Abstract
Dysregulation of microRNAs (miRNAs) is actively involved in the pathogenesis and tumorigenicity of hepatocellular carcinoma (HCC). miR-489 was found to play either oncogenic or tumor suppressive roles in human cancers. Recent study reported that the levels of miR-489 in late recurrent HCC patients were evidently higher than that in early recurrent cases, suggesting that miR-489 may function as a tumor suppressive miRNA in HCC. Yet, the clinical value and biological function of miR-489 remain rarely known in HCC. Here, we presented that miR-489 level in HCC tissues was notably reduced compared to matched non-cancerous specimens. Its decreased level was evidently correlated with adverse clinical parameters and poor prognosis of HCC patients. Accordingly, the levels of miR-489 were obviously down-regulated in HCC cells. Ectopic expression of miR-489 in HCCLM3 and MHCC97H cells prominently inhibits the migration and invasion of tumor cells and reduced lung metastases in vivo, while miR-489 knockdown increased these behaviors of HepG2 and MHCC97L cells. Mechanically, miR-489 negatively regulated matrix metalloproteinase-7 (MMP7) abundance in HCC cells. Herein, MMP7 was found to be a downstream molecule of miR-489 in HCC. An inversely correlation between miR-489 and MMP7 was confirmed in HCC specimens. MMP7 knockdown prohibited cell migration and invasion while MMP7 overexpression showed opposite effects on HCC cells. Furthermore, restoration of MMP7 expression could abrogate the anti-metastatic effects of miR-489 on HCCLM3 cells with enhanced cell migration and invasion. Altogether, miR-489 potentially acts as a prognostic predictor and a drug-target for HCC patients.
Introduction
microRNAs (miRNAs) inhibit the expression of target genes by contributing to the degradation or translational inhibition of target mRNAs [1]. They have been found to be actively involved in different cellular processes [2], [3] including proliferation, apoptosis, differentiation and movement. Emerging studies showed that abnormal expression and function of miRNAs play important roles in the pathogenesis and tumorigenicity of human malignancies [4], [5], [6]. Otherwise, miRNAs have been demonstrated to be hopeful diagnostic biomarkers and drug-targets of hepatocellular carcinoma (HCC) [7], [8], [9]. Investigating the expression and biological function of miRNAs in HCC will contribute to the discovery of new biomarkers and drug-targets for HCC patients.
miR-489 is found to be underexpressed in 4-hydroxytamoxifen-resistant and adriamycin-resistant human breast cancer cells compared to Tamoxifen-sensitive MCF-7 cells [10], [11], [12]. miR-489 overexpression reverses proliferation, migration and invasion of breast cancer cells by targeting Gse1 coiled-coil protein (GSE1) [13]. Restoration of miR-489 suppresses HER2-positive breast cancer cell growth and inhibits tumorigenesis and tumor growth in vivo [14]. Otherwise, miR-489 exerts a tumor suppressive role by inhibiting cell growth via inhibition of protein tyrosine phosphatase, non-receptor type 11 (PTPN11) [15] in hypopharyngeal squamous cell carcinoma. The level of miR-489 is reduced in cisplatin-resistant ovarian cancer (OC) cells and miR-489 overexpression inhibits OC cell survival, cell growth and induces apoptosis by targeting Akt3 [16]. In non-small cell lung cancer, knockdown of miR-489 facilitates cell invasion and epithelial mesenchymal transition (EMT) [17]. However, miR-489 is significantly overexpressed in oral squamous cell carcinoma [18], clear cell renal cell carcinoma [19], [20]. Recently, Yang Z et al. found that the levels of miR-489 in late recurrent HCC patients were notably lower than those in early recurrent cases [21]. Yet, the clinical value and biological role of miR-489 in HCC remain largely unknown.
Here, we confirmed that miR-489 was underexpressed in HCC specimens and cells. The low level of miR-489 associated with malignant clinical features of HCC patients and decreased survival rates. Our results showed that miR-489 inhibited the invasive ability of cancer cells in HCC. Moreover, matrix metalloproteinase-7 (MMP7) was recognized as a downstream molecule of miR-489 and possibly mediated the biological functions of miR-489 in HCC.
Material and Methods
Clinical Samples
Clinical specimens were obtained from 130 patients histologically diagnosed as HCC in the Department of Hepatobiliary Surgery at the Nanfang Hospital of Southern Medical University. Patients who received immunotherapy, chemotherapy or radiotherapy before surgical treatment were excluded. Informed consent was signed by each patient before clinical specimens collected and used. All specimens were stored in liquid nitrogen or fixed with formalin for further investigation. The protocol involved clinical specimens in this study was permitted by the Research Ethics Committee of Southern Medical University.
