Abstract
Intercellular heterogeneity, exacerbated by chromosomal instability (CIN), fosters tumor heterogeneity and drug resistance. However, extreme CIN correlates with improved cancer outcome, suggesting that karyotypic diversity required to adapt to selection pressures might be balanced in tumors against the risk of excessive instability. Here, we used a functional genomics screen, genome editing, and pharmacologic approaches to identify CIN-survival factors in diploid cells. We find partial anaphase-promoting complex/cyclosome (APC/C) dysfunction lengthens mitosis, suppresses pharmacologically induced chromosome segregation errors, and reduces naturally occurring lagging chromosomes in cancer cell lines or following tetraploidization. APC/C impairment caused adaptation to MPS1 inhibitors, revealing a likely resistance mechanism to therapies targeting the spindle assembly checkpoint. Finally, CRISPR-mediated introduction of cancer somatic mutations in the APC/C subunit cancer driver gene CDC27 reduces chromosome segregation errors, whereas reversal of an APC/C subunit nonsense mutation increases CIN. Subtle variations in mitotic duration, determined by APC/C activity, influence the extent of CIN, allowing cancer cells to dynamically optimize fitness during tumor evolution.
Significance
We report a mechanism whereby cancers balance the evolutionary advantages associated with CIN against the fitness costs caused by excessive genome instability, providing insight into the consequence of CDC27 APC/C subunit driver mutations in cancer. Lengthening of mitosis through APC/C modulation may be a common mechanism of resistance to cancer therapeutics that increase chromosome segregation errors.
Introduction
Chromosome missegregation leads to abrupt changes in gene expression and protein stoichiometry that result in a strong negative selection pressure when occurring in most diploid cell types, but which are tolerated in aneuploid cancer cells (reviewed in ref. 1). At least part of the selection against aneuploidy relies on p53, which limits cell propagation after chromosome missegregation and genome-doubling (2–4). Under selective pressure, however, chromosomal instability (CIN) enables cells to explore various karyotypic states, allowing the eventual emergence of subclones with improved fitness, a recurrent mode of adaptation observed in fungal pathogens, yeast, and mammalian cells, and a cause of treatment failure (5–17). Murine models largely support the notion that CIN favors tumor formation, but conversely excessive CIN appears to suppress tumorigenesis, analogous to mutational meltdown and error-prone catastrophe in bacterial and viral genetics (18–21). Although CIN has been generally associated with poor prognosis, patient stratification based on the degree of CIN has revealed that extremes of CIN are associated with improved prognosis, lending credence to the “just-right” threshold of genomic instability sufficient for tumor adaptation proposed by Cahill and colleagues (22–26). Excessive CIN appears deleterious for cell fitness, and, accordingly, enhancing chromosome missegregation has been proposed as an approach to target CIN cancer cells (18, 27, 28). Hence, selection could favor the attenuation of CIN in human cancer to prevent excessive genome instability while ensuring sufficient karyotypic instability to foster adaptation to a changing environment. Here, we explore cellular mechanisms contributing to the adaptation of excessive CIN in human cancer.
Results
Experimental Model for CIN Threshold and Tolerance
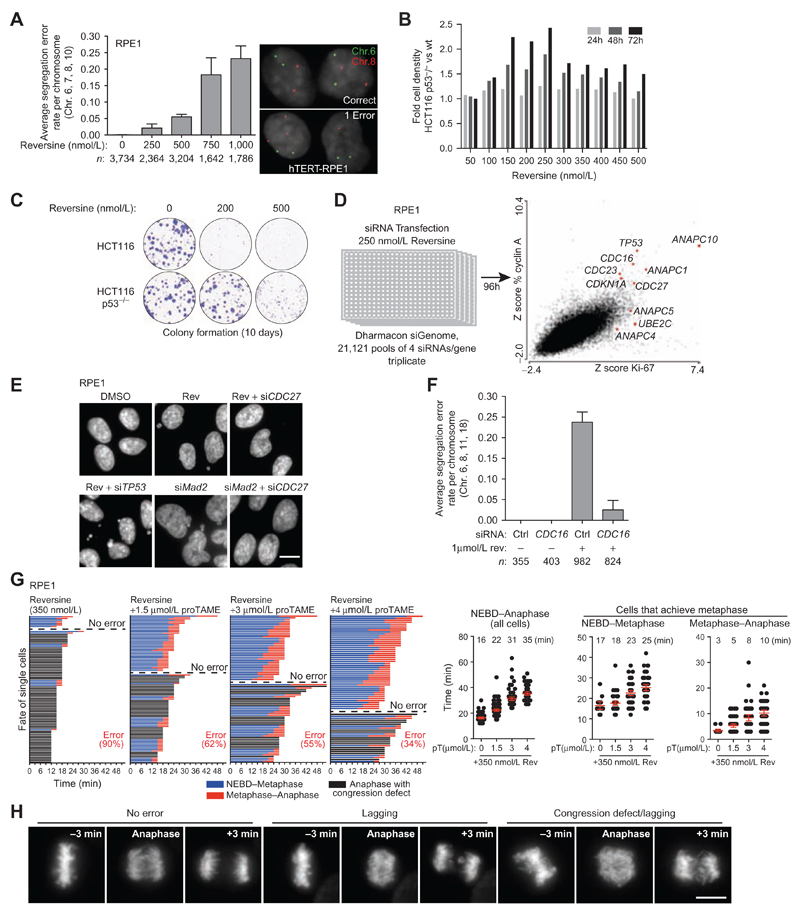
To investigate how cells respond and adapt to whole-chromosome missegregation, we sought a method to induce CIN in diploid cells that was amenable to high-throughput screening. Given the crucial role of the spindle assembly checkpoint (SAC) for chromosome segregation fidelity, we took advantage of reversine, an inhibitor of the SAC kinase MPS1 encoded by the gene TTK (29, 30). We investigated whether reversine titration could tune the frequency of segregation errors in cells. The hTERT-immortalized diploid epithelial cell line RPE1 and near-diploid HCT116 colorectal cancer cells were chosen for their karyotypic stability and refractoriness to CIN, which is largely due to their functional p53 pathway (2). We measured segregation error rates by centromeric FISH from RPE1 daughter cell pairs born during acute reversine exposure in mitosis. Reversine increased the error rate per chromosome pair per division in a dose-dependent manner, from 0.00027 in DMSO-treated cells (approximately 1 error per 165 divisions, assuming a diploid karyotype with equal error rates for all chromosomes) to 0.021 in 250 nmol/L (1 error per 2 divisions), 0.055 in 500 nmol/L (1.3 chromosome errors per division on average), 0.183 in 750 nmol/L (4.2 chromosomes per division), and 0.232 in 1 μmol/L reversine (5.3 chromosomes per division; Fig. 1A; Supplementary Table S1). Reversine treatment resulted in the p53-dependent expression of p21 (CDKN1A) and reduced proliferation in RPE1 and HCT116 cells, reflecting the activation of a cellular stress response to aneuploidy following a single passage through mitosis (Supplementary Fig. S1 A–S1F; ref. 2). TP53 RNAi increased the proliferation of RPE1 cells in reversine, reflecting greater tolerance to CIN and aneuploidy (Supplementary Fig. S1B; ref. 2). Phosphorylation of p53 at serine 15 was not detectable after reversine treatment, and the ATM inhibitor KU-55933 did not block p21 induction in reversine, suggesting that an ATM-mediated DNA-damage response was not the underlying cause for cell-cycle arrest (Supplementary Fig. S1G and S1H). We conclude that reversine titration allows control over the rate of chromosome segregation errors in otherwise diploid cells, thereby mimicking varying levels of CIN induction.
Figure 1. Identification of APC/C subunits in a genome-wide siRNA screen for CIN survival.
A, Chromosome segregation error rates determined by FISH in postmitotic daughter cells. RPE1 cells were treated with reversine for 2 hours, then mitotic cells were collected by shake-off and allowed to reattach in the presence of reversine, before fixation. Graph represents the average rate measured for chromosomes 6, 7, 8, and 10 (bars, average ± 95% CI). B, HCT116 wild-type and p53−/− isogenic lines were imaged for 72 hours by live-cell imaging. Cell density for each drug concentration was normalized to DMSO for wild-type and p53−/− cell separately. Fold difference in cell density of HCT116 p53−/− relative to wild-type is displayed for each drug concentration. C, Clonogenic assay using isogenic HCT116 cells grown for 10 days in the presence or absence or reversine, as indicated. D, Genome-wide RNAi screen for synthetic viability with MPS1 inhibition. RPE1 cells were synchronized in G0–G1 by contact inhibition, trypsinized, and reverse-transfected at low density in triplicate to allow uniform passage through mitosis. Cells were exposed to 250 nmol/L reversine for 96 hours and fixed. Automated image acquisition and analysis were performed for various parameters, and Z-scores were derived based on median plate normalization for each siRNA pool. E, DAPI staining of fixed cells 48 hours following the indicated siRNA treatments with 250 nmol/L reversine where indicated. F, Chromosome segregation error rates measured by FISH as in A. Cells were transfected with a nontargeting or a CDC16 siRNA pool, and 48 hours later, cells were synchronized with a single thymidine block. Reversine was added 10 hours after thymidine release, prior to mitotic entry. Mitotic cells were collected at 12 hours by shake-off, allowed to reattach on coverslips still in the presence of reversine, and fixed (bars, average ± 95% CI). G, Time-lapse fluorescence microscopy of RPE1 cells expressing H2B-mCherry imaged every 3 minutes, 1 hour following the addition of 350 nmol/L reversine +/− proTAME (pT), as indicated. The duration from NEBD to metaphase and metaphase to anaphase is shown for cells in which all chromosomes congressed to form a metaphase plate. Each row corresponds to a single cell (n > 60 cells each; bars, mean ± 95% CI). H, Sample images of segregation errors scored in G. Maximum intensity projections are shown for timeframes immediately preceding and following anaphase onset. Shown are examples of a correct division (no error), an example of a lagging chromosome following proper congression at metaphase (middle), and an example where anaphase occurred before all chromosomes congressed to the metaphase plate (congression defect, hence NEBD–metaphase could not be determined). Scale bar, 10 μm.
We next used reversine titration to examine the selective advantage of a genetic background permissive to CIN in response to increasing levels of CIN, by comparing HCT116 wild-type and TP53−/− isogenic cell lines (31). TP53 disruption provided a clear proliferative advantage in low concentrations of reversine (150–250 nmol/L) over a 72-hour period, but this difference was less pronounced when CIN levels were further increased (300–500 nmol/L; Fig. 1B). In a long-term assay, TP53 disruption enabled colony formation in 200 nmol/L reversine, whereas 500 nmol/L reversine severely impaired colony formation in both cell lines (Fig. 1C). These observations suggest that CIN tolerance caused by TP53 loss enables proliferation only when the segregation error rate is kept within a narrow window, beyond which excessive instability can no longer be tolerated (18, 27). We hypothesize that there might be selective pressures to prevent excessive CIN during cancer evolution in order to improve cell fitness.
