Abstract
The environmental bacterium Burkholderia pseudomallei causes an estimated 165,000 cases of human melioidosis per year worldwide, and is also classified as a biothreat agent. We used whole genome sequences of 469 B. pseudomallei isolates from 30 countries collected over 79 years to explore its geographic transmission. Our data point to Australia as an early reservoir, with transmission to Southeast Asia followed by onward transmission to South Asia, and East Asia. Repeated reintroduction was observed within the Malay Peninsula, and between countries bordered by the Mekong river. Our data support an African origin of the Central and South American isolates with introduction of B. pseudomallei into the Americas between 1650 and 1850, providing a temporal link with the slave trade. We also identified geographically distinct genes/variants in Australasian or Southeast Asian isolates alone, with virulence-associated genes being among those overrepresented. This provides a potential explanation for clinical manifestations of melioidosis that are geographically restricted.
Burkholderia pseudomallei is an environmental Gram-negative bacillus and the cause of melioidosis, a serious disease of humans and animals for which there is no licensed vaccine. Infection results from inoculation, ingestion or inhalation of B. pseudomallei, and is fatal in 10-40% of human cases1. To further understand the global dissemination of melioidosis, we sequenced 276 B. pseudomallei isolates cultured from humans with melioidosis or from the environment between 1935 and 2013. These originated from 30 countries across Australasia, Asia, Africa and Central and South America. We added to this whole genome data available for a further 193 B. pseudomallei isolates from Southeast Asia2 and Australia3, giving a total dataset comprising 469 isolates (See Supplementary Data 1 for details of isolates and references). The genetic diversity of these isolates was captured by mapping short-read genome sequences against a core genome created from the two chromosomes of B. pseudomallei K96243 (ref. 4), and by extracting both core and accessory coding sequences from the assembled genomes (see Methods). We used three different approaches to outline the population structure: phylogenetic reconstructions using single nucleotide polymorphisms (SNPs) called from core genome mapping (Figure 1a); SNPs from shared single-copy core genes (Supplementary Figure 1); and a tree-independent hierarchical Bayesian clustering (Supplementary Data 1).
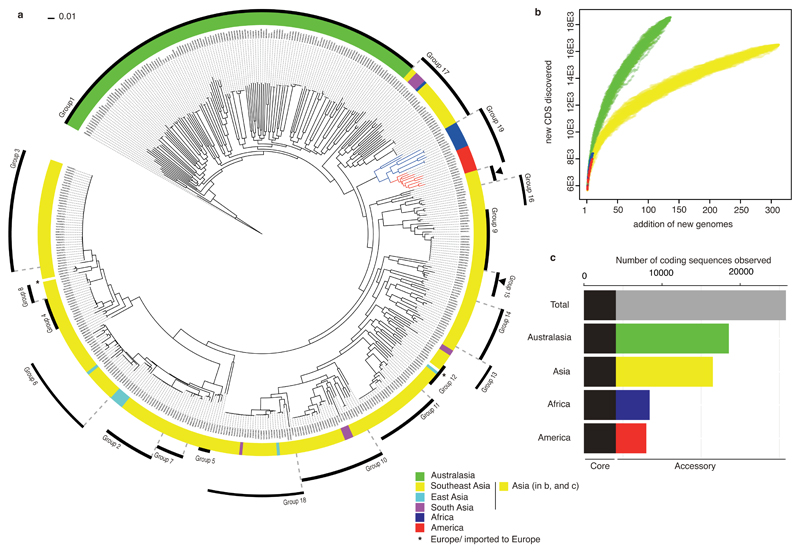
Figure 1. The phylogeny and pan-genome of B. pseudomallei.
Differences in level of bacterial diversity across different geographical origins: Australasia (green), Asia (yellow, cyan, and magenta for (a) and yellow for (b and c)), Africa (blue), America (red), and Europe (star). (a) A core SNP-based maximum likelihood phylogeny of 469 genomes with geographical origins highlighted. The tree was rooted on B. pseudomallei MSHR5619, the most genetically distant isolate based on pairwise SNP distance (see methods and Supplementary Figure 10). The outer ring represents population clusters based on BAPS hierarchical clustering (Group 1 – 19). Apart from Group 15, which is paraphyletic and marked by two black arrows, other groups each form a monophyletic branch. (b) Pan-genome accumulation curve representing rates of new gene discovery in isolates collected from different geographical origins. The order of new genome added was permutated 1,000 times to accommodate possible assortment. (c) Summary of core and accessory genomes of isolates grouped by geographical origins.
All three approaches demonstrated a clear genetic distinction between isolates from Australasia and Asia (two areas where melioidosis is endemic), supporting previous findings5,6. Isolates from Australasia had longer phylogenetic branches compared to isolates from other regions, indicative of greater genetic diversity (Figure 1a and Supplementary Figure 1). This was also observed from the pan-genome analysis7, which confirmed that the Australasian B. pseudomallei population had the highest rate of new gene discovery and the largest accessory genome (Figure 1b and 1c). Examination of data distribution confirmed that this finding was not related to different sampling periods or sequencing platforms used to generate the data (Supplementary Figure 2). These observations provide evidence for the hypothesis that Australia was an early reservoir for the current global B. pseudomallei population5,8, which is supported by the Australasian isolates being at the base of the tree (Supplementary Figure 1). An alternative explanation is that there have been repeated population bottlenecks outside Australia, but not within it. Figure 1a and Supplementary Figure 1 both delineated an apparent single transmission out of Australasia (consistent with previous findings9,10), and several independent transmission events from Southeast Asia to South Asia and East Asia. We also noted a monophyly and a single combined Bayesian cluster containing isolates from Africa and Central and South America, suggesting close ancestry (Figure 1a, Supplementary Figure 1 and Supplementary Data 1). The phylogenies also highlighted an African root for this group (100% bootstrap support), implying an African origin of the American isolates based on our sampling density.