Cell Culture and Transfection
Human HCC cell lines including HepG2, MHCC97L, MHCC97H and HCCLM3, a human immortalized normal hepatocyte cell line (LO2) and a human embryonic kidney (HEK293T) cell line were obtained from Chinese Academy of Sciences (Shanghai, China). All cells were cultured in DMEM (HyClone, Logan, UT, USA) along with fetal bovine serum (10%) (FBS; HyClone) and anti-biotics (Sigma-Aldrich, St. Louis, MO, USA). Cell cultures were kept in an incubator containing of 5% CO2 and humidified atmosphere at 37 °C.
miR-489 mimic (HmiR0107-MR04), miR-489 inhibitor (HmiR-AN0528-AM04) and the corresponding control vectors (CmiR0001-MR04; CmiR-AN0001-AM04) were bought from Genecopoeia (Guangzhou, China). MMP7 siRNA and scrambled siRNA were designed and synthesized by GenePharma (Shanghai, China). The above vectors were then tranduced into HCC cells with lippofectamine 2000 following manufactures' protocol. Retroviral vectors pMMP-MMP7 were constructed by inserting the corresponding cDNA into pMMP. The retroviruses were packaged and tranfected into HCC cells as previously described [22].
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA from HCC cells was isolated by miRNeasy Mini Kit (Qiagen, Hilden, Germany) and total RNA from HCC tissues were extracted with Trizon reagent (Invitrogen, Carlsbad, CA, USA). cDNAs were synthesized from 2 μg total RNA using TIANScript RT Kit (Tiangen biotech, Beijing, China). miR-489 levels in these samples were detected using TaqMan™ MicroRNA Assays based on the manufacturer's instructions (Applied Biosystems™, Carlsbad, CA, USA). Quantitative real-time PCR was performed using UltraSYBR Mixture (CW0957, CWBIO, Beijing, China) in a final volume of 10 μL in LC 480 PCR System (Roche, Indianapolis, IN, USA). The primers for miR-489 and U6, MMP7 and GAPDH were bought from Genecopoeia (Guangzhou, China). U6 was used as the control gene for the relative level of miR-489 while GAPDH served as internal control for MMP7. Data were presented as relative quantification based on the calculation of 2−ΔCt. ΔCt was derived from subtracting the Ct value of reference cDNA from the Ct value of the cDNA of interest. All primers for RT and PCR are shown as follows: miR-489 RT primer: 5′-CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG AGC TGC CGT-3′; miR-489 forward: 5′-ACA CTC CAG CTG GGG TGA CAT CAC ATA-3′; miR-489 reverse primer: 5′-TGG TGT CGT GGA GTC G-3′; MMP-7 primers were: 5′-GGT CAC CTA CAG GAT CGT ATC ATA T-3′(forward) and 5′-CAT CAC TGC ATT AGG ATC AGA GGA A-3′ (reverse); GAPDH primers were: 5′-CAA GCT CAT TTC CTG GTA TGA C-3′(forward) and 5′-CAG TGA GGG TCT CTC TCT TCC T-3′(reverse).
Luciferase Reporter Assay
To investigate whether miR-489 could interact with the 3′-UTRs of MMP7, wild type (wt) 3′-UTR of MMP7 predicted to interact with miR-489 or the mutant (mt) MMP7 3′-UTR was amplified. Then, the wt 3′-UTR of MMP7 or mt 3′-UTR of MMP7, and miR-489 mimic or miRNA scrambled control clones (NC) were co-transduced into HCC and HEK293T cells, respectively by lippofectamine 2000. 48 hours after co-transduction, the cells were lysed and detected using a Dual-Luciferase® Reporter Assay Kit (Promega, Madison, WI, USA) based on the manufacturer's protocols.
Wound Healing Assay
HCC cells transduced with corresponding vectors were subsequently seeded in 6-well plates to form the single confluent cell layer. The wound were made with 100ul tips in the confluent cell layer.0 and 24 hours after would scratching, the width of wound was photographed with phase-contrast microscope.
Transwell Migration and Invasion Assay
The migration and invasion ability of HCC cells were detected with Transwell chambers (BD Biosciences, Franklin Lakes, NJ, USA). 5–10 × 104 HCC cells suspended in 100ul medium without serum were seeded into the upper chamber, and lower chamber was full of 20% FBS to induce HCC cells migrating or invading through the membrane. Matrigel (1:6 dilution) was added on the upper chamber for invasion assay. Twenty-four hours later, cells with crystal violet staining that migrated or invaded across the Transwell membrane were numbered under optical microscope.
