Abstract
Oxidative stress is implicated in many diseases yet no simple, rapid, and robust measurement is available at the point-of-care to assist clinicians in detecting oxidative stress. Here, we report results from a discovery-based research approach in which a redox mediator is used to probe serum samples for chemical information relevant to oxidative stress. Specifically, we use an iridium salt (K2IrCl6) to probe serum for reducing activities that can transfer electrons to iridium and thus generate detectable optical and electrochemical signals. We show that this Ir-reducing assay can detect various biological reductants and is especially sensitive to glutathione (GSH) compared to alternative assays. We performed an initial clinical evaluation using serum from 10 people diagnosed with schizophrenia, a mental health disorder that is increasingly linked to oxidative stress. The measured Ir-reducing capacity was able to discriminate people with schizophrenia from healthy controls (p < 0.005), and correlations were observed between Ir-reducing capacity and independent measures of symptom severity.
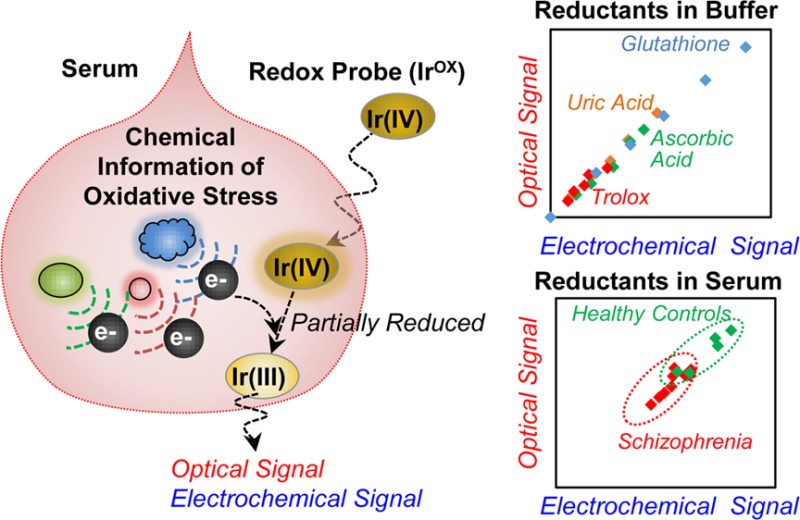
Increasing evidence links oxidative stress to the development of various diseases that include cancer, cardiovascular disease, neurodegenerative diseases, and neuropsychiatric diseases.1−4 The goal of this work is to develop a simple, rapid, objective measure of oxidative stress useful for both researchers and clinicians.5 To develop this method, we used a discovery-driven approach that is based on two underlying assumptions. First, we assume that chemical information on oxidative stress is present in serum and can be accessed by appropriate measurements. As suggested in Scheme 1a, chemical information on oxidative stress could include the oxidants believed to be responsible for damage (e.g., reactive oxygen and nitrogen species), the endogenously generated protective antioxidants (e.g., GSH and ascorbic acid), proteins involved in inflammation and protection (e.g., cytokines and defense enzymes),6,7 or the damage associated with oxidative stress (e.g., lipid peroxidation and protein carbonylation).8,9
Scheme 1. Redox Probing to Access Chemical Information of Oxidative Stress.
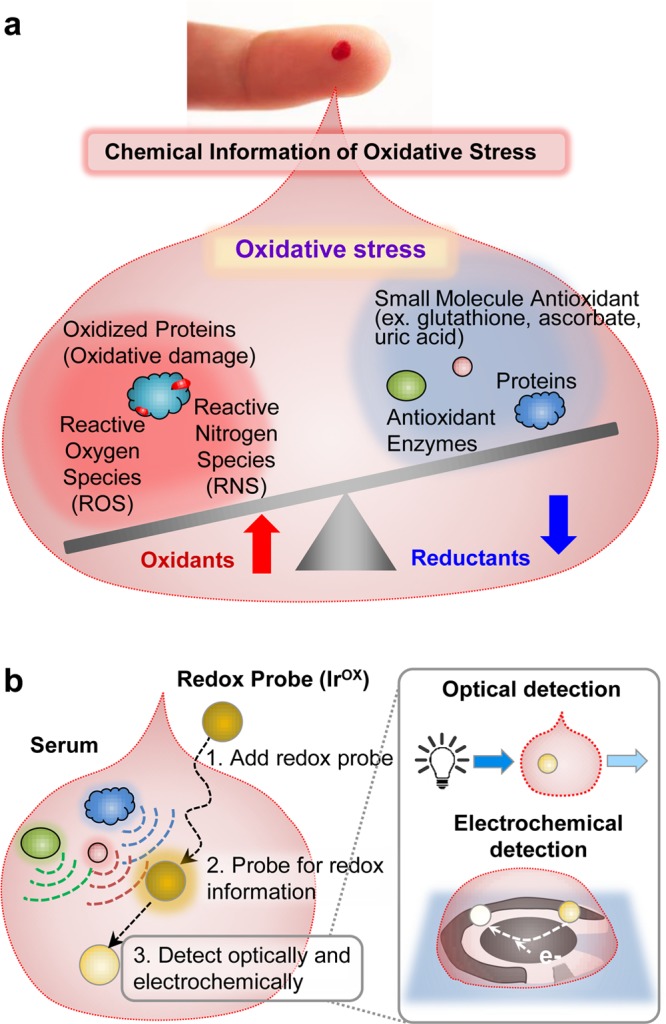
(a) Chemical information relevant to oxidative stress in blood. (b) The redox-mediator (K2IrCl6, IrOX) is used to probe for reducing activities and reports this information through optical and electrochemical modalities.
Traditional approaches to access chemical information on oxidative stress focus on chemically specific analytical methods (e.g., HPLC and mass spectrometry).8,9 Such instrument-intensive methods will likely be critical for researchers to unravel the detailed molecular mechanisms responsible for oxidative stress and to guide the development of future therapeutic interventions. However, these instrument-intensive measures do not lend themselves to the rapid, inexpensive, point-of-care analysis that could assist clinicians in diagnosing disease or tailoring treatments for today’s patients. Generally, clinicians rely on simpler tools that measure global (not chemically specific) indicators of a patient’s health. Examples of such global indicators come from physical measurements of body temperature, blood pressure, and heart rate. The goal of this work is to develop a simple, rapid method capable of probing the chemical information in serum to provide a global indicator of oxidative stress.10−12
The idea of probing a sample’s chemical information to provide a single integrated global measure is well-established in the food industry where antioxidant capacity assays are routinely used to compare the potential health beneficial activities of different foods.13−15 Variations of these antioxidant capacity assays are also being investigated for clinical analysis of oxidative stress, but each method has weaknesses when applied for serum analysis.10,13,16
The second assumption in our discovery-driven approach is that chemical signatures (i.e., biomarkers) of oxidative stress are accessible to measurements of redox activities. Historically, oxidative stress was believed to result from damage caused by reactive oxygen species (and other reactive species) and analytical measurements focused on detecting these chemical species, their generation, and consumption.7,17,18 However, emerging evidence indicates that oxidative stress is more nuanced than simply free radical damage, which may explain the general failure of therapeutic interventions that target free radicals.19−21 More recently, oxidative stress has been viewed as an imbalance between pro-oxidant and antioxidant activities,21−23 and sulfur-containing molecules (e.g., thiols) have emerged as integral chemical components in oxidative stress.22,24,25 Glutathione (GSH) is obviously linked to oxidative stress because oxidation of this sulfur-containing chemical species is a major physiological mechanism of antioxidant protection. However, the oxidation/reduction of the cysteine based sulfur switches in regulatory proteins provides the mechanisms for redox-based signals to be transduced into downstream cellular responses.26,27 Because free radicals are not obligatory intermediates in thiol oxidation, emerging theories characterize oxidative stress in terms of redox homeostasis and dysregulation.28−30 Assuming redox dysregulation is the underlying basis of oxidative stress, then we suggest: (i) redox-probing may be able to access relevant information on oxidative stress; (ii) redox-probing at a global level (vs measuring individual chemical components) may provide broader access to the relevant information on redox context; and (iii) a discovery-based approach is needed to identify relevant chemical signatures of oxidative stress (there are currently no validated hypotheses and biomarkers of oxidative stress). In essence, this second assumption states that while the chemical evidence of oxidative stress is of an unknown nature, it is accessible to a redox measurement.
