SUMMARY
Enhanced secretion of tumorigenic effector proteins is a feature of malignant cells. The molecular and cellular mechanisms underlying this feature are poorly defined. We identify PITPNC1 as a gene amplified in a large fraction of human breast cancer and over-expressed in metastatic breast, melanoma and colon cancer. Biochemical, molecular, and cell-biological studies reveal that PITPNC1 promotes malignant secretion by binding Golgi resident PI4P and localizing RAB1B to the Golgi. RAB1B localization to the Golgi allows for the recruitment of GOLPH3 to the trans-Golgi, which facilitates Golgi extension and enhanced vesicular release. PITPNC1-mediated vesicular release drives metastasis by increasing the secretion of pro-invasive and pro-angiogenic mediators HTRA1, MMP1, FAM3C, PDGFA, and ADAM10. We establish PITPNC1 as a PI4P-binding protein that enhances vesicular secretion capacity in malignancy.
INTRODUCTION
Phosphatidylinositols (PIs) are members of a large family of complex amphiphilic compounds comprised of a glycerol backbone, two fatty acids, a phosphate group, and an inositol head group. While these molecules constitute a minor component of eukaryotic cell membranes, they play key roles as transducers of signals within cells (Balla, 2013). PIs can be phosphorylated by various kinases, yielding mono, di, or tri-phosphorylated products. Specific phosphorylated PIs can be bound within lipid membranes by signal transducing proteins, can serve as docking sites for the selective recruitment of effector proteins to local cellular membrane compartments, or can be metabolized by specific PI phosphatases (Balla, 2013). The significance of this class of molecules to physiology and disease has been well-demonstrated through mechanistic studies on PI3-kinase and PTEN, which respectively positively and negatively regulate the formation of the tri-phosphorylated PIP3—a central regulator of growth and survival of cells (Wong et al., 2010). While earlier studies implicated PI3K and PTEN in the regulation of oncogenesis, subsequent work has revealed pervasive and fundamental roles for these PI modifying enzymes in the physiological and pathological function of many organelle systems (Balla, 2013).
The steps of the metastatic cascade require precise regulation of multiple cellular phenotypes and extracellular interactions. Understanding the molecular and cell-biological mechanisms by which genes execute these processes is a prerequisite for the development of effective therapies for treating and potentially preventing metastatic disease (Chiang and Massague, 2008; Hanahan and Weinberg, 2011; Huang et al., 2008; Kang et al., 2003; Minn et al., 2005; Pencheva and Tavazoie, 2013; Pencheva et al., 2012; Talmadge and Fidler, 2010; Tavazoie et al., 2008). A key feature of metastatic cells is their ability to impact various cell types within the tumor microenvironment through the release of secreted factors (Joyce and Pollard, 2009; Orimo et al., 2005). Yet the mechanisms that govern this phenotype are incompletely understood.
PITPNC1 was previously identified as a gene targeted by miR-126, a metastasis suppressor microRNA (Png et al., 2012). The cell-biological function of PITPNC1, however, was not defined. Here we reveal that PITPNC1 is genetically amplified in a large fraction of human breast cancers and that its over-expression significantly correlates with metastatic progression of breast, melanoma, and colon cancers. Through complementary biochemical and in vitro methods we have found that PITPNC1 binds PI4P–a previously unreported lipid substrate for PITP domain proteins–and that this interaction localizes PITPNC1 to the Golgi compartment within the cell. Golgi localization of PITPNC1 increases Golgi abundance of the PITPNC1 interacting protein RAB1B. RAB1B is a GTPase that cycles between a GDP bound inactive state bound to GDIalpha in the cytoplasm and a GTP bound active state in the Golgi where it is required for Golgi function (Barr, 2013; Wilson et al., 1996). PITPNC1-dependent RAB1B recruitment to the Golgi compartment enhances secretion by cancer cells. This optimized secretory state is accompanied by the extension of the Golgi morphology, which is known to occur in the context of enhanced vesicular release. Conversely, PITPNC1 depletion in malignant cells reduces the secretion of a number of secreted factors. Given that metastatic cells interact with various cell types within the tumor microenvironment through the release of secreted factors (Chiang and Massague, 2008; Hanahan and Weinberg, 2011; Huang et al., 2008; Kang et al., 2003; Minn et al., 2005; Pencheva et al., 2012; Talmadge and Fidler, 2010; Tavazoie et al., 2008), we sought to identify the proteins that exhibited enhanced secretion in metastatic cells as a result of PITPNC1-dependent events. By utilizing quantitative proteomics, we identified MMP1, HTRA1, FAM3C, ADAM10, and PDGFA as downstream mediators of the pro-metastatic and pro-angiogenic effects of PITPNC1 in breast cancer.
RESULTS
PITPNC1 promotes metastasis of multiple cancer types
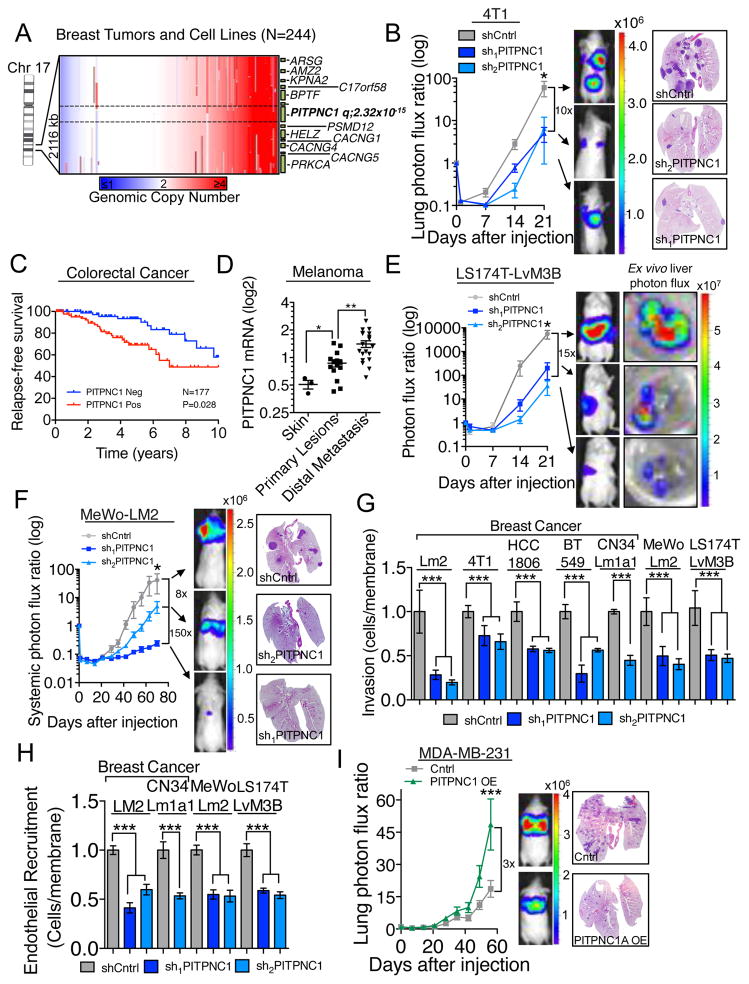
Analysis of genomic copy number data using the dataset derived by Beroukhim and colleagues (Beroukhim et al., 2010) revealed PITPNC1 to be significantly (q=2.32x10−15) amplified in 46% of 244 human breast cancer cells lines and tumors, with 15.9% of such amplifications being focal in nature (Figure 1A). Specific analysis of the breast cancer subtypes further revealed that triple negative breast cancer patients (TNBC) have increased levels of amplification relative to non-TNBC (Figure S1A). Expression of PITPNC1 was further increased in a large independent collection of human breast cancers relative to normal breast tissues (Figure S1B) and protein levels of PITPNC1 are elevated in MDA-MB-231 breast cancer cells compared to human mammary epithelial cells (Figure S1C). Furthermore, in vivo selected highly metastatic breast cancer cell populations LM2 and CN34Lm1a1 (Minn et al., 2005; Tavazoie et al., 2008) exhibited increased PITPNC1 protein abundance compared to their parental populations (MDA-MB-231 and CN34, Figure S1C). Interestingly, PITPNC1 knockdown reduced the ability of murine 4T1 as well as human CN34Lm1a1 and HCC1806 breast cancer cells to metastasize in immunocompetent and immunecompromised mice, respectively (Figure 1B and Figure S1D–G). These findings establish PITPNC1 as a pro-metastatic protein that is induced in metastatic breast cancer. Similar to breast cancer, we find that PITPNC1 protein levels are elevated in highly metastatic melanoma MeWo-LM2 (Pencheva et al., 2012) and colon cancer LS174T-LvM3 cells (Loo et al., 2015) relative to non-cancerous primary melanocytes and colon epithelial cells, respectively (Figure S1H). PITPNC1 expression in primary tumors was further significantly correlated with human metastatic progression outcomes in colorectal cancer and melanoma, suggesting that PITPNC1 may act centrally in the metastatic cascade to promote metastasis in multiple cancer types (Figures 1C,D). Knockdown of PITPNC1 in the highly metastatic colon cancer subline LS174T-LvM3 and the metastatic MeWo-LM2 melanoma line significantly reduced metastatic colonization by 15-fold and 8-fold, respectively (Figures 1E,F and Figures S1D). To further investigate the mechanism by which PITPNC1 promotes metastasis, we next sought to determine the metastatic phenotype(s) it governs. Specific depletion of PITPNC1 significantly inhibited invasion and endothelial recruitment (Png et al., 2012) by breast, colon and skin cancer cells, but did not affect in vitro migration or proliferation capacity (Figures 1G,H and Figures S1I–N). These findings reveal that the phenotypic effects of PITPNC1 depletion are selective and do not result from reduced cellular viability. Importantly, over-expression of PITPNC1 was sufficient to significantly enhance invasion, endothelial recruitment, and metastatic colonization by MDA-MB-231 breast cancer cells (Figures 1I, S1O–R). These findings establish PITPNC1 as a robust mediator of metastasis and pro-metastatic phenotypes in multiple cancer types.