Western Blotting
Two days post transfection, total protein was extracted, following by quantification with BCA protein assay kit (Pierce, Bonn, Germany). Protein were separated by 10% SDS-polyacrylamide gels and subsequently transferred to PVDF membranes. Then, 5% milk blocked membranes were incubated with MMP7 (10374–2-AP, Proteintech, Rosemont, IL, USA) antibody. The secondary antibodies were obtained from Cell signaling (Danvers, MA, USA). GAPDH (G8140, US Biological, Swampscott, MA, USA) was used as a loading control.
Immunohistochemistry (IHC)
The tumor tissues that were previously formalin-fixed and paraffin-embedded were sliced into 4 μm sections, and underwent deparaffination and then rehydration. Antigen retrieval, suppression of endogenous peroxidase activity and 10% skim milk blocking were performed before primary antibody incubation. MMP7 (Proteintech) antibody was used as primary antibody overnight at 4 °C. The following day, the slides were incubated with peroxidase conjugated secondary antibody (ZSGB BIO, Beijing China) for 90 min, and a peroxidase-labeled polymer, DAB solution was used for signal development for 5 min. The sections were counterstained with hematoxylin followed by dehydrating and mounting.
In Vivo Pulmonary Metastasis Assay
6 × 106 HCCLM3 cells that were transfected with miR-489 mimic or control vector were intravenously injected into nude mice. Five weeks after injection, all mice were euthanized, and the lungs were sectioned and stained by H&E to check if pulmonary metastatic foci formed. The protocol for these animal experiments were approved by the Ethics Review Committee of Southern Medical University.
Statistical Analysis
Data were presented as Mean ± SEM and analyzed by GraphPad Prism 5 software (GraphPad Software, Inc., San Diego, CA, USA). Chi-squared test was employed to explore the association between two variables. The Student’s t test and ANOVA were carried out to analyze continuous variable. Survival curves were constructed and differences among groups were calculated using the Kaplan–Meier method and Log-rank test. Spearman's rank correlation analysis was performed to reveal the correlation between two factors. The value of P less than .05 was considered to have statistical significance.
Results
miR-489 Expression is Decreased in HCC
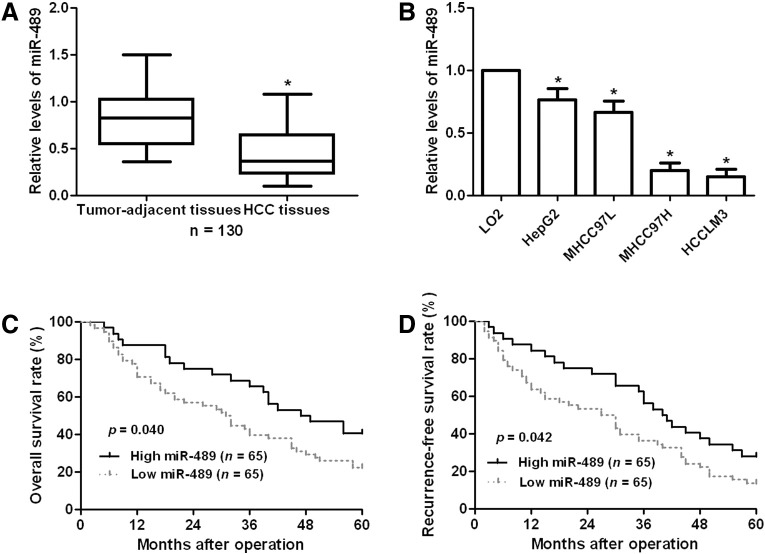
To examine the status of miR-489 in HCC, qRT-PCR was performed for 130 HCC cases. Our data disclosed that HCC tissues had significant decreased expression levels of miR-489 (P < .05, Figure 1A). Next, we compared the expression level of miR-489 between HCC cells lines and LO2 cells. Compared with LO2 cells, the levels of miR-489 in all HCC cells (HepG2, MHCC97L, MHCC97H, and HCCLM3) were significantly reduced, especially in high-metastatic cells (MHCC97H and HCCLM3) (P < .05, Figure 1B). These data indicate miR-489 probably plays a suppressive role in HCC.
Figure 1.
