Abstract
The contribution of microbiota in regulating multiple physiological and pathological host responses has been studied intensively in recent years. Evidence suggests that commensal microbiota can directly modulate different populations of cells of the immune system (e.g., Ivanov et al., 2008; Atarashi et al., 2011). Recently, we showed that protein extracts from gut commensal microbiota can activate retina-specific T cells, allowing these autoreactive T cells to then break through the blood-retinal barrier and trigger autoimmune uveitis in the recipient (Horai et al., 2015). The protocol below describes the method to prepare intestinal protein-rich extracts that can be used in various in vitro andin vivo immunological studies.
Background
Intestinal microbiota represent a complex community of microbes that provide a wide variety of innate and adaptive stimulants. Their isolation and purification from stool samples has been performed and protocols have been published (Mueller and Pan, 2013; Verberkmoes et al., 2009; Tanca et al., 2014; Xiong et al., 2015a; Xiong et al., 2015b). Most of these protocols have been developed with the aim of performing proteomic studies for characterization of the microbiota. Consequently, although they emphasize protein yield and purity, they are time consuming and may include a protein denaturating step that affects protein structure (Verberkmoes et al., 2009) or use reagents (i.e., sodium azide, SDS, phenol) that are incompatible with subsequent cell culture based assays (Tanca et al., 2014; Xiong et al., 2015a; Xiong et al., 2015b). These characteristics are not desired when functional immunological assays are intended to be performed with the extracted proteins.
We have developed a simple and fast method that can be used to obtain protein-rich extracts from different areas of the intestine as well as from stool samples. The protocol does not include denaturating steps and the protein-rich extracts can be used in different in vitro and in vivo immunological assays with live cells, including T cell stimulation for proliferation and for adoptive transfer (see Data analysis section).
Materials and Reagents
50 ml centrifuge tubes (Corning, Falcon®, catalog number: 352070)
15 ml centrifuge tubes (Corning, Falcon®, catalog number: 352095)
100 mm culture dish (Corning, Falcon®, catalog number: 353003)
Sterile 12 ml syringe (COVIDIEN, Monoject™, catalog number: 8881512878)
23 G × 1” needle (COVIDIEN, Monoject™, catalog number: 8881200383)
3 ml sterile syringe (COVIDIEN, Monoject™, catalog number: 8881513934)
Sterile 1 ml syringe (COVIDIEN, Monoject™, catalog number: catalog number: 8881501400)
1.5 ml microtubes (Eppendorf, catalog number: 022-36-411-1)
Syringe filter 0.22 µm, polyethersulfone, 33 mm, gamma sterilized (EMD Millipore, catalog number: SLGV033RS)
Sterile 1× PBS (Thermo Fisher Scientific, Gibco™, catalog number: 10010-023)
Pierce™Coomassie plus Bradford protein assay kit (Thermo Fisher Scientific, Thermo Scientific™, catalog number: 23236)
10 mg/ml aprotinin (Sigma-Aldrich, catalog number: A1153)
10 mg/ml leupeptin (Sigma-Aldrich, catalog number: L9783)
Phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: P7626) 15. Protease inhibitor cocktail (see Recipes)
Equipment
CO2 mouse euthanasia chamber (with flow meter) (Euthanex, model: E-20028)
−80 °C freezer (Thermo Fisher Scientific, Thermo Scientific™, model: Revco UXF)
Analytical balance (Mettler-Toledo International, model: AE50)
Refrigerated centrifuge (Thermo Fisher Scientific, Thermo Scientific™, model: Legend XFR)
Refrigerated microcentrifuge (Eppendorf, model: 5424R)
Vortex-Genie 2
Sonicator (Heat systems, model: XL2010)
Procedure
The following procedure describes the steps to collect the contents from the large intestine of one mouse. The protocol can be adapted to the collection of small intestine contents as well.
Euthanize mouse according to institutional guidelines using a CO2 euthanasia chamber.
