Abstract
Over the past decade, endoplasmic reticulum (ER) stress has emerged as an important mechanism involved in the pathogenesis of cardiovascular diseases including heart failure. Cardiac therapy based on ER stress modulation is viewed as a promising avenue toward effective therapies for the diseased heart. Here, we tested whether sirtuin-1 (SIRT1), a NAD+-dependent deacetylase, participates in modulating ER stress response in the heart. Using cardiomyocytes and adult-inducible SIRT1 knockout mice, we demonstrate that SIRT1 inhibition or deficiency increases ER stress-induced cardiac injury, whereas activation of SIRT1 by the SIRT1-activating compound STAC-3 is protective. Analysis of the expression of markers of the three main branches of the unfolded protein response (i.e., PERK/eIF2α, ATF6 and IRE1) showed that SIRT1 protects cardiomyocytes from ER stress-induced apoptosis by attenuating PERK/eIF2α pathway activation. We also present evidence that SIRT1 physically interacts with and deacetylates eIF2α. Mass spectrometry analysis identified lysines K141 and K143 as the acetylation sites on eIF2α targeted by SIRT1. Furthermore, mutation of K143 to arginine to mimic eIF2α deacetylation confers protection against ER stress-induced apoptosis. Collectively, our findings indicate that eIF2α deacetylation on lysine K143 by SIRT1 is a novel regulatory mechanism for protecting cardiac cells from ER stress and suggest that activation of SIRT1 has potential as a therapeutic approach to protect the heart against ER stress-induced injury.
The endoplasmic reticulum (ER) is the site for synthesis, folding and quality control of secreted and membrane proteins. Impairment of ER function in response to stresses such as disruption of calcium homeostasis or ischemia causes the accumulation of misfolded proteins in the ER lumen, resulting in ER stress.1 When ER stress occurs, the unfolded protein response (UPR) is activated to restore normal ER function. The UPR is initiated by activation of three proximal sensors, namely PERK, ATF6 and IRE1, which lead to eIF2α phosphorylation to attenuate global protein synthesis and to the transcription of UPR target genes (e.g., ATF4 and XBP1) to upregulate the expression of ER chaperones and proteins involved in ER-associated protein degradation. Activation of this self-protective pathway gives the cell a chance to restore normal ER function. However, in the case of severe or prolonged ER stress, the mitochondrial apoptotic process is triggered to eliminate damaged cells.2, 3, 4, 5
Over the past two decades, ER stress has emerged as an important mechanism involved in the pathogenesis of human diseases such as diabetes, obesity, neurodegenerative disorders and cancer. More recently, ER stress has also been implicated in cardiac diseases including myocardial infarction, ischemia, dilated cardiomyopathy and heart failure.6 Moderate induction of ER stress response is currently considered as protective, whereas prolongation of the response typically leads to apoptosis and contributes to the maladaptive response to injury. For example, overexpression of UPR-related proteins ATF6, XBP1s or PDI lowers apoptosis and protects the myocardium from I/R injuries.7, 8, 9 Furthermore, extinction of GRP78, the central regulator of UPR, induces more severe cardiac insults upon ischemia,10 whereas knockout of the pro-apoptotic factor CHOP protects mice against pressure overload-induced heart failure.11 Besides, chemical chaperones, such as 4-phenylbutyrate or tauroursodeoxycholic acid, protect the heart against isoproterenol (ISO)- or obesity-induced dysfunction by alleviating ER stress.12, 13 Therefore, cardiac therapy based on ER stress modulation could be promising to promote beneficial adaptations and avoid apoptosis.
Sirtuin-1 (SIRT1), a NAD+-dependent lysine deacetylase, is activated in response to various cellular stress14 and is cardioprotective in the context of aging, hypertrophy and myocardial infarction.15, 16, 17 However, whether and how SIRT1 modulates ER stress response in the heart has not been elucidated yet. Thus, we tested the role of SIRT1 in the modulation of the cardiac ER stress response in vitro and in vivo using adult-inducible SIRT1 knockout mice. We report that SIRT1 protects cardiomyocytes against ER stress-induced apoptosis by modulating the PERK/eIF2α pathway of the UPR at least through eIF2α deacetylation on lysine residue K143.
Results
SIRT1 deficiency increases cardiac dysfunction induced by ER stress
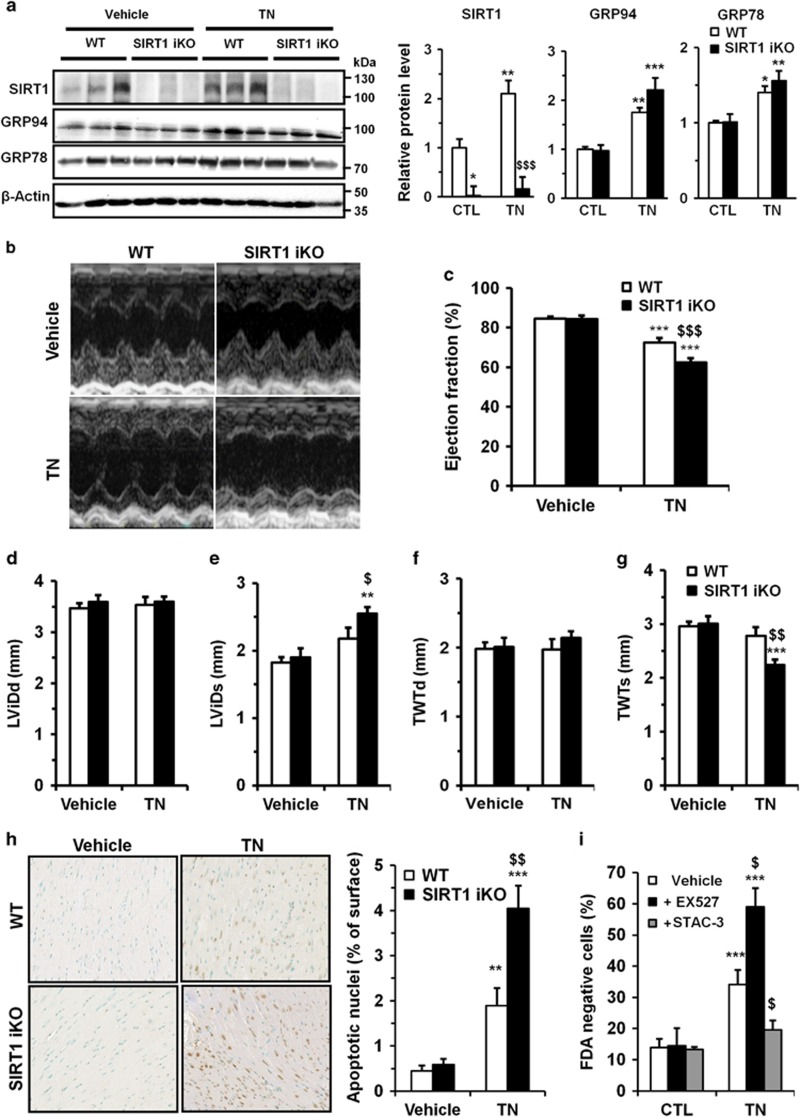
To investigate the functional role of SIRT1 in the context of cardiac ER stress, adult-inducible SIRT1 knockout (SIRT1 iKO) and control mice were injected with the ER stressor tunicamycin (TN, 2 mg/kg body weight). TN provoked an ER stress, as demonstrated by the upregulation of GRP94 and GRP78, which was not significantly modified by SIRT1 deletion (Figure 1a). Intraperitoneal challenge with TN did not affect organ weights except for liver (Supplementary Table S1). ER stress was accompanied by a decrease in ejection fraction (EF; Figures 1b and c) and fractional shortening (FS) (Supplementary Table S2). By contrast, the heart rate (Supplementary Table S2), left ventricular internal diameters (Figures 1d and e) or wall thickness (Figures 1f and g) were not affected. The deletion of SIRT1 exacerbated the deterioration of cardiac function induced by ER stress, with a significant decrease in EF and FS (Figures 1b, c and Supplementary Table S2), an increase of left ventricular systolic diameter (Figure 1e) and a decrease of systolic total wall thickness (Figure 1g). Given that ER stress is known to elicit myocardial damages through induction of apoptosis,18 the effects of SIRT1 deletion on ER stress-induced cardiomyocyte apoptosis was measured by terminal deoxyribonucleotidyl transferase-mediated dUTP-digoxigenin nick-end labeling (TUNEL) assay on mouse myocardium. The number of apoptotic cardiomyocytes induced by TN injection was significantly increased in myocardium of SIRT1 iKO mice (Figure 1h). Together, these data support a model in which activation of SIRT1 during ER stress is cardioprotective.
Figure 1.