A Synthetic Viability RNAi Screen for CIN Identifies APC/C Subunits
To identify genetic alterations that might promote survival following the induction of chromosome segregation errors, we performed a genome-wide siRNA screen in RPE1 cells exposed to 250 nmol/L reversine (Fig. 1D). Automated image acquisition and analysis was used to rank the effect of each siRNA pool on overall proliferation, based on cell number and the fraction of cells expressing the proliferation markers cyclin A2 and Ki-67. The screen identified seven subunits of the anaphase-promoting complex/cyclosome (APC/C) E3 ligase complex (CDC16, CDC23, CDC27, ANAPC1, ANAPC4, ANAPC5, ANAPC10), along with one of its E2 ligases UBE2C (UBCH10) whose knockdown conferred a proliferative advantage in reversine (Fig. 1D; Supplementary Table S2 for a gene ontology enrichment analysis). This phenotype could be validated with multiple siRNA duplexes targeting each subunit in RPE1 cells and was reproduced in HCT116 cells (Supplementary Fig. S2A–S2F). Collectively, the multitude of siRNA duplexes tested and subunits targeted suggest that our results are a consequence of on-target reduction in APC/C activity.
The SAC functions by inhibiting the APC/C during the early stage of mitosis until chromosome bi-orientation is achieved at metaphase, which in turn silences SAC signaling and triggers anaphase onset by allowing APC/C bound to its coactivator CDC20 to degrade securin and cyclin B (29). The duration from nuclear envelope breakdown to anaphase (NEBD–anaphase) therefore represents the time during which the SAC is active and monitors chromosome attachment to the mitotic spindle (32). Of note, complete pharmacologic inhibition of the APC/C or genetic ablation of the co-activator CDC20 robustly arrests cells at metaphase (33, 34), indicating that the RNAi conditions used here result in partial APC/C loss of function. APC/C subunit RNAi rescued cell-cycle arrest following MAD2 RNAi or reversine-induced SAC impairment, and reduced the number of micronuclei, suggesting that fewer segregation errors were occurring, in contrast to TP53 siRNA which did not reduce micronuclei formation (Fig. 1E; Supplementary Fig. S3A–S3C). FISH analyses confirmed that CDC16 RNAi or transient inhibition of proteasomal degradation with MG132 efficiently reduced the rate of segregation errors in reversine-treated cells (Fig. 1F; Supplementary Table S1; Supplementary Fig. S3D–S3F). These results are consistent with previous observations that imposing a long metaphase arrest (>80 minutes) using 12 μmol/L of the APC/C inhibitor proTAME can rescue segregation errors in MAD2-depleted cells (35). However, our initial analysis revealed that RNAi against various APC/C subunits, such as ANAPC10 or CDC16 RNAi, increased mitotic duration by less than 15 minutes in 500 nmol/L reversine (Supplementary Fig. S3G). To further explore how small changes in NEBD-anaphase duration affect chromosome segregation fidelity, we performed time-lapse fluorescence microscopy on RPE1/H2B-mCherry cells treated with 350 nmol/L reversine in combination with low doses of proTAME (1.5 μmol/L, 3 μmol/L, 4 μmol/L), which inhibits the APC/C by competing with the coactivator CDC20 (Fig. 1G and H; ref. 36). Under these conditions, we observed a mitotic delay of 6 to 19 minutes (from 16 minutes in reversine to 22, 31, and 35 minutes when combined with 1.5 μmol/L, 3 μmol/L, and 4 μmol/L proTAME, respectively), along with an increase in the number of cells that achieved bi-orientation (Fig. 1G). Longer NEBD-anaphase durations correlated with a progressive reduction in segregation errors from 90% to 34% (Fig. 1G). This indicates that partial APC/C dysfunction, resulting in mild delays in mitotic progression, can have profound effects in limiting the frequency of chromosome segregation errors.
CDC27 Haploinsufficiency Confers Resistance to CIN
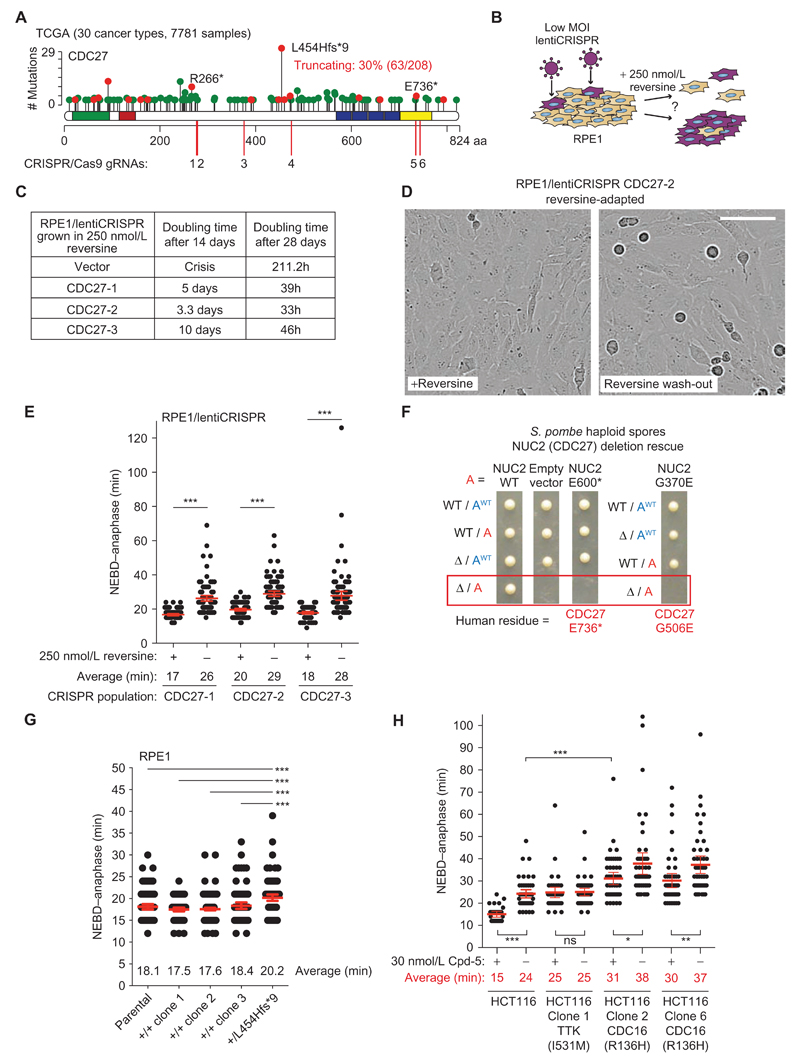
The observation that impaired APC/C activity could reduce the rate of segregation errors prompted us to investigate the relationship between APC/C mutations and CIN in cancer. We surveyed 30 cancer types from The Cancer Genome Atlas (TCGA) totaling 7,781 individual tumors. Depending on the cancer type, 0.7% to 22.9% of tumors harbored a nonsynonymous mutation in an APC/C subunit (median, 6%; average, 8%; Supplementary Table S3). Although this was not above the expected mutation rate for a group of 15 genes using randomization tests, it has already been established that the APC/C subunit CDC27 (also known as ANAPC3) is one of 23 cancer driver genes in colorectal cancer (37, 38), and was also identified in testicular germ cell tumors (ref. 39; these studies consider nonsynonymous point mutations). CDC27 is the most frequently mutated APC/C subunit (Supplementary Table S4), and, strikingly, it displays a high incidence of truncating mutations (30.3%) distributed widely across the gene (Fig. 2A; Supplementary Table S4). It is unusual to observe recurrent frameshift mutations such as the deletion creating L454Hfs*9, which may reflect an alignment error in the TCGA pipeline. Nevertheless, CDC27 would still bear a truncating mutation frequency of nearly 20% (34/179 = 19%) if those samples were excluded. CDC27 could be considered a candidate tumor suppressor according to the 20/20 rule, because truncating mutations represent close to 20% of all reported mutations (40). Truncating mutations are also predicted to have a clear detrimental impact on APC/C complex structural integrity based on the atomic structure (see details below; ref. 41), and heterozygous mutations could lead to fewer active complexes in the cell.
Figure 2. APC/C mutations in human cancer.
A, Lollipop plot representing the distribution, frequency, and type of mutations reported for CDC27 across 30 cancer types (see Supplementary Table S3). Green dots indicate missense mutations and red dots truncating mutations (nonsense, frameshift, and splice site). All lollipop plots were retrieved and adapted from cBioPortal (77, 78). The locations of the CRISPR/Cas9 guide RNA (gRNA) targeting sites used in the following experiments are indicated. B, Schematics describing the reversine adaptation assay. RPE1 populations were generated by infecting cells at low titre with lentiCRISPR vectors expressing either one of three different gRNAs targeting CDC27 (to control for possible off-target cleavage). Cells were grown in 250 nmol/L reversine by passaging cells when the dish became confluent or every 2 weeks, whichever came first. Approximate doubling times were derived from counting cells at every passage. For vector cells, additional plates were maintained in parallel to isolate rare resistant colonies (Supplementary Fig. S4). C, Doubling times of the RPE1/lentiCRISPR populations maintained in reversine as determined at days 14 and 28. D, Stills from time-lapse movies of the RPE1/lentiCRISPR/CDC27-2 population after 4 weeks in 250 nmol/L reversine. Note the actively dividing cells in reversine. Upon 24 hours following reversine washout, many round mitotic cells are visible, but these cells are delayed only in mitosis rather than arrested, as shown below in E (scale bar, 100 μm). E, Mitotic duration from NEBD–anaphase in three independent RPE1/lentiCRISPR CDC27 populations exposed to 250 nmol/L reversine for 4 weeks. Cells were imaged in parallel while still in 250 nmol/L reversine or 24 hours following reversine washout (bars, mean ± 95% CI; ***, P < 0.0001). F, Tetrad dissection of S. pombe strains and the genotype of each haploid spore bearing an extra allele (in red, mutated where indicated; see Supplementary Fig. S5A for mating scheme). Both the truncating mutation homologous to E736* (S. pombe E600*) and the missense mutant G506E (S. pombe G370E) failed to rescue NUC2 deletion lethality, indicating that they are deleterious mutations. WT, wild-type. G, Precise genome editing was performed in RPE1 cells to delete a single base pair at codon L454 (CTA) in one CDC27 allele, thereby creating a frameshift and stop codon 9 amino acids downstream. Mitotic duration from NEBD–anaphase was determined for the heterozygous L454Hfs*9 clone in parallel with the parental population and three nonedited clones isolated in parallel, all of which were never exposed to reversine. H, NEBD–anaphase duration of HCT116 clones isolated based on their resistance to the MPS1 inhibitor Cpd-5. Mitotic timings were either determined while cells were still maintained in 30 nmol/L Cpd-5 or following Cpd-5 washout (bars, mean ± 95% CI; *, P < 0.05; **, P < 0.01; ***, P < 0.0001; ns, nonsignificant).