Using these two assumptions and a discovery-based research approach, we developed a new method to probe for redox-information in serum samples. Scheme 1b illustrates that our method uses a redox-mediator to probe a serum sample for chemical information on oxidative stress.11 Low molecular weight, diffusible mediators are commonly used to access redox-active sites of insoluble macromolecules31,32 and internal redox centers of globular proteins (e.g., for mediated biosensing).33Scheme 1b shows that we selected K2IrCl6(IV) (designated IrOX) as our mediator. IrOx is a reasonably strong oxidant34,35 and has been shown to accept electrons from a broad range of biologically relevant reductants36 including GSH,37 ascorbate,38 and cysteine.39 The transfer of electrons from reducing species in serum to the IrOX mediator can generate both optical and electrochemical signals which are particularly convenient for rapid, point-of-care analysis.
Initially, we studied buffered solutions and observed that this Ir-reducing assay is sensitive to relevant bioreductants and especially to GSH. In subsequent studies with serum samples, we evaluated whether the Ir-reducing assay could provide clinically useful information for subjects diagnosed with a disease that is linked to oxidative stress. Specifically, we investigated correlations between the Ir-reduction values in serum and clinical measures of schizophrenia. Schizophrenia is a lifelong mental health disorder affecting about 1% of the world population,40 and increasing evidence links redox dysregulation and oxidative stress to the pathophysiology of schizophrenia.41−43 We show that the Ir-reducing assay can discriminate people diagnosed with schizophrenia from healthy controls (p < 0.005; AUC, 0.92) as well as correlate to subjective clinical measures of disease severity. We envision that the Ir-reducing assay could provide a simple, rapid tool that accesses chemical information on oxidative stress to guide both researchers and clinicians to better understand and manage oxidative stress disorders.
Results
Assay Development
Qualitative Validation of Redox Probe
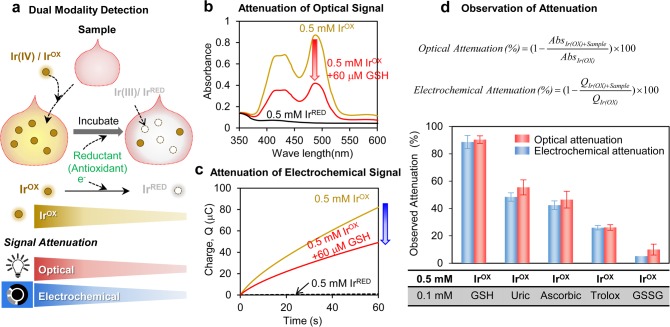
We used K2IrCl6(IV), designated IrOX, to probe our serum samples for redox information. As illustrated in Figure 1a, IrOX is a yellowish iridium(IV) complex that becomes colorless upon reduction to iridium(III), designated IrRED. As suggested in Figure 1a, the basis of our method is that we add IrOX to our serum sample to probe for reducing activities in the serum. Such reduction reactions can be detected by attenuations in either an optical signal associated with the loss of the yellow color or an electrical signal associated with a subsequent electrochemical titration of the remaining IrOX.
Figure 1.
Qualitative validation of Ir-reducing assay. (a) Scheme shows that the IrOX mediator reports reducing activities of a sample as attenuations in optical and electrochemical signals. (b) Optical signal (absorbance) attenuation and (c) electrochemical signal (reductive charge) attenuation of IrOX in the presence of a reduced glutathione (GSH). (d) Observed signal attenuations of IrOX when incubated with various reductants (measurements were performed in quadruplicate and error bars indicate standard deviation).
Attenuation of the optical signal is illustrated in Figure 1b, which shows IrOX (0.5 mM) has two strong absorbance peaks at 420 and 488 nm and these peaks are absent in the spectrum for IrRED. (Figure S1 of Supporting Information provides further details of this optical measurement.) Addition of the biological reductant glutathione (GSH; 60 μM) to the IrOX solution and incubation for 30 min was observed to attenuate this optical signal. (Figure S2 of Supporting Information shows attenuation is nearly complete after 30 min incubation.)
Attenuation of the electrochemical signal associated with the reduction of IrOX is illustrated in Figure 1c. For these measurements, a sample containing of IrOX (≈100 μL) was dropped onto the surface of a screen-printed 3 electrode system. Screen-printed electrodes were chosen because they are convenient, inexpensive, sensitive, and portable and thus are suitable for a point-of-care analysis.44 Reduction of IrOX is achieved using a constant imposed potential of 0 V vs Ag/AgCl. Figure 1c shows the reductive charge transfer for IrOX as measured by this chronocoulometry method. (Figure S3 of Supporting Information provides further details of this electrochemical measurement.) While solution containing the oxidized IrOX shows a high reductive charge transfer after 1 min (Q ≈ 80 mC), the solution containing the reduced IrRED shows minimal charge transfer (Q ≈ 1 mC). Figure 1c shows that the addition of GSH to the IrOX solution and incubation for 30 min leads to an attenuation of the reductive charge transfer.
The equations in Figure 1d show how we quantified attenuation of the optical signal (absorbance at 488 nm) and electrochemical signal (reductive charge transfer, Q, after 1 min at 0 V). Figure 1d also shows experimental results for the signal attenuation associated with various components. The reductant GSH shows the largest signal attenuation while the oxidized form of GSH, GSSG, shows the lowest signal attenuation (∼6% of GSH attenuation). Uric acid and ascorbic acid are common reductants in blood and they showed intermediate signal attenuation. Figure 1d also shows comparatively small signal attenuation for the commonly used antioxidant standard trolox.
In summary, Figure 1 provides initial evidence that IrOX can probe for redox information (e.g., the presence of reductants in a sample) and can report this information as an attenuation of signals through two separate modalities (optical and electrochemical). Importantly, the results in Figure 1d show good agreement in the measured attenuations between these two modalities.
Quantitative Validation
Intuitively, the reduction of IrOX and attenuation of the signals are expected to be linearly dependent on the concentration of reductants in the sample. To test this expectation, we mixed IrOX (0.5 mM) with varying concentrations of individual reductant, incubated for 30 min and measured signal attenuation. Figure 2a shows that attenuation of the optical signal increased linearly with concentration for various reductants. Similarly, Figure 2b shows attenuation of the electrochemical signal is linearly dependent on the reductant concentration. The slopes of the plots of Figure 2a,b provide a measure of the reductant’s ability to reduce IrOX and this Ir-reducing capacity follows the trend GSH ≫ uric acid ≈ ascorbic acid > trolox for both the optical and electrochemical measurements. (Figure S4 of Supporting Information provides further details of these measurements, and Figure S4f also shows that at higher reductant concentrations signal attenuation is complete and no longer sensitive to reductant levels).
Figure 2.