Figure 1. PITPNC1 promotes metastasis in multiple cancer types.
(A) Genomic copy number analysis of 244 breast tumors and cell lines. Data from Tumorscape (Beroukhim et al., 2010).
(B) Bioluminescence imaging plot of lung metastatic colonization by 50,000 4T1 breast cancer cells expressing a control or PITPNC1 targeting hairpins. n=5/group. Right, representative lung histology.
(C) Kaplan-Meier curve representing metastasis-free survival cohort of colorectal patients (n=177) as a function of their primary tumor’s PITPNC1 expression levels (Data from GSE17536). Patients whose primary tumors’ PITPNC1 expressions levels were greater or lower than the median for the population were classified as PITPNC1 positive (red) or negative (blue), respectively.
(D) PITPNC1 expression levels in normal skin, primary melanoma, and distal metastatic lesions of patients (n=37) (Haqq et al., 2005).
(E) Bioluminescence imaging quantification of liver colonization by 80,000 LS174T-LvM3 colon cancer cells expressing short hairpins targeting PITPNC1 or a control hairpin. Right, luciferase signal from ex vivo livers at day 21. n=6/group.
(F) Bioluminescence imaging plot of lung metastatic colonization by 40,000 MeWo control or PITPNC1 knockdown cells. n=5/group. Right, Lungs were extracted and stained by H&E.
(G) Matrigel invasion by 50,000 LM2, 4T1, HCC-1806, BT549, CN34Lm1a1, MeWo-LM2 or LS174T-LvM3 cells expressing PITPNC1 or control targeting hairpins. Data normalized to control group values. n=4/group.
(H) Trans-well recruitment of 80,000 human umbilical vein endothelial cells (HUVEC) by LM2, CN34LM1a1, MeWo-LM2 and LS174T-LvM3B cells. Data normalized to control group values. n=4/group.
(I) Bioluminescence imaging plot and histology of lung metastatic colonization by 40,000 MDA-MB-231cells transduced with PITPNC1 over-expression or a control vector. n=6/group.
Error bars represent S.E.M.
See also Figure S1.
PITPNC1 binds PI4P to promote metastatic progression
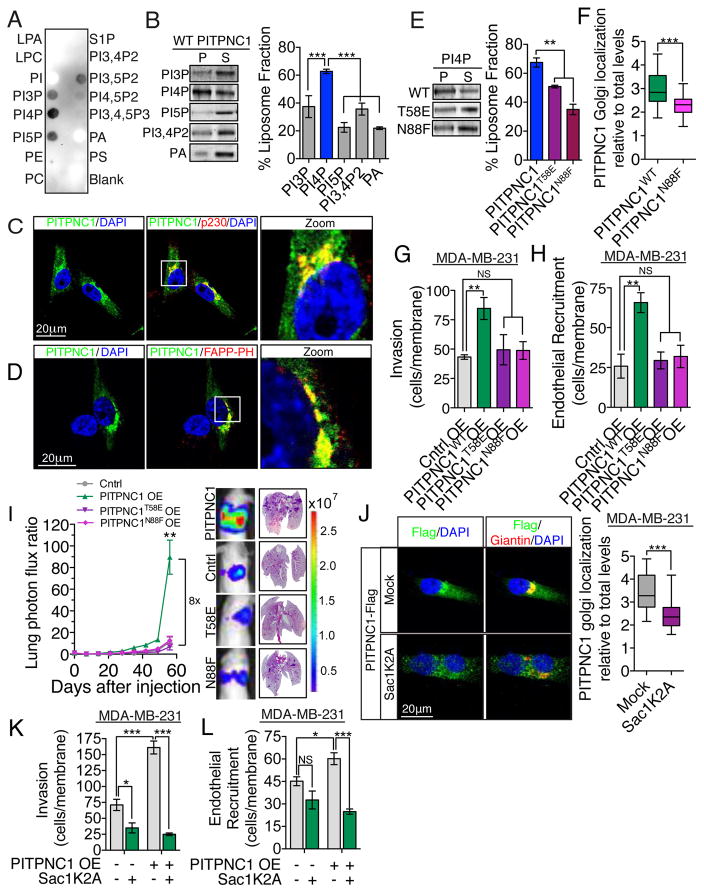
The N-terminal portion of PITPNC1 contains a PITP lipid-binding and transfer domain. To understand the molecular mechanism by which PITPNC1 regulates metastatic colonization, we sought to identify PITPNC1’s lipid substrate. We first performed a lipid-overlay assay in which a broad range of lipids were spotted on a membrane and blotted with purified recombinant PITPNC1 protein in order to identify its substrate. Lipid-blot assays revealed that PITPNC1 binds most strongly to PI4P with weaker binding to other PIs containing a single phosphate head group, namely PI3P and PI5P (Figure 2A and Figure S2A). To test these protein-lipid interactions in a more physiological context, we conducted vesicular pull-down experiments. Consistent with the robust lipid binding of PI4P binding by PITPNC1, recombinant PITPNC1 was found to bind to vesicles containing PI4P, but not those containing PI3P, PI5P, PI3,4P2 or phosphatidic acid (PA) (Figure 2B).
Figure 2. PITPNC1-mediated binding of PI4P drives metastasis.
(A) Lipid overlay assay. Lipid-bound recombinant PITPNC1-GST was detected with anti-GST antibody. The experiment was repeated 3 times.
(B) Vesicle pull-down assay of recombinant PITPNC1 using vesicles containing PE and PC in combination with PI3P, PI4P, PI5P, PI3,4P2 or PA respectively. n=3/group.
(C) MDA-MB-231 cells expressing GFP-tagged PITPNC1 (green) co-stained with anti-p230 antibody (red) and 4′,6-diamidino-2-phenylindole (DAPI, blue) were analyzed by immunofluorescence microscopy.
(D) MDA-MB-231 cells expressing GFP-tagged PITPNC1 (green) co-stained with DAPI (blue), and FAPP-PH for the detection of PI4P (red).
(E) Vesicle pull-down assay of recombinant wild-type PITPNC1 and PITPNC1 containing the N58E or N88F lipid binding mutations. n=3/group.
(F) MDA-MB-231 cells with stable expression of Flag-tagged wild-type or N88F mutant PITPNC1 co-stained with anti-Flag and anti-Giantin were analyzed by immunofluorescence microscopy. The intensity of Flag immunoreactivity in areas positive for Giantin was considered as the Golgi signal. The Golgi signal was normalized to total cellular levels to control for differences in expression levels between wild-type and N88F mutant PITPNC1. n=30/group.
(G,H) MDA-MB-231 cells expressing PIPTNC1WT, PITPNC1T58E, PITPNC1N88F, or control vector were subjected to the matrigel invasion assay (G) and endothelial recruitment assay (H). n=4/group.
(I) Bioluminescence imaging plot and lung histology of lung metastatic colonization by 40,000 MDA-MB-231 parental cells over-expressing PITPNC1, PITPNC1T58E, PITPNC1N88F or a control vector. n=6/group. H&E staining of representative lung sections.
(J) MDA-MB-231 cells with stable expression of Flag-tagged wild-type PITPNC1 where transfected with mock or Sac1K2A and co-stained with anti-Flag and anti-Giantin. The intensity of Flag immunoreactivity in areas positive for Giantin was considered as the Golgi signal. The Golgi signal was normalized to total cellular levels. n=30/group.
(K,L) MDA-MB-231 cells expressing PITPNC1 or control vector and cells expressing PITPNC1 and that was transfected with Sac1K2A were subjected to matrigel invasion assay (K) and the endothelial recruitment assay (L). n=4/group.
For B,E,G,H, I, K,L error bars represent S.E.M. Box and whiskers plots represent the data in figure F and J with the upper and lower bars showing minimum and maximum data points
See also Figure S2.