In order to test if CDC27 truncating mutations confer a selective advantage during the onset of CIN, we used CRISPR/Cas9 to engineer three RPE1 populations carrying CDC27 frameshift mutations by infecting cells with a lentiCRISPR vector at a low multiplicity of infection (MOI; Fig. 2A guide RNAs 1-2-3 and Fig. 2B). We reasoned that each polyclonal population would contain a mixture of mostly wild-type cells with some CDC27 heterozygous cells (because CDC27 is an essential subunit; ref. 42). CRISPR-targeted populations were exposed to 250 nmol/L reversine continuously for over 4 weeks, which was lethal to RPE1/lentiCRISPRVector cells except for rare clones. In contrast, RPE1/lentiCRISPRCDC27 populations rapidly adapted and could be continuously passaged in reversine (Fig. 2C and D; Supplementary Fig. S4A). This procedure resulted in a strong enrichment for clones with heterozygous CDC27 disruption after reversine treatment compared with untreated populations (CDC27-1: 100% vs. 8.7%; CDC27-2: 100% vs. 26%; CDC27-3: 93% vs. 4.3%; Supplementary Fig. S4B). Reversine washout after selection caused a delay of 9 to 10 minutes in anaphase onset, resulting in an apparent accumulation of mitotic cells, consistent with the selection of cells with impaired APC/C activity (Fig. 2D and E). CDC27 haploinsufficiency is therefore sufficient to confer a selective advantage under conditions that would otherwise lead to extreme CIN and lethality.
Deleterious CDC27 Mutations Are Selected in Cancer
To further explore if CDC27 mutations found in cancer disrupt APC/C activity, we took advantage of the sequence similarity between human and Schizosaccharomyces pombe (fission yeast) proteins, and the possibility to assay mutations in a haploid context. We tested a subset of mutations for which a homologous residue is found in S. pombe (NUC2 for CDC27), and assayed their ability to rescue lethality of NUC2 deletion in haploid spores (Supplementary Fig. S5A). Truncating NUC2 at position E600, which corresponds to one of the most C-terminal CDC27 nonsense mutations in human cancers (E736*), did not support growth (Fig. 2A and F). Similarly, substituting the homologous residue in NUC2 to mimic CDC27 G506E caused lethality, whereas the R629K mutation resulted in slow growth with delayed cyclin B degradation in mitosis (Fig. 2F; Supplementary Fig. S5B). CDC27 forms a homodimer within the APC/C complex and mediates the interaction with the Ile-Arg motif (IR tail) of a co-activator and the IR tail of APC10 in a symmetrical manner, which is required for substrate recognition by the APC/C (refs. 41, 43; Supplementary Fig. S5C). Examination of the human APC/C atomic structure (41, 43) revealed that the CDC27 G506E mutation would disrupt the IR tail binding site required for the interaction with the coactivators (CDH1 and CDC20) and with APC10. In addition, truncating CDC27 at E736 would also reduce contact with the IR tail and WD40 domain of APC/C coactivators, with the IR tail of APC10, and with the CDC16 subunit. E736* and any upstream CDC27 truncating mutations often found in cancer would therefore severely impair APC/C activity and likely behave as null alleles, in line with the lethality of NUC2 E600* in S. pombe. To test this in human cells, we repeated the reversine selection assay described in Fig. 2B using three new CDC27 lentiCRISPR populations, including two that targeted the region around residue E736 and one designed to disrupt CDC27 near E468 (Fig. 2A, guides 4-5-6). We found that truncating human CDC27 near E736 was just as efficient in conferring resistance to prolonged reversine treatment as upstream truncating mutations (Supplementary Fig. S5D). This confirms that all CDC27 truncating mutations reported in TCGA are likely to be functionally relevant (except Q793*, which has not been tested). We also applied the same protocol to disrupt ANAPC1 and ANAPC4 near their respective most frequent truncation site reported in TCGA, also revealing rapid adaptation to reversine (Supplementary Fig. S5D). Deleterious mutations in CDC27 and other APC/C subunits can therefore be selected for in cancer and impair APC/C activity.
To examine the consequence of a CDC27 truncating mutation in unperturbed conditions, we engineered RPE1 cells in which one allele contained the single base deletion that creates the L454Hfs*9 truncation (Fig. 2A and G). Monoallelic disruption of CDC27 was sufficient to significantly delay anaphase onset by less than 3 minutes compared with wild-type clones or the parental population (P ≤ 0.0006), revealing that mitotic duration is sensitive to APC/C gene dosage even without selective pressure imposed by sustained MPS1 inhibition.
Impact of Cancer-Associated Missense Mutations on APC/C Structure
To gain further insight into the consequence of APC/C missense mutations, we analyzed the published APC/C structure (41) to assess the impact of a subset of mutations on the structural integrity of the complex (Supplementary Table S5). We restricted our analysis to APC/C mutations reported in nine cancer types (bladder, breast, colon, head and neck, glioblastoma multiforme, clear cell kidney carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, and melanoma). A total of 132 missense mutations were examined in various subunits, 93 of which affected residues that could be mapped onto the APC/C structure (ref. 41; Supplementary Table S5). Fifteen mutations occurred in the catalytic and substrate recognition subunits, and may compromise E2 recruitment and substrate binding, respectively. In total, 40 mutations are predicted to disrupt the structural integrity of the APC/C (Supplementary Table S5). At least 60 of 132 mutations were also predicted to be deleterious based on MutationAssessor (44) or PolyPhen-2 (45). In summary, potentially deleterious missense mutations in several APC/C subunits are found in cancer.
APC/C Dysfunction Confers Resistance to MPS1 Inhibitors
The pharmacologic induction of CIN has been proposed as a therapeutic approach to target cancer cells (27), and several MPS1 inhibitors have been developed for this purpose, some of which are currently in drug development (clinicaltrials.gov identifiers NCT02366949, NCT02138812, and NCT02792465). Understanding the resistance mechanisms to such inhibitors and predicting how this might occur mechanistically is essential to improve drug design and potentially identify exploitable synthetic lethal pathways. In a previous study, recovery of HCT116 clones resistant to a lethal dose of the MPS1 inhibitor Compound-5 (Cpd-5) led to the identification of mutations in the MPS1 kinase domain that conferred resistance to various MPS1 inhibitors (46). However, six resistant clones did not carry a TTK mutation based on cDNA sequencing (46). Here, we performed targeted exon sequencing of these six clones for TTK, TP53, UBE2C, and all APC/C subunits and found two of the six clones carried a CDC16R136H heterozygous mutation, which is a highly conserved arginine residue. Interestingly, R136H clones displayed a longer mitotic duration while grown in Cpd-5 compared with untreated parental cells, and Cpd-5 washout prolonged NEBD-anaphase duration, suggestive of disrupted APC/C activity (Fig. 2H). In contrast, as expected, mitotic duration was unchanged with or without Cpd-5 in a clone bearing a mutation in the MPS1 kinase domain (I531M, Fig. 2H). Altered APC/C activity might therefore represent a resistance mechanism to MPS1 inhibitors distinct from the acquisition of somatic mutations in TTK itself.
APC/C Dysfunction Attenuates CIN after Genome Doubling
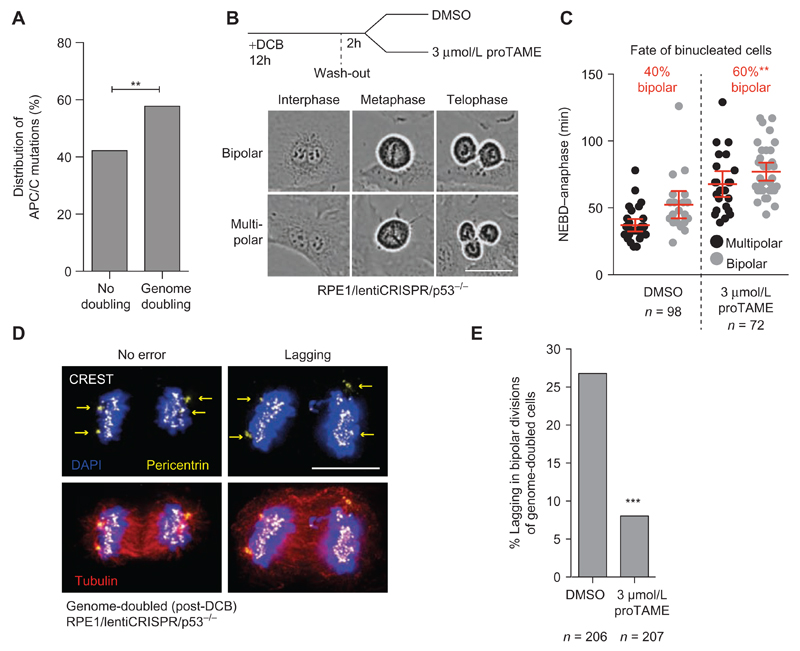
Next, we sought to examine whether APC/C dysfunction might also reduce CIN resulting from causes other than SAC defects. Genome doubling constitutes a precursor of CIN in several cancer types (47, 48), and experimental models have also shown that genome-doubled (tetraploid) cells are tumorigenic and prone to CIN (3, 4, 47). We found that APC/C subunit nonsynonymous point mutations occurred significantly more frequently in genome-doubled compared with non–genome-doubled tumors (58%; P = 0.005, Fisher exact test; Fig. 3A). Genome-doubling events, which can arise from cell division failure, inherently increase CIN due to the presence of extra centrosomes (3, 4). Extra centrosomes increase the formation of merotelic attachment errors, where a chromosome is attached to opposite poles (Supplementary Fig. S6A). If uncorrected, merotely may result in lagging chromosomes during chromosome separation and is considered to constitute a major cause of CIN in cancer (3, 4, 49–51). Efficient clustering of supernumerary centrosomes also prevents multipolar divisions, which result in extreme karyotypic changes and are most often lethal (Supplementary Fig. S6B; refs. 4, 52).