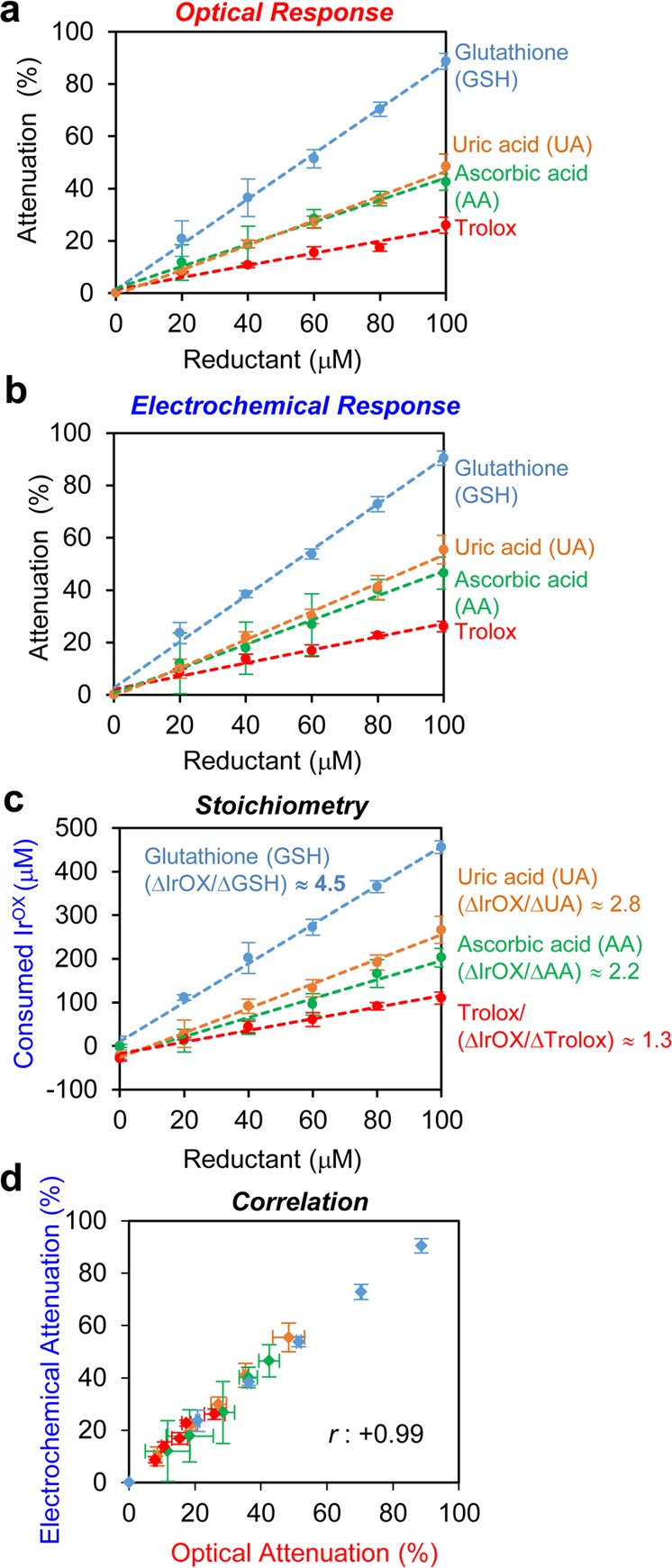
Quantitative validation of Ir-reducing assay. (a) Optical signal attenuation and (b) electrochemical signal attenuation relative to the concentration of individual reductants. (c) Consumed IrOX to oxidize individual reductant versus concentration of reductants. (d) Correlation between optical signal attenuation and electrochemical signal attenuation (N = 17, r = +0.99). Measurements were performed in quadruplicate and error bars indicate standard deviation.
Table 1 lists proposed reactions associated with the reduction of IrOX by the various reductants. In the absence of O2, it has been reported that the predominant IrOX oxidation of GSH is a 6 electron transfer to generate sulfonate (GSO3–), while minor amounts of the oxidized disulfide (GSSG) are formed.37,45−47 (Figure S5 of Supporting Information shows GSH oxidation reactions that have been proposed to explain these stoichiometries). In the presence of O2, experimental measurements showed the transfer of 4.2 electrons from GSH to IrOX although no reactions were proposed.37 This stoichiometric value of 4.2 is similar to the value of ∼4.5 observed in Figure 2c (Figure S6 of Supporting Information provides further details of these calculations).37 For the case of ascorbic acid (AA), Table 1 shows a 2 electron transfer to IrOX was reported,38 which is also consistent with our calculated value (∼2.2) observed in Figure 2c.
Table 1. Reaction Stoichiometries with IrOX/Ir(IV).
| reductants | reaction stoichiometry with IrOX/Ir(IV) | ref |
|---|---|---|
| glutathione (GSH) | 6 Ir(IV) + GSH + 3H2O → 6 Ir(III) + GSO3– + 7H+ | (37) |
| 2 Ir(IV) + 2GSH → 2 Ir(III) + GSSG + 2H+ | (37) | |
| ascorbate | 2 Ir(IV) + H2A → 2 Ir(III) + A + 2H+ | (38) |
| cysteine | 6 Ir(IV) + HSCH2CHNH3COO– + 3H2O → 6 Ir(III) + HO3SCH2CHNH2COO– + 7H+ | (39,45) |
| quinols | 2 Ir(IV) + H2Q → 2 Ir(III) + Q + 2H+ | (73) |
The correlation between the optical and electrochemical signals is shown in Figure 2d, which shows a cross-plot of the attenuation percentages for the two modalities. As expected, there is a strong linear correlation in the attenuation of these two signals (correlation coefficient, r = 0.995).
Comparison with Other Methods
Several commercial methods have been developed to measure the global reducing capacity of a sample.14,15 These methods are based on the electron transfer from reductants in a sample to an added oxidant (probe), which causes a color change of the probe. We performed measurements with one standard commercial method, the cupric reducing antioxidant capacity (CUPRAC) assay, to compare with our Ir-reduction method because the CUPRAC method has recently been used to measure total antioxidant activities in serum.48 As shown in Figure 3a, this commercial assay is based on a sample’s ability to transfer electrons to a colorless Cu(II) (CuOX) solution to generate a purple-colored Cu(I) (CuRED). The color change associated with this reaction is monitored by measuring the absorbance at 570 nm. In contrast to our Ir-reducing assay where the signals are attenuated in the presence of reductant, the optical signals for the Cu-reducing assay increase in the presence of reductants.
Figure 3.
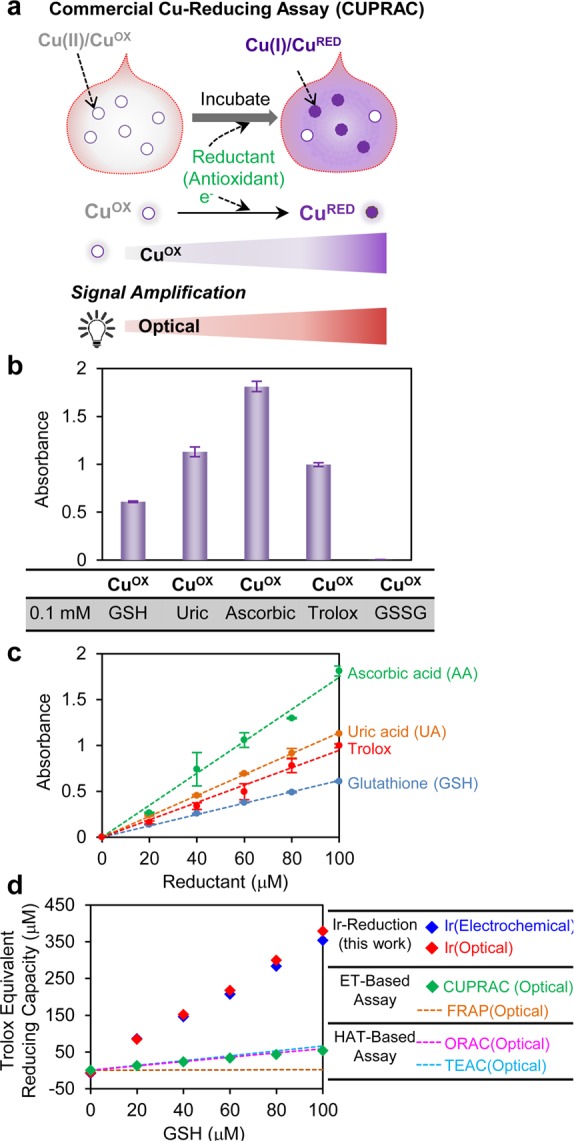
Comparison of Ir-reducing assay with other methods. (a) Scheme illustrating commercial Cu-reducing assay. (b) Optical signal (absorbance) of CuOX when incubated with various reductants. (c) Optical signal increase of CuOX relative to the concentration of individual reductants. (d) GSH sensitivity of Ir-reduction assay compared with other methods (data from Ir-reduction and Cu-reduction assays were experimentally measured while the best fit lines were from Cao et al. (1998).49 Measurements in parts b–d were performed in triplicate (all error bars indicate standard deviation).