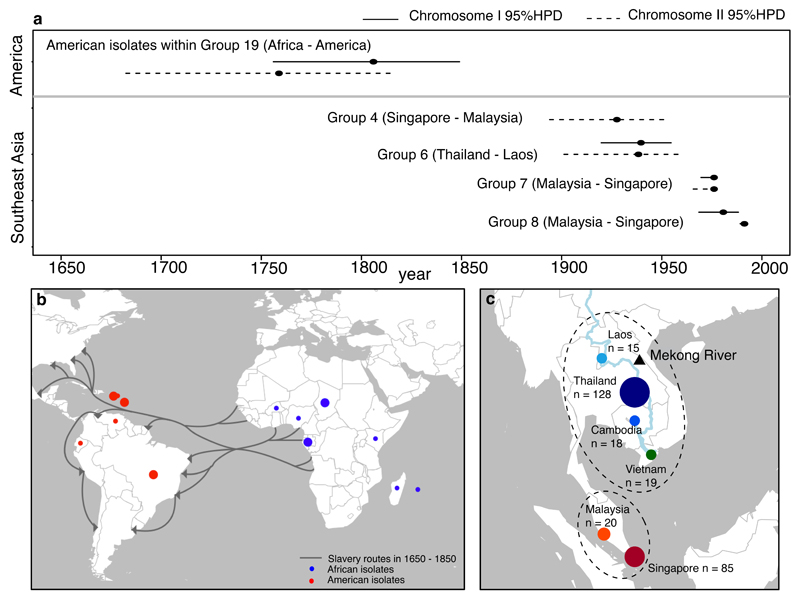
We then estimated a timeline for the intercontinental and regional spread of B. pseudomallei by identifying and analysing 19 separate Bayesian clusters comprising isolates from Australia and Oceania (group 1), Asia (groups 2 to 18), and Africa and America (group 19). To improve our sensitivity to detect genetic variants, we remapped sequence reads from each cluster against a closely related reference genome (Supplementary Figure 3). After removing sequences that had been horizontally acquired by recombination11, temporal signals were determined for each cluster with the timeline estimated by BEAST12 (Supplementary Figs 4, 5 and 6). Clock signals were captured for American isolates within the African-American cluster, and for four Asian clusters. The most recent common ancestor for the American isolates was estimated to be 1806 or 1759 based on either chromosome I or II, respectively (combined 95% highest posterior density (HPD) interval of both chromosomes, 1682-1849) (Figure 2a). The introduction of B. pseudomallei into the Americas overlaps with the height of the slave trade between 1650 - 1850, during which an estimated 10-15 million people and related cargoes including environmentally contaminated food and water were transported from Africa to the Americas (Figure 2b)13,14. Dating of Asian clusters showed that recent common ancestors could be defined for three Malaysian-Singaporean clusters and one Thai – Laos cluster, all of which dated to the 20th century (Figure 2a). The most recent common ancestor of other Asian and Australasian clusters is very likely to pre-date these estimates, but dating of these deeper evolutionary events is less reliable.
Figure 2. Timeline of trans-continental and sub-regional spread of B. pseudomallei.
(a) Estimated time when the most recent common ancestor (MRCA) of each cluster emerged. Time (black dots) and 95% highest posterior density (horizontal line) were estimated by BEAST for those clusters with temporal signals. Estimations were performed separately for chromosome I (solid lines), and II (dotted lines). Overlapping estimations between the two chromosomes provide further confidence in the time interval in which the MRCA emerged. The estimation for chromosome I of group 4 did not reach a credible effective sample size and was excluded. (b) Transatlantic slave trade routes and sampling locations of African and American isolates. Each dot represents the geographical origin of isolates used for the time estimation with the size proportional to the number of isolates. (c) The geographical landscape and isolates used to determine sub-regional connectivity. Isolates representing six Southeast Asian countries were plotted on the map, highlighting the geographical proximity of the Mekong group, and the Malay group. The number of isolates from each country was annotated.
Within the Asian isolates, the majority of Southeast Asian clusters either contained isolates from the Malay Peninsula (Malaysia and Singapore – here termed “the Malay sub-region”), or from countries bordered by the Mekong river (Thailand, Laos, Cambodia and Vietnam – here termed “the Mekong sub-region”) (Supplementary Figure 7a, Supplementary Data 1). To further examine this pattern, we estimated the number of times B. pseudomallei transitioned between Southeast Asian countries. This revealed a greater number of transitions within the same sub-regions than between sub-regions (two-tailed Mann-Whitney U test, p-value < 2.2x10-16) (Supplementary Figure 7b). The connectivity observed within sub-regions may be explained by geographical proximity, cultural links or trading networks associated with the Mekong river15,16 (Figure 2c). In addition to an unequal number of transitions, B. pseudomallei may have spent different amounts of evolutionary time in these countries (total branch lengths of multiple sub-sampling phylogenetic trees) (Supplementary Figure 7c). Assuming a homogenous mutation rate, our results are indicative of a higher proportion of evolutionary time spent in the Mekong versus the Malay sub-region (two-tailed Mann-Whitney U test, p-value < 2.2x10-16), and possibly suggests that the Mekong sub-region has been a hotspot for B. pseudomallei evolution in the Southeast Asian endemic zone. It is possible that this observation may be influenced by evolutionary rate variation on each branch, but the local clock cannot be reliably assessed across this dataset.