Having observed lipid binding towards PI4P, we questioned if this activity is required for the cellular localization of PITPNC1. GFP-tagged PITPNC1 displayed diffuse cytoplasmic localization as previously described (Takano et al., 2003), but showed most intense staining in perinuclear regions that co-stained with p230–marker of the trans Golgi network (Figure 2C). The Golgi is the sorting hub for vesicles destined for the plasma membrane and endosomal compartments, and these functions are dependent on resident PI4P (Mayinger, 2012). Conversely, co-staining of PITPNC1 and PI4P using the PI4P-specific PH-domain of FAPP1 confirmed that PITPNC1 accumulates in PI4P-positive areas of the Golgi (Figures 2D, Figure S2B). Previous studies have shown that T59E and N90F mutations of PITPα, another PITP domain family member that binds PI and phosphatidylcholine in vitro, abrogates lipid binding (Milligan et al., 1997; Tilley et al., 2004; Yoder et al., 2001). Homology analysis of PITPα and PITPNC1 revealed these amino acids to be conserved in PITPNC1 (T58 and N88). We therefore produced recombinant forms of both single amino acid mutations and tested their capacities for binding PI4P. Both mutations abrogated the PI4P binding activity of PITPNC1 (Figure 2E). To determine if PI4P binding mediates Golgi localization of PITPNC1, we expressed Flag-tagged wild-type or N88F lipid binding mutant PITPNC1 in breast cancer cells and performed immunofluorescence analysis between PITPNC1-Flag and the Golgi marker Giantin. These studies revealed that PITPNC1 localization to the Golgi is dependent upon PI4P binding (Figure 2F, Figure S2C). We then asked if such PI4P binding and Golgi localization is required for the metastasis promoting function of PITPNC1. To this end, we over-expressed either wild-type or the two lipid binding mutant forms of PITPNC1 in MDA-MB-231 cells and found that wild-type PITPNC1, but neither of the lipid binding mutants, was sufficient to promote invasion, endothelial recruitment, and metastatic colonization (Figures 2G–I and Figure S2D,E). Finally, in complimentary experiments, we reduced trans-Golgi PI4P levels via expression of the trans-Golgi localized PI4P phosphatase mutant Sac1K2A and asked if this manipulation would affect PITPNC1’s ability to promote metastatic phenotypes (Figure S2F,G) (Mayinger, 2012). Consistent with a required role for trans-Golgi PI4P to recruit PITPNC1 to the Golgi compartment, reduction of trans-Golgi-localized PI4P by Sac1K2A reduced PITPNC1 localized in the Golgi (Figure 2J). Further, PITPNC1 over-expression failed to promote invasion and endothelial recruitment phenotypes following Sac1K2A mediated Golgi PI4P reduction in MDA-MB-231 cells (Figure 2K,L). Overall, these findings reveal that the interaction between PI4P and PITPNC1 in the trans-Golgi is required for its ability to promote metastasis.
PITPNC1 forms a protein complex with 14-3-3 isoforms and RAB1B
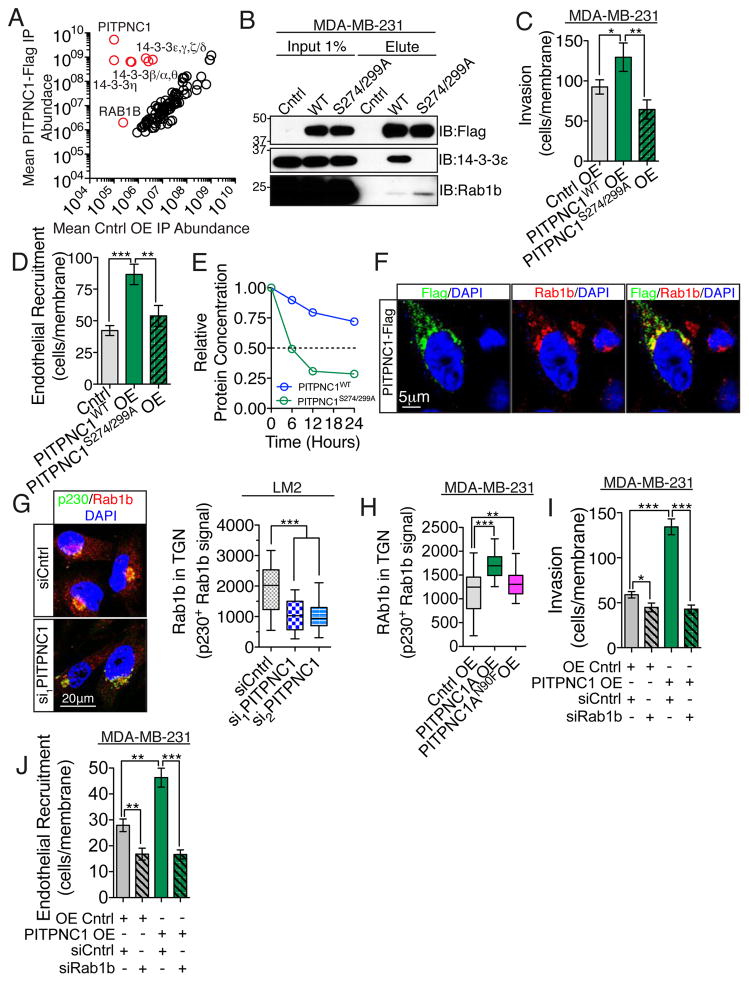
To gain further mechanistic insights into the cellular function of PITPNC1, we next sought to identify potential PITPNC1 interacting proteins. To this end, we expressed Flag-tagged PITPNC1 in MDA-MB-231 breast cancer cells and performed immunoprecipitation of the Flag epitope followed by in-solution trypsin digestion and label free mass spectrometry quantification. This revealed that PITPNC1 binds significantly to six isoforms of the 14-3-3 protein family and to the GTPase RAB1B (Figure 3A). 14-3-3 proteins are scaffolding proteins that bind phosphorylated serine residues. Previously, 14-3-3 was found to bind phosphorylated PITPNC1 serine-274 and serine-299 in a manner that protects PITPNC1 from degradation (Garner et al., 2011). Western blot analysis of immunoprecipitated protein from wild-type and S274/299A mutant PITPNC1 confirmed that the interaction with 14-3-3 is dependent on serine 274 and 299 phosphorylation (Figure 3B). We next asked if binding to 14-3-3 is required for the pro-metastatic function of PITPNC1 by performing invasion and endothelial recruitments assays with both wild-type and the S274/299A serine mutant of PITPNC1. We found 14-3-3 binding is necessary for PITPNC1-mediated pro-metastatic phenotypes (Figure 3C,D). As expected, this effect is likely explained by the shorter half-life of the S274/299A mutant (Figure 3E). In addition to 14-3-3, we identified a novel binding partner for PITPNC1, the small GTPase RAB1B (Figure 3A,B). To further elucidate the cellular function of the interaction between PITPNC1 and RAB1B, we performed co-localization experiments and found that the vast majority of the overlap between the two proteins is detected in the perinuclear Golgi region (Figure 3F). Given our finding that PITPNC1 localization to the Golgi is required for its ability to promote metastasis and that RAB1B regulates Golgi function (Dugan et al., 1995; Haas et al., 2007; Plutner et al., 1991), we next investigated whether PITPNC1 promotes metastasis by enhancing the abundance of RAB1B in the Golgi. Immunofluorescence analysis revealed a 1.8-fold reduction of endogenous RAB1B in the trans-Golgi upon depletion of PITPNC1 (Figure 3G, Figure S3A,B) without any accompanying change in total cellular RAB1B protein levels as determined by western blotting (Figure S3C). Conversely, over-expression of wild-type PITPNC1 led to a significant increase in Golgi-localized RAB1B (Figure 3H, Figure S3D). Consistent with the effect of PITPNC1 on Golgi-localization of RAB1B being dependent on its PI4P-binding capacity, over-expression of the lipid-binding mutant of PITPNC1 minimally impacted RAB1B abundance in the Golgi (Figure 3H).
Figure 3. PITPNC1 forms a protein complex with 14-3-3 isoforms and RAB1B.
(A) Lysates from MDA-MB-231 cells expressing Flag-tagged PITPNC1 or empty control vector were subjected to immunoprecipitation by anti-Flag beads. The eluate was trypsin digested in solution and the liquid chromatography-tandem mass spectrometry (LC-MS/MS) spectra were analyzed by label free quantification. Comparison of eluted proteins by the empty vector (horizontal axis) and PITPNC1-Flag (vertical axis) revealed PITPNC1, several 14-3-3 protein forms and RAB-1B to be significantly different (p<0.05) between the two samples. n=3/group.
(B) Western blot analysis of input and immunoprecipitated Flag-tagged wild-type and S274/299A mutant forms of PITPNC1 from MDA-MB-213 cells using anti-Flag, anti-14-3-3ε and anti-RAB1B antibodies.
(C,D) MDA-MB-231 cells expressing empty control vector, wild-type or S274/299A mutant PITPNC1 were subjected to invasion (C) and endothelial recruitment assays (D). n=4/group.
(E) The half life of wild-type and S274/299 mutant PITPNC1 in MDA-MB-231 cells were determined by treating cells with 100ug/ml cyclohexamide over 24 hours and analyzing PITPNC1 abundance in cellular lysates by western blotting at the given time points. The experiment was repeated twice.
(F) Immunofluorescence analysis of Flag-tagged PITPNC1 and endogenous RAB1B by staining with anti-Flag (green) and anti-RAB1B (red) in MDA-MB-231 breast cancer cells.
(G) Golgi levels of endogenous RAB1B in LM2 breast cancer cells treated with control or two independent siRNAs targeting PITPNC1. Areas of the trans-Golgi compartment were defined by anti-p230 (green) immunoreactivity. Analysis of the anti-RAB1B signal intensity (red) in p230 positive areas defined the Golgi specific RAB1B signal. n=30/group.
(H) MDA-MB-231 cells expressing an empty control vector, wild-type or N88F lipid binding mutant PITPNC1 were subjected to the same analysis as in (G). n=46/group.
(I,J) Matrigel invasion (I) and endothelial recruitment assay (J) of MDA-MB-231 cells expressing empty control vector or PITPNC1. 48 hours prior to the experiments both the control and PITPNC1 overexpressing cells were transfected with control siRNA or siRNA targeting RAB1B. n=4/group.