Figure 3b shows the optical response (absorbance at 570 nm) when the CuOX probe was mixed with various reductants and incubated for 30 min. As expected, the reduced glutathione (GSH) shows a positive response in this assay while the oxidized glutathione (GSSG) shows no response. Figure 3b also shows the optical response of CuOX varied depending on the reductant tested. For instance, ascorbic acid showed the highest response in this Cu-reduction assay.
To quantitatively measure the reducing capacity of various reductants, we mixed the CuOX probe with varying concentrations of an individual reductant, incubated for 30 min and measured the absorbance at 570 nm. As expected, Figure 3c shows the optical response is proportional to the concentration of reductant being tested. (Figure S7 of Supporting Information provides further details of these measurements.) As observed in Figure 2, the slopes in the plot of Figure 3c can be related to a reductant’s reducing capacity. With the Cu-reduction assay, ascorbic acid has the highest reducing capacity and GSH has the lowest with the following trend: ascorbic acid > uric acid ≈ trolox > GSH. This trend is different than that observed with our Ir-reduction assay (Figure 2). Thus, despite the fact that both assays are based on an electron-transfer reduction mechanism, the redox probes (IrOX or CuOX) have differing sensitivities for accepting electrons from reductants.
In addition to the Cu-reduction assay, several other global assays have been developed to assess a sample’s total antioxidant activities. These methods have been prominently applied to foods to provide a single-value measure of antioxidant activities for the purpose of understanding and comparing health beneficial properties of foods.14,15 These methods have also been extended to clinical samples in an effort to provide a quantitative measure useful for characterizing oxidative stress.10,49,50 Typically, these assays are based on either a hydrogen atom transfer (HAT) or electron transfer (ET) mechanism.14,15,48 HAT-based assays measure the ability of an antioxidant to scavenge free radicals by hydrogen donation and these methods include oxygen radical absorbance capacity (ORAC) and total radical-trapping antioxidant parameter (TRAP). In ET-based assays, the reducing capacity of reductants in a sample is measured by transferring an electron from the reductant to an oxidant probe that could be metals, carbonyls, and radicals.15 The widely used ET-based assays are the trolox equivalent antioxidant capacity (TEAC) assay, the ferric ion reducing antioxidant power (FRAP) assay, the N,N-dimethyl-p-phenylenediamine (DMPD) assay, and the Cu-reduction (CUPRAC) assay of Figure 3a.
Figure 3d shows the sensitivity of these various antioxidant assays to GSH. The Ir-reducing assay, using either the optical or electrochemical signals, shows comparatively high sensitivity to GSH compared to commercial Cu-reducing (CUPRAC) assay. Previous literature reports provided a comparison of the GSH-sensitivity for the ORAC, TEAC, and FRAP methods, and the best-fit lines from these studies are also shown in Figure 3d.49 These lines show that these standard methods have a comparatively low sensitivity for GSH, which is also consistent with reports that the FRAP assay has low sensitivity for detecting thiols in biological fluids.15 One possible explanation for the greater sensitivity of the Ir-reducing assay for GSH is the more oxidative redox potential of the IrOX mediator. Table 2 lists the redox potentials for each redox probe (i.e., oxidant) for the various reducing capacity assays. It is important to note however that thermodynamic explanations based on redox potentials may not be sufficient to explain differences in these methods because there can be significant kinetic barriers to electron transfer reactions. For instance, Figure 3d shows the FRAP assay is unable to detect GSH despite the fact that the Fe(III) oxidant has a more oxidative redox potential compared to that for the Cu(II) oxidant of the CUPRAC method that is able to detect GSH.
Table 2. Redox Potentials of Various Redox Probes.
In summary, the Ir-reducing assay uses IrOX as an oxidative probe and reports information through either optical or electrochemical modalities. This method can detect reducing-activities from various reductants and is especially sensitive to GSH. GSH (and thiols in general) are believed to be important endogenous biological antioxidants yet these compounds are rather sluggish in transferring electrons and thus methods to detect biothiols often require special mediators (i.e., oxidative probes) or nanoparticles for their oxidation.51−55 Not surprisingly, conventional antioxidant capacity assays developed for food applications are rather insensitive to GSH: phenolics and ascorbate (not thiols) are considered to be the important food antioxidants and thus special attention to GSH was not required for developing antioxidant measures for food analysis. For clinical applications, however, the high GSH-sensitivity of the Ir-reducing assay may be an especially important asset when probing serum samples for redox information on oxidative stress.
Clinical Testing
The underlying hypotheses of this study are that (i) blood serum contains chemical information on oxidative stress, and (ii) this chemical information can be accessed by a global (i.e., chemically nonspecific) method of redox probing.10 Directly testing these hypotheses is currently impossible because of the ill-defined nature of oxidative stress, as well as uncertainties of which individual chemical species are the best markers of oxidative stress. Initial support for these hypotheses is provided by an experiment in which serum was treated with an oxidative stressor (i.e., H2O2) and the change of its reducing capacity was measured. Figure S8 of Supporting Information shows that the addition of oxidative stressor (0.5 mM H2O2) decreased the reducing capacity of serum by up to 50%, which might be associated with the oxidation of amino acids by this stressor.56−58 The focus of our study is a less direct, but potentially more important, test of these hypotheses by evaluating correlations between measurements from our Ir-reduction assay and independent clinical measures of disease. For this, we measured serum samples from ten people diagnosed with schizophrenia and five healthy controls, and we evaluated possible correlations with clinical measures of disease (note: as in previous measurements, O2 was not excluded during serum analysis). Growing evidence suggests oxidative stress plays an important role in schizophrenia.43,59−61 Importantly, no independent blood tests are currently widely used by clinicians to assist in diagnosing or evaluating the treatment response of schizophrenia.62
Comparison of Schizophrenia and Healthy Control Groups
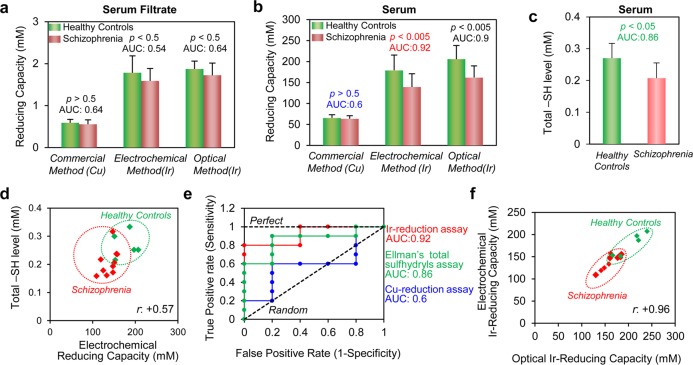
In initial studies, we removed proteins from serum by filtering the serum using a centrifugal membrane filter (molecular weight cutoff = 10 kDa), diluted the filtrate 10-fold with phosphate buffered saline (PBS), and then analyzed the diluted filtrate by both the commercial Cu-reduction assay and the Ir-reduction assay (with both optical and electrochemical detection). Analysis of serum filtrates is expected to detect reducing contributions from low molecular weight components of serum such as ascorbic acid (AA), α-tocopherol, β-carotene, ubiquinol, glutathione (GSH), uric acid (UA), and bilirubin.49,63 For comparison purposes, we normalized reducing capacity in terms of trolox equivalents, which is the common standard used for antioxidant reducing assays.15Figure 4a compares the serum filtrate’s reducing capacity between the schizophrenia and control groups. The commercial Cu-reducing assay shows no differences between these serum filtrates (p = 0.54), while the Ir-reducing assay shows serum filtrates from healthy controls have nonsignificant but higher average reducing capacity compared to those from the schizophrenia group (p = 0.23).
Figure 4.