Knockout of SIRT1 sensitizes mice to ER stress-induced cardiac injury. (a) WT and SIRT1 iKO mice were injected i.p. with TN (2 mg/kg) or vehicle (150 mM dextrose) for 16 h and the levels of SIRT1 and ER stress markers GRP94 and GRP78 were analyzed by western blot in heart tissue. β-Actin was used as loading control. Relative expression of proteins (normalized to WT CTL) is presented in bar graphs. Error bars, S.E.M. (n=3). (b-g) Echocardiographic data were recorded 72 h after vehicle or TN treatment. (b) Representative images obtained by transthoracic echocardiography. (c) EF of WT and SIRT1 iKO mice 72 h after vehicle or TN treatment. (n=9). (d) Left ventricular internal dimension (diastole), LVIDd, in vehicle or TN-treated mice (n=9). (e) Left ventricular internal dimension (systole), LVIDs, in vehicle or TN-treated mice (n=9). (f) Total wall thickness (diastole), TWTd, in vehicle or TN-treated mice (n=9). (g) Total wall thickness (systole), TWTs, in vehicle or TN-treated mice (n=9). Results are presented as mean±S.E.M. *P<0.05, **P<0.01, ***P<0.005 versus respective control. $P<0.05, $$P<0.01, $$$P<0.005 versus WT TN. (h) WT and SIRT1 iKO mice were injected i.p. with TN (2 mg/kg) or with vehicle (150 mM dextrose) for 72 h and subjected to TUNEL assay. Representative micrographs showing apoptotic cardiomyocyte nuclei (brown) are presented (scale bar=40 μm). Quantification of apoptotic nuclei is showed in bar graph. Error bars, S.E.M. (n=6). (i) Freshly isolated ARVMs were left untreated or treated with 10 μg/ml TN for 24 h±10 μM EX527 or 1 μM STAC-3 pretreatment and cell viability was determined. Results presented in bar graph are expressed as mean±S.E.M. of percentages of dead cells (FDA-negative cells, n=6). **P<0.01, ***P<0.005 versus control. $P<0.05, $$P<0.01 versus TN alone
Activation of SIRT1 protects adult cardiomyocytes from ER stress-induced cell death
To establish that TN directly targets cardiomyocytes to induce cell death, we tested the effect of this ER stressor on the viability of adult rat ventricular cardiomyocytes (ARVMs) with or without the SIRT1-specific inhibitor EX527. TN treatment (10 μg/ml, 24 h) induced an increase of cell death (Figure 1i and Supplementary Figure S1), which was higher when SIRT1 was inhibited, corroborating the data obtained in vivo. By contrast, pretreatment of ARVM with STAC-3, a SIRT1-specific activator,19 almost totally abrogated cell death (Figure 1i), indicating that SIRT1 activation protects adult cardiomyocytes from ER stress-induced cell death. These results, combined with those obtained in vivo, show that activation of SIRT1 has a protective role against ER stress-induced cardiomyocyte death.
SIRT1 regulates ER stress-induced mitochondrial apoptosis in cardiac cells
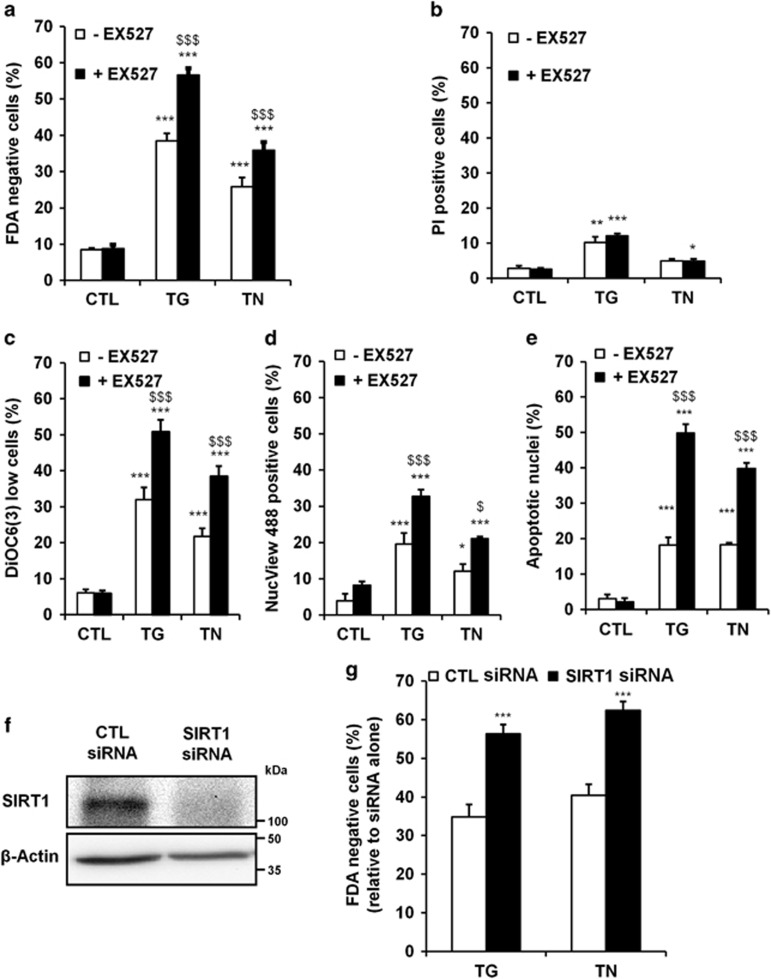
To deeply investigate the mechanism underlying the modulating activity of SIRT1 against ER stress-induced apoptosis, we developed a cell culture model of ER stress induction using H9c2 cardiac cell line. As expected, in response to the ER stressors thapsigargin (TG) or TN, an increase in the level of the ER stress markers GRP78 and CHOP was observed (Supplementary Figure S2a). Inhibition of SIRT1 by EX527 resulted in sensitization of cells to ER stress-induced cell death as shown in Figure 2a and Supplementary Figure S3. We then characterized the mode of cell death triggered by SIRT1 inhibition upon ER stress. In response to TG and TN, with or without SIRT1 inhibitor, the percentage of cells with compromised plasma membrane integrity remained very low (Figure 2b), excluding the involvement of necrosis. By contrast, ΔΨm loss (Figure 2c), caspase-3 activation (Figure 2d) and DNA fragmentation (Figure 2e and Supplementary Figure S2b) induced by ER stress were markedly increased by SIRT1 inhibitor. The general caspase inhibitor ZVAD-fmk and specific caspase-9 or caspase-3 inhibitors (Ac-LEHD-cmk and Ac-DEVD-cmk, respectively) conferred protection against TG- and TN-induced cell death both in the absence or the presence of EX527 (Supplementary Figures S2c and d), confirming that caspase are involved in our model. Moreover, depletion of SIRT1 with siRNA (Figure 2f) significantly enhanced cell death induced by TG and TN (Figure 2g). Therefore, SIRT1 inhibition or knockdown exacerbates the mitochondrial-regulated apoptotic pathway triggered by ER stress. In addition, TN-treated cells exhibited ultrastructural features of apoptosis and ER stress, including nuclear condensation/fragmentation, multiple contact sites between ER and mitochondria, ER dilatation and mitochondria enlargement (Supplementary Figures S5a and b). In cells incubated with the combination of the ER stressor and the SIRT1 inhibitor, the ER appeared swollen and mitochondria were rounded, considerably enlarged and devoid of cristae, indicating that organelle morphology was dramatically altered. Taken together, our results show that inhibition of SIRT1 enhanced ER stress-induced apoptosis in H9c2 cells. As these observations are similar to those observed in vivo, H9c2 cardiomyoblasts constitute a valuable cell model to study the mechanism of SIRT1-mediated protection against severe ER stress. We also tested whether the protective role of SIRT1 in response to ER stress can be mediated through autophagy modulation. TG and TN enhanced the autophagic flux as evidenced by the accumulation of LC3-II (Supplementary Figure S4). However, no further modulation of autophagy was observed when SIRT1 was inhibited.
Figure 2.