The most common presentation of human melioidosis in both Asia and Australia is one or more of bacteremia, pneumonia and liver and/or splenic abscesses. By contrast, some of the less common clinical manifestations show geographical segregation, including encephalomyelitis in Australia. Moreover, mortality is lower in Australasia than Southeast Asia (10% versus 40%, respectively)17. Differences in human genetics and access to medical care including intensive care facilities are likely to contribute to different outcomes, but bacterial factors could also contribute to disease severity or to specific clinical manifestations. To investigate the genetic basis that might explain clinical differences between Australasia and Southeast Asia, we systematically screened for particular kmers (DNA words) that were enriched in Australasian isolates alone, or in Southeast Asian isolates alone using a kmer based GWAS18 (see methods and Supplementary Data 2, 3 and 4). The strong link between the population structure and the geographical origin described above led us to omit population stratification in the GWAS analysis. Kmers were then clustered into loci based on their genetic proximity. This resulted in the identification of 468 and 14 loci that were specific to the Australasian and Southeast Asian population, respectively. Australasia- and Southeast Asia-specific loci were each distributed across multiple phylogenetic branches of their respective population (Supplementary Figure 8), suggesting that these were not solely driven by clonality in the population structure but may have been independently acquired and/or lost on multiple occasions. The mechanisms that have driven these patterns will be the subject of further investigation.
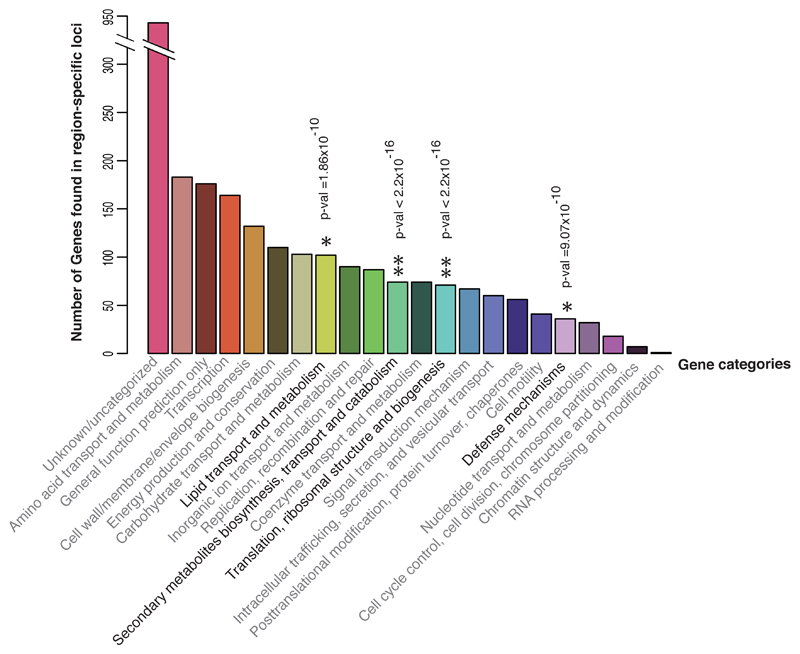
Region-specific loci included those that may enhance survival and inter-bacterial competition in specific niches. They may also reflect virulence factors that contribute to the documented regionally distinct clinical manifestations. To facilitate the biological interpretation of these data, loci were categorised by the function of genes (COG), gene ontology (GO) and pathway terms. Some genes had no functional match in the curated database, but 64.3% could be assigned which revealed that region-specific genes were widely dispersed across multiple functions (Figure 3). Functional enrichment analyses highlighted elevated frequencies of the terms “secondary metabolite biosynthesis”, “translation”, “lipid transport and metabolism” and “defense mechanisms” among region-specific genes compared to random expectation from a reference genome (one-sided Fisher test p-value < 2.2 x 10-16, < 2.2 x 10-16, 1.86 x 10-10 and 9.07 x 10-10 respectively, Supplementary Data 5). The latter contained several virulence genes involved in disease pathogenesis. Our results highlighted several virulence loci with known region-specific variations, including Burkholderia thailandensis-like flagellum and chemotaxis cluster (BTFC), and Burkholderia mallei-like BimA (BmBimA)19,20. Both BTFC and BmBimA facilitate bacterial motility inside host cells21,22, with the latter frequently detected in isolates associated with encephalomyelitis in Australia19. These findings validate our analytic approach and the ability to detect genetic variations based on geographical origin. The GWAS also identified unappreciated regional variations in well and less well characterised virulence loci (Supplementary Data 4), some examples of which are described below.
Figure 3. Region-specific genetic signatures.
Functional categories of genes (COG) localised in region-specific loci. One-sided Fisher’s exact test was used to search for terms that showed significant departure from random expectation in the reference genome. Asterisks highlight terms with heightened frequency following Bonferroni correction for multiple testing. *denotes terms with p-value <10-9, while ** denotes terms with p-value <2.2x10-16.
Filamentous hemagglutinin (fha) is a surface exposed and secreted protein that functions as an adhesin and immunomodulator across different bacterial species. In B. pseudomallei, the number of fha genes varies between isolates, and different combinations of fha genes have been observed between Australia and Thailand23. Furthermore, patients infected by B. pseudomallei with a specific fha variant are more likely to have infection associated with positive blood cultures19. We identified alternative adhesins/filamentous hemagglutinin variants in the Australasian population (Supplementary Data 4). For example, the BURPS668_RS04895 variant in Australasian isolates differed from its non-Australasian ortholog by a group of kmers that clustered in an extended signal peptide for the Type V secretion system, and in hemagglutinin repeat domains (Supplementary Figure 9a). Such variation may alter protein secretion, binding affinity and specificity.
Intracellular pathogens have evolved various mechanisms for macrophage and immune evasion. Experimental evidence has shown that B. pseudomallei is capable of subverting antigen presentation and macrophage killing via polysaccharide capsule (CPS) and a type III secretion system (T3SS)24. We identified an Australasian-variant in CPS I (Supplementary Figure 9b), marked by kmers clustered in genes coding for two capsular polysaccharide export ABC transporter transmembrane proteins and putative sulfotransferase. We also identified variation in T3SS between the Australasian and Southeast Asian population (Supplementary Figure 9c). B. pseudomallei carries at least three clusters of T3SS, including T3SS-3 which is considered a virulence factor in mammalian infection. We noted genetic variants in T3SS-3 proteins bsaU, bsaR, bsaP, bsaO, an upstream region of a transcription factor bprR known to activate genes encoding structural components of T3SS-325, and an oxygen-regulated invasion protein orgA in the Australasian population. Infection assays using a macrophage cell line have shown reduced bacterial escape and lower intracellular bacterial survival of a bsaU mutant26, although the phenotype of geographical variants has not been established.