In figure C,D, I and J error bars represents S.E.M. Box and whiskers plots represent the data in figure G,H with the upper and lower bars showing minimum and maximum data points.
See also Figure S3.
These results suggest that PITPNC1 regulates RAB1B Golgi localization to mediate its metastatic effects. To functionally test this relationship, we performed epistasis experiments by assessing the ability of PITPNC1 to promote invasion and endothelial recruitment in the absence of RAB1B. RAB1B depletion completely abrogated the PITPNC1-mediated enhancement of invasion and endothelial recruitment capacity (Figure 3I,J, Figure S3E,F). Importantly, the reduction in invasion and endothelial recruitment was more pronounced in PITPNC1 over-expressing cells relative to control cells. These finding are consistent with RAB1B acting as a downstream effector of PITPNC1-mediated metastasis. Collectively, these findings reveal that PITPNC1 interacts with RAB1B in the Golgi to drive metastasis.
PITPNC1-dependent RAB1B localization to the Golgi affects Golgi structure and optimizes secretory function
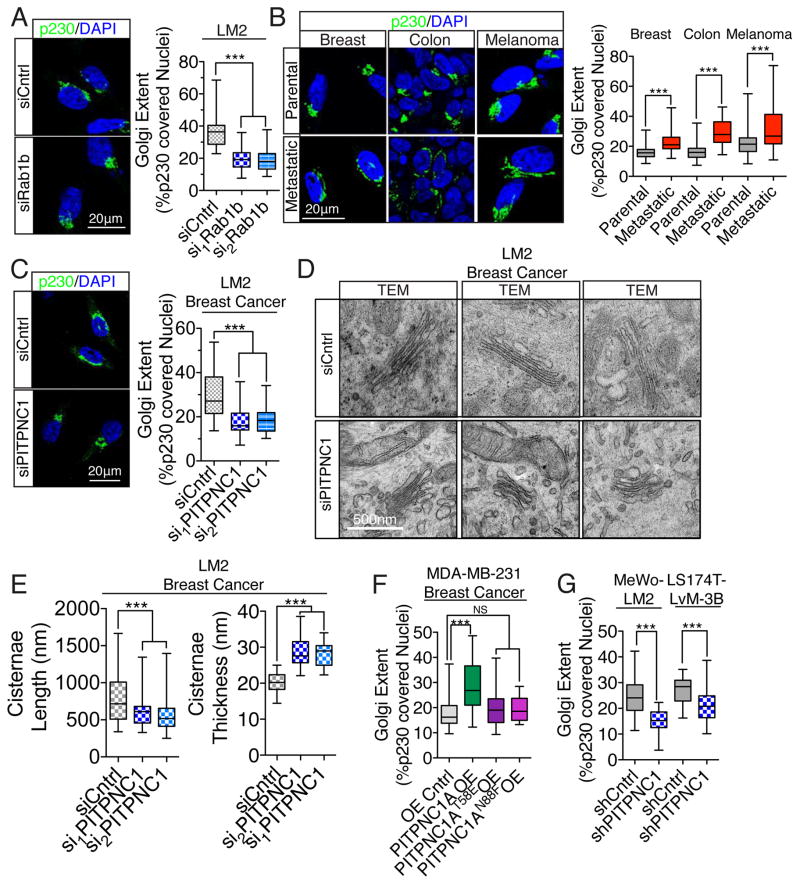
Our findings suggest a critical role for Golgi-localized RAB1B in PITPNC1-mediated metastasis. Previous studies have described a crucial role for RAB1B in maintaining Golgi structure and function (Romero et al., 2013). The Golgi network consists of stacks of membrane-bound structures. Proteins destined for secretion are transported to the trans face of the Golgi network—from which vesicular budding and release occurs (Kepes et al., 2005). In actively secreting cells, enhanced vesicular budding results in extension of the trans-Golgi network, which is ‘stretched’ by tensile forces from actin cytoskeletal proteins interfacing with proteins bound to vesicular membranes mediating vesicular release (Dippold et al., 2009; Lazaro-Dieguez et al., 2006). While investigating the phenotypic effect of RAB1B depletion, we noted a clear impact on Golgi morphology. We found that acute depletion of RAB1B led to the condensation of the Golgi (Figure 4A, Figure S3E). We next asked if changes in Golgi structure is a morphological feature that correlates with metastatic capacity in breast, colon and skin cancers by comparing the Golgi morphology between poorly metastatic parental cell populations and their in vivo-selected highly metastatic derivative sublines (Loo et al., 2015; Minn et al., 2005; Pencheva et al., 2012). Interestingly, we found that in these cancers, highly metastatic sub-populations consistently exhibited more extended Golgi structure compared to their poorly metastatic parental cell population (Figure 4B). This suggests that altered Golgi structure and function may contribute to metastatic capacity. We next wondered if PITPNC1 regulates such Golgi morphology. Indeed, PITPNC1 depletion caused the Golgi to condense as analyzed by both immunofluorescence and transmission electron microscopy (TEM) in breast cancer cells (Figure 4C,D). In addition to cisternae length, PITPNC1 depletion resulted in increased cisternae thickness, but did not significantly affect the number of Golgi stacks or the number of Golgi associated vesicles (Figure 4E, Figure S4A). Conversely, gain-of-function experiments revealed that PITPNC1 expression promotes Golgi cisternae length and reduces cisternae thickness, consistent with PITPNC1 functioning as a metastasis promoter (Figure S4B). Additionally, the ability of PITPNC1 to extend the Golgi was dependent on its binding to PI4P, as the N88F lipid-binding mutant was unable to alter Golgi structure (Figure 4F, Figure S4C). Consistent with our in vivo metastasis results, PITPNC1 also promoted Golgi extension in melanoma and colon cancer cells, suggesting that PITPNC1 may impact the metastatic phenotypes of these cancer types via its effect on Golgi function (Figure 4G, Figure S4D).
Figure 4. PITPNC1 promotes Golgi extension, a phenotype associated with metastasis.
(A) LM2 cells transfected with either control or siRNAs targeting RAB1B for 24 hours were immuno-stained for p230 and DAPI. The Golgi extent was quantified as the fraction of the nucleus circumference that was covered by p230 positive Golgi signal. n= 40/group.
(B) Highly metastatic derivates of the poorly metastatic MDA-MB-231 breast-, Ls714 colon-, and MeWo melanoma cancer cells developed previously (Minn et al., 2005; Pencheva et al., 2012) as well as their parental cell populations were analyzed for Golgi extent as in (A). n=40/group.
(C) Golgi extent was analyzed by immunohistochemistry of LM2 breast cancer cells transfected with siRNAs targeting PITPNC1 or a control siRNA. n=50/group.
(D) Golgi structure was analyzed in TEM images of LM2 cells transfected with siRNAs targeting PITPNC1 or a control siRNA. n=50/group.
(E) Images from (D) were quantified for cisternae length and thickness.
(F) Golgi extent was analyzed in MDA-MB-231 breast cells expressing empty control vector, wild-type, T58E or N88F mutant forms of PITPNC1. n=20/group.
(G) Golgi extent analyzed in highly metastatic MeWo and Ls714 subpopulations expressing either a control or PITPNC1 targeting hairpin. n=25/group.
Box and whiskers plots represent the data in figure A–G with the upper and lower bars showing minimum and maximum data points.
See also Figure S4.