SIRT1 protects cardiac cells from ER stress-induced apoptosis. (a) The percentage of dead cells (FDA-negative cells) was assessed by flow cytometry after treatment of H9c2 cells for 48 h with TG (5 μM) or TN (10 μg/ml) in the absence or presence of SIRT1 inhibitor EX527 (10 μM). (b-d) Same as (a) except that cells were stained with (b) PI to determine the percentage of necrosis/late apoptosis (PI high cells), (c) DiOC6(3) to estimate ΔΨm loss (DiOC6(3) low cells), (d) DEVD-NucView™ 488 to measure active caspase-3 (NucView 488-positive cells), (e) DAPI to analyze apoptotic nuclei. (f) H9c2 cells were transfected with SIRT1 or control siRNA and SIRT1 expression was assessed after 24 h by western blot. (g) H9c2 cells were transfected with SIRT1 or control siRNA for 24 h then treated for 48 h with TG or TN and cell death (FDA-negative cells) was measured by flow cytometry. Data in the bar graphs represent mean±S.E.M. (n=4). *P<0.05, **P<0.01, ***P<0.005 versus control. $P<0.05, $$$P<0.005 versus TG or TN alone
SIRT1 regulates the UPR by modulating PERK/eIF2α pathway signaling
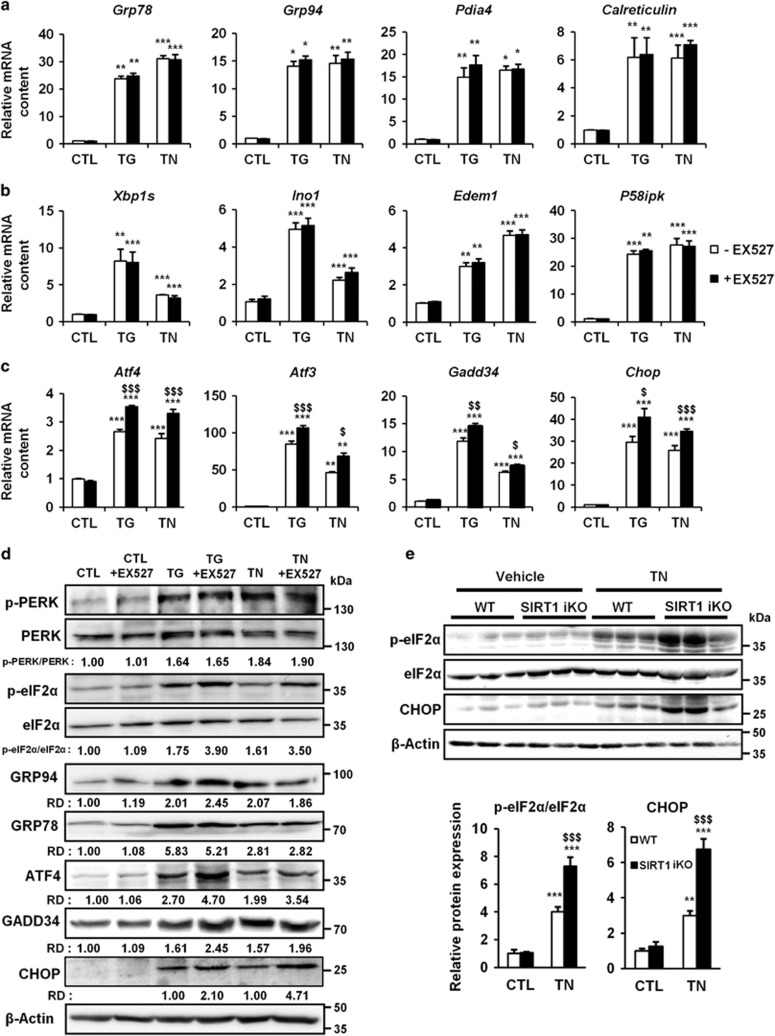
To elucidate the mechanisms underlying the cardioprotective effects of SIRT1 in severe ER stress, we measured the expression of different markers of the three branches of the UPR (i.e., PERK, ATF6 and IRE1) in H9c2 cells. As expected, TG and TN treatments induced a robust increase in the expression of all members of the UPR tested (Figures 4a–c). The inhibition of SIRT1 did not significantly affect the expression of the members of the ATF6 branch (Figure 4a) or of the IRE1 branch (Figure 4b) induced by ER stressors. By contrast, SIRT1 inhibition further stimulated TG- and TN-mediated transcription of ATF4, ATF3, GADD34 and CHOP genes (Figure 4c), suggesting a hyperactivation of the PERK/eIF2α pathway. To consolidate our results, we measured the protein level of members of the PERK/eIF2α/ATF4/CHOP branch. TG and TN induced an increase in GRP94 and GRP78 protein levels and a rapid phosphorylation of PERK, which were not modified by SIRT1 inhibitor (Figure 4d), suggesting a mechanism in which SIRT1 acts downstream of PERK to regulate this pathway. Interestingly, the phosphorylation of eIF2α and the level of the downstream effectors ATF4, GADD34 and CHOP were higher when SIRT1 was inhibited. In addition, the induction of eIF2α phosphorylation and the upregulation of CHOP expression (Figure 4e, WT mice) were also increased in the myocardium of SIRT1 knockout mice. Collectively, these results indicate that SIRT1 regulates the UPR and attenuates the activation of the PERK/eIF2α pathway in response to ER stress in cardiac cells.
SIRT1 deficiency increases PERK/eIF2α pathway activation and cardiac dysfunction induced by ISO
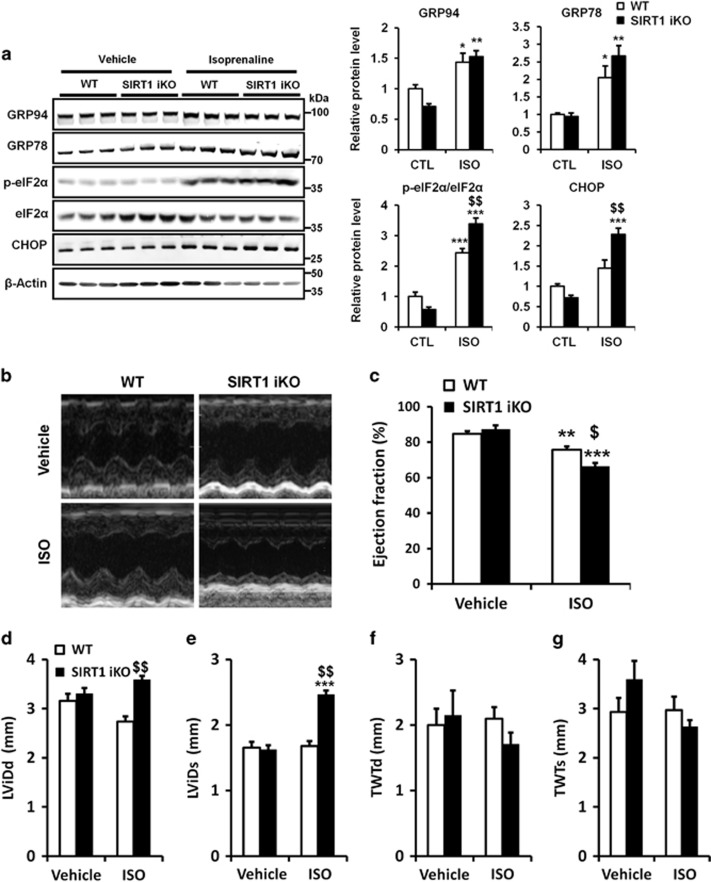
We next investigated whether SIRT1 also modulates the PERK/eIF2α pathway in a pathophysiological model of cardiac injury. SIRT1 iKO mice were injected with ISO, to elicit a catecholaminergic stress, which has been well documented to induce cardiac dysfunction associated with ER stress.12, 20, 21, 22, 23 WT and SIRT1 iKO mice were subcutaneously injected with ISO (150 mg/kg body weight) and ER stress and heart function were analyzed. ISO injection triggered an ER stress, as demonstrated by the upregulation of GRP94 and GRP78. However, no difference in GRP protein levels were observed when comparing WT with SIRT1 iKO samples (Figure 4a), whereas eIF2α phosphorylation and CHOP expression levels were increased. As the expression of the ER chaperones GRP78 and GRP94 are induced by the ATF6 pathway,24 these results suggest that SIRT1 regulates the PERK/eIF2α pathway but not the ATF6 pathway in response to ISO, as observed for TN-induced ER stress (Figures 1a and 3). Catecholaminergic stress was accompanied by a significant impairment of cardiac function as judged by decrease in EF (Figures 4b, c) and FS (Supplementary Table S3) induced by ISO. The heart rate (Supplementary Table S3), left ventricular internal diameters (Figures 4d and e) or wall thickness (Figures 4f and g) were not affected. After SIRT1 deletion, we observed a more important decrease in EF and FS (Figures 4b, c and Supplementary Table S3) and an increase of left ventricular internal diameters (Figures 4d and e), showing that SIRT1 deficiency exacerbates the cardiac dysfunction induced by ISO. Similarly to what we described above for TN-induced ER stress, the activation of SIRT1 is cardioprotective against ISO-induced cardiac injury at least in part by attenuating PERK/eIF2α pathway activation.
Figure 3.
SIRT1 regulates UPR by modulating PERK/eIF2α pathway activation. (a–c) H9c2 cells were left untreated or treated with TG (5 μM) or TN (10 μg/ml) ±EX527 (10 μM) and the relative mRNA levels of UPR target genes were quantified by qPCR and expressed as fold change over untreated controls. Values represent mean±S.E.M. (n=6). **P<0.01, ***P<0.005 versus control. $P<0.05, $$P<0.01, $$$P<0.05 versus TG or TN alone. Members of the (a) ATF6 pathway, (b) IRE1 pathway and (c) PERK/eIF2α pathway. (d) Cells were left untreated or treated with TG (5 μM) or TN (10 μg/ml) for 15 min (p-PERK and PERK), 90 min (p-eIF2α and eIF2α) or 24 h (GADD34, ATF4 and CHOP) ±EX527 (10 μM). Representative western blots of the level of members of the PERK/eIF2α axis are presented. β-Actin was used as loading control. (e) WT and SIRT1 iKO mice were injected i.p. with TN (2 mg/kg) or with vehicle (150 mM dextrose) for 72 h, proteins from the left ventricle of the heart were analyzed. Representative western blot of the level of members of the PERK/eIF2α axis are presented. Bar graphs summarize changes in protein expression from three independent measurements. Data are represented as fold change over untreated WT mice. **P<0.01, ***P<0.005 versus WT CTL, $$$P<0.05 versus WT TN. Error bars, S.E.M.
Figure 4.