A distinctive feature of B. pseudomallei infection is the formation of multi-nucleated giant cells (MNGC), which results from cell membrane fusion between infected and uninfected host cells. This enables bacterial cell-to-cell spread while avoiding detection by host immunity. One of the key requirements for MNGC formation is a functional Type 6 secretion system cluster 1 (T6SS-1)24. We detected regional variation that extended from a known Australasian BmBimA variant to an upstream region of virAG regulator. This locus contains variations in hemolysin-coregulated protein (hcp), type VI secretion lysozyme-like protein (tssE), and ATP-dependent clp protease located on T6SS-1 (Supplementary Figure 9d). It remains to be seen whether region-specific variations in components of T6SS-1 and upstream of the virA regulator could affect disease pathogenesis.
In conclusion, our results indicate that movement of people and cargo has led to the dissemination of B. pseudomallei, a finding with implications for our increasingly globalised lifestyle. The carrier could have been contaminated soil, water or plants, or humans and other animals with clinical or sub-clinical disease. Given the frequency of B. pseudomallei transmission within Asia, it is striking that there appears to have been only one transmission event out of a diverse Australasian population into another geographical location. This might suggest that simple transmission is not sufficient, and that an adaptive bacterial event may also have been necessary. This could reflect the fact that the fauna of Australia and Southeast Asia are significantly different (the Wallace Line27). Identification of numerous bacterial genes or gene variants that are geographically segregated provides a rich resource for biological studies of the basis for region-specific clinical syndromes in melioidosis.
Methods
Bacterial collection and DNA sequencing
The global B. pseudomallei collection sequenced for this study contained 276 isolates from the environment and human disease. The rationale underpinning isolate selection from available global collections was to maximise distribution over time and geography, with representatives from each continent (see Supplementary Figure 2a). A very limited number of isolates had been stored and were available in areas where melioidosis is either uncommon or under-reported based on lack of microbiology infrastructure, which resulted in an unequal geographic representation. DNA libraries were prepared according to the Illumina protocol and sequenced on an Illumina HiSeq2000 or Miseq with paired-end runs to give a mean coverage of 84 reads per nucleotide (range 35 – 450). Publicly available sequence data for a further 193 isolates (16 reference genomes, 76 Australasian isolates3 and 101 Southeast Asian isolates2) and their accession numbers are also tabulated in Supplementary Data 1.
Genome Assembly and Annotation
To control for potential contamination in each sample with other closely related species, taxonomic identity was assigned to all short reads and assemblies using Kraken28. Multilocus sequence typing (MLST) was derived from Illumina read data by mapping against the MLST sequence archive (http://bpseudomallei.mlst.net/). Unless previously assembled3, de novo assembly of short read data was performed using Velvet.29 The kmer size was varied between 60% and 90% of the read length, and the assembly with the best N50 selected. Contigs shorter than the insert size length were filtered out. The sequence data were then used to further improve the assembly. Contigs were iteratively scaffolded using the process described in Chewapreecha et al.30. As a QC step, reads were mapped back to the assembly using SMALT v. 0.7.4. (http://www.sanger.ac.uk/resources/software/smalt/). The assembly pipeline gave an average total length of 7,139,337 bp (range 6,744,467 – 7,536,799) from 101 contigs (range 72 - 356) with an average contig length of 84,361 bp (range 20,098– 192,188 bp) and an N50 of 223,075 (range 37,455 – 1,142,362). Gene predictions and annotations of draft reference genomes as well as other assemblies were performed using Prokka31. On average, 5,980 predicted coding sequences were assigned onto each genome (range 5,701 to 6,671 per each genome), falling within the similar range of a predicted 6,332 coding sequences in the first reference genome K96243 of 7.2 Mb.4,32
Pan-genome analysis
Based on annotated assemblies, a pan-genome was calculated for all 469 isolates using Roary7. An all-against-all comparison was performed using BLASTP and sequences clustered using a percentage identity of 92%, which was found to be a threshold that optimised specificity and sensitivity in this dataset (Supplementary Figure 10c and 10d). We identified a total of 25,812 predicted coding sequences (CDS), with 4,064 and 21,748 genes assigned to the core (present in 99% of isolates), and accessory (variably present) genome, respectively, which is comparable to that reported previously33. We used rarefaction curves to compare the number of predicted coding sequences as a function of the number of samples detected at different geographies (Figure 1b and Supplementary Figure 2c and 2d). A randomisation scheme with 1,000 permutations were employed to test our hypotheses about geographical diversity in gene contents. We also tested whether a greater rate of new gene discovery per number of samples sequenced in Australasia was biased by different sampling timeframes or because the sequence data obtained from elsewhere were generated by different sequencing platforms. After sub-sampling the data (Supplementary Figure 2) to have equal representatives by year and sequencing quality, neither showed a change in the plot trajectory.
Phylogeny based on shared single-copy core genes between B. pseudomallei and B. thailandensis
We repeated the pan-genome analysis described above with the inclusion of Burkholderia thailandensis genome E264 (accession numbers: NC_007651.1 and NC_007650.1), a closely related species that was used as an outgroup to root the tree. This demonstrated that 1,605 single-copy core genes were shared between B. thailandensis and B. pseudomallei. An approximate maximum likelihood phylogenetic tree was estimated by FastTree version 2.1.334 using GTR+CAT (General Time Reversible with per-site rate CATegories) model of approximation for site rate variation and was resampled 1,000 times (Supplementary Figure 1). The total number of single nucleotide polymorphic sites (SNPs) called was 127,421, of which 69,473 SNPs (54.53%) represented differences between B. thailandensis and B. pseudomallei. This left 57,948 SNPs to resolve the B. pseudomallei population structure.