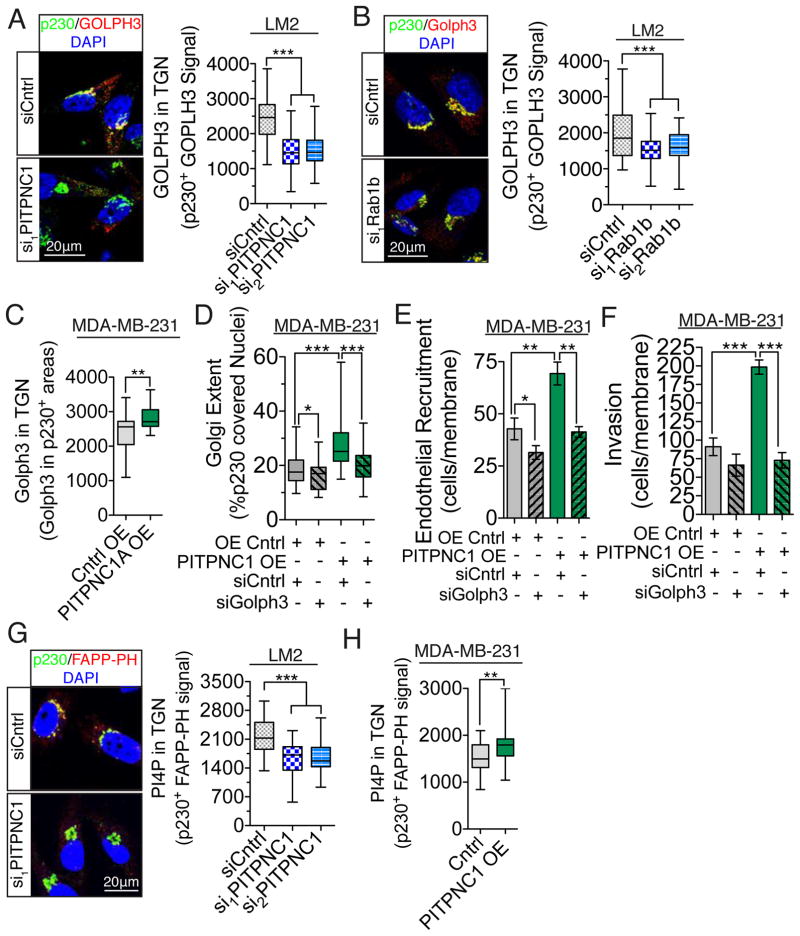
Having demonstrated that PITPNC1 promotes Golgi extension and that Golgi extension correlates with increased metastatic capability, we next sought to examine how the PITPNC1-RAB1B complex regulates Golgi structure. The Golgi structure condensation (decreased cisternae length and increased thickness) we observed upon acute depletion of either PITPNC1 or RAB1B phenocopies depletion of another protein, GOLPH3, which bridges Golgi PI4P and the actomyosin MYO18A to promote vesicle release and associated Golgi extension (Dippold et al., 2009). Increased GOLPH3 abundance in the trans-Golgi would therefore allow for increased Golgi extension and vesicular release. To this end, we performed immunofluorescence analysis of GOLPH3 and the trans-Golgi marker p230 in the setting of either PITPNC1 or RAB1B depletion. Indeed, we found that depletion of either PITPNC1 or RAB1B led to an approximate 2-fold reduction in GOLPH3 Golgi abundance, without changing the total cellular GOLPH3 protein content (Figures 5A,B; Figure S5A). Over-expression of PITPNC1 caused an increase in GOLPH3 Golgi levels (Figure 5C, Figure S5B). We then asked if PITPNC1-dependent recruitment of GOLPH3 to the Golgi is required for PITPNC1-mediated phenotypes. We found that PITPNC1-mediated Golgi extension is dependent on Golgi GOLPH3 expression, as depletion of GOLPH3 reversed the extended Golgi phenotype induced by PITPNC1 overexpression by 63% (Figure 5D, Figure S5C–E). Furthermore, in accordance with the findings of Dippold et al., (Dippold et al., 2009) we found that depletion of GOLPH3 in malignant cells condensed the Golgi in breast cancer cells (Figure 5D). Additional epistasis experiments confirmed that GOLPH3 is functionally downstream of PITPNC1-mediated invasion and endothelial recruitment phenotypes (Figure 5E,F). As GOLPH3 is recruited to the Golgi through PI4P, we next wondered if the PITPNC1-RAB1B complex recruits GOLPH3 to the Golgi complex through regulating the local Golgi PI4P abundance. Interestingly, co-immunofluorescence imaging using the trans-Golgi marker p230 with the FAPP1-PH domain and an anti-PI4P antibody revealed that PITPNC1 knockdown significantly reduced trans-Golgi PI4P abundance, while PITPNC1 over-expression significantly increased trans-Golgi PI4P levels (Figures 5G,H; Figure S5F–H). This indicates that PI4P abundance in the trans-Golgi is governed by PITPNC1 expression in cancer cells and leads to a model whereby PITPNC1 recruits RAB1B to the Golgi and augments Golgi PI4P levels. Increased Golgi PI4P abundance enhances the binding of GOLPH3 to the Golgi—allowing for Golgi extension and vesicular release. PITPNC1 could enhance Golgi PI4P abundance through multiple mechanisms such as recruitment of a specific PI4-kinase to the Golgi or by enhancing the activity of Golgi-resident PI4-kinases. To identify a potential PI4-kinase that could mediate this effect, we performed invasion experiments analyzing the epistatic relationship between PITPNC1 and each of the four PI4-kinases. This revealed that only depletion of PI4KIIIalpha abolished PITPNC1-induced invasion, while not affecting baseline invasion capacity (Supplemental Figure 5I). While these data suggest that PI4KIIIalpha could represent a downstream mediator of PITPNC1 enhanced Golgi PI4P abundance, further studies are required to mechanistically establish this relationship.
Figure 5. PITPNC1 facilitates recruitment of GOLPH3 to the Golgi.
(A,B) LM2 cells transfected with either control siRNA or siRNAs targeting PITPNC1 (A) or RAB1B (B) were immunocytochemically stained for endogenous GOLPH3 and p230. GOLPH3 levels in the trans-Golgi were quantified as mean fluorescence intensity of GOLPH3 in p230-positive regions. n=40/group.
(C) MDA-MB-231 cells expressing PITPNC1 or control vector were analyzed for trans-Golgi GOLPH3 levels as in (A).
(D) Golgi extent analyzed in MDA-MB-231 cells transfected with GOLPH3 siRNA or control siRNA in the setting of PITPNC1 or control over-expression. n=30/group.
(E,F) MDA-MB-231 cells were transfected with GOLPH3 siRNA or control siRNA in the setting of control or PITPNC1 over-expression and subjected to the invasion (G) and endothelial recruitment (H) assays. n=4/group.
(G,H) LM2 cells transfected with siRNAs targeting PITPNC1 or a control siRNA (G) or MDA-MB-231 cells over-expressing PITPNC1 or control vector (H) were stained for PI4P using FAPP-PH domain (red), p230 (green) and DAPI (blue). PI4P levels in the trans-Golgi were quantified as mean fluorescence intensity of FAPP1-PH signal in p230-positive regions. n=50/group.
Box and whiskers plots represent the data in figure A–D and G,H with the upper and lower bars showing minimum and maximum data points. In figure E and F, error bars represents S.E.M.
See also Figure S5.
The above findings reveal a pathway wherein metastatic cancer cells up-regulate PITPNC1, which mediates GOLPH3 recruitment to the trans-Golgi. This recruitment extends Golgi morphology and leads to the enhancement of pro-metastatic phenotypes of invasion and endothelial recruitment.
PITPNC1 drives metastasis by regulating the secretion of pro-invasive and pro-angiogenic genes
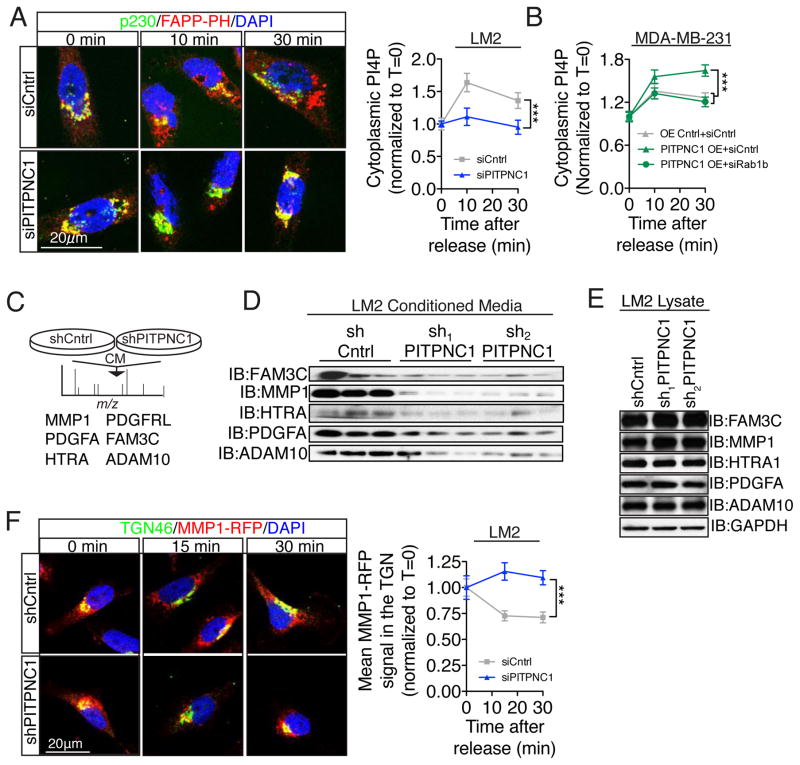
Multiple steps in the metastatic cascade are orchestrated by the secretion of regulatory proteins from cancer cells in the microenvironment. We have thus far demonstrated that PITPNC1 in concert with RAB1B promotes metastasis by recruiting GOLPH3 to extend the Golgi morphology. Given that PITPNC1 regulates invasion and endothelial recruitment (Figures 1G,H) which are both mediated by secretion of proteins, that RAB1B is critical for Golgi function (Dugan et al., 1995; Plutner et al., 1991), and that Golgi extension reflects enhanced PI4P/GOLPH3/MYO18A activity to promote vesicular release (Dippold et al., 2009), we hypothesized that the pro-metastatic effects of PITPNC1 are mediated through enhanced secretory capacity. This model is also consistent with our previous finding that PITPNC1 acts upstream of secreted IGFBP2 in miR-126 regulated metastasis (Png et al., 2012). To directly test this, we followed Golgi vesicular release by tracking vesicular PI4P in the cytoplasm in a Golgi exit assay (Figure S6A). Interestingly, PITPNC1 depletion significantly reduced the release of PI4P-containing vesicles from the Golgi (Figure 6A). Conversely, PITPNC1 over-expression increased the secretory capacity of the Golgi in a RAB1B-dependent manner in MDA-MB-231 cells (Figure 6B, Figure S6B).
Figure 6. PITPNC1 facilitates malignant secretion.
(A) Golgi exit assay analysis of LM2 cells transfected with control or PITPNC1-targeting siRNA. Following a 2 hour incubation at 23°C in the presence of 100μg/mL cyclohexamide, the cells were returned to 37°C and prepared for immunofluorescence for PI4P (FAPP-PH) and p230 analysis at time 0, 10, and 30 min. The abundance of PI4P containing vesicles released to the cytoplasm was determined by subtracting Golgi localized PI4P from the total cellular PI4P signal. All measurements of cytoplasmic PI4P were blinded. n=10/time-point/group. Significance test between groups was performed with Fisher’s method.