The status and prognostic value of miR-489 expression in HCC. (A) The alterative expression of miR-489 between HCC tissues and normal tumor-adjacent tissues. n = 130, *P < .05 by t test. (B) The differences expression of 4 different HCC cells lines (HepG2, MHCC97L, MHCC97H and HCCLM3) and human immortalized hepatocyte LO2. n = 3 repeats with similar results, *P < .05 by ANOVA. (C) and (D) The “low” or “high” of miR-489 level was defined according to the cut-off value, which was defined as the median value of the cohort of patients tested (0.83). Compared with those of high miR-489 level (n = 65), miR-489 low-expressing patients (n = 65) had significantly reduced overall survival and recurrence-free survival rates. P < .05 by log-rank test.
Underexpression of miR-489 Correlates with Adverse Clinical Parameters and Prognosis of HCC Patients
To clarify the clinical value of miR-489 in HCC, all patients were grouped into miR-489 low group and miR-489 high group according to the cut-off value, which was defined as the median value of the cohort of patients tested (0.83). As shown in Table 1, HCC patients with low expression of miR-489 had more tumor nodes (P = .010), venous infiltration (P = .046) and advanced tumor-node-metastasis (TNM) stage (P = .009). Furthermore, survival analyses indicated that patients with low expression showed significantly reduced 5-year overall and recurrence-free survival (P = .040 and P = .042, respectively, Figure 1C and D). We suggest that miR-489 is a possible prognostic biomarker for HCC patients.