Figure 3. APC/C dysfunction reduces CIN caused by merotely and lagging chromosomes in cancer cell lines.
A, Genome-doubling status of tumors bearing a mutation in one of the APC/C subunits. Derived from 2,694 tumors from 9 cancer types for which mutational and copy-number data were available to determine genome-doubling status (**, P = 0.0014, Fisher exact test). B and C, Genome doubling (tetraploidization) was induced in RPE1/p53−/− cells by a 12-hour treatment with 4 μmol/L DCB to block cytokinesis. DCB was washed off, and cells were allowed to recover for 2 hours, after which 3 μmol/L proTAME or DMSO was added to the media and cells were imaged every 3 minutes to capture the first mitosis following tetraploidization. Mitotic duration from NEBD–anaphase was measured exclusively for binucleated cells, and the fraction of bipolar or multipolar divisions was scored (scale, 50 μm; bars, mean ± 95% CI; **, P = 0.01 for division outcome, Fisher exact test). D and E, Cells were grown on glass coverslips, treated with DCB using the same protocol as described in C, and fixed for immunostaining. Genome-doubled cells were identified on the basis of having four centrosomes (identified using pericentrin staining), and the frequency of anaphase lagging chromosomes was scored (***, P < 0.0001, Fisher exact test).
To directly assess the importance of APC/C activity in genome-doubled cells, we transiently blocked cytokinesis using Dihydrocytochalasin B (DCB) and examined the first division of binucleated cells with and without partial APC/C inhibition using 3 μmol/L proTAME (Fig. 3B). Because cell-cycle progression beyond G1 is limited by p53 after tetraploidization (53), we performed these experiments in RPE1 cells where TP53 was disrupted using CRISPR/Cas9 (Supplementary Fig. S7A and S7B). Time-lapse microscopy revealed 40% of binucleated cells underwent a bipolar division, and bipolarity was associated with a longer NEBD-anaphase duration, concordant with previous observations (refs. 52, 54, 55; Fig. 3C, DMSO). Partial APC/C inhibition using pro-TAME delayed anaphase onset and significantly increased the frequency of bipolar divisions in binucleated cells to 60% (P = 0.01), suggesting that APC/C dysfunction provides more time for centrosome clustering to take place (Fig. 3C). Pro-TAME also caused a significant reduction in the frequency of lagging chromosomes among tetraploid cells undergoing bipolar divisions (27% to 8%, P < 0.0001, Fig. 3D and E). These results suggest that APC/C dysfunction can stabilize genome-doubled cells by preventing potentially catastrophic multipolar division, but also by reducing the extent of CIN during bipolar divisions, likely by allowing more time for endogenous error-correction mechanisms to function.
APC/C Dysfunction Rescues Merotely
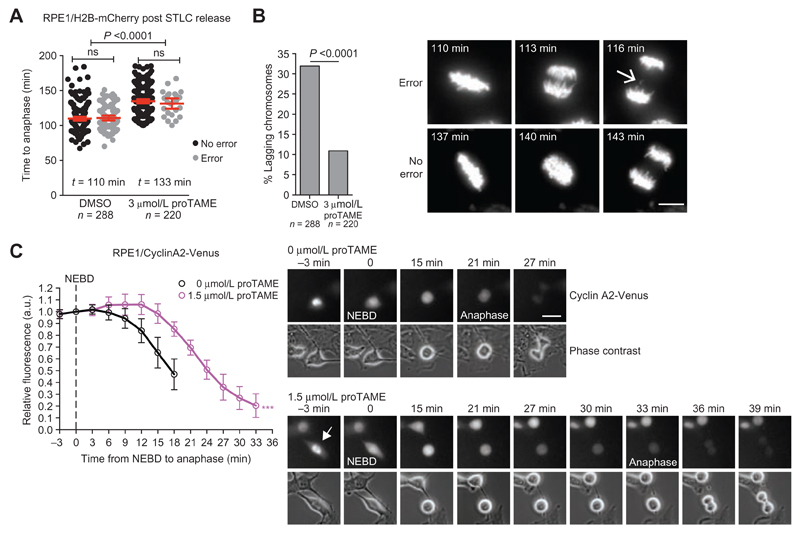
Merotelic attachment errors also occur in non–genome-doubled cells and compromise genome stability more than other types of attachment error because they do not activate the SAC and consequently do not delay mitotic progression to allow error correction. We induced merotely in RPE1/H2B-mCherry diploid cells by transiently arresting cells in mitosis with the Eg5 inhibitor S-Trityl-L-cysteine (STLC) and examined the importance of APC/C activity on merotelic attachment correction by live-cell imaging (Fig. 4A; Supplementary Fig. S6A; ref. 30). APC/C inhibition upon STLC release using 3 μmol/L proTAME delayed anaphase onset after STLC recovery by 23 minutes (110 to 133 minutes) and reduced the frequency of lagging chromosomes by 3.5-fold (39% to 11%, P < 0.0001; Fig. 4A and B). Within each condition (DMSO or 3 μmol/L proTAME), the time of anaphase onset was identical regardless of whether a segregation error occurred, confirming that these attachment errors have evaded SAC surveillance. Hence, APC/C dysfunction can reduce CIN caused by both SAC-impairment–dependent and –independent mechanisms. As expected, APC/C inhibition in mitosis did not reduce the frequency of structural aberrations (acentric and dicentric chromosomes) induced by premitotic replicative stress following aphidicolin treatment (Supplementary Fig. S8A and S8B). We conclude that although merotely cannot activate the SAC and in turn inhibit the APC/C, impaired APC/C activity creates a context favorable for endogenous error correction mechanisms to take place by delaying mitotic progression.
Figure 4. APC/C dysfunction results in CIN buffering and adaptation to extreme CIN.
A and B, RPE1/H2B-mCherry cells were arrested in mitosis using 40 nmol/L STLC, collected by shake-off, and released into media containing 3 μmol/L proTAME or DMSO (t = 0h). Stacks were acquired every 3 minutes to determine anaphase onset (A) and the presence of lagging chromosomes (B; error bars, mean ± 95% CI). Sample images from movies are shown, and the arrow indicates a lagging chromosome in a cell without proTAME. Representative images are shown (scale bar, 10 μm). C, Degradation kinetics of cyclin A2-Venus fluorescence quantified from unsynchronized single cells as they progress through mitosis. 1.5 μmol/L proTAME or DMSO was added 2 hours before imaging. Total cell fluorescence was quantified and normalized to the level at NEBD. Curves end at anaphase onset (n = 24 cells per condition; error bars indicate SD; ***, P < 0.0001 Student t test). Time to reach 50% maximum intensity was used for statistical analysis.
To further investigate how mild APC/C inhibition could facilitate error correction, we considered cyclin A2, a known APC/C substrate, whose degradation is not inhibited by SAC signaling in prometaphase, in contrast to cyclin B (32, 56). Sustained cyclin A2 levels were shown to create a permissive condition for error correction to take place during prometaphase by preventing premature stabilization of kinetochore–microtubule attachments, and its degradation below a permissive threshold might contribute to the step-wise increases in stability observed from prometaphase to metaphase and anaphase (57). Given our previous observation that proTAME delays prometaphase progression (NEBD-metaphase; Fig. 1G), we investigated whether a slight reduction in APC/C activity delayed cyclin A2 degradation in early mitosis. We used RPE1 cells in which an endogenous cyclin A2 allele has been tagged with the yellow-fluorescent protein Venus, allowing the quantification of its decay prior to anaphase (56). We observed that the decline of cyclin A2 levels below 100% following NEBD was delayed from 6 to 15 minutes in the presence of 1.5 μmol/L proTAME (the lowest dose used in our assays; Fig. 4C). Maintaining cyclin A2 levels in early mitosis for a longer period of time might thus facilitate merotelic attachment correction and reduce segregation errors when APC/C is pharmacologically or mutationally impaired. Determination of spindle microtubule stability during prometaphase using a mCherry-photoactivatable-GFP-Tubulin construct in RPE1 cells revealed similar half-lives with or without proTAME (Supplementary Fig. S9 A and S9B). This supports a model whereby impaired APC/C simply prolongs the time during which cyclin A2 is maintained at a level permissive for efficient error correction to take place.
CIN Attenuation through APC/C Dysfunction in Aneuploidy-Tolerant p53-Null Cells
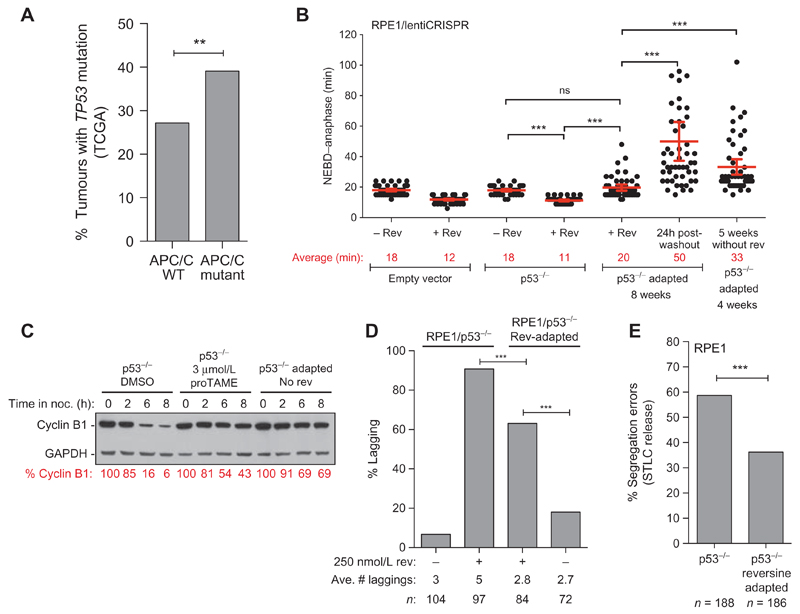
Although TP53 is a major determinant of cellular proliferation following chromosome missegregation and its disruption is widely associated with CIN, aneuploidy tolerance mechanisms are probably unable to cope with the deleterious effects of excessive CIN that create extreme changes in gene dosage in cells suffering gain or loss of chromosomes (Fig. 1B and C; Supplementary Figs. S1–S3). A larger proportion of APC/C-mutated than APC/C–wild-type tumors also carry a TP53 mutation in the cohort of nine cancer types analyzed (39% vs. 28%, P = 0.004; Fig. 5A, considering nonsynonymous point mutations exclusively), a correlation independent of genome-doubling status (P = 0.00263), but borderline significant when considering the higher mutational burden of TP53-mutated tumors (P = 0.079). We subjected RPE1/p53−/− populations to 250 nmol/L reversine for up to 8 weeks, and cells readily became resistant (Supplementary Fig. S10A). RPE1/p53−/− cells grown in reversine displayed a mitotic duration significantly longer than RPE1/p53−/− cells acutely treated with reversine (20 and 11 minutes, respectively, for NEBD-anaphase), but similar to RPE1/p53−/− cells that have never been exposed to reversine (18 minutes; Fig. 5B). Drug washout significantly delayed NEBD-anaphase timing in reversine-adapted cells (50 minutes), and these cells maintained a longer NEBD-anaphase for several weeks after reversine removal, indicative of a transmissible and viable phenotype (Fig. 5B). Similar results were obtained in two RPE1-independent p53-null populations following drug adaptation and washout using 2 μmol/L AZ3146, an MPS1 inhibitor structurally unrelated to reversine (Supplementary Fig. S10B). RPE1/p53−/− reversine-adapted cells displayed greater cyclin B1 stability during a prolonged mitotic arrest in nocodazole compared with RPE1/p53−/− cells that have never been exposed to reversine, confirming that APC/C activity is indeed reduced in the adapted population (ref. 58; Fig. 5C). Importantly, adaptation of RPE1/p53−/− cells in reversine was associated with a lower chromosome segregation error rate compared with RPE1/p53−/− cells acutely treated with reversine, with fewer lagging chromosomes when errors did occur (63% error with 2.8 lagging chromosomes on average vs. 91% with 5 lagging chromosomes, respectively; Fig. 5D). In addition, RPE1/p53−/− cells after adaptation (but grown without reversine for over 4 weeks) displayed fewer lagging chromosomes than treatment-naïve RPE1/p53−/− cells after STLC washout, indicative of fewer uncorrected merotelic attachments in adapted cells (Fig. 5E). Targeted exon sequencing on the RPE1/p53−/− adapted population did not reveal the clonal expansion of an APC/C subunit mutation, suggesting APC/C attenuation through parallel pathways that may involve transcriptional or epigenetic deregulation. Mitotic duration lengthening through attenuation of APC/C activity might therefore represent a common resistance mechanism to MPS1 inhibitors. We propose that even though p53-null cells display greater tolerance to segregation errors, which will undoubtedly contribute to CIN propagation, there is still a selective pressure to reduce CIN within a sustainable range, and impaired APC/C activity is one such mechanism to reach this equilibrium.