Clinical testing of reducing capacity of healthy control and schizophrenia groups. (a) Reducing capacity of filtered serum and (b) serum for healthy control (N = 5) and schizophrenia (N = 10) groups. Reducing capacity was measured by the commercial Cu-reduction method and the Ir-reduction method with both electrochemical and optical detection. (c) Measurement of total sulfhydryl groups (−SH) in serum samples of healthy control and schizophrenia groups. (d) Correlation between total sulfhydryl groups and Ir-reducing capacity (electrochemical detection) of serum sample (N = 15, r = +0.57, p = 0.026). (e) Receiver operating characteristic (ROC) curves for electrochemical Ir-reduction method, Cu-reduction method, and Ellman’s total sulfhydryls assay for diagnosis of the schizophrenia group from the healthy control group. (f) Correlation between Ir-reducing capacities measured electrochemically and optically (N = 15, r= +0.96). Measurements in parts a–c were performed in quadruplicate (error bars indicate standard deviation).
In addition to measuring serum filtrates, we performed measurements on serum after diluting the serum 1000-fold with PBS (Figure S9 of the Supporting Information shows that after 1000-fold dilution, the serum absorbance approaches that of the buffer background). Figure 4b shows that when these serum samples were evaluated by the commercial Cu-reducing assay, no significant differences were observed between schizophrenia and control groups (p = 0.63). In contrast, results from the Ir-reducing assay show considerably higher reducing capacities for serum from control group compared to serum from the schizophrenia group (p < 0.005). Also, the Ir-reducing assay with serum (Figure 4b) showed greater discriminating abilities compared to results with filtered serum (Figure 4a).
One possible explanation for the ability of the Ir-reducing assay to detect differences between the control and schizophrenia groups (compared to the commercial Cu-reducing assay), is the greater sensitivity of the Ir-reducing assay to sulfhydryl groups as observed in Figure 3d. To evaluate this possibility, we assayed serum for total sulfhydryl groups (e.g., GSH and protein sulfhydryls) using a modified Ellman’s method.64−66Figure 4c shows that the serum from control group has higher sulfhydryl values compared to serum from the schizophrenia group. Figure 4d shows a modest positive correlation (N = 15, r = +0.57, p = 0.026) between total sulfhydryl group assay and the Ir-reducing capacity as measured electrochemically. [Note: for clarity, error bars are not shown in Figure 4d but are shown in Figure S10 of the Supporting Information.] This correlation indicates that the higher Ir-reducing capacity is modestly related to higher levels of total sulfhydryls. Thus, it appears that the higher sulfhydryl content in the serum from healthy controls is partially responsible for the higher measured Ir-reducing capacity.
We characterized the clinical diagnostic performance of our assay using a receiver operating characteristic (ROC) curve analysis to determine if the measurements could discern the schizophrenia group from the healthy controls.62,67,68 In this method, the area under the ROC curve (AUC) for a perfect diagnostic test would be 1.0 while a random test would yield a value of 0.5. As shown in Figure 4e, the calculated AUC values for the Ir-reducing capacity assay (with electrochemical detection) for serum was determined to be 0.92 (95% confidence interval (CI): 0.76–1.08; p = 0.01), which compares to the value of 0.6 for the Cu-reducing serum assay (95% CI, 0.26–0.94; p = 0.54) and 0.86 for the Ellman’s free sulfhydryl group assay of serum (95% CI, 0.63–1.09; p = 0.03). Thus, this analysis provides additional support that the Ir-reducing assay accesses clinically useful chemical information. (Figure S11 of the Supporting Information provides further details of ROC curves.)
To further evaluate the Ir-reducing assay results for serum samples (Figure 4b), we prepared a cross-plot between optical and electrochemical measurements. Figure 4f shows a strong correlation between these two independent measurement modalities (N = 15, r = +0.96) even in serum analysis. [Note: for clarity, error bars are not shown in Figure 4f but are shown in Figure S12 of the Supporting Information]
In summary, the results in Figure 4 indicate that the Ir-reducing capacity measurements with diluted serum can distinguish the schizophrenia group from the control group. The lower observed reducing activities in the serum of the schizophrenia group is consistent with suggestions that oxidative stress is linked to schizophrenia.43,59 While this initial test of Ir-reducing assay is promising, a larger sized sample will be required to support clinical conclusions.
Ir-Reducing Capacity Correlations to Age and Disease Severity
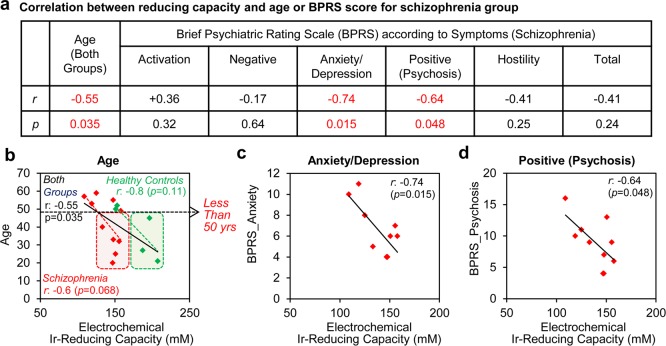
The original free radical theory of aging hypothesized that aging results from cumulative damage associated with free radicals69 and several studies have established correlations between age and various markers of oxidative damage.70,71 If the Ir-reducing assay accesses important chemical information on oxidative stress, then we would expect correlations between age and Ir-reducing capacity. As expected, Figure 5a,b shows inverse correlations between age and Ir-reducing capacity for the individual schizophrenia and control groups as well as for the overall population of both groups. Importantly, the boxed regions in Figure 5b show that if an age cutoff of 50 years is applied to our data, the Ir-reducing assay can fully distinguish the schizophrenia group from healthy controls (p < 0.05). Specifically, the two overlapping data points in Figure 4f are for the oldest healthy controls.
Figure 5.
Correlation of Ir-reducing capacity with age and symptom severity. (a) Correlations between Ir-reducing capacity assay (electrochemical detection) and age or symptoms as measured by the brief psychiatric rating scale (BPRS). (b) Correlation between of Ir-reducing capacity and age for healthy control and schizophrenia groups. (c) Correlation between Ir-reducing capacity and anxiety/depression symptom. (d) Correlation between Ir-reducing capacity and positive (psychosis) symptom.
Potentially, the Ir-reducing assay is accessing chemical information on oxidative stress that is related to the severity of symptoms in people with schizophrenia, and thus correlations might be expected between Ir-reduction capacity and independent clinical measures of symptom severity. The most widely used scale for measuring psychotic symptoms is the brief psychiatric rating scale (BPRS), which is based on a clinician’s interview and observations of the patient.72 The BPRS scale considers several items, and higher scores indicate more severe symptoms. Figure 5a shows statistical information for correlations between these composite psychotic symptoms and Ir-reducing capacity for the 10 persons in the schizophrenia group. As expected, most symptoms show a negative correlation between symptom severity and Ir-reduction capacity (i.e., greater symptom severity is correlated to greater oxidative stress). The strongest correlation was observed between anxiety/depression and Ir-reducing capacity (N = 10, r = −0.74, p = 0.015) in Figure 5c, while the weakest correlation was observed for the negative symptom. The positive symptom (psychosis) also showed a high correlation between symptom severity and Ir-reducing capacity (N = 10, r = −0.64, p = 0.048) in Figure 5d.
In summary, the initial clinical results in Figure 5 further support a conclusion that the Ir-reduction assay accesses chemical information that could be useful for understanding and managing diseases that are believed to be linked to oxidative stress.
Discussion
Clinicians routinely assess patients using simple physical measurements that provide global information in a timely manner (e.g., measurements of temperature, pulse and blood pressure). Blood contains valuable chemical information on a patient’s health, and blood tests are routinely used to access specific chemical information (e.g., of individual metabolites, antibodies, or biomarkers). For the case of oxidative stress, a focus on specific (vs global) chemical information may be less helpful to clinicians for two reasons. First, acquiring specific chemical information often requires specialized instrumentation in centralized laboratories which generally means this chemical information is not available in a timely manner. Second, for the case of oxidative stress, it is not clear what specific chemical information is most relevant. As a result, for diseases such as schizophrenia, clinicians do not even use chemical information for diagnosis or assessment. Here, we report a method to access global chemical information on oxidative stress. While the development of this method was guided by chemical/medical intuition (e.g., a requirement for high GSH sensitivities), this method is not chemically specific but more broadly probes for redox-information. Initial clinical evaluations indicate this method may access valuable chemical information while the speed and simplicity of the method suggests this information could be available at the point-of-care.