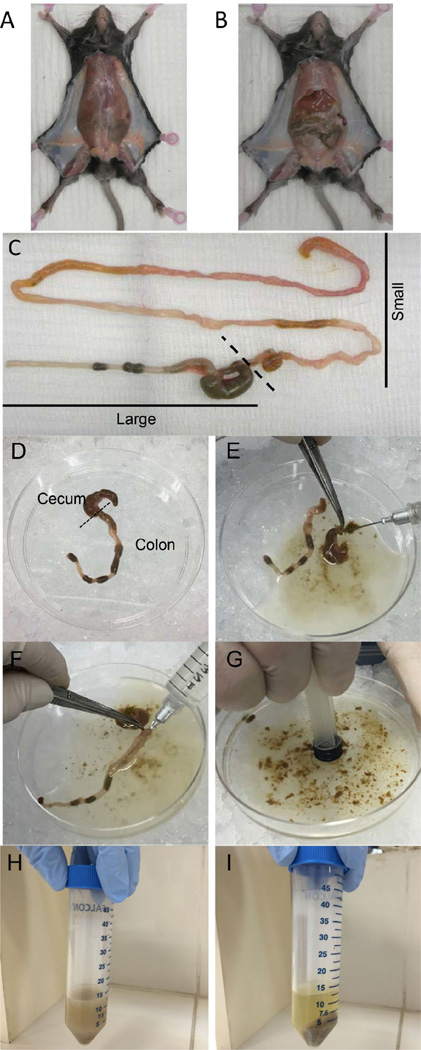
Cut the skin of the abdomen without disrupting the peritoneum (Figure 1A).
Cut the peritoneum to expose the small and large intestines (Figure 1B).
Carefully push the intestines out of the abdomen and isolate the complete small and large intestines by cutting just after the stomach and as low as possible of the colon (Figure 1C).
Remove large intestine from the beginning of the cecum to the end of the colon. Transfer intestine to a 100 mm culture dish placed on ice (Figure 1D).
Flush out intestinal contents with up to 10 ml of ice-cold PBS using a 12 ml syringe + 23 G×1” needle (Figure 1E, F).
Disperse intestinal contents in the PBS by applying pressure with a 3 ml syringe plunger (Figure 1G). Transfer contents to a 50 ml tube (Figure 1H).
Pellet the contents by centrifugation at 2,000 × g for 10 min at 4 °C (Figure 1I).
Pour off and discard the supernatant by gentle inversion of the tube.
Tare analytical balance with empty 50 ml tube and adjust to zero. Weigh the tube with pelleted intestinal contents.
Resuspend the pellet in freshly prepared protease inhibitor cocktail (see Recipes) at a concentration of 2 g/ml.
Freeze pellet in −80 °C freezer for 20 min, then thaw for 20 min and vortex at maximum speed for 5 sec to disrupt bacterial cells. Repeat the freeze/thaw/vortex cycle two additional times.
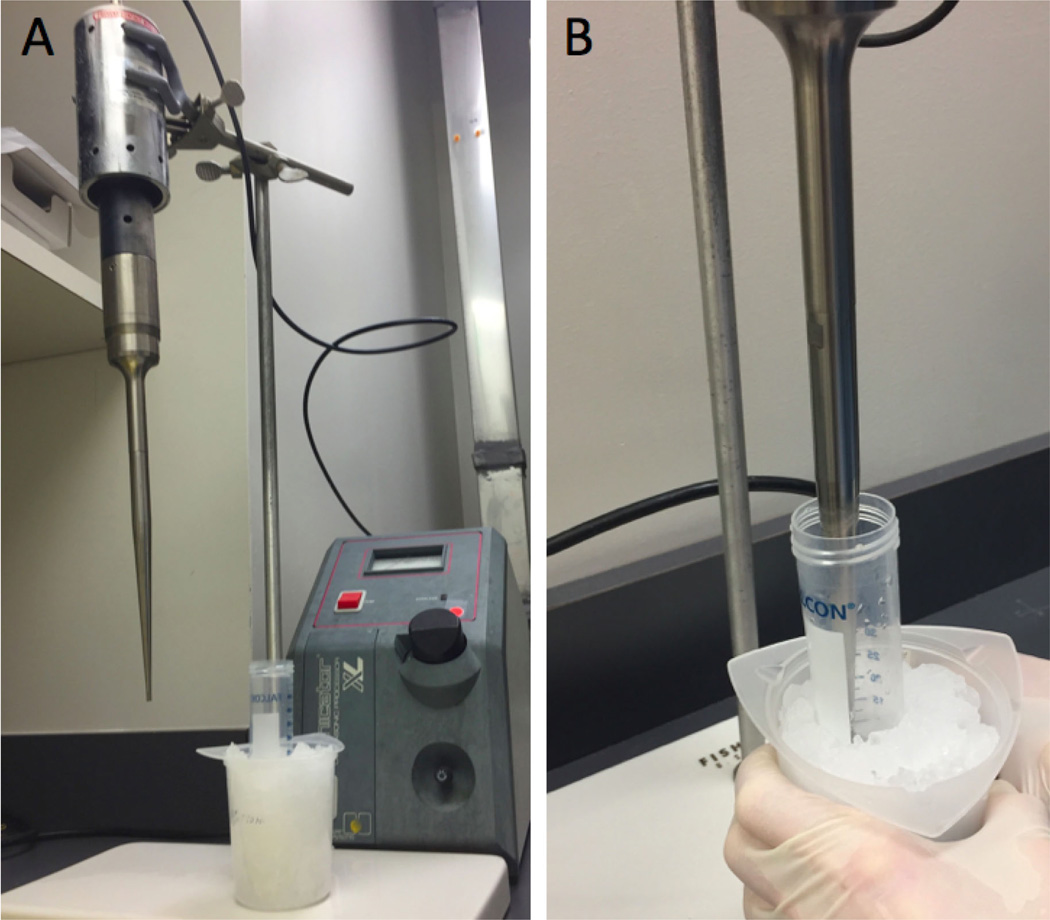
To complete cell lysis, sonicate the contents for 30 sec on ice, followed by 30 sec rest on ice. Sonicate 5 times in total (use power setting #2 for up to 2 ml of sample with fixed vibration frequency at 20 KHz) (Figure 2).