Phylogeny based on core genome mapping of B. pseudomallei
A tree was constructed by mapping Illumina sequenced short reads to references using SMALT 0.7.4 (Figure 1a). Fully sequenced chromosomes and long reads sequenced by other platforms3 were shredded to create 100 bp paired-end reads before mapping. Reads were mapped against the core genome of B. pseudomallei strain K96243 (accession numbers BX571965 and BX571966) with bases called and aligned using a method previously described in Harris et al.35 and Page et al.36. Genetic divergence compared with the K96243 core genome ranged from 0.73 to 5.61%, and variants were identified at 324,637 SNPs (range 5,650 to 43,221 sites per isolate). A maximum-likelihood phylogeny was estimated with RAxML37 using a general time reversible nucleotide substitution model with four gamma categories for rate heterogeneity and 100 bootstrap support.
Hierarchical Bayesian clustering
A tree-independent hierarchical Bayesian clustering with hierBAPS38,39 was employed to determine the population structure generated from the core genome mapping alignment. This method allows the population to be sub-divided into groups with closely related genetic backgrounds and allows the recombination detection tool (Gubbins) to operate within its best performing range40. Except for the Australasian cluster (Group 1), which contained the highest amount of diversity for each isolate and could not be further sub-clustered, we continued the hierarchical clustering until the diversity observed in secondary or tertiary clusters fell within the limit of recombination detection (Supplementary Figure 10b). This resulted in 19 groups (Supplementary Data 1) for subsequent lineage-specific analyses. Except for Group 15 and a bin cluster (35 isolates), Group 1 - 14 and 16 - 19 each formed a monophyletic group in the phylogeny (Figure 1a).
Analysis of individual lineages
Evolutionary parameters and date of most recent common ancestors were determined for 19 clusters. For each cluster, closely related reference genomes were chosen for mapping to increase variant calling sensitivity (Supplementary Figure 3). Where relevant reference genomes were not available as complete chromosomal contigs, draft reference genomes were created from de novo assemblies. One isolate within each of these clusters was selected, assembled and ordered relative to its closest reference using ABACAS v2.5.141 and ACT42 followed by manual curation. Short reads from all members of each cluster were then mapped against this lineage-specific reference using SMALT 0.7.4. Bases were called and aligned with short insertions and deletions included using the method described in Harris et al.43. Recombination fragments were called and removed from the alignment using Gubbins11. A lineage-specific phylogeny was reconstructed using the remaining variants (Supplementary Figure 4).
Timeline reconstruction
We first tested for a positive correlation between date of isolation and root-to-tip distance obtained from a lineage-specific phylogeny with recombination removed using Path-O-Gen v1.4 (Supplementary Figure 5). Of 19 clusters, a consistent clock-like behaviour across both chromosomes was observed in a group of American isolates within the African-American cluster and five other Asian clusters (groups 4, 5, 6, 7 and 8). Except for group 5 where the number of isolates were too low (n=4) to allow credible estimations, other clusters were analysed by BEAST v1.712 to determine the clock rate and the time when the most recent common ancestor emerged. We performed model selection on combinations of strict, relaxed log-normal, relaxed exponential, and random clock models and constant, exponential, logistic and skyline population models. For each, three independent chains were run for 50 million iterations, and sampled at every 1,000 generations. Models that failed to converge based on visual inspection44 of the trace files or had effective sampling size (ESS) values < 200 for key parameters were discarded. Stepping-stone and path-sampling analyses did not show appreciable differences between clock models, potentially suggesting that there may be insufficient rate variation within each group to warrant the use of a complex clock model. Thus, the strict clock with fewest parameters was employed to avoid over-fitting of parameters as suggested in 45. We used the Bayesian skyline model as the tree prior to describing demographic history. Except for chromosome I of group 4 which did not achieve a credible ESS, the time calibrated phylogenetic trees, clock rates and time since most recent common ancestor (TMRCA) of estimated clusters are reported in Supplementary Figure 6.
Due to a small sample size used for each estimated cluster (American isolates within group 19: 9 isolates, group 4: 11 isolates, group 6: 24 isolates, group 7: 9 isolates, and group 8: 6 isolates), we also performed a date-randomised test as described in Murray et al.46 to estimate the rigour of the true temporal signals compared to noise. For each tested cluster, we performed 1,000 permutations with the true date, but randomised root-to-tip distance. Regression coefficient R2 of the true data was ranked and compared to R2 of the randomised data (Supplementary Figure 5). Ranks of the true signals ranged from 34th (group 6 chromosome II) to 97th (group 8 chromosome II), suggesting that noise had an effect on a small dataset. Aside from small sample size, our clock rate on each chromosome for the clusters estimated by BEAST is consistent with previous estimates in Burkholderia species47 and other bacteria35,48–50. This suggests that the results generated here are non-random.
Ancestral state reconstruction on geographic locations of Southeast Asian isolates
Ancestral reconstruction was performed on the maximum likelihood global core genome phylogeny to assess the connectivity of isolates, and infer which population might act as source versus sink in Southeast Asia. To avoid sampling bias, we sub-sampled the phylogeny so that there were equal numbers of isolates from Thailand, Laos, Cambodia, Vietnam, Malaysia and Singapore (n=15 for each country), and resampled 1,000 times. Countries containing less than 15 isolates were excluded. We treated countries as discrete geographic characters. For each sub-sampled tree, we used stochastic character mapping make.simmap available in R package phytools v0.5-10 51,52 to estimate both the transitions between different geographical characters and the total time spent in each geographical character. Stochastic mapping was performed under an asymmetric model of character change for 1,000 simulations.