(B) Golgi exit assay in MDA-MB-231 cells expressing either a control vector or PITPNC1 overexpressing cells that was transfected with control siRNA or RAB1B targeting siRNA. n=20/time point. Significance test between groups was performed with Fisher’s method.
(C) Conditioned media was collected from SILAC-labeled LM2 control and PITPNC1 knockdown cells and subjected to liquid chromatography-mass spectrometry (LC-MS/MS) to identify proteins underrepresented in PITPNC1 knockdown media.
(D) Western blot analysis for PDGFA, HTRA1, MMP1, ADAM10, and FAM3C in conditioned media from LM2 cells expressing short hairpins targeting PITPNC1 or a control hairpin.
(E) Western blot analysis for PDGFA, HTRA1, MMP1, ADAM10, and FAM3C in cellular lysates from LM2 cells expressing short hairpins targeting PITPNC1 or a control hairpin.
(F) Golgi exit assay of control and PITPNC1 knockdown LM2 cells tracking RFP-labeled MMP1. TGN localization was defined as TGN46-postive regions. n=25/timepoint/group. Significance test between groups was performed with Fisher’s method.
Error bars represent S.E.M.
See also Figure S6.
These findings suggest that the PITPNC1-RAB1B complex promotes the metastatic potential of cancer cells by enhancing the Golgi secretion capacity. To identify the set of secreted proteins that mediate the pro-metastatic effects of PITPNC1, we conducted stable isotope labeling by amino acids in cell culture (SILAC) on media conditioned by control versus PITPNC1-depleted cells. Proteomic analysis revealed 6 proteins (MMP1, PDGFA, HTRA1, PDGFRL, FAM3C, and ADAM10) that are all known to be expressed by breast cancer cells that were at least two-fold less abundant in conditioned media from PITPNC1-depleted cells relative to control cells (Figure 6C). Western blot analysis of conditioned media confirmed that the secreted protein levels of all these factors except PDGFRL decreased significantly upon PITPNC1 knockdown (Figure 6D, Figure S6C). These effects were not secondary to reduced protein production as no differences in total cellular protein abundance were observed (Figure 6E). Conversely, overexpression of PITPNC1 leads to the increased secretion of all five proteins whitout changing the overall intracellular protein levels (Figure S6D,E). These findings are consistent with a model wherein these secreted factors are not released from the Golgi upon PITPNC1 depletion as a consequence of impaired PI4P/GOLPH3 function. To further test this, we used MMP1-RFP as a marker for metastatic secretion in a Golgi exit assay. Consistent with our model, PITPNC1 depletion significantly reduced MMP1-RFP release from the TGN (Figure 6F).
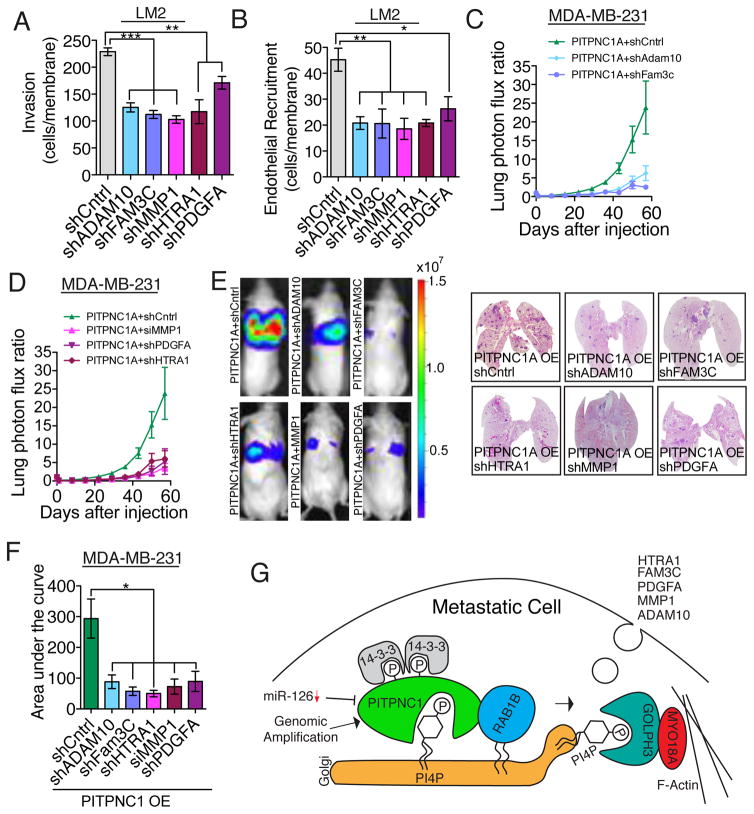
Our model thus predicts that FAM3C, MMP1, HTRA1, PDGFA and ADAM10 are pro-metastatic and pro-angiogenic genes that may drive metastasis. To test this, we first depleted each gene in highly metastatic LM2 cells and assessed their ability to invade and to recruit endothelial cells. Both invasion and endothelial recruitment phenotypes by the cancer cells were highly dependent on each of these proteins to varying degrees (Figure 7A,B, Figure S7A). This suggests that in vivo-selected metastatic cells are optimized for maximal invasion and endothelial recruitment capacity through the enhanced secretion of multiple factors that cooperate in these processes. Moreover, these findings reveal that these secreted factors similarly impact the movement of both cancer cells and endothelial cells through extracellular matrix.
Figure 7. PITPNC1 promotes metastasis by facilitating secretion of pro-invasive and pro-angiogenic genes.
(A,B) LM2 cells transduced with short hairpins targeting ADAM10, FAM3C, MMP1, HTRA1, PDGFA or a control, were subjected to the matrigel invasion assay (A) and endothelial recruitment assay (B).
(C–E) Bioluminescence imaging plot of lung metastatic colonization by 40,000 MDA-MB-231 cells transduced with a control shRNA or shRNAs targeting ADAM10, FAM3C, HTRA, PDGFA or a siRNA targeting MMP1 in the setting of PITPNC1 over-expression. n=6/group. Right, representative lung bioluminescence images.
(F) Area under the curve quantification of (C and D).
(G) Model of PITPNC1-mediated metastatic secretion through recruitment of RAB1B to the trans-Golgi compartment of the cell.
Error bars represent S.E.M.
See also Figure S7.
Given the individual importance of each of these secreted factors, we next investigated whether they each contributed to PITPNC1-mediated metastatic colonization. Consistent with the cooperative impact of these secreted factors on metastatic progression, we found all five of these genes to be required for the optimized metastatic colonization activity induced by PITPNC1 over-expression (Figures 7C–F). These epistatic experiments reveal PITPNC1 to drive metastasis, in part, through augmenting the cellular release of FAM3C, MMP1, HTRA1, PDGFA and ADAM10 secreted pro-metastatic proteins.
DISCUSSION
Our findings reveal PITPNC1 to be genetically amplified in nearly half of human breast tumors, over-expressed in multiple prevalent cancer types, and its expression levels to correlate with metastatic progression in melanoma, breast, and colon cancers. PITPNC1 promotes metastasis by enhancing vesicular secretion, leading to the augmented cellular release of a set of pro-metastatic genes consisting of MMP1, PDGFA, HTRA1, FAM3C, and ADAM10 (Figure 7G). PITPNC1 accomplishes this by increasing the abundance of RAB1B in the Golgi compartment of the cell. Through a complementary set of biochemical and cell-biological experiments, we have identified PI4P as the lipid substrate of PITPNC1. PITPNC1 was originally named Phosphoinositide transfer protein, cytoplasmic 1 based on its homology to the phosphoinositol transfer proteins, however it is less well characterized than other members of the PITP family. While an in vivo lipid substrate of PITPNC1 has not been previously identified, a recent study demonstrated in vitro binding and transfer of phosphatidic acid (PA) by recombinant PITPNC1 (Garner et al., 2012). In addition to our biochemical analyses, the localization of PITPNC1 to the TGN, a compartment that is enriched in PI4P, as well as our SAC1-K2A epistasis experiments, support the conclusion that PI4P is the predominant physiological in vivo substrate for PITPNC1 in the cells studied herein. Our findings expand on the molecular, cellular, and organismal role of the PITP family by suggesting that PITPNC1 acts to coordinate the morphology of the Golgi by the recruitment of RAB1B through binding to Golgi resident PI4P.
Our findings furthermore identify extended Golgi morphology as a key morphological feature of highly metastatic cells. We find that PITPNC1 in complex with RAB1B regulates Golgi PI4P levels. Enhanced Golgi PI4P levels increase the abundance of the PI4P binding protein GOLPH3–a key regulator of Golgi morphology. Our work thus identifies GOLPH3 and its function in secretion as a key component of the metastatic cascade. This is corroborated clinically by the association of GOLPH3 over-expression with poor prognosis in multiple cancer types (Scott et al., 2009; Zeng et al., 2012; Zhou et al., 2012). By enhancing the levels of PITPNC1, highly metastatic cells are able to target GOLPH3 to its site of action in the trans-Golgi.