Knockout of SIRT1 exacerbates PERK/eIF2α pathway activation and cardiac injury induced by ISO. (a) WT and SIRT1 iKO mice were injected subcutaneously with ISO (150 mg/kg) or vehicle (NaCl 0,9%) for 6 h and the levels of ER stress markers GRP94, GRP78, p-eIF2α, eIF2α and CHOP were analyzed by western blot in heart tissue. β-Actin was used as loading control. Relative expression of proteins (normalized to WT CTL) is presented in bar graphs. Error bars, S.E.M. (n=3). (b-g) Echocardiographic data were recorded 48 h after vehicle (NaCl 0,9%) or ISO treatment. (b) Representative images obtained by transthoracic echocardiography. (c) EF of WT and SIRT1 iKO mice after vehicle or ISO treatment. (n=5). (d) Left ventricular internal dimension (diastole), LVIDd, in vehicle or ISO-treated mice (n=5). (e) Left ventricular internal dimension (systole), LVIDs, in vehicle or ISO-treated mice (n= 5). (f) Total wall thickness (diastole), TWTd, in vehicle or ISO-treated mice (n=5). (g) Total wall thickness (systole), TWTs, in vehicle or ISO-treated mice (n=5). Results are presented as mean±S.E.M. **P<0.01, ***P<0.005 versus respective control. $P<0.05, $$P<0.01 versus WT ISO
SIRT1 interacts with and deacetylates eIF2α
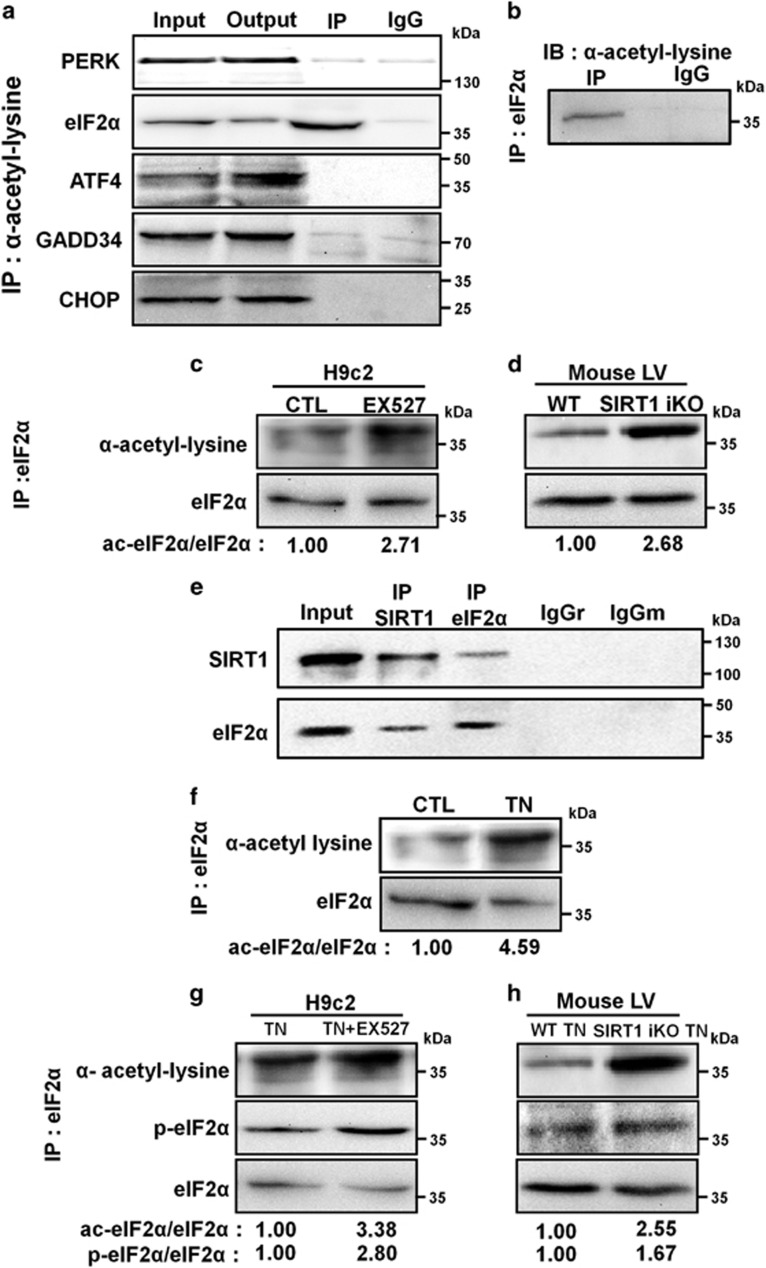
SIRT1 is known to regulate protein activity through deacetylation on lysine residues. To investigate the mechanism by which SIRT1 attenuates the PERK/eIF2α pathway in response to ER stress, proteins acetylated on lysine residues were pulled down from H9c2 cell lysates. eIF2α was present in anti-acetyl lysine immunoprecipitates, whereas PERK, ATF4, GADD34 and CHOP were not detected, suggesting that among the members of the PERK/eIF2α axis tested, eIF2α is the only lysine-acetylated protein (Figure 5a). Reciprocal immunoprecipitation confirmed the acetylation of eIF2α on lysine residues (Figure 5b). These data identify acetylation as a previously unknown post-translational modification of eIF2α. The inhibition of SIRT1 greatly increased the acetylation of eIF2α as shown in Figure 5c. The acetylation level of eIF2α was also evaluated in heart tissue from WT or SIRT1 iKO mice, and we found a more pronounced acetylation of eIF2α when SIRT1 was deleted (Figure 5d). To determine whether SIRT1 and eIF2α physically interact with each other in cardiac cells, co-immunoprecipitation experiments were carried out. Immunoprecipitation of endogenous SIRT1 from H9c2 cell lysates co-precipitated eIF2α (Figure 5e). The reverse experiment, immunoprecipitating endogenous eIF2α and immunoblotting for SIRT1, confirmed their physical interaction. We next investigated whether acetylation of eIF2α was modulated by SIRT1 in response to ER stress. ER stress induced by TN increased the level of eIF2α acetylation (Figure 5f). This lysine acetylation was more important when SIRT1 was inhibited by EX527 or genetically deleted (Figures 5g and h), indicating that SIRT1 limits the acetylation level of eIF2α in response to ER stress in vitro and in vivo. Interestingly, in parallel to acetylation, the level of phosphorylation of eIF2α was also increased upon ER stress when SIRT1 was inhibited or deleted, suggesting a dynamic interplay between these two post-translational modifications. Collectively, these results showed that eIF2α is acetylated on one or more lysine residues and that this acetylation is regulated, at least in part, by SIRT1 during ER stress response.
Figure 5.
SIRT1 physically interacts with and deacetylates eIF2α. (a) Immunoprecipitation of acetylated proteins from H9c2 lysate followed by immunoblotting with the indicated antibodies. Input, input fraction; Output, supernatant after immunoprecipitation; IP, immunoprecipitate; IgG: negative control. (b) eIF2α was immunoprecipitated from H9c2 lysate and its level of acetylation was analyzed by immunoblotting with anti-acetyl-lysine antibody. (c) eIF2α was immunoprecipitated from control or H9c2 cells treated with EX527 (10 μM, 4 h) and its level of acetylation was determined by immunoblotting with anti-acetyl-lysine antibody. The ratio of acetylated versus total eIF2α is presented relative to control. (d) The level of acetylation of eIF2α was determined from immunoprecipitates of left ventricle extracts of WT or SIRT1 iKO mice. The ratio of acetylated versus total eIF2α is presented relative to WT mice. (e) The physical interaction between endogenous SIRT1 and eIF2α was demonstrated by co-immunoprecipitation. SIRT1 was precipitated from H9c2 lysate with anti-SIRT1 antibody and blotted with anti-eIF2α antibody, and vice versa. Negative controls: IgGr, rabbit IgG, IgGm, mouse IgG. (f) eIF2α was immunoprecipitated from control or H9c2 cells treated with TN (10 μg/ml, 4 h) and its level of acetylation was determined by immunoblotting with anti-acetyl-lysine antibody. Ratios of acetylated versus total eIF2α are presented relative to control. (g) H9c2 cells were treated with TN (10 μg/ml, 4 h) ±EX527 (10 μM), eIF2α was immunoprecipitated and its levels of acetylation and phosphorylation were analyzed. Ratios of acetylated and phosphorylated versus total eIF2α are presented relative to TN alone. (h) WT and SIRT1 iKO mice were injected with TN (2 mg/kg), eIF2α was immunoprecipitated from left ventricle and its levels of acetylation and phosphorylation were analyzed. Ratios of acetylated or phosphorylated versus total eIF2α are presented relative to WT mice
SIRT1 deacetylates eIF2α on lysine residues K141 and K143
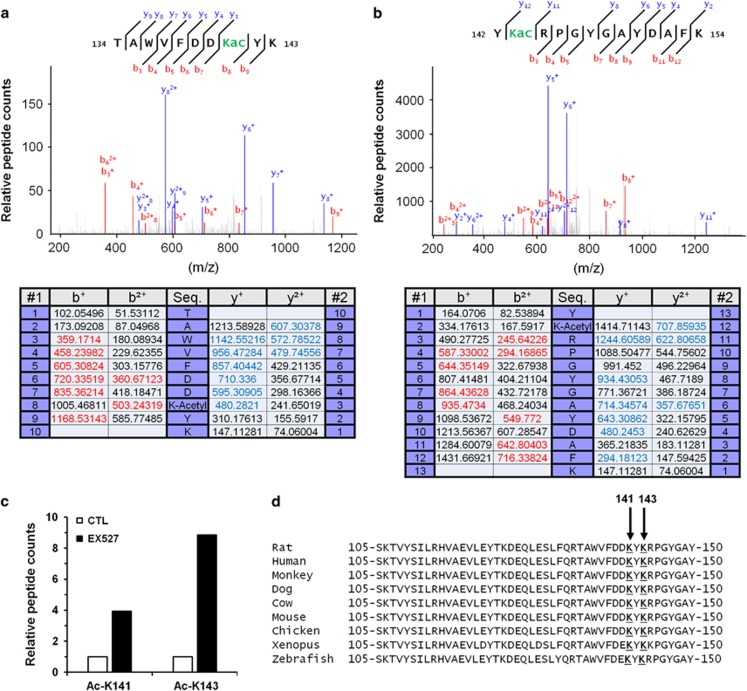
To identify eIF2α acetylation sites, eIF2α was immunoprecipitated from H9c2 cell lysate and analyzed by nanoLC-MS/MS. Analysis revealed different trypsin-cleaved peptides near K141 (peptides TAWVFDDK and TAWVFDDKYK, Figure 6a) and K143 (peptides RPGYGAYDAFK and YKRPGYGAYDAFK, Figure 6b) residues. Moreover, on each peptide an increase of 42 011 Dalton, corresponding to the addition of one acetyl group, was found in the mass of lysine K141 and K143 (Figures 6a and b). These results indicated that eIF2α is acetylated at least on K141 and K143 residues. In addition, inhibition of SIRT1 by EX527 increased the quantity of acetylated peptides on K141 and K143 residues, suggesting that SIRT1 deacetylates these two residues (Figure 6c). As shown in Figure 6d, the protein sequence of eIF2α is highly conserved among species and specifically the K141 and K143 residues.