To assess the connectivity of isolates, we categorised geographical characters into two groups based on geographical proximity. The Mekong sub-region represents countries bordered by the Mekong river including Thailand, Laos, Cambodia and Vietnam; the Malay sub-region comprises Malaysia and Singapore. Changes between geographical characters were counted after grouping into two categories: 1) transitions within the same sub-region, and 2) transitions between sub-regions. The occurrence of transitions within and between the two sub-regions was compared using a two-tailed Mann-Whitney U test (Supplementary Figure 7b). To infer which population might act as the source, we compared the time spent in the Mekong and Malay sub-regions and compared this using a two-tailed Mann-Whitney U test (Supplementary Figure 7c). The choice of non-parametric Mann-Whitney U test over parametric test was due the violation of normally distributed data.
Identification of distinct genes/variants in Australasian and Southeast Asian populations
Kmer-based GWAS without correction for population structure
We first considered the optimal approach to perform a GWAS for B. pseudomallei. Given the high level of genomic plasticity and large accessory genomes (Figure 1c), we concluded that a GWAS based on core genome SNPs as used elsewhere53,54 would be sub-optimal as this fails to capture the extent of genetic variation. Instead, we used kmers (DNA words of length k) as an alternative to a SNP-based analysis. Unlike a traditional GWAS where genetic causes of particular phenotypes were identified while adjusting for population stratification, we employed GWAS to search for genetic markers in the Australasian and Southeast Asian populations, some of which may intrinsically define population structure. A control for population structure was thus omitted. Two independent GWAS runs were performed to search for variable kmers in the Australasia population alone (Australasia GWAS), and the Southeast Asian population alone (SEA GWAS). For both GWAS runs, the data were randomly divided into a discovery and a validation dataset. The Australasia GWAS comprised a set of 80 Australasia and 200 non-Australasian isolates, and was validated with 57 Australasian and 132 non-Australasian isolates. Similarly, the SEA GWAS comprised a random set of 180 Southeast Asian and 105 non-Southeast Asian isolates, and was confirmed using 114 Southeast Asian and 65 non-Southeast Asian isolates. We used the reference-independent GWAS pipeline Seer by Lees et al.18 to search for kmers with region-specific patterns. All kmers of length 9-100 bp were scanned from all assembled reads using fsm-lite (https://github.com/nvalimak/fsm-lite). Only kmers seen in 5-95% of the total population were retained to reduce false positives from testing underpowered kmers. Seer18 was performed on the discovery data using geographical origin of isolates (Australasia/ non-Australasia or Southeast Asia/ non-Southeast Asia) as binary phenotype (y) and the presence/absence of each kmer as tested genotype X:
The direction of association (positive or negative) is described by β. Kmers with a conservative cut-off p-value < 10-8 in the logistic regression were considered further as suggested in Lees et al.18. Australasia and SEA GWAS yielded 77,787 and 43,663 kmers, respectively, that were positively or negatively associated with Australasia or Southeast Asia populations (Supplementary Data 2). Among these, 42,521 kmers that were positively associated with Australasia were negatively associated with Southeast Asia (Supplementary Figure 11). Kmers that reached significance in the discovery data were confirmed in the validation data. To aid visualisation, the frequencies of 5,000 randomly chosen kmers from the Australasia and SEA GWAS in the validation data have been plotted in Supplementary Figure 11.
Mapping and kmer clustering
Significant kmers were searched for an exact match in de novo assemblies and fully sequenced chromosomes using BLAT v. 3455 with minimum match and score adjusted to cater for low complexity kmers as below.
blat assembly kmers.query –minMatch=1 –minScore=10 output
To facilitate biological interpretation, kmers were grouped into clusters based on their genetic distance. We defined the size of operons based on the length of transcription fragments reported in Ooi et al.56. Any kmers located within 7.68 kb (the size of an operon covering 95th percentile of transcription fragments) were grouped together into a locus. On average, each locus had a median of 66 kmers (range 2 -11,072 kmers), with the size of the loci ranging from 40 – 70,684 bp. The binary patterns in size of region-specific loci (Supplementary Figure 12) likely reflect different scales of variation, with smaller and larger peaks corresponding to small-scale differences (including SNPs) and large-scale differences (including regions of mobile genetic elements incorporated via homologous recombination or site specific recombination), respectively. As the GWAS was not corrected for population structure, we further tested whether the predicted loci were subjected to clonality. The presence and absence of each locus (measured by % of detected kmers) were plotted against the phylogeny. Their scattering patterns across multiple branches suggested that region-specific loci were not strictly driven by a clonal population structure (Supplementary Figure 8).
COG, GO and pathway terms found in region-specific loci
We annotated the biological properties of kmers within coding regions using information from the functional categories (COG term), Gene Ontology (GO term), and pathway data (KEGG, InterPro and UniPathway), available from the Burkholderia Genome Database57. The reference genome Bp668 contained 40,986 out of 78,929 region-specific kmers, of which 23,565 overlapped with coding regions. One-sided Fisher’s exact test was used to search for COG, GO and pathway terms in kmers that showed significant departure from random expectation in the Bp668 genome. We tested kmers enrichment in 22 COG terms, 1,485 GO terms, and 408 pathway terms using a strict Bonferroni correction with a required p-value of 0.01/1,915 = 5.22x10-6. Significant COG terms were marked in Figure 3. Additional GO and pathway enrichment analyses are discussed in the supplementary note. Terms with significant deviation are tabulated in Supplementary Data 5.
Statistics and visualisation
Visualisation of phylogenetic trees and statistical analyses were performed in R58, iToL59, and FigTree v 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Data availability
New sequence data for the study isolates have been deposited in the ENA under study accession number ERP001193 and ERP002658, with the accession numbers for individual isolates listed in Supplementary Data 1. Supplementary Data 2-5 provide information that supports the data presented.