Transfer the contents to 1.5 ml microtubes and centrifuge at 14,000 × g for 30 min at 4 °C.
The protein-rich supernatant is aspirated (typically 1 to 2 ml) without disturbing the pellet and collected into a 15 ml tube placed on ice.
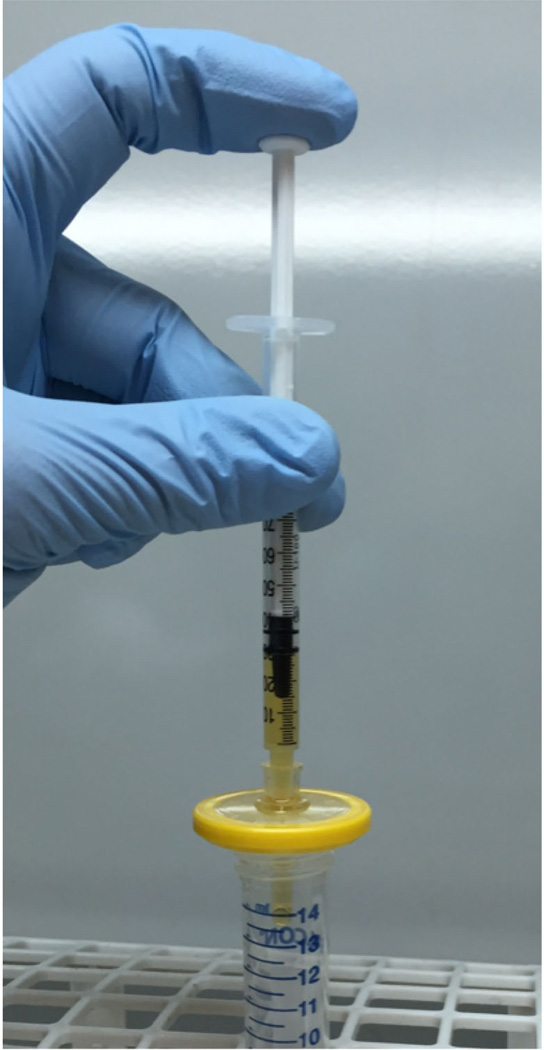
Sterilize the extract using a 1 ml syringe and a 0.22 µm syringe filter (Figure 3).
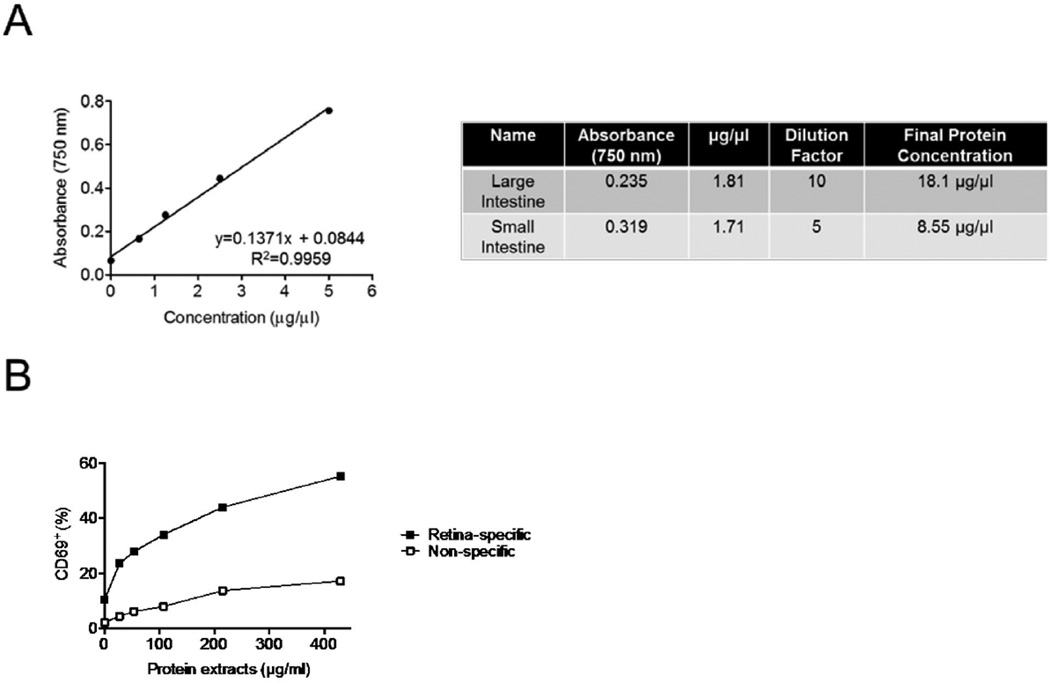
Determine protein concentration of extract using Bradford protein assay kit or equivalent, following manufacturer’s protocol. We suggest a neat, 1:10, 1:100, and 1:1,000 dilution of the extract for the assay to ensure that the result falls on the standard curve.
