical for the regulation of PKB/Akt stability and activation phosphorylation. Oncogene 2009; 28: 2436–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ray PS, Sim M-S, Zhou XZ, Lu KP, Lee a V et al. FOXC1 regulates the functions of human basal-like breast cancer cells by activating NF-κB signaling. Oncogene 2012; 31: 4798–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 2013; 13: 714–726. [DOI] [PubMed] [Google Scholar]
- Nassar D, Blanpain C. Cancer Stem Cells: Basic Concepts and Therapeutic Implications. Annu Rev Pathol Mech Dis 2016; 11: 47–76. [DOI] [PubMed] [Google Scholar]
- Prat A, Baselga J. The role of hormonal therapy in the management of hormonal-receptor-positive breast cancer with co-expression of HER2. Nat Clin Pract Oncol 2008; 5: 531–542. [DOI] [PubMed] [Google Scholar]
- Khanal P, Yun HJ, Lim SC, Ahn SG, Yoon HE, Kang KW et al. Proyl isomerase Pin1 facilitates ubiquitin-mediated degradation of cyclin-dependent kinase 10 to induce tamoxifen resistance in breast cancer cells. Oncogene 2012; 31: 3845–3856. [DOI] [PubMed] [Google Scholar]
- Lee KY, Lee JW, Nam HJ, Shim JH, Song Y, Kang KW. PI3-Kinase/p38 kinase-dependent E2F1 activation is critical for pin1 induction in tamoxifen-resistant breast cancer cells. Mol Cells 2011; 32: 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes BT, Bailey ML, Brandl CJ, Shilton BH, Litchfield DW. Non-catalytic participation of the pin1 peptidyl-prolyl isomerase domain in target binding. Front Physiol 2013; 4: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JD, Potter A. Pin1 inhibitors: Pitfalls, progress and cellular pharmacology. Bioorganic Med. Chem. Lett 2013; 23: 4283–4291. [DOI] [PubMed] [Google Scholar]
- Kiran Aithal B, Sunil Kumar MR, Nageshwar Rao B, Udupa N, Satish Rao BS. Juglone, a naphthoquinone from walnut, exerts cytotoxic and genotoxic effects against cultured melanoma tumor cells. Cell Biol Int 2009; 33: 1039–1049. [DOI] [PubMed] [Google Scholar]
- Wei S, Kozono S, Kats L, Nechama M, Li W, Guarnerio J et al. Active Pin1 is a key target of all-trans retinoic acid in acute promyelocytic leukemia and breast cancer. Nat Med 2015; 21: 457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Greenwood A, Binder L, Bigio EH, Denial S, Nicholson L et al. Proline isomer-specific antibodies reveal the early pathogenic tau conformation in Alzheimer's disease. Cell 2012; 149: 232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]