In addition to providing timely chemical information at the point-of-care, we believe there is a second potential advantage of the Ir-reducing assay. If this measurement proves to be a reliable indicator to assist in the diagnosis and assessment of symptom severity, then these measurements could become an important investigational tool. For instance, this measurement could provide clinical researchers with a readily measurable objective target to assess therapeutic interventions. Alternatively, experimental research to unravel the chemical basis of the Ir-reduction signal could discover clues of the molecular mechanisms important in oxidative stress. Such a “reverse engineering” of the Ir-reduction signal could provide a complementary approach to alternative, instrument-intensive discovery approaches (i.e., -omic based methods). In summary, we believe the Ir-reduction assay could be important because it provides simple near-real-time access to important global chemical information in serum.
Conclusions
Here we report a simple, rapid, and robust method to probe serum for chemical information relevant to oxidative stress. This iridium-reducing assay uses K2IrCl6 (IrOX) as a redox mediator to detect the serum’s reducing activities and can detect this activity by independent optical and electrochemical modalities. Compared to alternative global reducing assays, the Ir-reducing assay has a high sensitivity to GSH which is an especially important attribute for probing serum for information on oxidative stress. Initial clinical evaluations show that the Ir-reducing assay can discern a schizophrenia group (N = 10) from healthy controls (N = 5, p < 0.005) and showed an inverse correlation between reducing activities and the severity of the anxiety/depression (N = 10, r = −0.74, p = 0.015) and psychosis symptoms (N = 10, r = −0.64, p = 0.048) for the schizophrenia group. In conclusion, the Ir-reducing assay accesses global chemical information on oxidative stress with the sensitivity, speed, and simplicity required for point-of-care measurement. Potentially, this chemical measurement could complement other global physical measures (e.g., temperature and blood pressure) used routinely for the rapid clinical evaluation of a patient’s status.
Experimental Section
Chemicals
The following were purchased from Sigma-Aldrich: K2IrCl6 (IV), K3IrCl6 (III), glutathione (reduced, GSH), glutathione (oxidized, GSSG), ascorbic acid, uric acid, 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB), phosphate buffered saline (PBS). The water (>18 MΩ) used in this study was obtained from a Super Q water system (Millipore). A stock solution of 0.5 mM K2IrCl6(IV) was prepared in PBS (pH 7.4).
Serum Samples and Symptom Assessment
Recruitment of people to participate in a clinical study designed to collect blood samples occurred between May 2015 and August 2016. Blood samples were collected from the Maryland Psychiatric Research Center, University of Maryland School of Medicine. We recruited two populations, people with a DSM-IV diagnosis of schizophrenia or schizoaffective disorder and a population of individuals without a major psychiatric diagnosis. All participants completed data collection procedures in a single 1–2 h study visit. Additionally, participants provided detailed clinical information. Blood samples (45 mL) were collected using 6 tubes of BD Vacutainers and centrifuged at 3000 rpm. The resulting supernatant was removed using disposable plastic 1 mL pipets. It was apportioned into 1 mL aliquots and stored at −80 °C in a freezer before analysis. To assay serum, the frozen serum was thawed at room temperature in the air for 30 min and then it was kept in an ice bath before the measurement.
The study to collect human serum from people with schizophrenia and healthy controls was approved by the University of Maryland School of Medicine IRB, and informed consent was obtained from all study participants prior to the research procedure.
A trained interviewer interviewed the people with schizophrenia to assess the Brief Psychiatric Rating Scale (BPRS), a clinical evaluation that rapidly provides measurement of clinical symptoms. The BPRS total score as well as five domain scores are calculated (positive, negative, anxiety/depression, hostility, and activation).
Research staff tasked with the collection of biological samples and protected health information have completed the requisite training and implemented standard procedures as required by The State of Maryland Department of Mental Health and Hygiene (DHMH) and the University of Maryland School of Medicine. Informed consent was obtained from each participant after reviewing relevant risks and benefits for the project. Successful completion of the Evaluation to Sign Consent was also required to demonstrate participant understanding of the voluntary nature of research, study tasks, and risks. The respective Institutional Review Boards for the University of Maryland and DHMH have approved this project and specified its conduct as having minimal risk to research participants.
Instrumentation
Electrochemical measurements (cyclic voltammetry (CV)) and chronocoulometry (CC)) were performed to measure the electrochemical signal (CHI420a electrochemical analyzer, CHInstruments, TX). For the electrochemical assay, we used a screen-printed carbon paste electrode (CHInstruments, TX) with carbon working and counter electrodes, and a Ag/AgCl reference electrode. The optical signal was recorded using a microplate reader (SperctraMax M2, Molecular Devices, CA).
Ir-Reducing Capacity Assay in Buffer
A stock solution of 10 mM K2IrCl6 and stock solutions of ascorbic acid (1 mM) and glutathione (GSH, 1 mM), oxidized glutathione (GSSG, 1 mM), trolox (1 mM) and uric acid (0.4 mM) were prepared in 0.1 M PBS (pH 7.4). A portion of each antioxidant stock solution was added into a 96 well-plate to generated 0 (blank), 20 μM, 40 μM, 60 μM, 80 μM, 100 μM antioxidant solutions. The 0.1 M PBS was added to each well to bring the volume to 95 μL. In each well, 5 μL of 10 mM K2IrCl6 (final concentration 0.5 mM) was added, mixed by pipetting and incubated for 30 min at room temperature. After that, the absorbance was measured at 488 nm using a microplate reader (optical measurement). For an electrochemical measurement, 100 μL of the mixture from each well was dropped onto a screen-printed electrode by covering all of the electrodes (working, counter, and reference electrodes). A constant potential of 0 V was applied to the electrode, and the charge was measured for 1 min using a chronocoulometry technique. All data shown in Figure 1 and Figure 2 were averaged from the measurements in quadruplicate, and the error bar indicates standard deviation (s.d.).
Ir-Reducing Capacity Assay in Filtered Serum
For a reducing capacity assay in filtered serum, microcon centrifugal filter device (EMD Millipore, MA) was used to remove biomacromolecues (MW > 10 kDa) from serum. Serum was pipetted into the device and the assembly was placed in a centrifuge (Centrifuge 5415c, Eppendorf) and spun at 14 000g. The filtrate was used for reducing capacity assay. A volume of 10 μL of filtered serum was added into a 96 well plate containing 85 μL of 0.1 M PBS, and then 5 μL of 10 mM K2IrCl6 was added to each well (this procedure results in a 10-fold dilution of the filtered serum). After adding the filtrate, buffer, and mediator, the solution was mixed by pipetting and incubated for 30 min at room temperature. The optical and electrochemical responses were measured as described above.
Ir-Reducing Capacity Assay in Serum
For a reducing capacity assay in serum, serum was first diluted 20-fold with 0.1 M PBS and 2 μL of diluted serum was added into a 96 well plate containing 93 μL of 0.1 M PBS, and then 5 μL of 10 mM K2IrCl6 solution was added to each well (this procedure results in an overall 1000-fold dilution of the filtered serum). After adding the solutions, mixing by pipetting and incubating for 30 min at room temperature, the optical response and electrochemical response were measured as described above. All data shown in Figure 4 were averaged from the quadruplicate measurements and the error bar indicates standard deviation.
Cu-Reducing Capacity Assay
To compare the reducing capacity measured with our method, a commercial reducing capacity assay was performed (cupric reducing antioxidant capacity (CUPRAC) assay, MAK187 from Sigma-Aldrich). To measure the reducing capacity of the sample, a portion of sample was added into a 96 well-plate and water was added to each well to bring the volume to 50 μL. In each well, 50 μL of Cu(II) working solution provided in an assay kit was added, mixed by pipetting and incubated for 30 min at room temperature. After incubation, the absorbance was measured at 570 nm using a microplate reader (optical measurement). All data shown in Figure 3 were averaged from the triplicate measurements, and the error bar indicates standard deviation.