Figure 6.
Lysines K141 and K143 are the acetylation sites on eIF2α targeted by SIRT1. (a) NanoLC-MS/MS spectrum of acetylated eIF2α peptides containing lysine K141. (b) NanoLC-MS/MS spectrum of acetylated eIF2α peptides containing lysine K143. (c) Relative acetylated K141 or K143 peptide count in control or EX527 (10 μM) treated cells. (d) Sequence alignment of the region surrounding K141 and K143 residues of eIF2α
Loss of eIF2α acetylation at lysine K143 reduces ER stress-induced apoptosis
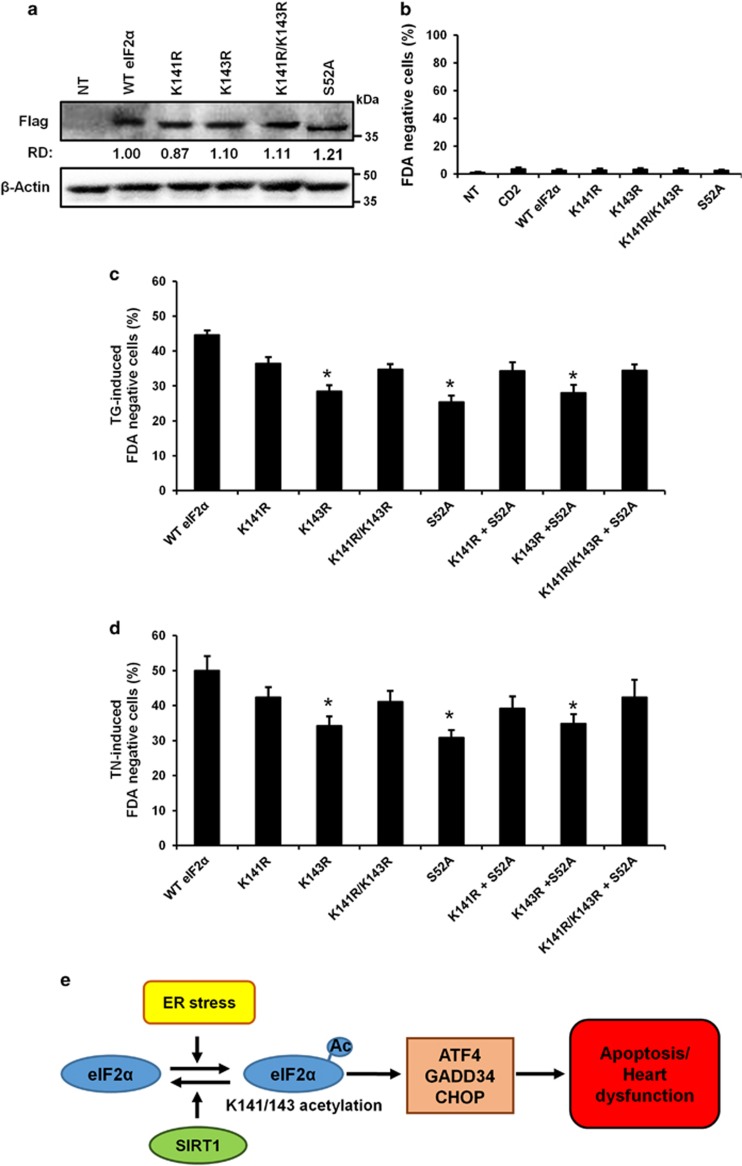
To investigate a causative role of eIF2α acetylation in ER stress-induced apoptosis, we generated acetylation-defective mutants of eIF2α in which either lysine 141 (K141R) or 143 (K143R) or both residues (K141R/K143R) were replaced with arginine. As shown in Figure 7, Flag-tagged eIF2α wild-type or mutants were expressed at similar levels and showed no discernible effect on basal cell viability (Figures 7a and b). Mutation of lysines to deacetylation-mimic arginines reduced the level of TG- and TN-induced cell death when compared with wild-type eIF2α (Figures 7c and d). However, only the K143R mutant significantly protected cells from ER stress-induced cell death. Interestingly, the level of protection conferred by the K143R mutation was remarkably similar to that provided by the phosphorylation mutant S52A, suggesting a potential relationship between the level of eIF2α phosphorylation at serine 52 and acetylation at lysine 143. This hypothesis is reinforced by the observation that co-transfection of acetylation mutants with the phosphorylation mutant S52A did not show any synergistic or adverse effects on cell death. Altogether, these data demonstrate that acetylation of eIF2α on the K143 residue has a role in the regulation of ER stress-induced cell death in cardiac cells.
Figure 7.
Acetylation of eIF2α on lysine K143 regulates ER stress-induced cell death. (a–c) Wild-type and mutant eIF2α expression plasmids were transfected into H9c2 cells for 24 h, ER stress was induced or not by TG (5 μM) or TN (10 μg/ml) and cell death was analyzed in transfected cells by flow cytometry after 48 h by FDA assay. For acetylation-defective mutants, either lysine 141 (K141R) or 143 (K143R) or both residues (K141R/K143R) were replaced with arginine. For eIF2α phosphorylation-defective mutant, serine 52 (S52A) was replace by alanine. (a) Expression levels of the Flag-tagged eIF2α mutants. β-Actin was used as loading control. (b) Effect of wild-type and mutant eIF2α expression on cell death. (c and d) Effect of wild-type and mutant eIF2α on (c) TG-induced cell death (FDA-negative cells), (d) TN-induced cell death (FDA-negative cells). Results are expressed as mean±S.E.M. (n=6). *P<0.05, versus WT eIF2α. (e) Model of SIRT1 protection against ER stress-induced injury in cardiac cells. Upon ER stress, eIF2α is acetylated on K141/K143 residues leading to induction of cardiac injury through cardiomyocyte apoptosis. The deacetylation of eIF2α by SIRT1 protects cardiomyocytes from ER stress-induced apoptosis and heart dysfunction
Discussion
Recent studies have demonstrated the importance of ER stress in the development of cardiac diseases and have suggested that modulation of ER stress response could be cardioprotective.11, 12, 25 Moreover, increasing evidence suggest that SIRT1, as a master regulator of cellular stress, may be a therapeutic target for the prevention of cardiovascular diseases.26, 27 The main focus of this study was thus to investigate the possible role of SIRT1 as a regulator of ER stress in cardiac cells and the mechanisms by which it regulates ER stress response. We show that ER stress impairs heart function and that SIRT1 inhibition or deletion results in increased ER stress-induced apoptosis and cardiac dysfunction. Conversely, SIRT1 activation by the SIRT1-activating compound STAC-3 protects cardiomyocytes from ER stress-induced cell death. We also provide evidence that SIRT1 protects the heart by modulating the PERK/eIF2α pathway of the UPR at least through deacetylation of eIF2α on lysine K143 (Figure 7e).
It has been shown that a primary function of SIRT1 is to protect against different cardiac stresses.28, 29, 30 Our results show that SIRT1 also has a protective role against acute ER stress induced by TN or ISO in the heart. The increase in SIRT1 expression observed after TN treatment may help protect cardiac cells from excessive ER stress-induced cell death. It has been reported that overexpression of ATF4 increases SIRT1 expression in cancer cell lines,31 suggesting that the upregulation of ATF4 observed in our model in response to ER stress may increase SIRT1 expression.
SIRT1 was previously reported to regulate the IRE1 branch of the UPR through deacetylation of XBP1s in embryonic fibroblasts and its deficiency resulted in a decrease of ER stress-induced apoptosis. The authors proposed that SIRT1 exerts deleterious effects and sensitizes these cells to ER stress-induced apoptosis via repressing XBP1s signaling.32 In our experiments, we did not detect any effect of SIRT1 inhibition on the IRE1/XBP1s axis, but rather on the PERK/eIF2α pathway. In addition, we demonstrated that inhibition, knockdown or genetic deletion of SIRT1 increases cell death triggered by ER stress, indicating that SIRT1 protects cardiomyocytes from severe ER stress. Such discrepancies may reflect cell-type specificities including differentiation state. However, to overcome the shortcomings often associated with cell culture models, we further confirmed the cardioprotective effect of SIRT1 activation against ER stress-induced cell death in ventricular adult cardiomyocytes and in in vivo mouse models of TN- and ISO-induced cardiac dysfunction. Very recently, Guo et al.33 have observed that resveratrol-mediated activation of SIRT1 reduced cardiomyocyte apoptosis induced in a diabetic cardiomyopathy model. They suggested that the protection conferred by SIRT1 may involve attenuation of the PERK, ATF6 and IRE1 pathways of ER stress, but no mechanistic insights were provided. Here, using specific pharmacological and genetics tools, we show in a model in which ER stress is specifically induced, that the protection afforded by SIRT1 occurs through regulation of the PERK/eIF2α pathway of the UPR. Activation of PERK/eIF2α pathway contributes to cell adaptation after initiation of ER stress, mostly by limiting protein translation through eIF2α phosphorylation. Nevertheless, in cells with irrecoverable levels of ER stress, the PERK/eIF2α pathway can induce apoptosis by sustaining the level of expression of the pro-apoptotic factor CHOP.5 This study indicates that inhibition or inactivation of SIRT1 results in hyperactivation of PERK/eIF2α pathway, which is associated with increased expression of CHOP and cardiomyocyte apoptosis. In addition, pharmacological activation of SIRT1 by the selective activator STAC-3,19 provides effective protection against ER stress-induced cell death. These findings suggest that SIRT1, by attenuating PERK/eIF2α pathway activation, reduces the initiation of apoptosis and promotes cell survival thus supporting the therapeutic potential of SIRT1 activators for the treatment of cardiac pathologies associated with ER stress.