Figure 5. Acquired CIN attenuation through APC/C dysfunction in aneuploidy-tolerant p53-null cells.
A, Fraction of TP53-mutated tumors among samples that are either wild-type for all APC/C subunits or in which at least one APC/C subunit is mutated. Derived from 2,694 tumors from 9 cancer types (bladder, breast, colon, and head and neck cancers, glioblastoma multiforme, kidney renal clear cell carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, and melanoma; **, P = 0.005, Fisher exact test). See Methods for the number of samples per tumor types. B, NEBD–anaphase duration of RPE1/p53−/− cells after acute treatment with 250 nmol/L reversine (added 2 hours before imaging). In parallel, RPE1/p53−/− grown for 8 weeks in reversine (adapted) were imaged while still in 250 nmol/L reversine and 1 hour after reversine washout, as indicated. The last column represents RPE1/p53−/− grown for 4 weeks in reversine, followed by 5 weeks without reversine (bars, average ± 95% CI; ***, P < 0.0001). C, Cyclin B degradation kinetics determined by western blot from cells that were maintained in mitosis for the indicated duration with 50 nmol/L nocodazole (a microtubule poison). RPE1/p53−/− cells were maintained in nocodazole ± 3 μmol/L proTAME, whereas reversine-adapted RPE1/p53−/− cells (growing over 4 weeks in the absence of reversine) were maintained in nocodazole only. ImageQuant was used to normalize cyclin B levels to GAPDH, and % cyclin B is expressed relative to t = 0h for each cell line. D, The fraction of anaphase cells with lagging chromosomes was determined for RPE1/p53−/− cells acutely treated with 250 nmol/L reversine, and RPE1/p53−/− reversine-adapted cells while maintained in reversine or grown without reversine 24 hours before fixation (***, P < 0.0001, Fisher exact test). E, H2B-mCherry was introduced in RPE1/p53−/− cells and RPE1/p53−/− reversine-adapted that had been cultured at least 5 weeks without reversine. Merotelic attachments were induced using the STLC arrest/washout protocol, and the frequency of segregation errors was determined by time-lapse fluorescence microscopy (from three experiments; ***, P < 0.0001, Fisher exact test).
APC/C Inhibition or Frameshift Mutation Limits Chromosome Segregation Errors in CIN Cell Lines
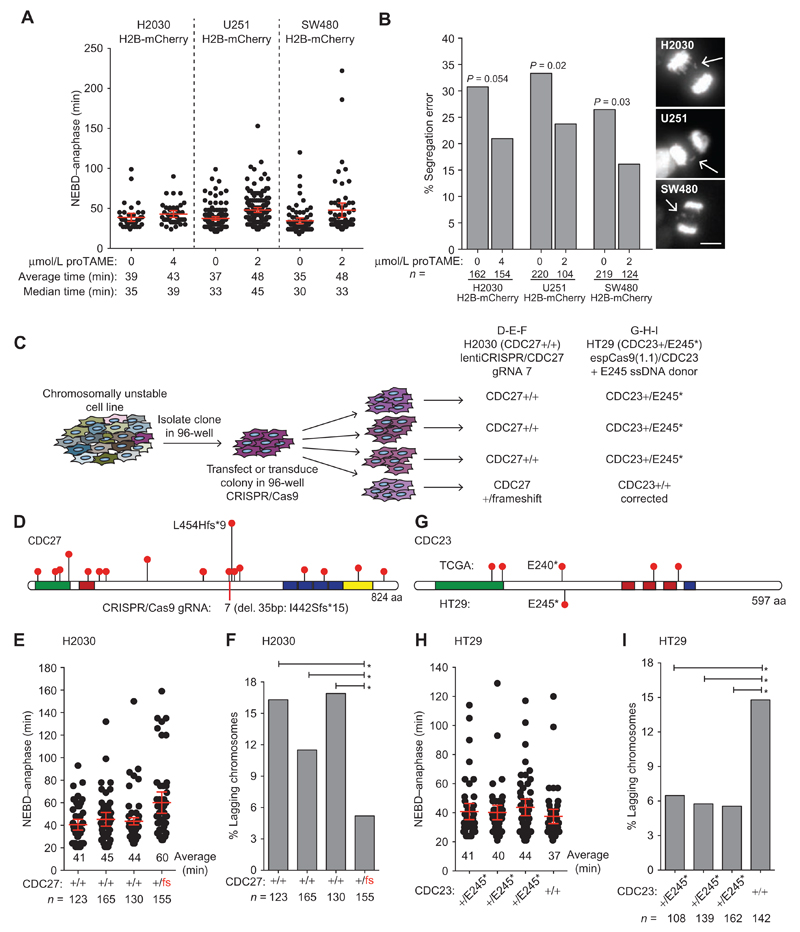
To test whether partial APC/C inhibition could reduce the frequency of naturally occurring segregation errors in cancer cell lines, we chose three cell lines with a high frequency of anaphase lagging chromosomes (H2030 lung adenocarcinoma, U251 glioblastoma, and SW480 colon carcinoma). We introduced H2B-mCherry to perform live-cell imaging using concentrations of proTAME that caused mild delays in anaphase onset (range of 4–13 minutes) and measured the frequency of divisions with lagging chromosomes (Fig. 6A and B). Although each cell line showed a greater variation in basal NEBD-anaphase duration compared with untreated RPE1 or HCT116 cells, which likely reflects the phenotypic variability symptomatic of CIN, we observed a consistent reduction in lagging chromosomes with proTAME (Fig. 6B). These results suggest that impairing APC/C function can also reduce the rate of endogenous segregation errors in established CIN cancer cell lines.
Figure 6. APC/C subunit mutational status affects CIN in cancer cells.
A and B, H2B-mCherry was introduced in H2030, U251, and SW480 cell lines and NEBD–anaphase duration (A) as well as the frequency of anaphase segregation errors (B) with and without proTAME was determined by time-lapse fluorescence microscopy. Stacks were acquired every 3 minutes, and an example of segregation error is shown for each cell line (scale bar, 10 μm; in A, bars, mean ± 95% CI of a representative experiment; in B, P values from Fisher exact test). C, Experimental procedure used to generate CRISPR/Cas9 edited H2030 and HT29 cells used for plots D to I. In each case, a single clone was infected (lentiCRISPR/CDC27 for H2030) or transfected (espCas9(1.1)/CDC23 + ssDNA donor) in a well of a 96-well plate before the colony reached confluency. Following transfection or transduction, the colony was dispersed by limiting dilution into 96-well plates. Clones were then screened for heterozygous disruption of CDC27 in H2030 cells or correction of the heterozygous E245 nonsense mutation in HT29 cells. Nonedited clones identified during screening were used as controls. Phenotypic analysis of all newly derived cell lines was performed following minimal clonal expansion to limit phenotypic diversity that may be acquired due to ongoing CIN. D, Lollipop plot of CDC27 showing only truncating mutations reported in TCGA and the location of the guide RNA used to disrupt CDC27 in H2030 cells. The clone isolated contained a heterozygous 35-bp deletion creating the truncation I442Sfs*15. E, NEBD–anaphase duration was determined for the H2030 clones using phase–contrast time-lapse microscopy (3 minutes/frame; bars, average ± 95% CI). F, The frequency of anaphase lagging chromosomes was determined on fixed cells by indirect IF microscopy (*, P < 0.05; Fisher exact test). G, Lollipop plot of CDC23 showing only truncating mutations reported in TCGA and the HT29 nonsense mutation from HT29 cells. H, NEBD–anaphase duration was determined for HT29 clones using phase–contrast timelapse microscopy (3 minutes/frame; bars, average ± 95% CI). I, The frequency of anaphase lagging chromosomes was determined on fixed cells by indirect IF microscopy (*, P < 0.05; Fisher exact test).
To substantiate this finding, we generated H2030 cells bearing a frameshift mutation in one allele of CDC27 (Fig. 6C). H2030 cells were chosen because they efficiently formed colonies upon limiting dilution (unlike U251 or SW480). We first isolated a single H2030 colony to avoid clonal variation due to CIN within the population and performed in-well transduction with a lentiCRISPR/CDC27 vector using a low viral titre. A second round of limiting dilution was then performed to identify rare clones in which gene editing took place in a single allele (Fig. 6C). We chose a guide RNA (gRNA) to disrupt CDC27 near the middle of the protein (Fig. 6D guide 7, having demonstrated previously that all CDC27 truncating mutations disrupt APC/C activity). Disruption of a single CDC27 allele significantly delayed mitosis compared with three wild-type clones isolated in parallel, and was also associated with a significant reduction in lagging chromosomes (Fig. 6E and F). Monoallelic disruption of an APC/C subunit is therefore sufficient to reduce naturally occurring chromosome segregation errors.