Measurement of Total Sulfhydryl Groups of Serum Samples
Total sulfhydryl groups of serum samples were assayed according to manufacturer’s instructions (Quantification of Sulfhydryls, Uptima, Interchim). A dilution buffer (30 mM Tris HCl, 3 mM EDTA, pH 8.2) and DTNB working solution (3 mM in methanol) were prepared. As a standard for the free sulfhydryl group (−SH) assay, we prepared GSH solutions (0.1 mM to ∼1 mM in dilution buffer). A volume of 20 μL of sample or standard solution, 75 μL of dilution buffer, 25 μL of DTNB reagent, and 400 μL of methanol were added into a microcentrifuge tube. After 5 min incubation, the mixture was centrifuged at 3000g for 5 min at room temperature. The supernatant was transferred into a microplate. The optical absorbance was measured at 412 nm. All data shown in Figure 4c were averaged from the quadruplicate measurements, and the error bar indicates standard deviation.
Statistical Analysis
The calculation of p-values in Figure 4 was performed using a mixed design analysis of variance (SPANOVA). Receiver operating characteristic (ROC) curves for the antioxidant assays in Figure 4e and Figure S11 were determined utilizing OriginPro (OriginLab Corporation), and values were determined for area under curve (AUC) as shown in Figure 4, the 95% confidence intervals, and the p-values. Relationships between reducing capacity and age or BPRS scores in Figure 5 were assessed using Pearson’s correlation coefficient. p-values in Figure 5 were obtained from regression analysis (ANOVA). All of the clinical data in Figure 4 and Figure 5 are provided in Tables S1–S8 of the Supporting Information.
Acknowledgments
We gratefully acknowledge financial support from the United States: National Science Foundation (Grant CBET-1435957), Defense Threat Reduction Agency (Grant HDTRA1-13-1-0037), and the National Institutes of Health (Grant R56 MH105571).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.6b03620.
Additional experimental results, statistical analysis, clinical data, and suggested reaction mechanism (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Valko M.; Leibfritz D.; Moncol J.; Cronin M. T. D.; Mazur M.; Telser J. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Pohanka M. Curr. Med. Chem. 2014, 21, 356–364. 10.2174/09298673113206660258. [DOI] [PubMed] [Google Scholar]
- Finkel T.; Holbrook N. J. Nature 2000, 408, 239–247. 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Emiliani F. E.; Sedlak T. W.; Sawa A. Curr. Opin Psychiatry 2014, 27, 185–190. 10.1097/YCO.0000000000000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohen R.; Nyska A. Toxicol. Pathol. 2002, 30, 620–650. 10.1080/01926230290166724. [DOI] [PubMed] [Google Scholar]
- Elmarakby A. A.; Sullivan J. C. Cardiovasc. Ther. 2012, 30, 49–59. 10.1111/j.1755-5922.2010.00218.x. [DOI] [PubMed] [Google Scholar]
- Schieber M.; Chandel; Navdeep S. Curr. Biol. 2014, 24, R453–R462. 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraibar M. A.; Ladouce R.; Friguet B. J. Proteomics 2013, 92, 63–70. 10.1016/j.jprot.2013.05.008. [DOI] [PubMed] [Google Scholar]
- Fedorova M.; Bollineni R. C.; Hoffmann R. Mass Spectrom. Rev. 2014, 33, 79–97. 10.1002/mas.21381. [DOI] [PubMed] [Google Scholar]
- Ghiselli A.; Serafini M.; Natella F.; Scaccini C. Free Radical Biol. Med. 2000, 29, 1106–1114. 10.1016/S0891-5849(00)00394-4. [DOI] [PubMed] [Google Scholar]
- Kim E.; Liu Y.; Ben-Yoav H.; Winkler T. E.; Yan K.; Shi X.; Shen J.; Kelly D. L.; Ghodssi R.; Bentley W. E.; Payne G. F. Adv. Healthcare Mater. 2016, 5, 2595–2616. 10.1002/adhm.201600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford F. P.; Whitehead T. P. Ann. Clin. Biochem. 1998, 35, 48–56. 10.1177/000456329803500105. [DOI] [PubMed] [Google Scholar]
- Serafini M.; Del Rio D. Redox Rep. 2004, 9, 145–152. 10.1179/135100004225004814. [DOI] [PubMed] [Google Scholar]
- Huang D.; Ou B.; Prior R. L. J. Agric. Food Chem. 2005, 53, 1841–1856. 10.1021/jf030723c. [DOI] [PubMed] [Google Scholar]
- Prior R. L.; Wu X.; Schaich K. J. Agric. Food Chem. 2005, 53, 4290–4302. 10.1021/jf0502698. [DOI] [PubMed] [Google Scholar]
- Erel O. Clin. Biochem. 2004, 37, 112–119. 10.1016/j.clinbiochem.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Apel K.; Hirt H. Annu. Rev. Plant Biol. 2004, 55, 373–399. 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Nathan C.; Cunningham-Bussel A. Nat. Rev. Immunol. 2013, 13, 349–361. 10.1038/nri3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura F. A.; de Andrade K. Q.; dos Santos J. C. F.; Araújo O. R. P.; Goulart M. O. F. Redox Biol. 2015, 6, 617–639. 10.1016/j.redox.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter S.; Gupta S. C.; Chaturvedi M. M.; Aggarwal B. B. Free Radical Biol. Med. 2010, 49, 1603–1616. 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies H. Redox Biol. 2015, 4, 180–183. 10.1016/j.redox.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. P. Antioxid. Redox Signaling 2006, 8, 1865–1879. 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- Jones D. P. American Journal of Physiology - Cell Physiology 2008, 295, C849. 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer F. Q.; Buettner G. R. Free Radical Biol. Med. 2001, 30, 1191–1212. 10.1016/S0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- Levonen A.-L.; Hill B. G.; Kansanen E.; Zhang J.; Darley-Usmar V. M. Free Radical Biol. Med. 2014, 71, 196–207. 10.1016/j.freeradbiomed.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klomsiri C.; Karplus P. A.; Poole L. B. Antioxid. Redox Signaling 2011, 14, 1065–1077. 10.1089/ars.2010.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieyal J. J.; Chock P. B. Antioxid. Redox Signaling 2012, 16, 471–475. 10.1089/ars.2011.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursini F.; Maiorino M.; Forman H. J. Redox Biol. 2016, 8, 205–215. 10.1016/j.redox.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigelius-Flohé R.; Flohé L. Antioxid. Redox Signaling 2011, 15, 2335–2381. 10.1089/ars.2010.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmstrom K. M.; Finkel T. Nat. Rev. Mol. Cell Biol. 2014, 15, 411–421. 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- Kim E.; Liu Y.; Leverage W. T.; Yin J.-J.; White I. M.; Bentley W. E.; Payne G. F. Biomacromolecules 2014, 15, 1653–1662. 10.1021/bm500026x. [DOI] [PubMed] [Google Scholar]
- Kim E.; Panzella L.; Micillo R.; Bentley W. E.; Napolitano A.; Payne G. F. Sci. Rep. 2015, 5, 18447. 10.1038/srep18447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Electroanalysis 2001, 13, 983–988. . [DOI] [Google Scholar]