The translation initiation factor eIF2α has a critical role in translation and its phosphorylation on serine 51/52 by PERK is considered as a rate-limiting step in translation in response to ER stress.1 Although eIF2α phosphorylation inhibits general protein translation initiation, it is required for selective translation of several mRNAs of stress response proteins including ATF4, which in turn activates the expression of the pro-apoptotic factor CHOP.34 Although phosphorylation of eIF2α has been intensively studied, the understanding of the molecular events regulating its phosphorylation is poorly documented. Biochemical studies have highlighted the importance of the conformational state of eIF2α for phosphorylation,35 raising the hypothesis that an undescribed post-translational modification may regulate eIF2α conformation and its subsequent phosphorylation.36 We demonstrate that eIF2α is acetylated on lysines K141 and K143, that eIF2α physically interacts with SIRT1 and that SIRT1 deacetylates eIF2α on these lysine residues. SIRT1 interaction with eIF2α has been shown in HeLa cancer cells,37 but no data on eIF2α acetylation/deacetylation was reported. In response to ER stress, inhibition of SIRT1 promotes both hyperacetylation of eIF2α and cardiomyocyte death, suggesting that eIF2α acetylation on lysine K141 or K143 may regulate ER stress-induced apoptosis. We demonstrated that blocking lysine K143 acetylation by conservative mutation to arginine reduces ER stress-induced cell death, suggesting that deacetylation of eIF2α on lysine K143 by SIRT1 may be an important novel regulatory mechanism for protecting cardiac cells from severe ER stress.
The activity of numerous targets of SIRT1 is known to be regulated both by deacetylation and by phosphorylation (e.g., PGC-1α, p53, FOXO and Beclin1),14, 38 even if the interplay between these two post-translational modifications is not fully understood. Our observations that acetylation and phosphorylation of eIF2α increase in parallel in response to ER stress when SIRT1 is inhibited or knocked out and that acetylation (K143R) and phosphorylation (S52A) mutants protect cells from ER stress-induced cell death at a similar level suggest that SIRT1, by regulating the level of acetylation of eIF2α, may regulate its level of phosphorylation and thus its activity. Although additional studies are needed to clarify the relationship between acetylation and phosphorylation of eIF2α, our data provide a clue as to how SIRT1 limits PERK/eIF2α pathway activation and ER stress-induced apoptosis.
It has been demonstrated that ER stress and SIRT1 are involved in the development of numerous pathologies such as cardiovascular diseases, diabetes, cancer, neurodegenerative disorders and obesity.27, 39, 40, 41, 42 Hence, it will be interesting to test whether the regulation of ER stress response through SIRT1-mediated deacetylation of eIF2α is also beneficial in these diseases. Finally, the demonstration that SIRT1 activation by STAC-3 protects cardiomyocytes from ER stress-induced cell death provides grounds for the development of therapies based on SIRT1-activating compounds in ER stress-associated cardiac diseases.
Materials and Methods
Animals
C57BL/6J mice carrying both the UBC-Cre-ERT2 and the SIRT1floxΔE4/floxΔE4 alleles (UBC-Cre-ERT2 × SIRT1floxΔE4/floxΔE4) called herein SIRT1 iKO mice were described previously.19, 43 Excision of the exon 4 of SIRT1 was induced by tamoxifen administration at 2–4 months of age (25 mg/kg i.p. daily × 4 days). To study ER stress in vivo, 4 weeks after tamoxifen treatment mice (n=9) were given i.p. injection of TN 2 mg/kg body weight or the equivalent volume of vehicle (Dextrose 150 mM) and were killed 16 h after injection for molecular studies. Echocardiographic parameters were assessed 72 h after injection and mice were then killed for heart histochemical and molecular analysis. To study the role of SIRT1 in a myocardial injury model, mice (n=5 for each group) were given subcutaneous injection of ISO 150 mg/kg body weight or the equivalent volume of vehicle (NaCl 0.9%) twice (at T0 and T5h30) and were killed after 6 h for molecular studies. Echocardiographic parameters were assessed after 2 days of treatment and mice were then killed for heart molecular analysis. All experiments were performed in conformity with the European Community guiding principles in the care and use of animals (Directive 2010/63/EU of the European Parliament). Authorizations to conduct animal experiments were obtained from the French Ministère de l'Agriculture, de la Pêche et de l'Alimentation (no. 92-284, 27 June 2007).
Cell culture and reagents
H9c2 rat cardiomyoblast cell line was purchased from ATCC (Manassas, VA, USA). Cells were cultured in DMEM medium supplemented with 100 U/ml penicillin, 100 mg/ml streptomycin and 10% FBS (Lonza, Levallois, France), at 37 °C under 5% CO2/95% air. ER stress was induced by treatment with TG (5 μM) or TN (10 μg/ml). EX527 was from Tocris (Bristol, UK), STAC-3 was from Santa Cruz (Dallas, TX, USA). Caspase inhibitors were from Bachem AG (Bubendorf, Switzerland). All other chemicals were purchased from Sigma-Aldrich (Lyon, France). DEVD-NucView™ 488 caspase-3 substrate was from Interchim (Montluçon, France). All other fluorescent probes were from Fischer Scientific (Illkirch, France).
siRNA transfection
A mixture of five SIRT1 siRNA provided as a single reagent was purchased from Santa Cruz. H9c2 cells were plated overnight at 25 × 104 cells in 24-well plate and were transfected with 16 pg of SIRT1 siRNA oligonucleotides or non-target control oligonucleotides according to manufacturer's instructions. Twenty-four hours after transfection, medium was removed and cells were treated or not with TG or TN for 48 h.
Echocardiography
Transthoracic echocardiography was performed using a 15 MHz transducer (Vivid 9, General Electric Healthcare, GE Medical Systems, Buc, France) under 2.5% isoflurane gas anesthesia. Two-dimensional (2D)-guided M-mode echocardiography was used to determine wall thickness and left ventricular chamber volume at systole and diastole and contractile parameters, such as FS or EF.
Histochemical analysis
Hearts were fixed in 10% formalin and embedded in paraffin. Apoptosis was detected in tissue sections (4 μm) using the TUNEL assay (Apoptag Kit, peroxidase staining, Merck Millipore, Molsheim, France) according to the manufacturer's recommendations. The slides were counterstained with methyl green. Apoptotic cells were quantified using a computer-based morphogenic analysis software (TRIBVN ICS Framework, Chatillon, France) in a blinded manner. For each mouse, an entire heart cross section was analyzed. The surface of the positive area (stained in brown) was measured for each specimen and expressed as a percentage of the total tissue area.
Flow cytometry analysis
The fluorescent probe fluorescein diacetate (FDA) was used to assess cell viability. After treatment, cells were incubated for 5 min at 37 °C with 0.2 μg/ml FDA and analyzed by flow cytometry. Mitochondrial transmembrane potential (ΔΨm) was analyzed by cell staining with 10 nM DiOC6(3) for 20 min at 37 °C. Necrosis was estimated by adding 10 μg/ml of propidium iodide (PI) just before flow cytometric analysis. Caspase-3 activation was assessed using DEVD-NucView 488 caspase-3 substrate. After treatment, cells were incubated with 5 μM DEVD-NucView 488 for 30 min at RT before cytometric analysis. Fluorescence of cells was analyzed on a Cell Lab Quanta MPL cytometer (Beckman Coulter, Villepinte, France).
Sample preparation for transmission electron microscopy
H9c2 cells were fixed with 2% glutaraldehyde/0.1 M sodium cacodylate for 1 h at RT, pelleted and rinsed with 0.1 M sodium cacodylate/0.2 M sucrose for 1 h. Samples were epon-embedded, sliced, mounted on slides and the images were obtained at the electron microscopy facility of INRA (Jouy-en-Josas, France).
Adult cardiomyocyte isolation and ER stress induction
Retrograde heart perfusion according to the Langendorff method was performed to isolate adult rat ventricle cardiomyocytes as previously described.44 Cells were plated onto dishes coated with laminin and were kept at room temperature for 2 h. Cells were then treated with 10 μg/ml TN with or without EX527 (20 μM) or STAC-3 (1 μM) for 24 h at 37 °C under 5% CO2/95% air.