The amount of extract to be used to stimulate cells in vitro may vary according to the experimental design. In the original article (Horai et al., 2015), 10–500 µg/ml of extract was used to stimulate retina-specific autoreactive T cells in vitro.
Figure 1. Intestinal contents collection.
A. Opening the skin of the abdomen without disrupting the peritoneum. B. Opening peritoneum to expose intestine. C. Isolation of complete small and large intestine. D. Large intestine. E and F. Flushing out cecum (E) and colon (F) contents with PBS. G. Dispersing intestinal contents by using a plunger. H. Transfer contents into a 50 ml tube. I. Pellet contents after centrifugation.
Figure 2. Cell lysis of the intestinal contents by sonication.
Following three freeze/thaw/vortex cycles, contents are sonicated on ice. The sonicator apparatus (A) and the procedure (B).
Figure 3. Sterilization of the extract.
After centrifugation, bacteria-rich protein supernatant/extract is collected and passed through a 0.22 µm syringe filter.
Data analysis
Recipes
-
Protease inhibitor cocktail
PBS supplemented with:
10 µg/ml aprotinin
10 µg/ml leupeptin
0.5 mM phenylmethylsulfonyl fluoride (PMSF)
Figure 4. Representative results.
A. Standard curve generated from protein assay kit (left) and typical final protein concentrations of bacteria-rich protein extracts obtained from large and small intestines (right). B. Induction of CD69 expression in retina-specific versus non-specific T cells from R161H mice after 20 h of stimulation with the extracts (adapted from Figure 6C, Horai et al., 2015).
Acknowledgments
This work was supported by National Eye Institute Intramural funding NEI/NIH (Project #EY000184).
References
- 1.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horai R, Zarate-Blades CR, Dillenburg-Pilla P, Chen J, Kielczewski JL, Silver PB, Jittayasothorn Y, Chan CC, Yamane H, Honda K, Caspi RR. Microbiota-dependent activation of an autoreactive T cell receptor provokes autoimmunity in an immunologically privileged site. Immunity. 2015;43(2):343–353. doi: 10.1016/j.immuni.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4(4):337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mueller RS, Pan C. Sample handling and mass spectrometry for microbial metaproteomic analyses. Methods Enzymol. 2013;531:289–303. doi: 10.1016/B978-0-12-407863-5.00015-0. [DOI] [PubMed] [Google Scholar]
- 5.Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J, Lefsrud MG, Apajalahti J, Tysk C, Hettich RL, Jansson JK. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3(2):179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 6.Tanca A, Palomba A, Pisanu S, Deligios M, Fraumene C, Manghina V, Pagnozzi D, Addis MF, Uzzau S. A straightforward and efficient analytical pipeline for metaproteome characterization. Microbiome. 2014;2(1):49. doi: 10.1186/s40168-014-0049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong W, Abraham PE, Li Z, Pan C, Hettich RL. Microbial metaproteomics for characterizing the range of metabolic functions and activities of human gut microbiota. Proteomics. 2015;15(20):3424–3438. doi: 10.1002/pmic.201400571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong W, Giannone RJ, Morowitz MJ, Banfield JF, Hettich RL. Development of an enhanced metaproteomic approach for deepening the microbiome characterization of the human infant gut. J Proteome Res. 2015;14(1):133–141. doi: 10.1021/pr500936p. [DOI] [PMC free article] [PubMed] [Google Scholar]