- George P.; Irvine D. H. Biochem. J. 1954, 58, 188–195. 10.1042/bj0580188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J. G.; Duarte V.; Hickerson R. P.; Burrows C. J. Nucleic Acids Res. 1998, 26, 2247–2249. 10.1093/nar/26.9.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthje S.; Böttger M. Biochim. Biophys. Acta, Bioenerg. 1989, 977, 335–340. 10.1016/S0005-2728(89)80089-1. [DOI] [Google Scholar]
- Bhattarai N.; Stanbury D. M. Inorg. Chem. 2012, 51, 13303–13311. 10.1021/ic301955y. [DOI] [PubMed] [Google Scholar]
- Drury W. D.; Dekorte J. M. Inorg. Chem. 1983, 22, 121–125. 10.1021/ic00143a026. [DOI] [Google Scholar]
- Kottapalli K. K.; Adari K. K.; Vani P.; Govindan S. K. Transition Met. Chem. 2005, 30, 773–777. 10.1007/s11243-005-4827-3. [DOI] [Google Scholar]
- van Os J.; Kapur S. Lancet 2009, 374, 635–645. 10.1016/S0140-6736(09)60995-8. [DOI] [PubMed] [Google Scholar]
- Gilca M.; Piriu G.; Gaman L.; Delia C.; Iosif L.; Atanasiu V.; Stoian I. Psychopharmacology 2014, 231, 4703–4710. 10.1007/s00213-014-3624-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak A.; Steullet P.; Cabungcal J.-H.; Werge T.; Ingason A.; Cuenod M.; Do K. Q. Antioxid. Redox Signaling 2013, 18, 1428–1443. 10.1089/ars.2012.4858. [DOI] [PubMed] [Google Scholar]
- Yao J. K.; Keshavan M. S. Antioxid. Redox Signaling 2011, 15, 2011–2035. 10.1089/ars.2010.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metters J. P.; Kadara R. O.; Banks C. E. Analyst 2011, 136, 1067–1076. 10.1039/c0an00894j. [DOI] [PubMed] [Google Scholar]
- Bhattarai N.; Stanbury D. M. J. Phys. Chem. B 2014, 118, 1097–1101. 10.1021/jp4116723. [DOI] [PubMed] [Google Scholar]
- Mailloux R. J.; McBride S. L.; Harper M.-E. Trends Biochem. Sci. 2013, 38, 592–602. 10.1016/j.tibs.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Loi V. V.; Rossius M.; Antelmann H. Front. Microbiol. 2015, 6, 1. 10.3389/fmicb.2015.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apak R.; Özyürek M.; Güçlü K.; Çapanoğlu E. J. Agric. Food Chem. 2016, 64, 997–1027. 10.1021/acs.jafc.5b04739. [DOI] [PubMed] [Google Scholar]
- Cao G.; Prior R. L. Clin. Chem. 1998, 44, 1309–1315. [PubMed] [Google Scholar]
- Rice-Evans C.; Miller N. J. In Oxygen Radicals in Biological Systems, Part D; Methods in Enzymology, Vol. 234; Academic Press: San Diego, CA, 1994; pp 279–293. [DOI] [PubMed] [Google Scholar]
- Ran X.; Sun H.; Pu F.; Ren J.; Qu X. Chem. Commun. 2013, 49, 1079–1081. 10.1039/c2cc38403e. [DOI] [PubMed] [Google Scholar]
- Ge J.; Huang Z.-M.; Xi Q.; Yu R.-Q.; Jiang J.-H.; Chu X. Chem. Commun. 2014, 50, 11879–11882. 10.1039/C4CC05309E. [DOI] [PubMed] [Google Scholar]
- Niu L.-Y.; Chen Y.-Z.; Zheng H.-R.; Wu L.-Z.; Tung C.-H.; Yang Q.-Z. Chem. Soc. Rev. 2015, 44, 6143–6160. 10.1039/C5CS00152H. [DOI] [PubMed] [Google Scholar]
- Jung H. S.; Chen X.; Kim J. S.; Yoon J. Chem. Soc. Rev. 2013, 42, 6019–6031. 10.1039/c3cs60024f. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Liu Y.; Kim E.; Bentley W. E.; Payne G. F. Anal. Chem. 2016, 88, 7213–7221. 10.1021/acs.analchem.6b01394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson B. C.; Chang C. J. Nat. Chem. Biol. 2011, 7, 504–511. 10.1038/nchembio.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasil M.; Halliwell B.; Hutchison D. C. S.; Baum H. Biochem. J. 1987, 243, 219. 10.1042/bj2430219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballal S.; Radi R.; Kirk M. C.; Barnes S.; Freeman B. A.; Alvarez B. Biochemistry 2003, 42, 9906–9914. 10.1021/bi027434m. [DOI] [PubMed] [Google Scholar]
- Albayrak Y.; Ünsal C.; Beyazyüz M.; Ünal A.; Kuloğlu M. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2013, 45, 144–149. 10.1016/j.pnpbp.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Zhang X. Y.; Chen D. C.; Xiu M. H.; Tang W.; Zhang F.; Liu L.; Chen Y.; Liu J.; Yao J. K.; Kosten T. A.; Kosten T. R. Schizophr. Res. 2012, 139, 66–72. 10.1016/j.schres.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Flatow J.; Buckley P.; Miller B. J. Biol. Psychiatry 2013, 74, 400–409. 10.1016/j.biopsych.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E.; Izmailov R.; Spain M.; Barnes A.; Mapes J. P.; Guest P. C.; Rahmoune H.; Pietsch S.; Leweke F. M.; Rothermundt M.; Steiner J.; Koethe D.; Kranaster L.; Ohrmann P.; Suslow T.; Levin Y.; Bogerts B.; van Beveren N.; McAllister G.; Weber N.; Niebuhr D.; Cowan D.; Yolken R. H.; Bahn S. Biomark. Insights 2010, 5, 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B. P. Physiol. Rev. 1994, 74, 139–162. [DOI] [PubMed] [Google Scholar]
- Costa C. M. d.; Santos R. C. C. d.; Lima E. S. J. Bras. Patol. Med. Lab. 2006, 42, 345–350. 10.1590/S1676-24442006000500006. [DOI] [Google Scholar]
- Sedlak J.; Lindsay R. H. Anal. Biochem. 1968, 25, 192–205. 10.1016/0003-2697(68)90092-4. [DOI] [PubMed] [Google Scholar]
- Gordonov T.; Kim E.; Cheng Y.; Ben-Yoav H.; Ghodssi R.; Rubloff G.; Yin J.-J.; Payne G. F.; Bentley W. E. Nat. Nanotechnol. 2014, 9, 605–610. 10.1038/nnano.2014.151. [DOI] [PubMed] [Google Scholar]
- Laksanasopin T.; Guo T. W.; Nayak S.; Sridhara A. A.; Xie S.; Olowookere O. O.; Cadinu P.; Meng F.; Chee N. H.; Kim J.; Chin C. D.; Munyazesa E.; Mugwaneza P.; Rai A. J.; Mugisha V.; Castro A. R.; Steinmiller D.; Linder V.; Justman J. E.; Nsanzimana S.; Sia S. K. Sci. Transl. Med. 2015, 7, 273re1. 10.1126/scitranslmed.aaa0056. [DOI] [PubMed] [Google Scholar]
- Bewick V.; Cheek L.; Ball J. Critical Care 2004, 8, 46. 10.1186/cc2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekimi S.; Lapointe J.; Wen Y. Trends Cell Biol. 2011, 21, 569–576. 10.1016/j.tcb.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J. K.; Leonard S.; Reddy R. Dis. Markers 2006, 22, 83–93. 10.1155/2006/248387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J. K.; Reddy R.; van Kammen D. P. Psychiatry Res. 2000, 97, 137–151. 10.1016/S0165-1781(00)00230-4. [DOI] [PubMed] [Google Scholar]
- Overall J. E.; Gorham D. R. Psychol. Rep. 1962, 10, 799–812. 10.2466/pr0.1962.10.3.799. [DOI] [Google Scholar]
- Pelizzetti E.; Mentasti E.; Baiocchi C. J. Phys. Chem. 1976, 80, 2979–2982. 10.1021/j100908a014. [DOI] [Google Scholar]
- Apak R.; Güçlü K.; Özyürek M.; Karademir S. E. n.; Altun M. Free Radical Res. 2005, 39, 949–961. 10.1080/10715760500210145. [DOI] [PubMed] [Google Scholar]
- Apak R.; Güçlü K.; Özyürek M.; Çelik S. E. Microchim. Acta 2008, 160, 413–419. 10.1007/s00604-007-0777-0. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.