Evaluation of cell death by fluorescence microscopy
To detect apoptosis in H9c2 cells, nuclei were stained with 2.5 μg/ml Hoechst 33348 for 5 min. Condensed or fragmented nuclei were considered as apoptotic. To analyze cell death, cardiomyocytes were stained with 0.2 μg/ml of FDA for 5 min. FDA-negative cardiomyocytes were considered as dead cells. More than 1000 cells were counted for each condition and representative micrographs were taken on a Leica fluorescence microscope (Leica Microsystems, Nanterre, France).
RNA isolation and quantitative RT-PCR
RNA was isolated from cultured cells using Zymo Research (Freiburg, Germany) Quick RNA MiniPrep according to the manufacturer's instructions or from the left ventricle of the heart using Trizol reagent (Fisher Scientific, Illkirch, France). Total RNA (1 μg) was reverse transcribed using BioRad iScript reverse transcription kit. For real-time PCR, cDNA was amplified by the 'two-step' SsoFast EvaGreen supermix (Bio-Rad, Marnes-la-Coquette, France), heated at 95 °C for 30 s, followed by 50 cycles of denaturation at 95 °C during 2 s and hybridization/elongation at 60 °C during 5 s. The PCR primers were obtained from Eurofins (Les Ulis, France) and are listed in table EV4. The results were quantified according to the Cq value method, where Cq is defined as the quantification cycle of PCR at which the amplified product is detected. The ratio (1+Etarget gene)^−(Cq sample−Cq control)targetgene/(1+Ereference gene)^−(Cq sample−Cq control) gene was calculated, where E represents the efficiency of the quantitative PCR reaction. With this calculation, expression of control was equivalent to one (Supplementary Table 4).
Western blot analysis
Cells and tissues were lysed in RIPA lysis buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 1% Triton, 1 mM EDTA, 0.1% SDS, 0.5% deoxycholic acid) plus a cocktail of protease and phosphatase inhibitors (Roche, Boulogne-Billancourt, France), PMSF and a cocktail of deacetylase inhibitors (Santa Cruz, Heidelberg, Germany) for 30 min at 4 °C. Proteins (50 μg) were separated by SDS-polyacrylamide gel electrophoresis and transferred to PVDF membranes (Merck Millipore, Molsheim, France). Membranes were incubated overnight at 4 °C with the following antibodies: anti-GRP94, anti-GRP78, anti-phospho-PERK, anti-PERK, anti-GAPDH, anti-phospho-eIF2α, anti-eIF2α, anti-acetylated lysine, anti-p53, anti-LC3-II and anti-CHOP from Cell Signaling (Leiden, The Netherlands), anti-Flag from Novus (Lille, France), anti-β-actin (sc-47778), anti-ATF4 (sc-200), anti-histone H1 (sc-10806) and anti-GADD34 (sc-8327) from Santa Cruz, anti-SIRT1 anti-acetylated p53 from Abcam (Paris, France), anti-acetylated histone H1 from Sigma-Aldrich (Lyon, France). Proteins were detected on a ChemiDoc XRS (Bio-Rad) by using the ECL method according to the manufacturer's instructions (Merck Millipore). To calculate the relative density (RD), ImageJ software was used and the intensity of each protein was normalized to β-actin or GAPDH. The data obtained were then expressed as the ratio of the intensity of the protein in treated cells or mice to that of the corresponding protein in untreated cells or vehicle-treated mice (RD). The level of protein phosphorylation/acetylation was expressed as the ratio of the phosphorylated/acetylated protein versus the total protein.
Native and denaturing immunoprecipitations
To analyze the level of eIF2α acetylation, cells were lysed in a denaturing IP buffer (1% SDS, 50 mM Tris-HCl, 5 mM EDTA, 10 mM DTT, 1 mM PMSF, 15 U/ml DNase, pH 7.4) supplemented with a protease and phosphatase inhibitors cocktail (Roche) and a cocktail of deacetylase inhibitors (Santa Cruz). Lysate were heat at 90 °C for 5 min and then diluted in non-denaturing buffer to trap SDS (1% Triton X-100, 50 mM Tris-HCl, 5 mM EDTA, 300 mM NaCl, 0.02% azide de sodium, 1 mM PMSF, pH 7.4) supplemented with a cocktail of protease and deacetylase inhibitors. Lysine-acetylated proteins or eIF2α protein were immunoprecipitated using anti-acetylated lysine antibody or anti-eIF2α antibody coated on A/G magnetic beads (Merck Millipore). Mouse anti-IgG (Santa Cruz) were used as control. To examine physical interaction between SIRT1 and eIF2α, cell were lysed in non-denaturing lysis buffer (20 mM Tris-HCl, 137 mM NaCl, 1% Triton X-100, 2 mM EDTA, pH 8) supplemented with a protease and phosphatase inhibitors cocktail (Roche) for 30 min at 4 °C. SIRT1 or eIF2α proteins were immunoprecipitated using anti-SIRT1 of anti-eIF2α antibodies coated on A/G magnetic beads (Merck Millipore). Mouse and rabbit anti-IgG (Santa Cruz) were used as control. Imunoprecipitated proteins were run on SDS-PAGE and were revealed with anti-eIF2α, anti-PERK, anti-ATF4, anti-CHOP, anti-GADD34, anti-acetylated lysine or anti-SIRT1 antibodies.
Mass spectrometry analysis
Immunoprecipitated eIF2α protein was separated by SDS-PAGE and In-gel trypsin digestion was carried out as previously described.45 The gel slices corresponding to the bands between 25 and 50 kDa from H9c2 cells treated or not with EX527 were excised. After reduction and alkylation, the proteins were digested with a modified sequencing grade trypsin (Sigma-Aldrich). The digestion was carried out overnight at 37 °C and the fragmented peptides were extracted from the gel with 5% formic acid/50% acetonitrile. Mass spectrometric analyses were performed using nanoflow liquid chromatography tandem mass spectrometry (LC-MS/MS). LC-MS/MS was carried out on an RSLC Ultimate-3000 nanoflow LC system (ThermoScientific, Waltham, MA, USA) coupled to an Hybrid LTQ-Orbitrap Velos mass spectrometer instrument (ThermoScientific). A capillary peptide trap (1 mm 300 μm ID) was used to desalt and concentrate the tryptic peptides before separation on a reverse-phase 15 cm 75 μm ID pepMap column, packed with C18-resin (ThermoScientific). The column was directly mounted in the nanoelectrospray ion sources and the peptides were eluted by applying a linear gradient program (5–50% B 30 min, 50% B 30–35 min and 50–95% B 35–60 min where mobile phase B was 0.1% formic acid/95% HPLC grade acetonitrile/5% HPLC grade water and mobile phase A was 0.1% formic acid/5% HPLC grade acetonitrile/95% HPLC grade water) at a constant flow of 170 nl/ml. The analysis using the Hybrid LTQ-Orbitrap Velos type of instrument was performed as previously described.46 The instrument was operated in data-dependent acquisition mode to automatically switch between MS and MS/MS. MS/MS spectra of these ions were obtained by fragmenting the ions using collision-induced dissociation with normalized collision energy of 30% in the ion trap. The data files generated by the LC-MS/MS were converted to Sequest generic format files and were searched using Proteome Discoverer Search Engine (ThermoScientific) against the human NCBI non-redundant database. Peptides searches were performed using carbamidomethyl as fixed modification and oxidized methionine and acetylated lysine as variable modification.
Vectors and site-directed mutagenesis
Plasmids encoding human CD2 (pCDNA3-hCD2), WT eIF2α (eIF2a 1) and eIF2α S52A phosphorylation mutant (eIF2a 2) were a gift from D Ron (Addgene, Teddington, UK, plasmid #21810, #21807 and #21808, respectively). eIF2α mutants of acetylation were generated by site directed mutagenesis (Genscript antibodies-online GmbH, Paris, France) by converting either lysine 141 or 143 or both lysines to an arginine residue (codon change from TCA to TTA). In addition, a FLAG-tag was added in the C-terminal domain of the mutants and WT eIF2α. For transfection experiments, H9c2 cells were plated overnight at 75 × 104 cells in 24-well plate and were co-transfected using Turbofect (Life Technologies, Fisher Scientific, Illkirch, France) with a mix of 0.2 μg tRFP-encoding vector/0.8 μg vector of interest to allow cell death analysis only in transfected cells (tRFP-positive cells), according to manufacturer's instructions. Twenty-four hours post-transfection, cells were treated with TG or TN for 48 h and cell death was analyzed by FDA assay. The FDA fluorescence was determined by cytometric analysis of samples gated on tRFP-positive cells.
Statistical analysis
All data are presented as mean±S.E.M. Data were analyzed using Sigma Stat (Sigma Stat, version 3.0, Systat Software, San Jose, CA, USA). One-way ANOVA was used to assess differences among groups followed by Newman–Keuls or Dunn's post hoc test. When two groups were compared, differences were assessed by Student's t-test. Differences between groups were considered significant if the P-value was <0.05. *P<0.05; **P<0.01; ***P<0.005 versus CTL. $P<0.05; $$P<0.01; $$$P<0.005 versus EX527 or STAC-3 untreated cells or WT mice.