Supplementary Material
Acknowledgements
The authors thank the Wellcome Trust Sanger Institute library construction, sequence and core informatics teams, and Elizabeth Blane for their technical support. We thank Drs Tanistha Nandi and Patrick Tan at Genome Institute of Singapore; and Drs Erin Price and Derek Sarovich at Menzies School of Health Research, Australia for providing access to publically available WGS data. We thank the following people who provided isolates or DNA: Professor Nicholas Day, MORU, Faculty of Tropical Medicine, Mahidol University; Drs Paul Newton, Manivanh Vongsouvath, Mayfong Mayway, Viengmon Davong, Olay Lattana, Catrin Moore, Sayaphet Rattanavong and the directors and staff of Mahosot Hospital, Vientiane, Lao PDR; Dr Varun Kumar, Ankor Hospital for Children, Siem Reap, Cambodia; Dr James Campbell, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam; Dr Hui Suk Wai, Ocean Park Corporation, Hong Kong SAR, China; Mr Chun Kham and Dr Thong Phe, Sihanouk Hospital Centre of Hope, Phnom Penh, Cambodia; Dr Joost W. Wiersinga, Academic Medical Center (AMC), Amsterdam, the Netherlands; Professor Jan Jacobs, ITM, Antwerp, Belgium; Dr Julie E. Russell, National Collection of Type Cultures, UK; Dr Ty Pitt, NHS Blood and Transplant, UK; Mr Daniel Godoy, Imperial College, UK; Dr Stephane Emonet, Geneva University Hospitals, Switzerland; Dr Susan Morpeth, Middlemore Hospital, New Zealand; and Dr Jay Gee, CDC, USA. C.C. is a Sir Henry Wellcome post-doctoral Fellow (grant ref: 107376/Z/15/Z). J.C., M.V., Z.Y. were supported by the COIN Centre of Excellence and Z.Y. by a HIIT post-doctoral fellowship. A.E.M. is supported by Biotechnology and Biological Sciences Research Council grant BB/M014088/1. B.G.S. was supported by the Wellcome Trust grant WT089472. D.A.B.D and R.P. are supported by the Wellcome Trust grants 106698/Z/14 and B9R00760. D.L. and V.W. are supported by the Wellcome Trust grant 089275/Z/09/Z. M.M. and B.J.C are supported by the Australian National Health and Medical Research Council through project grants #1046812 and #1098337. This publication presents independent research supported by the Health Innovation Challenge Fund (WT098600, HICF-T5-342), a parallel funding partnership between the Department of Health and Wellcome Trust. The views expressed in this publication are those of the author(s) and not necessarily those of the Department of Health or Wellcome Trust. This project was also funded by a grant awarded to the Wellcome Trust Sanger Institute (098051).
Footnotes
Author contributions
A.T., B.D.S., S.L.H., C.B., M.M., V.W., D.L., R.P., B.G.S., P.K., D.A.B.D. and B.J.C. collected and provided the samples for the study. C.C. designed and performed the analyses. M.T.G.H, S.R.H., A.E.M., J.C., J.P. and G.D. designed and contributed materials and analysis tools. M.V., N.V., Z.Y., and J.C. performed the kmer based analyses in the first draft. C.C. performed the kmer based analysis in the revised draft. Z.Y. and J.C. performed cluster analyses. S.J.P. was responsible for management of the study. S.J.P. and C.C. wrote the paper with input from all authors. All authors approved the manuscript prior to submission.
References
Note: number 1 - 27 are in text references
- 1.Limmathurotsakul D, et al. Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2015.8. [DOI] [PubMed] [Google Scholar]
- 2.Nandi T, et al. Burkholderia pseudomallei sequencing identifies genomic clades with distinct recombination, accessory, and epigenetic profiles. Genome Res. 2015;25:608. [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson SL, et al. Whole-Genome Sequences of 80 Environmental and Clinical Isolates of Burkholderia pseudomallei. Genome Announc. 2015;3 doi: 10.1128/genomeA.01282-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holden MT, et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci U S A. 2004;101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price EP, et al. Large-scale comparative genomics identifies unprecedented melioidosis cases in northern Australia caused by an Asian Burkholderia pseudomallei strain. Appl Environ Microbiol. 2015 doi: 10.1128/AEM.03013-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearson T, et al. Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biol. 2009;7:78. doi: 10.1186/1741-7007-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dale J, et al. Epidemiological tracking and population assignment of the non-clonal bacterium, Burkholderia pseudomallei. PLoS Negl Trop Dis. 2011;5:e1381. doi: 10.1371/journal.pntd.0001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gee JE, Allender CJ, Tuanyok A, Elrod MG, Hoffmaster AR. Burkholderia pseudomallei type G in Western Hemisphere. Emerg Infect Dis. 2014;20:682–684. doi: 10.3201/eid2004.130960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarovich DS, et al. Phylogenomic Analysis Reveals an Asian Origin for African Burkholderia pseudomallei and Further Supports Melioidosis Endemicity in Africa. mSphere. 2016;1 doi: 10.1128/mSphere.00089-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Croucher NJ, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolchin P. American Slavery: 1619-1877. Penguin; 1995. [Google Scholar]
- 14.Thomas H. The Slave Trade, The Story of the Atlantic Slave Trade:1440-1870. Simon & Schuster Paperbacks; 1997. [Google Scholar]
- 15.Nguyen TD. The Mekong River and the Struggle for Indochina: Water, War and Peace. Praeger Publishers; 1999. [Google Scholar]
- 16.Liu JH, Lawrence B, Ward C. Social representations of history in Malaysia and Singapore: On the relationship between national and ethnic identity. Asian J Soc Psychol. 2002;5:3–20. [Google Scholar]
- 17.Currie BJ. Melioidosis: evolving concepts in epidemiology, pathogenesis, and treatment. Semin Respir Crit Care Med. 2015;36:111–125. doi: 10.1055/s-0034-1398389. [DOI] [PubMed] [Google Scholar]