Restoring APC/C Function Increases CIN
Finally, we took advantage of the CIN colorectal cancer cell line HT29, which carries an endogenous heterozygous nonsense mutation in the APC/C subunit CDC23 (also known as ANAPC8, amino acid position E245; Fig. 6G). We used a similar protocol as described above to limit clonal variation caused by CIN and transfected a single HT29 colony with a CRISPR/Cas9 construct together with a wild-type CDC23 single-stranded DNA (ssDNA) donor. Correction of the CDC23 E245* truncating mutation back to wild-type caused a modest decrease in mitotic duration, and a significant increase in the frequency of lagging chromosomes compared with three clones bearing a monoallelic E245* mutation (+/E245*; Fig. 6H and I).
Taken together, the mutational status of APC/C subunits can therefore directly affect chromosome segregation fidelity in cancer cells. These data suggest that several APC/C subunits, most notably CDC27, represent haploinsufficient tumor suppressors, owing to the delicate equilibrium required to maintain stoichiometry of the APC/C and its activity, with a role in preventing excess CIN thereby improving cell fitness.
Discussion
Mounting evidence from cancer genomics studies and experimental systems suggest that the degree of CIN determines its tumor-promoting or tumor-suppressive biological output, with excessive CIN associated with improved clinical outcome across several tumor types (18, 19, 23, 24, 26, 27, 59, 60). This paradoxical relationship between CIN and cancer raises the question of whether the optimal level of CIN in cancer cells is reached (and perhaps passed) in response to the stochastic acquisition of genetic alterations, or whether secondary alterations might be selected to prevent excessive CIN. The stochastic model implies that cancer cell fitness could decrease during tumor evolution, as cancer cells would likely accumulate more defects, increasing mitotic or premitotic chromosome missegregation events. The results presented here provide a potential mechanism through which cancer cells reduce their segregation error rate and therefore avoid the detrimental consequences of excessive CIN. Such a mechanism might have important consequences for tumor evolution because an intermediate level of CIN is more likely to be efficiently propagated by limiting cell-autonomous lethality, while still driving tumor cell diversification and heterogeneity, the substrate for selection and evolutionary fitness.
In support of this hypothesis, we show that cell populations can adapt to excessive CIN over a matter of weeks, by subtly lengthening mitotic duration. Mutations in APC/C subunits that would otherwise have very mild consequences on overall proliferation (in the range of 3 to 15 minutes over an 18- to 24-hour cell cycle) might thus efficiently reduce segregation errors following cellular perturbations that have been recognized as some of the most potent CIN-inducing defects, such as genome-doubling, merotelic attachment errors, and SAC dysfunction. APC/C mutations were also enriched in genome-doubled tumors, whereas our experimental evidence shows that APC/C inhibition increases the frequency of bipolar spindle formation and reduces CIN after genome doubling. This provides support for the selection of APC/C dysfunction to balance the beneficial consequences of rising cellular diversity against the fitness costs of excessive CIN. In addition, we provide functional evidence for the role of truncating mutations in the colorectal cancer driver CDC27 in rescuing cells from excessive CIN when segregation errors are experimentally elevated using MPS1 inhibitors. Importantly, monoallelic truncation of CDC27 in the CIN cell line H2030 significantly reduced the frequency of lagging chromosomes. Conversely, restoring the heterozygous CDC23 E245* mutation naturally occurring in the CIN colorectal cell line HT29 back to wild-type caused an increase in lagging chromosomes, further indicating that APC/C mutations have a direct impact on chromosome segregation fidelity in cells with CIN. Our detailed analysis of cancer-derived APC/C mutations suggests that although the mutation frequency of the complex as a whole is not higher than would be expected by chance, many mutations are very likely to have a functional impact. This highlights key statistical issues with the derivation of driver gene status when considering large molecular complexes containing several different proteins, any one of which when mutated could potentially disrupt the function of the entire complex.
Limiting CIN might also be important to facilitate long-term survival in genetic backgrounds permissive for CIN, such as in TP53-disrupted cells, a relevant observation because 39% of tumors with an APC/C mutation also carry a TP53 mutation (2, 61). This likely reflects the limits that aneuploidy tolerance mechanisms can provide in response to overwhelming CIN that cells may experience during cancer evolution. We propose that genetic and epigenetic alterations affecting the expression or integrity of several APC/C subunits could affect the complex processivity, offering parallel avenues to select for impaired activity following the onset of CIN. Our findings also shed light on resistance mechanisms to therapeutics such as TTK inhibitors, which serve to increase chromosome missegregation. A number of cancer drugs targeting mitotic processes, including paclitaxel, can lead to extreme CIN (62) and cell death, raising the possibility that APC/C mutations might also be a resistance mechanism to other anticancer drugs that function by precipitating chromosome segregation errors. Compensatory mechanisms may be important to mitigate several types of genomic instabilities during cancer development and restrict the generation of structural chromosomal aberrations. For example, telomere crisis early during cancer development followed by telomerase reactivation (hence stabilization) has been described for a number of cancer types (63–66). DNA-repair processes are upregulated in choroid plexus carcinoma, a type of brain tumor harboring extensive somatic DNA copy-number alterations, and focal amplification of syntenic genes with roles in DNA repair found to be important for tumor maintenance was proposed to help stabilize cancer cells following the acquisition of extensive rearrangements (67). Restoration of homologous recombination has also been described as a mechanism of resistance to PARP inhibitors in BRCA1/2-deficient tumors (67, 68). Numerous compensatory mechanisms might thus be selected during tumorigenesis and the acquisition of drug resistance in order to stabilize the various sources of genomic instability and optimize fitness.
The delicate nature of the evolutionary trade-off between the advantages of increased cellular diversity for subsequent generations, balanced against the fitness costs of excessive CIN, could offer therapeutic opportunities to exploit the CIN paradox in cancer medicine. Targeting mechanisms that limit CIN in tumors, forcing cells past a threshold of CIN tolerance, might provide a compelling strategy to limit tumor evolution and adaptation.
Methods
Cell Lines
All cell lines used in this study were obtained from the ATCC unless stated otherwise and reauthenticated by the London Research Institute’s Cell Services core facility (now Francis Crick Institute) by short tandem repeat DNA profiling (hTERT-RPE-1, June 2015; HCT116 from ECACC, February 2015; HCT116 wild-type (+/+) and p53−/−, June 2010, a generous gift from Bert Vogelstein; H2030, October 2015; U251, January 2015; SW480, October 2015; HT29, June 2016) and regularly tested to confirm absence of Mycoplasma. H2B-mCherry was introduced by lentiviral delivery (Addgene plasmid 21217, a gift from Mark Mercola). RPE1/Cyclin A2-Venus heterozygous cells were a generous gift from Jonathon Pines (The Institute of Cancer Research, London, UK).
RNAi Screen
The primary screen was performed using the human siRNA Smart-pool siGenome library from Dharmacon, whereas the secondary screen was performed using the corresponding deconvoluted siRNA duplexes (where four siRNAs included in the pool were tested independently). Contact-inhibited RPE1 cells were reverse-transfected in triplicate 384-well plates using Lullaby transfection reagent [OZ Biosciences, 800 cells/well, 0.2 μL Lullaby/well, 18.75 nmol/L siRNA, 250 nmol/L reversine (Sigma)] using a Biomek FX liquid handling robot (Beckman). Cells were cultured for 96 hours and fixed with 4% formaldehyde (ThermoFisher, no. 28908), except for the deconvolution experiment, where cells were pulsed for 1 hour with 10 μmol/L EdU before fixation to label cells in S phase. Cells were subsequently permeabilized and blocked in PBS + 3% BSA + 0.2% Triton-X100 for 30 minutes and processed for immunofluorescence (IF) and EdU detection (LifeTechnology) according to the manufacturer’s protocol. Images were acquired and analyzed using an ArrayScan VTi-automated microscope (Cellomics), except for total cell counts, which were acquired on an Acumen Explorer eX3 laser scanning microplate cytometer (TTPLabtech). For each parameter, a median Z-score was derived using plate normalization [(well value – plate median)/plate Median Absolute Deviation].
FISH
Unsynchronized RPE1 cells were treated with the indicated concentrations of reversine for 2 hours. Mitotic cells were collected by mitotic shake-off and seeded on glass coverslips, while still in reversine, for 2 hours until daughter cells reattached. Cells were allowed to swell in 0.56% KCl for 5 minutes, fixed in ice-cold 3:1 methanol:acetic acid for 15 minutes, and dehydrated with successive 5-minute incubations in 70%, 85%, and 99.9% ethanol. Hybridization to centromeric probes for chromosomes 6, 7, 8, 10, 11, or 18 was performed following the manufacturer’s protocol (Cytocell). Daughter cells where the centromeric signal was within a micronucleus were excluded because it could belong to either cell. Measured rates are therefore likely an underestimate of the actual rate.
Immunofluorescence Microscopy
For detection of interphase nuclear proteins, cells were fixed in 4% formaldehyde (ThermoFisher, cat. 28908). For segregation errors, RPE1 cells were fixed for 40 minutes in ice-cold MeOH, and HCT116 cells for 15 minutes in 0.2% Triton X-100, 10 mmol/L PIPES, 5 mmol/L EGTA, 1 mmol/L MgCl2, and 4% formaldehyde. Primary and secondary antibodies were incubated in 3% BSA and 0.02% Triton X-100. FISH and interphase images were acquired on a Zeiss Axio Imager M2 microscope using a Plan-Neofluar 340/1.3 numerical aperture (NA) oil objective lens (Zeiss) or Plan Apochromat 363/1.4 NA oil objective lens (Zeiss) equipped with an ORCA-ER camera (Hamamatsu) and controlled by Volocity 6.0.1. software (Improvision). For segregation error analyses and fluorescence live-cell microscopy, images were acquired using an Olympus DeltaVision RT microscope equipped with 40×, 63×, and 100× Olympus 3100 or 360 1.4 NA UPlanSApo oil immersion objectives (Applied Precision, LLC) and a Coolsnap HQ2 camera. For segregation error analyses, cells were stained with antitubulin, anticentromere, and antipericentrin antibodies, along with DAPI to stain DNA. Anaphase cells were identified based on the spindle morphology using the tubulin channel (hence without prior knowledge of whether an error occurred or not). Note that 0.2-μm stacks across the whole cell were acquired, and deconvolution of image stacks was performed with SoftWorx Explorer (Applied Precision, LLC). For fluorescence live-cell imaging of H2B-mCherry, stacks of 2 μm were acquired. Long-term imaging and bright-field NEBD-anaphase analysis were done using an Incucyte FLR device equipped with a 20x objective (Essen BioScience).