The importance of phosphoinositides in cancer development and progression has previously mainly focused on PI(3,4,5)P3 levels, which are regulated by PI3-kinase and phosphatase tensin homolog (PTEN). PI3-kinase and PTEN constitute two of the most frequently mutated genes in cancer. The signaling pathway downstream of PI(3,4,5)P3 has also been well documented in human cancer through mechanistic and pathologic studies (Wong et al., 2010). However there exist a large number of other phosphoinositides with largely uncharacterized roles in cancer progression. Each of these PIs demonstrates spatial and temporal restriction within the cell, serving as markers of distinct cellular organelles or as platforms for signaling pathways. While previous studies have focused on the phosphorylation state of the phosphoinositides, our studies underscore the importance of phosphoinositide-mediated recruitment of effector proteins to malignant phenotypes such as metastatic invasion and endothelial recruitment.
Our unbiased proteomic analysis of metastatic cells revealed that PITPNC1 depletion impairs secretion of multiple of proteins—many of which have been previously implicated in cancer invasion and angiogenesis. These five secreted factors found to be downstream of PITPNC1 in augmenting metastasis were all found to promote both invasion and endothelial recruitment, indicating that each secreted factor impacts both breast cancer cell invasion and endothelial cell migration through interactions with the microenvironment or through activation of receptors expressed by these cell types. Several of these secreted factors have been previously implicated in cancer cell invasion. MMP1, a matrix metalloprotease, has been implicated in the early steps of metastatic progression in multiple cancer types through its role in degradation of the extracellular matrix to facilitate invasion of cancer cells (Page-McCaw et al., 2007). Cancer cell-induced remodeling of the microenvironment may also be exploited to facilitate migration of endothelial cells, thereby further enhancing cancer cells’ metastatic capacity. Two of the other secreted factors identified are also proteases: ADAM10, another metalloprotease, and HTRA1, a serine protease. These factors may also engage in a role similar to MMP1 by modifying the extracellular environment for cancer cells and endothelial cells to promote metastasis. PDGFA is an established pro-angiogenic factor that has been previously shown to be essential for tumor angiogenesis in multiple solid tumor types, including breast and colon cancers (Anan et al., 1996; Nakagawa et al., 2004). While the endothelial role of PDGF proteins is well characterized, breast cancer cells have also been found to up-regulate PDGFR (Heldin, 2013), indicating a potential cell-autonomous role for PDGF proteins in cancer progression. FAM3C, also called ILEI, is the least characterized of the PITPNC1 regulated secreted proteins. While the mechanistic basis of its action is still uncharacterized, FAM3C has been shown to associate with human breast cancer progression (Waerner et al., 2006).
PITPNC1’s clinical correlation with metastatic outcomes in breast, colon, and melanoma cancers is surprising given the diverse tissue origins of these cancer types. While both breast cancer and melanoma predominantly metastasize to the lung and brain, colon cancer metastasizes mainly to the liver and to a lesser extent the lung. However, successful metastasis of all these cancers requires invasion, extravasation, and angiogenesis—processes that require secreted factors acting on the microenvironment or on other cell types. PITPNC1’s role as a general regulator of secretion highlights the importance of the secretory pathway in the metastatic progression of these cancer types. Given its over-expression in a broad set of human cancers and its regulatory control of a large number of pro-metastatic genes, our findings suggest promise for therapeutic targeting of PITPNC1.
EXPERIMENTAL PROCEDURES
Detailed descriptions of methods are provided in the Extended Experimental Procedures.
Animal experiments
All animal experiments were conducted in accordance to a protocol approved by the Institutional Animal Care and Use Committee at Rockefeller University. Lung and liver metastasis assays by MDA-MB-231, LM2, CN34Lm1a1, MeWo-LM2 and LS174T-LvM3 were performed as described previously (Pencheva et al., 2012; Png et al., 2012). Lung colonization by triple reporter-labeled 4T1 cells was performed in 7–8 weeks old Balb/c mice. 50,000 cells were injected intravenously through tail vein administration. Lung colonization by triple reporter-labeled HCC-1806 cells were performed in NOD-SCIDgamma mice by injecting 100,000 cells intravenously through the tail vein.
Cell Lines
All breast and melanoma cell lines were propagated as described previously (Pencheva et al., 2012; Png et al., 2012). The liver metastatic LS174T-LvM3 subline was derived from the human LS174 colon cancer line through rounds of in vivo selection (Loo et al., 2015). LS174T-LvM3 colorectal cancer and 4T1 breast cancer cells were propagated in DMEM supplemented with 10% FBS.
Matrigel Invasion Assay
Cancer cells were conditioned in 0.2% FBS DMEM-based media for 16 hr. 50,000 cells in starvation medium were seeded per well into matrigel-coated trans-well inserts (BD Biosciences) and incubated at 37°C for 20 hr. The inserts were rinsed in PBS, the apical side of each insert was scraped to remove cells, and inserts were fixed in 4% paraformaldehyde for 15 min. The inserts were excised and mounted onto slides using VectaShield mounting medium containing DAPI (Vector Laboratories). The basal side of each insert was imaged using an inverted fluorescence microscope (Zeiss Axiovert 40 CFL) at 10X magnification. Five representative images were obtained for each insert. The number of invaded cells was quantified using ImageJ software (NIH).
Endothelial Recruitment Assay
50,000 cancer cells were seeded into 24-well plates approximately 24 hr prior to the start of the assay. HUVEC cells were serum-starved in EGM-2 media (Lonza) supplemented with 0.2% FBS for 20 hr. Cancer cells were washed with PBS and 1 mL 0.2% FBS EGM-2 medium was added to each well. Each well was then fitted with a 3.0μm HTS Fluoroblock trans-well migration insert (BD Falcon). 80,000 HUVECs, resuspended in 0.5 mL of starvation media, were seeded into each insert and incubated at 37°C for 20 hr. The inserts were processed and analyzed as described for the matrigel invasion assay.
Immunofluorescence Imaging
Immunofluorescence detection of PI4P was performed as described in (Hammond et al., 2009). Briefly, cells were fixed and permeabilized with 20μM digitonin before staining using recombinant FAPP-PH or anti-PI4P (Echelon Biolscience). A more detailed description can be found in the Extended Experimental Procedures.
Golgi Exit Assay
The Golgi Exit Assay was performed with slight modifications as previously described (Bonazzi et al., 2005). Cells were incubated at 23°C for 2 hours in the presence of 100μg/mL cyclohexamide in DMEM supplemented with 0.2%FBS before being transferred to 37°C and fixed at the given time points. The cells were stained using FAPP-PH and p230 and analyzed as described in the Extended Experimental Procedures.
Stable Isotope Labeling of Amino acids in Culture (SILAC)
Secreted proteins from control and PITPNC1 depleted cells were analyzed in conditioned media using SILAC. Briefly, cells were grown in DMEM-Flex media contained in the SILAC Protein ID & Quantitiation Kit (Invitrogen) and changed to serum-free DMEM-Flex media 16 hours prior to collection of conditioned media. Proteins were separated by 1D gel electrophoresis and subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS). Further details may be found in the Extended Experimental Procedures.
Data analysis
The prognostic power of PITPNC1 to predict colorectal cancer metastasis-free survival was tested for significance using the Mantel–Cox log-rank test performed using Graphpad Prism 5 software. Fishers combined probability test were calculated to compare groups for the Golgi exit assays using Graphpad Prism 5 software. For all other comparisons, the one-tailed t-test was used. Symbols were used as follows * P<0.05, ** P<0.01, *** P<0.001. P≤0.05 was considered statistically significant.
Supplementary Material
SIGNIFICANCE.
Secretion of pro-tumorigenic proteins is a required feature for efficient metastatic colonization. We find that PITPNC1–a protein whose expression correlates with tumor progression in breast cancer, colon cancer and melanoma–regulates metastatic secretion. PITPNC1 does this by recruiting the small GTPase RAB1B to the trans-Golgi compartment of the cell by binding to the Golgi resident lipid PI4P. In the Golgi, the PITPNC1/RAB1B complex enhances metastatic secretion capacity by recruiting the protein GOLPH3 to the Golgi. This enables the enhanced release of vesicles containing effector proteins.
Acknowledgments
We thank members of the Tavazoie laboratory for comments on previous versions of this manuscript and Seth Field (University of California, San Diego) for insightful discussions. The Sac1-K2A construct was a kind gift from Dr. Peter Mayinger, University of Oregon (Rohde et al., 2003). We thank S. Chandarlapaty at MSKCC, USA for providing HCC-1806 and BT549 breast cancer cells. N.H. is a Rockefeller University Anderson Center for Cancer Research Postdoctoral Fellow and recipient of the Bergen Research Foundation Recruitment Program. C.A.S. was supported by a National Science Foundation Graduate Research Fellowship. K.N was supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program. S.F.T is a Department of Defense Era of Hope Scholar and a Rita Allen Foundation Scholar.
Footnotes
AUTHOR CONTRIBUTIONS
S.F.T. conceived the project and supervised all research. C.A.S., N.H., and S.F.T. wrote the manuscript. C.A.S., N.H., and K.N. designed, performed, and analyzed the experiments. H.M. performed mass spectrometry and analyzed results. K.U. performed electron microscopy.