- 18.Lees JA, et al. Sequence element enrichment analysis to determine the genetic basis of bacterial phenotypes. Nat Commun. 2016;7:12797. doi: 10.1038/ncomms12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarovich DS, et al. Variable virulence factors in Burkholderia pseudomallei (melioidosis) associated with human disease. PLoS One. 2014;9:e91682. doi: 10.1371/journal.pone.0091682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuanyok A, et al. A horizontal gene transfer event defines two distinct groups within Burkholderia pseudomallei that have dissimilar geographic distributions. J Bacteriol. 2007;189:9044–9049. doi: 10.1128/JB.01264-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.French CT, et al. Dissection of the Burkholderia intracellular life cycle using a photothermal nanoblade. Proc Natl Acad Sci U S A. 2011;108:12095–12100. doi: 10.1073/pnas.1107183108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benanti EL, Nguyen CM, Welch MD. Virulent Burkholderia species mimic host actin polymerases to drive actin-based motility. Cell. 2015;161:348–360. doi: 10.1016/j.cell.2015.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuanyok A, et al. Genomic islands from five strains of Burkholderia pseudomallei. BMC Genomics. 2008;9:566. doi: 10.1186/1471-2164-9-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willcocks SJ, Denman CC, Atkins HS, Wren BW. Intracellular replication of the well-armed pathogen Burkholderia pseudomallei. Curr Opin Microbiol. 2016;29:94–103. doi: 10.1016/j.mib.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, et al. Characterization and analysis of the Burkholderia pseudomallei BsaN virulence regulon. BMC Microbiol. 2014;14:206. doi: 10.1186/s12866-014-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bast A, et al. Caspase-1-dependent and -independent cell death pathways in Burkholderia pseudomallei infection of macrophages. PLoS Pathog. 2014;10:e1003986. doi: 10.1371/journal.ppat.1003986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallace AR. On the Physical Geography of the Malay Archipelago. Journal of the Royal Geographical Society of London. 1863;7:205–212. [Google Scholar]
- 28.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chewapreecha C, et al. Dense genomic sampling identifies highways of pneumococcal recombination. Nat Genet. 2014;46:305–309. doi: 10.1038/ng.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 32.Nandi T, et al. A genomic survey of positive selection in Burkholderia pseudomallei provides insights into the evolution of accidental virulence. PLoS Pathog. 2010;6:e1000845. doi: 10.1371/journal.ppat.1000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spring-Pearson SM, et al. Pangenome Analysis of Burkholderia pseudomallei: Genome Evolution Preserves Gene Order despite High Recombination Rates. PLoS One. 2015;10:e0140274. doi: 10.1371/journal.pone.0140274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price MN, Dehal PS, Arkin AP. FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLoS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Page AJ, et al. SNP-sites: rapid efficient extraction of SNPs from multiFASTA alignments. Microbial Genomics. 2016;2 doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and postanalysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corander J, Marttinen P, Siren J, Tang J. Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics. 2008;9:539. doi: 10.1186/1471-2105-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng L, Connor TR, Siren J, Aanensen DM, Corander J. Hierarchical and Spatially Explicit Clustering of DNA Sequences with BAPS Software. Mol Biol Evol. 2013;30:1224–1228. doi: 10.1093/molbev/mst028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Croucher NJ, et al. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat Genet. 2013;45:656–663. doi: 10.1038/ng.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Assefa S, Keane TM, Otto TD, Newbold C, Berriman M. ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics. 2009;25:1968–1969. doi: 10.1093/bioinformatics/btp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carver TJ, et al. ACT: the Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 43.Harris SR, et al. Genome specialization and decay of the strangles pathogen, Streptococcus equi, is driven by persistent infection. Genome Res. 2015;25:1360–1371. doi: 10.1101/gr.189803.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rambaut A, Suchard MA, Xie D, Drummond AJ. Tracer v1.6. 2014 < http://beast.bio.ed.ac.uk/Tracer>.
- 45.Ho SY, Duchene S. Molecular-clock methods for estimating evolutionary rates and timescales. Mol Ecol. 2014;23:5947–5965. doi: 10.1111/mec.12953. [DOI] [PubMed] [Google Scholar]
- 46.Murray GGR, et al. The effect of genetic structure on molecular dating and tests for temporal signal. Methods in Ecology and Evolution. 2016;7:80–89. doi: 10.1111/2041-210X.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lieberman TD, et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet. 2011;43:1275–1280. doi: 10.1038/ng.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathers AJ, et al. Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae at a single institution: insights into endemicity from wholegenome sequencing. Antimicrob Agents Chemother. 2015;59:1656–1663. doi: 10.1128/AAC.04292-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young BC, et al. Evolutionary dynamics of Staphylococcus aureus during progression from carriage to disease. Proc Natl Acad Sci U S A. 2012;109:4550–4555. doi: 10.1073/pnas.1113219109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Revell LJ. phytools: An R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3:217–223. [Google Scholar]
- 52.Bollback JP. SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics. 2006;7:88. doi: 10.1186/1471-2105-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laabei M, et al. Predicting the virulence of MRSA from its genome sequence. Genome Res. 2014;24:839–849. doi: 10.1101/gr.165415.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chewapreecha C, et al. Comprehensive identification of single nucleotide polymorphisms associated with beta-lactam resistance within pneumococcal mosaic genes. PLoS Genet. 2014;10:e1004547. doi: 10.1371/journal.pgen.1004547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ooi WF, et al. The condition-dependent transcriptional landscape of Burkholderia pseudomallei. PLoS Genet. 2013;9:e1003795. doi: 10.1371/journal.pgen.1003795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winsor GL, et al. The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics. 2008;24:2803–2804. doi: 10.1093/bioinformatics/btn524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2008. http://www.R-project.org. [Google Scholar]
- 59.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
New sequence data for the study isolates have been deposited in the ENA under study accession number ERP001193 and ERP002658, with the accession numbers for individual isolates listed in Supplementary Data 1. Supplementary Data 2-5 provide information that supports the data presented.