Acknowledgments
We thank N Viet Hung for Mass Spectrometry analysis (SFR A Lwoff Platform). We also thank F Lefebvre for helpful technical assistance and Valérie Domergue-Dupont and the animal core facility of IPSIT for efficient handling and preparation of the animals. We gratefully acknowledge C Longin and S Chat for electron microscopy work (TEM platform of INRA). We thank Dr R Fischmeister for continuous support and careful reading of the manuscript. WT and mutant (S52A) eIF2α encoding plasmids were kindly provided by D Ron (University of Cambridge). This work was supported by grants from LabEx LERMIT, FRM (Fondation pour la Recherche Médicale) to A Garnier (#DPM20121125546) and Région Ile de France CODDIM to R Ventura-Clapier (#cod110153). A Prola received a fellowship from Groupe de Réflexion sur la Recherche Cardiovasculaire (GRRC). A Guilbert received a fellowship from Région Ile de France CORDDIM. D Sinclair is supported by the Glenn Foundation for Medical Research, a gift from the Schulak Family, the Juvenile Diabetes Foundation, and a MERIT award from the NIH/NIA.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by A Villunger
DAS consults for GlaxoSmithKline, Metrobiotech, Ovascience and BigDataBio. The remaining authors declare no conflict of interest.
Supplementary Material
References
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 2007; 8: 519–529. [DOI] [PubMed] [Google Scholar]
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 2012; 13: 89–102. [DOI] [PubMed] [Google Scholar]
- Csordas G, Thomas AP, Hajnoczky G. Quasi-synaptic calcium signal transmission between endoplasmic reticulum and mitochondria. EMBO J 1999; 18: 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath H, O'Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell 2007; 129: 1337–1349. [DOI] [PubMed] [Google Scholar]
- Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol 2011; 13: 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Kitakaze M. ER stress in cardiovascular disease. J Mol Cell Cardiol 2010; 48: 1105–1110. [DOI] [PubMed] [Google Scholar]
- Martindale JJ, Fernandez R, Thuerauf D, Whittaker R, Gude N, Sussman MA et al. Endoplasmic reticulum stress gene induction and protection from ischemia/reperfusion injury in the hearts of transgenic mice with a tamoxifen-regulated form of ATF6. Circ Res 2006; 98: 1186–1193. [DOI] [PubMed] [Google Scholar]
- Severino A, Campioni M, Straino S, Salloum FN, Schmidt N, Herbrand U et al. Identification of protein disulfide isomerase as a cardiomyocyte survival factor in ischemic cardiomyopathy. J Am Coll Cardiol 2007; 50: 1029–1037. [DOI] [PubMed] [Google Scholar]
- Thuerauf DJ, Marcinko M, Gude N, Rubio M, Sussman MA, Glembotski CC. Activation of the unfolded protein response in infarcted mouse heart and hypoxic cultured cardiac myocytes. Circ Res 2006; 99: 275–282. [DOI] [PubMed] [Google Scholar]
- Shintani-Ishida K, Nakajima M, Uemura K, Yoshida K. Ischemic preconditioning protects cardiomyocytes against ischemic injury by inducing GRP78. Biochem Biophys Res Commun 2006; 345: 1600–1605. [DOI] [PubMed] [Google Scholar]
- Fu HY, Okada K, Liao Y, Tsukamoto O, Isomura T, Asai M et al. Ablation of C/EBP homologous protein attenuates endoplasmic reticulum-mediated apoptosis and cardiac dysfunction induced by pressure overload. Circulation 2010; 122: 361–369. [DOI] [PubMed] [Google Scholar]
- Ayala P, Montenegro J, Vivar R, Letelier A, Urroz PA, Copaja M et al. Attenuation of endoplasmic reticulum stress using the chemical chaperone 4-phenylbutyric acid prevents cardiac fibrosis induced by isoproterenol. Exp Mol Pathol 2012; 92: 97–104. [DOI] [PubMed] [Google Scholar]
- Ceylan-Isik AF, Sreejayan N, Ren J. Endoplasmic reticulum chaperon tauroursodeoxycholic acid alleviates obesity-induced myocardial contractile dysfunction. J Mol Cell Cardiol 2011; 50: 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol 2010; 5: 253–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X et al. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res 2007; 100: 1512–1521. [DOI] [PubMed] [Google Scholar]
- Hsu CP, Zhai P, Yamamoto T, Maejima Y, Matsushima S, Hariharan N et al. Silent information regulator 1 protects the heart from ischemia/reperfusion. Circulation 2010; 122: 2170–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planavila A, Iglesias R, Giralt M, Villarroya F. Sirt1 acts in association with PPARalpha to protect the heart from hypertrophy, metabolic dysregulation, and inflammation. Cardiovasc Res 2011; 90: 276–284. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xia Z, La Cour KH, Ren J. Activation of Akt rescues endoplasmic reticulum stress-impaired murine cardiac contractile function via glycogen synthase kinase-3beta-mediated suppression of mitochondrial permeation pore opening. Antioxid Redox Signal 2011; 15: 2407–2424. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hubbard BP, Gomes AP, Dai H, Li J, Case AW, Considine T et al. Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science 2013; 339: 1216–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks WW, Conrad CH. Isoproterenol-induced myocardial injury and diastolic dysfunction in mice: structural and functional correlates. Comp Med 2009; 59: 339–343. [PMC free article] [PubMed] [Google Scholar]
- Dalal S, Foster CR, Das BC, Singh M, Singh K. Beta-adrenergic receptor stimulation induces endoplasmic reticulum stress in adult cardiac myocytes: role in apoptosis. Mol Cell Biochem 2012; 364: 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichtova Z, Novotova M, Kralova E, Stankovicova T. Morphological and functional characteristics of models of experimental myocardial injury induced by isoproterenol. Gen Physiol Biophys 2012; 31: 141–151. [DOI] [PubMed] [Google Scholar]
- Wallner M, Duran JM, Mohsin S, Troupes CD, Vanhoutte D, Borghetti G et al. Acute catecholamine exposure causes reversible myocyte injury without cardiac regeneration. Circ Res 2016; 119: 865–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell 2007; 13: 365–376. [DOI] [PubMed] [Google Scholar]
- Castillero E, Akashi H, Pendrak K, Yerebakan H, Najjar M, Wang C et al. Attenuation of the unfolded protein response and endoplasmic reticulum stress after mechanical unloading in dilated cardiomyopathy. Am J Physiol Heart Circ Physiol 2015; 309: H459–H470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Onofrio N, Vitiello M, Casale R, Servillo L, Giovane A, Balestrieri ML. Sirtuins in vascular diseases: emerging roles and therapeutic potential. Biochim Biophys Acta 2015; 1852: 1311–1322. [DOI] [PubMed] [Google Scholar]
- Winnik S, Auwerx J, Sinclair DA, Matter CM. Protective effects of sirtuins in cardiovascular diseases: from bench to bedside. Eur Heart J 2015; 36: 3404–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 2005; 434: 113–118. [DOI] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Dominy JE Jr, Blattler SM, Jedrychowski MP, Banks AS, Lim JH et al. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+). Mol Cell 2011; 44: 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrin N, Kaushik VK, Fortier E, Wall D, Pearson KJ, de Cabo R et al. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS One 2009; 4: e8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Xia L, Zhang Y, Wang H, Xu W, Hu H et al. Activating transcription factor 4 confers a multidrug resistance phenotype to gastric cancer cells through transactivation of SIRT1 expression. PloS one 2012; 7: e31431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FM, Chen YJ, Ouyang HJ. Regulation of unfolded protein response modulator XBP1s by acetylation and deacetylation. Biochem J 2011; 433: 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Liu W, Liu B, Zhang B, Li W, Xu Y. SIRT1 suppresses cardiomyocyte apoptosis in diabetic cardiomyopathy: an insight into endoplasmic reticulum stress response mechanism. Int J Cardiol 2015; 191: 36–45. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 2000; 6: 1099–1108. [DOI] [PubMed] [Google Scholar]
- Dey M, Velyvis A, Li JJ, Chiu E, Chiovitti D, Kay LE et al. Requirement for kinase-induced conformational change in eukaryotic initiation factor 2alpha (eIF2alpha) restricts phosphorylation of Ser51. Proc Natl Acad Sci USA 2011; 108: 4316–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2alpha kinases: their structures and functions. Cell Mol Life Sci 2013; 70: 3493–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh HS, Reizis B, Robbins PD. SIRT1 associates with eIF2-alpha and regulates the cellular stress response. Sci Rep 2011; 1: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Li X, Zhang P, Chen WD, Zhang HL, Li DD et al. Acetylation of beclin 1 inhibits autophagosome maturation and promotes tumour growth. Nat Commun 2015; 6: 7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Chevet E, Harding HP. Targeting the unfolded protein response in disease. Nat Rev Drug Discov 2013; 12: 703–719. [DOI] [PubMed] [Google Scholar]
- Cominacini L, Mozzini C, Garbin U, Pasini A, Stranieri C, Solani E et al. Endoplasmic reticulum stress and Nrf2 signaling in cardiovascular diseases. Free Radic Biol Med 2015; 88(Pt B): 233–242. [DOI] [PubMed] [Google Scholar]
- Giblin W, Skinner ME, Lombard DB. Sirtuins: guardians of mammalian healthspan. Trends Genet 2014; 30: 271–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes SA, Papa FR. The role of endoplasmic reticulum stress in human pathology. Annu Rev Pathol 2015; 10: 173–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C et al. The NAD(+)/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell 2013; 154: 430–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde I, Vandecasteele G, Lezoualc'h F, Fischmeister R. Characterization of the cyclic nucleotide phosphodiesterase subtypes involved in the regulation of the L-type Ca2+ current in rat ventricular myocytes. Br J Pharmacol 1999; 127: 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 1996; 68: 850–858. [DOI] [PubMed] [Google Scholar]
- Pachl F, Ruprecht B, Lemeer S, Kuster B. Characterization of a high field Orbitrap mass spectrometer for proteome analysis. Proteomics 2013; 13: 2552–2562. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.