Photoactivation experiments were performed on RPE1 cells stably expressing GPAC-tubulin +/− 3 μmol/L proTAME. GPAC variant 1 (69) was cloned N-terminally of human alpha 1b tubulin into pLVX. Cells were monitored until a bipolar mitotic spindle formed in early mitosis (using the mCherry channel). Two sequential photoactivations were done 5 minutes apart in each cell to compare spindle stability as cells progressed during prometaphase, using a focused 405-nm laser on a 1-μm/9-μm region on one side of the spindle. Images were then acquired every 5 seconds for 3 minutes to monitor GFP fluorescence dissipation. These conditions did not cause photobleaching nor prevent progression into anaphase. Fluorescence intensity profiles for each activation event were thresholded to remove background noise and normalized. A nonlinear regression analysis was applied to the resulting data, to fit an exponential curve using Prism 6 software (GraphPad software) to measure the rate of tubulin turnover.
Antibodies and Dyes
The following antibodies were used for IF or western blot (WB): cyclin A [AT10 1:500-IF, gift from Julian Gannon (Francis Crick Institute)], Ki-67 (Abcam ab15580, 1:5,000-IF), p21 (MH9, 1:2,000-IF; AC8, 1:1,000-WB, gift from J. Gannon), p53 (OP43 Calbiochem, 1:1,000-IF/WB), p53-pS15 (Cell Signaling Technology, 1:1,000), CDC27 (AF3.1, 2 μg/mL, gift from J. Gannon), alpha-tubulin (Sigma, clone B512, 1:10,000-IF), anti-centromere antibodies (HCT-0100, 1:2,000; Immunovision), MKLP1 (BD Biosciences 612682, 1:1,000-IF), CDC16 (Santa Cruz K-16, 1:1,000-WB), and GAPDH (Abcam ab8245, 1:80,000-WB). Secondary antibodies conjugated to Alexa Fluor 488, 568, 594, or 647 (Invitrogen/ThermoFisher) were used for IF detection. EdU was detected by incubating cells for 30 minutes in 100 mmol/L Tris-HCl pH8.5, 4 mmol/L CuSO4, 500 nmol/L Alexa Fluor Azide-488 or -647 (Invitrogen), and 100 mmol/L ascorbic acid. DNA was stained with 1 μg/mL 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes).
Sequencing
An IonTorrent panel (Thermo Fisher Scientific) was designed to amplify all coding exons of TTK, TP53, UBE2C, CDC20, and the 14 core APC/C subunits (ANAPC1, 4, 5, 7, 10, 11, 12, 13, 15, 16, CDC16, CDC23, CDC26, CDC27). Next-generation sequencing was performed, and mutation calling was performed as described previously (70).
CRISPR/Cas9
The following gRNA sequences were cloned into the lentiCRISPR vector (gift from Feng Zhang, Addgene plasmid 49535), as described (71):
CTCTTAGTCCATTAACCCCA (CDC27-gRNA 1)
TTGACTTACCTTGGGGTTAA (CDC27-gRNA 2)
CTTCTTCGAGGCAGTGCGTT (CDC27-gRNA 3)
TGAGCCTTCTTCGTGAAATG (CDC27-gRNA 4)
AAGTAAACGAGGGATTCTTT (CDC27-gRNA 5)
TCCTATTAAGAAGTAAACGA (CDC27-gRNA 6)
AGATTAAAGGCCTGAATCTG (CDC27-gRNA 7)
TCGACGCTAGGATCTGACTG (TP53-1)
CGTCGAGCCCCCTCTGAGTC (TP53-2)
AGCAAAGGATGCAGCACCAG (ANAPC1-gRNA 1)
ACTGCCTTCCAAGACCAGCA (ANAPC1-gRNA 2)
CTTGGTATCAGCAAGAGCAA (ANAPC4-gRNA).
For precise genome editing, CDC27 guide 7 or the CDC23 gRNA below was cloned into eSpCas9(1.1) (gift from Feng Zhang, Addgene #71814) which expresses a mutant Cas9 with improved specificity compared with wild-type Cas9 (72). Cells were cotransfected with the corresponding ssDNA donor vectors which also carried a silent mutation to facilitate identification and prevent Cas9 editing of the donor template.
GAGATTCTGATACTTTTGCA (CDC23-gRNA)
CDC23 wild-type ssDNA donor
GGATGAAAGAGTTTTTTCTGGCTCATATATACACAGAGTTGCAGTTGATAGAGGAGGCTCTGCAAAAGTATCAGAATCTCATTGATGTGGGCTTCTCTAAGAGCTCGTA
CDC27 L454Hfs*9 ssDNA donor
AATACAAAGATAATATTAAGCATTTAAAACAGACCTGCTGCTGCTTTTTGTGATTAAAGGCCTGAATTTGAGGTGTGATTGTGGATATTTTCCCTTCTGAAATGATGGAAGAGTCCAA.
Virus particles were produced in 293T cells by cotransfecting lentiCRISPR vectors with psPAX2 and pMD2.G (Addgene Plasmid #12260 and #12259, gifts from Didier). RPE1 cells were infected at low MOI to favor single viral integrants, followed by puromycin (5 μg/mL) selection for 7 days.
Compounds
proTAME was obtained from BostonBiochem (I-440). Reversine (cat. no. R3904), STLC (cat. no. 164739), DCB (cat. no. D1641), Z-Leu-Leu-Leu-al (MG132, cat. no. C2211), and doxorubicin hydrochloride (cat. no. D1515) were obtained from Sigma.
APC/C Mutation Analysis
For the correlation between APC/C mutations and genome doubling, we used data from 2,694 TCGA tumors, across nine cancer types for which both copy-number data and mutational status were available (see ref. 73): bladder urothelial carcinoma (n = 130), breast invasive carcinoma (n = 903), colon adenocarcinoma (n = 190), glioblastoma multiforme (n = 250), head and neck squamous cell carcinoma (n = 289), kidney renal clear cell carcinoma (n = 303), lung adenocarcinoma (n = 257), lung squamous cell carcinoma (n = 131), skin cutaneous melanoma (n = 241).
Amino acid positions reported are based on the following UniProt IDs: CDC27 (P30260); ANAPC1 (Q9H1A4); ANAPC2 (Q9UJX6); CDC27 (P30260); ANAPC4 (Q9UJX5); ANAPC5 (Q9UJX4); ANAPC7 (Q9UJX3); ANAPC10 (Q9UM13); ANAPC11 (Q9NYG5); ANAPC13 (Q9BS18); ANAPC15 (P60006); ANAPC16 (Q96DE5); CDC16 (Q13042); CDC20 (Q12834); CDC23 (Q9UJX2); CDC26 (Q8NHZ8). Ninety-three of the 132 APC/C subunit mutations affected residues that were resolved on the APC/C atomic structure (PDB: 4UI9). Each one was analyzed individually, and the predicted impact on the APC/C structure or function is detailed in Supplementary Table S5.
Pombe Strain Handling and Time-Lapse Imaging
Cell culture conditions and growth media were as previously described by Moreno and colleagues (74). In brief, cells were cultured in YE4S, and crosses were performed on MEA4S. The heterozygous NUC2 deletion strain was derived from the Bioneer collection (75). We used an internally tagged cdc13-GFP allele (ref. 76; kind gift from S. Hauf) for our single-cell cyclin B degradation analysis. Timelapse imaging was performed at 25°C on a DeltaVision Elite system (GE Healthcare). During imaging, cells were cultured in YE4S using a Cellasics microfluidics system coupled to an Y04C plate (EMD Millipore). Image analysis was performed using custom Matlab scripts.
Supplementary Material
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Acknowledgments
We would like to thank Frank Uhlmann and members of the Swanton laboratory for suggestions and for critically reading the manuscript, David Barford for insightful comments regarding the APC/C structure, and Ming Jiang and Rebecca E. Saunders for their expert assistance with high-throughput screening and analysis.
The results published here are in part based on data generated by TCGA project established by the National Cancer Institute and National Human Genome Research Institute. The data were retrieved through dbGaP (Database of Genotypes and Phenotypes) authorization (accession number phs000178.v9.p8). Information about TCGA and the investigators and institutions that constitute the TCGA research network can be found at http://cancergenome.nih.gov/.