Information supplementing this article consists of seven figures, four tables, and extended experimental procedures
References
- Anan K, Morisaki T, Katano M, Ikubo A, Kitsuki H, Uchiyama A, Kuroki S, Tanaka M, Torisu M. Vascular endothelial growth factor and platelet-derived growth factor are potential angiogenic and metastatic factors in human breast cancer. Surgery. 1996;119:333–339. doi: 10.1016/s0039-6060(96)80120-6. [DOI] [PubMed] [Google Scholar]
- Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FA. Review series: Rab GTPases and membrane identity: causal or inconsequential? J Cell Biol. 2013;202:191–199. doi: 10.1083/jcb.201306010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonazzi M, Spano S, Turacchio G, Cericola C, Valente C, Colanzi A, Kweon HS, Hsu VW, Polishchuck EV, Polishchuck RS, et al. CtBP3/BARS drives membrane fission in dynamin-independent transport pathways. Nat Cell Biol. 2005;7:570–580. doi: 10.1038/ncb1260. [DOI] [PubMed] [Google Scholar]
- Chiang AC, Massague J. Molecular basis of metastasis. N Engl J Med. 2008;359:2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippold HC, Ng MM, Farber-Katz SE, Lee SK, Kerr ML, Peterman MC, Sim R, Wiharto PA, Galbraith KA, Madhavarapu S, et al. GOLPH3 bridges phosphatidylinositol-4- phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139:337–351. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan JM, deWit C, McConlogue L, Maltese WA. The Ras-related GTP-binding protein, Rab1B, regulates early steps in exocytic transport and processing of beta-amyloid precursor protein. J Biol Chem. 1995;270:10982–10989. doi: 10.1074/jbc.270.18.10982. [DOI] [PubMed] [Google Scholar]
- Garner K, Hunt AN, Koster G, Somerharju P, Groves E, Li M, Raghu P, Holic R, Cockcroft S. Phosphatidylinositol transfer protein, cytoplasmic 1 (PITPNC1) binds and transfers phosphatidic acid. J Biol Chem. 2012;287:32263–32276. doi: 10.1074/jbc.M112.375840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner K, Li M, Ugwuanya N, Cockcroft S. The phosphatidylinositol transfer protein RdgBbeta binds 14-3-3 via its unstructured C-terminus, whereas its lipid-binding domain interacts with the integral membrane protein ATRAP (angiotensin II type I receptor-associated protein) Biochem J. 2011;439:97–111. doi: 10.1042/BJ20110649. [DOI] [PubMed] [Google Scholar]
- Haas AK, Yoshimura S, Stephens DJ, Preisinger C, Fuchs E, Barr FA. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J Cell Sci. 2007;120:2997–3010. doi: 10.1242/jcs.014225. [DOI] [PubMed] [Google Scholar]
- Hammond GR, Schiavo G, Irvine RF. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2) Biochem J. 2009;422:23–35. doi: 10.1042/BJ20090428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Haqq C, Nosrati M, Sudilovsky D, Crothers J, Khodabakhsh D, Pulliam BL, Federman S, Miller JR, 3rd, Allen RE, Singer MI, et al. The gene expression signatures of melanoma progression. Proc Natl Acad Sci U S A. 2005;102:6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin CH. Targeting the PDGF signaling pathway in tumor treatment. Cell communication and signaling: CCS. 2013;11:97. doi: 10.1186/1478-811X-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang QH, Gumireddy K, Schrier M, Le Sage C, Nagel R, Nair S, Egan DA, Li AP, Huang GH, Klein-Szanto AJ, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nature Cell Biology. 2008;10:202–U283. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Kepes F, Rambourg A, Satiat-Jeunemaitre B. Morphodynamics of the secretory pathway. Int Rev Cytol. 2005;242:55–120. doi: 10.1016/S0074-7696(04)42002-6. [DOI] [PubMed] [Google Scholar]
- Lazaro-Dieguez F, Jimenez N, Barth H, Koster AJ, Renau-Piqueras J, Llopis JL, Burger KN, Egea G. Actin filaments are involved in the maintenance of Golgi cisternae morphology and intra-Golgi pH. Cell motility and the cytoskeleton. 2006;63:778–791. doi: 10.1002/cm.20161. [DOI] [PubMed] [Google Scholar]
- Loo JM, Scherl A, Nguyen A, Man FY, Weinberg E, Zeng Z, Saltz L, Paty PB, Tavazoie SF. Extracellular metabolic energetics can promote cancer progression. Cell. 2015;160:393–406. doi: 10.1016/j.cell.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayinger P. Phosphoinositides and vesicular membrane traffic. Biochim Biophys Acta. 2012;1821:1104–1113. doi: 10.1016/j.bbalip.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan SC, Alb JG, Jr, Elagina RB, Bankaitis VA, Hyde DR. The phosphatidylinositol transfer protein domain of Drosophila retinal degeneration B protein is essential for photoreceptor cell survival and recovery from light stimulation. J Cell Biol. 1997;139:351–363. doi: 10.1083/jcb.139.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Liyanarachchi S, Davuluri RV, Auer H, Martin EW, Jr, de la Chapelle A, Frankel WL. Role of cancer-associated stromal fibroblasts in metastatic colon cancer to the liver and their expression profiles. Oncogene. 2004;23:7366–7377. doi: 10.1038/sj.onc.1208013. [DOI] [PubMed] [Google Scholar]
- Nile AH, Tripathi A, Yuan P, Mousley CJ, Suresh S, Wallace IM, Shah SD, Pohlhaus DT, Temple B, Nislow C, et al. PITPs as targets for selectively interfering with phosphoinositide signaling in cells. Nat Chem Biol. 2014;10:76–84. doi: 10.1038/nchembio.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15:546–554. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, Tavazoie SF. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151:1068–1082. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plutner H, Cox AD, Pind S, Khosravi-Far R, Bourne JR, Schwaninger R, Der CJ, Balch WE. Rab1b regulates vesicular transport between the endoplasmic reticulum and successive Golgi compartments. J Cell Biol. 1991;115:31–43. doi: 10.1083/jcb.115.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481:190–194. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- Rohde HM, Cheong FY, Konrad G, Paiha K, Mayinger P, Boehmelt G. The human phosphatidylinositol phosphatase SAC1 interacts with the coatomer I complex. J Biol Chem. 2003;278:52689–52699. doi: 10.1074/jbc.M307983200. [DOI] [PubMed] [Google Scholar]
- Romero N, Dumur CI, Martinez H, Garcia IA, Monetta P, Slavin I, Sampieri L, Koritschoner N, Mironov AA, De Matteis MA, et al. Rab1b overexpression modifies Golgi size and gene expression in HeLa cells and modulates the thyrotrophin response in thyroid cells in culture. Mol Biol Cell. 2013;24:617–632. doi: 10.1091/mbc.E12-07-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KL, Kabbarah O, Liang MC, Ivanova E, Anagnostou V, Wu J, Dhakal S, Wu M, Chen S, Feinberg T, et al. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature. 2009;459:1085–1090. doi: 10.1038/nature08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano N, Owada Y, Suzuki R, Sakagami H, Shimosegawa T, Kondo H. Cloning and characterization of a novel variant (mM-rdgBbeta1) of mouse M-rdgBs, mammalian homologs of Drosophila retinal degeneration B gene proteins, and its mRNA localization in mouse brain in comparison with other M-rdgBs. J Neurochem. 2003;84:829–839. doi: 10.1046/j.1471-4159.2003.01591.x. [DOI] [PubMed] [Google Scholar]
- Talmadge JE, Fidler IJ. AACR centennial series: the biology of cancer metastasis: historical perspective. Cancer Res. 2010;70:5649–5669. doi: 10.1158/0008-5472.CAN-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilley SJ, Skippen A, Murray-Rust J, Swigart PM, Stewart A, Morgan CP, Cockcroft S, McDonald NQ. Structure-function analysis of human [corrected] phosphatidylinositol transfer protein alpha bound to phosphatidylinositol. Structure. 2004;12:317–326. doi: 10.1016/j.str.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Waerner T, Alacakaptan M, Tamir I, Oberauer R, Gal A, Brabletz T, Schreiber M, Jechlinger M, Beug H. ILEI: a cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell. 2006;10:227–239. doi: 10.1016/j.ccr.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Wilson AL, Erdman RA, Maltese WA. Association of Rab1B with GDP-dissociation inhibitor (GDI) is required for recycling but not initial membrane targeting of the Rab protein. J Biol Chem. 1996;271:10932–10940. doi: 10.1074/jbc.271.18.10932. [DOI] [PubMed] [Google Scholar]
- Wong KK, Engelman JA, Cantley LC. Targeting the PI3K signaling pathway in cancer. Curr Opin Genet Dev. 2010;20:87–90. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder MD, Thomas LM, Tremblay JM, Oliver RL, Yarbrough LR, Helmkamp GM., Jr Structure of a multifunctional protein. Mammalian phosphatidylinositol transfer protein complexed with phosphatidylcholine. J Biol Chem. 2001;276:9246–9252. doi: 10.1074/jbc.M010131200. [DOI] [PubMed] [Google Scholar]
- Zeng Z, Lin H, Zhao X, Liu G, Wang X, Xu R, Chen K, Li J, Song L. Overexpression of GOLPH3 promotes proliferation and tumorigenicity in breast cancer via suppression of the FOXO1 transcription factor. Clin Cancer Res. 2012;18:4059–4069. doi: 10.1158/1078-0432.CCR-11-3156. [DOI] [PubMed] [Google Scholar]
- Zhou J, Xu T, Qin R, Yan Y, Chen C, Chen Y, Yu H, Xia C, Lu Y, Ding X, et al. Overexpression of Golgi phosphoprotein-3 (GOLPH3) in glioblastoma multiforme is associated with worse prognosis. J Neurooncol. 2012;110:195–203. doi: 10.1007/s11060-012-0970-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.