Grant Support
This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001169, FC001121, FC001155, and FC001209), the UK Medical Research Council (FC001169, FC001121, FC001155, and FC001209), and the Wellcome Trust (FC001169, FC001121, FC001155, and FC001209). Support was also provided by Cancer Research UK (TRACERx), the CRUK Lung Cancer Centre of Excellence, the Rosetrees Trust, Novo- Nordisk Foundation (ID 16584), the Prostate Cancer Foundation, the Breast Cancer Research Foundation (BCRF), the European Research Council (THESEUS), and the Marie Curie Network/European Commission PloidyNet. Research was also supported by a Stand Up To Cancer Laura Ziskin Prize. Stand Up To Cancer is a program of the Entertainment Industry Foundation. L. Sansregret was supported by fellowships from Cancer Research UK, the Canadian Institutes of Health Research, the European Molecular Biology Organization, and NovoNordisk Foundation. J.O. Patterson was supported by a Boehringer Ingelheim Fonds PhD fellowship. M. Petronczki was supported by Cancer Research UK and the EMBO Young Investigator Programme. C. Swanton is Royal Society Napier Research Professor. Support was provided to C. Swanton by the National Institute for Health Research, the University College London Hospitals Biomedical Research Centre, and the Cancer Research UK University College London Experimental Cancer Medicine Centre.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
Conception and design: L. Sansregret, P. Nurse, M. Petronczki, C. Swanton
Development of methodology: L. Sansregret, J.O. Patterson, C. Swanton
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): L. Sansregret, J.O. Patterson, S. Dewhurst, C. López-García, A. Koch, A. Rowan, R. Instrell, M. Howell
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): L. Sansregret, J.O. Patterson, A. Koch, N. McGranahan, W.C.H. Chao, S. Horswell, M. Way, M. Howell, M.R. Singleton, C. Swanton
Writing, review, and/or revision of the manuscript: L. Sansregret, S. Dewhurst, C. López-García, R.H. Medema, P. Nurse, M. Petronczki, C. Swanton
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): D.J. Barry, M. Howell
Study supervision: L. Sansregret, R.H. Medema, P. Nurse, Petronczki, C. Swanton
References
- 1.Santaguida S, Amon A. Short- and long-term effects of chromosome missegregation and aneuploidy. Nat Rev Mol Cell Biol. 2015;16:473–85. doi: 10.1038/nrm4025. [DOI] [PubMed] [Google Scholar]
- 2.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–81. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–7. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 4.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–70. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen G, Mulla WA, Kucharavy A, Tsai HJ, Rubinstein B, Conkright J, et al. Targeting the adaptability of heterogeneous aneuploids. Cell. 2015;160:771–84. doi: 10.1016/j.cell.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen G, Bradford WD, Seidel CW, Li R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature. 2012;482:246–50. doi: 10.1038/nature10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–5. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee AJ, Endesfelder D, Rowan AJ, Walther A, Birkbak NJ, Futreal PA, et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 2011;71:1858–70. doi: 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sotillo R, Schvartzman JM, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464:436–40. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li R, Hehlman R, Sachs R, Duesberg P. Chromosomal alterations cause the high rates and wide ranges of drug resistance in cancer cells. Cancer Genet Cytogenet. 2005;163:44–56. doi: 10.1016/j.cancergencyto.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Rutledge SD, Douglas TA, Nicholson JM, Vila-Casadesus M, Kantzler CL, Wangsa D, et al. Selective advantage of trisomic human cells cultured in non-standard conditions. Sci Rep. 2016;6:22828. doi: 10.1038/srep22828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Millet C, Ausiannikava D, Le Bihan T, Granneman S, Makovets S. Cell populations can use aneuploidy to survive telomerase insufficiency. Nat Commun. 2015;6:8664. doi: 10.1038/ncomms9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan AW, Hanlon Newell AE, Bi W, Finegold MJ, Olson SB, Beaudet AL, et al. Aneuploidy as a mechanism for stress-induced liver adaptation. J Clin Invest. 2012;122:3307–15. doi: 10.1172/JCI64026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ly P, Eskiocak U, Kim SB, Roig AI, Hight SK, Lulla DR, et al. Characterization of aneuploid populations with trisomy 7 and 20 derived from diploid human colonic epithelial cells. Neoplasia. 2011;13:348–57. doi: 10.1593/neo.101580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selmecki AM, Dulmage K, Cowen LE, Anderson JB, Berman J. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet. 2009;5:e1000705. doi: 10.1371/journal.pgen.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon-Chung KJ, Chang YC. Aneuploidy and drug resistance in pathogenic fungi. PLoS Pathog. 2012;8:e1003022. doi: 10.1371/journal.ppat.1003022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silk AD, Zasadil LM, Holland AJ, Vitre B, Cleveland DW, Weaver BA. Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc Natl Acad Sci U S A. 2013;110:E4134–41. doi: 10.1073/pnas.1317042110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Duijf PH, Benezra R. The cancer biology of whole-chromosome instability. Oncogene. 2013;32:4727–36. doi: 10.1038/onc.2012.616. [DOI] [PubMed] [Google Scholar]
- 21.Lynch M, Burger R, Butcher D, Gabriel W. The mutational meltdown in asexual populations. J Hered. 1993;84:339–44. doi: 10.1093/oxfordjournals.jhered.a111354. [DOI] [PubMed] [Google Scholar]
- 22.Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and Darwinian selection in tumours. Trends Cell Biol. 1999;9:M57–60. [PubMed] [Google Scholar]
- 23.Roylance R, Endesfelder D, Gorman P, Burrell RA, Sander J, Tomlinson I, et al. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol Biomarkers Prev. 2011;20:2183–94. doi: 10.1158/1055-9965.EPI-11-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birkbak NJ, Eklund AC, Li Q, McClelland SE, Endesfelder D, Tan P, et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011;71:3447–52. doi: 10.1158/0008-5472.CAN-10-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jamal-Hanjani M, A’Hern R, Birkbak NJ, Gorman P, Gronroos E, Ngang S, et al. Extreme chromosomal instability forecasts improved outcome in ER-negative breast cancer: A prospective validation cohort study from the TACT trial. Ann Oncol. 2015;26:1340–6. doi: 10.1093/annonc/mdv178. [DOI] [PubMed] [Google Scholar]
- 26.Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2016;22:105–13. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janssen A, Kops GJ, Medema RH. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc Natl Acad Sci U S A. 2009;106:19108–13. doi: 10.1073/pnas.0904343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Godek KM, Venere M, Wu Q, Mills KD, Hickey WF, Rich JN, et al. Chromosomal instability affects the tumorigenicity of glioblastoma tumor-initiating cells. Cancer Discov. 2016;6:532–45. doi: 10.1158/2159-8290.CD-15-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–93. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 30.Santaguida S, Tighe A, D’Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 32.Sivakumar S, Gorbsky GJ. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat Rev Mol Cell Biol. 2015;16:82–94. doi: 10.1038/nrm3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sackton KL, Dimova N, Zeng X, Tian W, Zhang M, Sackton TB, et al. Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature. 2014;514:646–9. doi: 10.1038/nature13660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manchado E, Guillamot M, de Carcer G, Eguren M, Trickey M, Garcia-Higuera I, et al. Targeting mitotic exit leads to tumor regression in vivo: Modulation by Cdk1, Mastl, and the PP2A/B55alpha,delta phosphatase. Cancer Cell. 2010;18:641–54. doi: 10.1016/j.ccr.2010.10.028. [DOI] [PubMed] [Google Scholar]
- 35.Zeng X, Sigoillot F, Gaur S, Choi S, Pfaff KL, Oh DC, et al. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 2010;18:382–95. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang S, Chang L, Alfieri C, Zhang Z, Yang J, Maslen S, et al. Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature. 2016;533:260–4. doi: 10.1038/nature17973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Litchfield K, Summersgill B, Yost S, Sultana R, Labreche K, Dudakia D, et al. Whole-exome sequencing reveals the mutational spectrum of testicular germ cell tumours. Nat Commun. 2015;6:5973. doi: 10.1038/ncomms6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature. 2015;522:450–4. doi: 10.1038/nature14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schreiber A, Stengel F, Zhang Z, Enchev RI, Kong EH, Morris EP, et al. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470:227–32. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- 43.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature. 2014;513:388–93. doi: 10.1038/nature13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koch A, Maia A, Janssen A, Medema RH. Molecular basis underlying resistance to Mps1/TTK inhibitors. Oncogene. 2016;35:2518–28. doi: 10.1038/onc.2015.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dewhurst SM, McGranahan N, Burrell RA, Rowan AJ, Gronroos E, Endesfelder D, et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov. 2014;4:175–85. doi: 10.1158/2159-8290.CD-13-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–40. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol. 2011;27:585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- 50.Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–27. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol. 2009;19:1937–42. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ganem NJ, Cornils H, Chiu SY, O’Rourke KP, Arnaud J, Yimlamai D, et al. Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell. 2014;158:833–48. doi: 10.1016/j.cell.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Z, Loncarek J, Khodjakov A, Rieder CL. Extra centrosomes and/or chromosomes prolong mitosis in human cells. Nat Cell Biol. 2008;10:748–51. doi: 10.1038/ncb1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sansregret L, Vadnais C, Livingstone J, Kwiatkowski N, Awan A, Cadieux C, et al. Cut homeobox 1 causes chromosomal instability by promoting bipolar division after cytokinesis failure. Proc Natl Acad Sci U S A. 2011;108:1949–54. doi: 10.1073/pnas.1008403108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collin P, Nashchekina O, Walker R, Pines J. The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nat Cell Biol. 2013;15:1378–85. doi: 10.1038/ncb2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kabeche L, Compton DA. Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation. Nature. 2013;502:110–3. doi: 10.1038/nature12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brito DA, Rieder CL. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr Biol. 2006;16:1194–200. doi: 10.1016/j.cub.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kops GJ, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci U S A. 2004;101:8699–704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Janssen A, Kops GJ, Medema RH. Targeting the mitotic checkpoint to kill tumor cells. Horm Cancer. 2011;2:113–6. doi: 10.1007/s12672-010-0059-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burds AA, Lutum AS, Sorger PK. Generating chromosome instability through the simultaneous deletion of Mad2 and p53. Proc Natl Acad Sci U S A. 2005;102:11296–301. doi: 10.1073/pnas.0505053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zasadil LM, Andersen KA, Yeum D, Rocque GB, Wilke LG, Tevaarwerk AJ, et al. Cytotoxicity of paclitaxel in breast cancer is due to chromosome missegregation on multipolar spindles. Sci Transl Med. 2014;6:229ra43. doi: 10.1126/scitranslmed.3007965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meeker AK, Hicks JL, Platz EA, March GE, Bennett CJ, Delannoy MJ, et al. Telomere shortening is an early somatic DNA alteration in human prostate tumorigenesis. Cancer Res. 2002;62:6405–9. [PubMed] [Google Scholar]
- 64.Rudolph KL, Millard M, Bosenberg MW, DePinho RA. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat Genet. 2001;28:155–9. doi: 10.1038/88871. [DOI] [PubMed] [Google Scholar]
- 65.Feldmann G, Beaty R, Hruban RH, Maitra A. Molecular genetics of pancreatic intraepithelial neoplasia. J Hepatobiliary Pancreat Surg. 2007;14:224–32. doi: 10.1007/s00534-006-1166-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, et al. In situ analyses of genome instability in breast cancer. Nat Genet. 2004;36:984–8. doi: 10.1038/ng1409. [DOI] [PubMed] [Google Scholar]
- 67.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–5. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 68.Jaspers JE, Kersbergen A, Boon U, Sol W, van Deemter L, Zander SA, et al. Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer Discov. 2013;3:68–81. doi: 10.1158/2159-8290.CD-12-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Welman A, Serrels A, Brunton VG, Ditzel M, Frame MC. Two-color photoactivatable probe for selective tracking of proteins and cells. J Biol Chem. 2010;285:11607–16. doi: 10.1074/jbc.M110.102392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murugaesu N, Wilson GA, Birkbak NJ, Watkins TB, McGranahan N, Kumar S, et al. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov. 2015;5:821–31. doi: 10.1158/2159-8290.CD-15-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–8. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, Swanton C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med. 2015;7:283ra54. doi: 10.1126/scitranslmed.aaa1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 75.Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–23. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kamenz J, Mihaljev T, Kubis A, Legewie S, Hauf S. Robust ordering of anaphase events by adaptive thresholds and competing degradation pathways. Mol Cell. 2015;60:446–59. doi: 10.1016/j.molcel.2015.09.022. [DOI] [PubMed] [Google Scholar]
- 77.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.