Table 1.
Correlation between the Clinicopathologic Features and miR-489 Expression in HCC
| Characteristics | Total No. of Patients, n = 130 | miR-489 Expression Status |
P | ||
|---|---|---|---|---|---|
| Low | High | ||||
| Age (y) | <50 | 54 | 26 | 28 | .722 |
| ≥50 | 76 | 39 | 37 | ||
| Sex | Male | 98 | 50 | 48 | .684 |
| Female | 32 | 15 | 17 | ||
| HBV | Absent | 42 | 17 | 25 | .134 |
| Present | 88 | 48 | 40 | ||
| Serum AFP level (ng/mL) | <400 | 48 | 23 | 25 | .716 |
| ≥400 | 82 | 42 | 40 | ||
| Tumor size (cm) | <5 | 49 | 23 | 26 | .587 |
| ≥5 | 81 | 42 | 39 | ||
| No. of tumor nodules | 1 | 102 | 45 | 57 | .010* |
| ≥2 | 28 | 20 | 8 | ||
| Cirrhosis | Absent | 54 | 28 | 26 | .722 |
| Present | 76 | 37 | 39 | ||
| Venous infiltration | Absent | 96 | 43 | 53 | .046* |
| Present | 34 | 22 | 12 | ||
| Edmondson-Steiner grading | I + II | 98 | 45 | 53 | .103 |
| III + IV | 32 | 20 | 12 | ||
| TNM tumor stage | I + II | 97 | 42 | 55 | .009* |
| III + IV | 33 | 23 | 10 | ||
HCC, hepatocellular carcinoma; HBV, hepatitis B virus; AFP, alpha-fetoprotein; TNM, tumor-node-metastasis.
Statistically significant.
miR-489 Inhibits the Mobility of HCC Cells
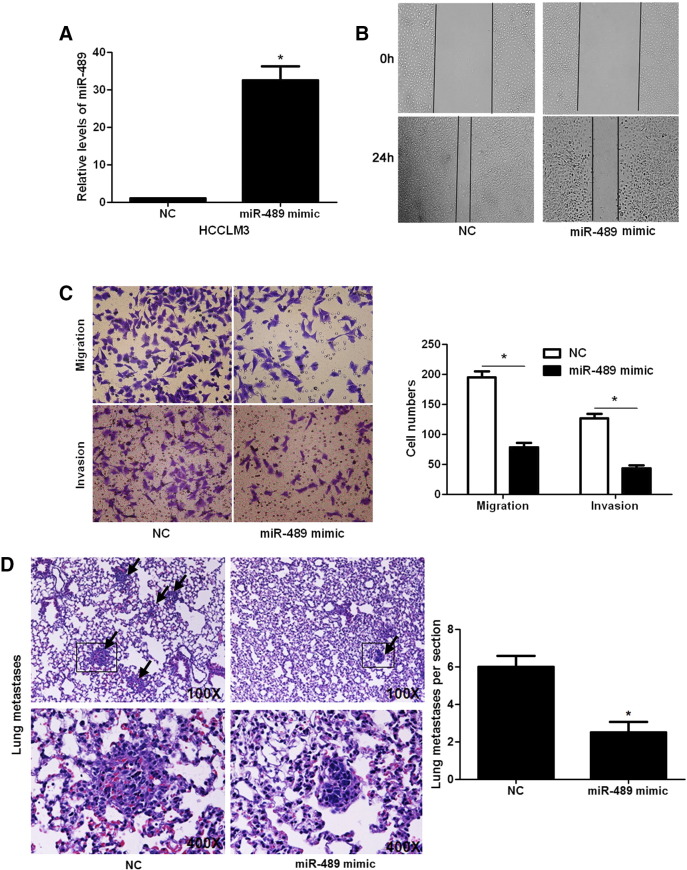
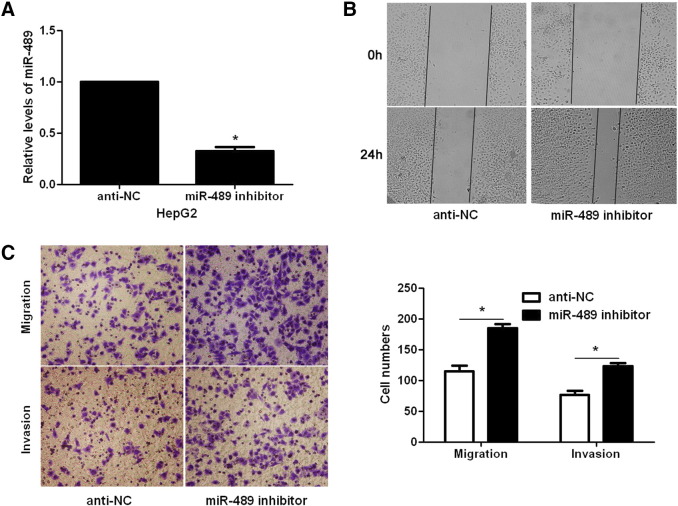
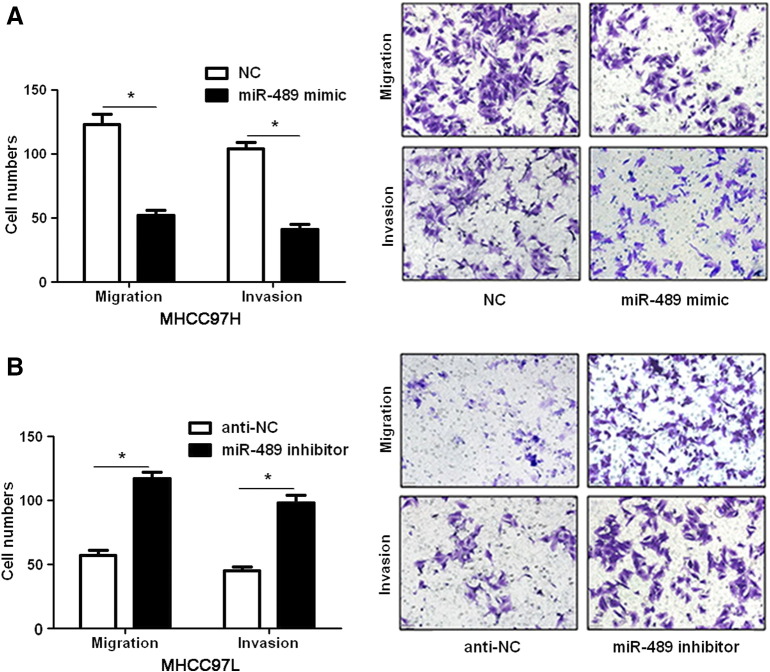
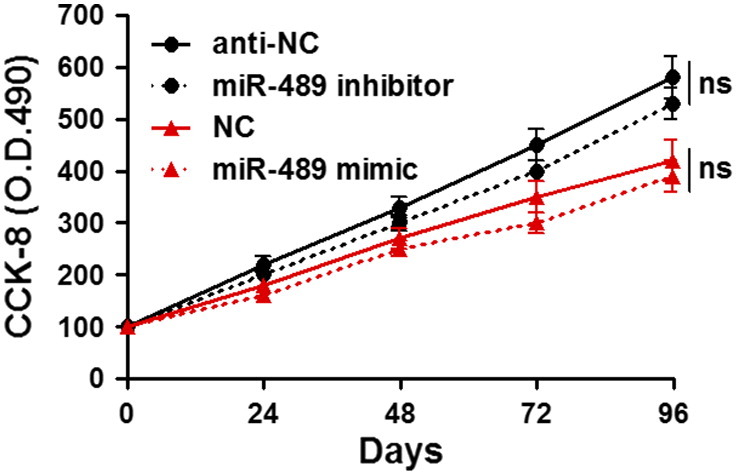
Since increased cancer cell mobility is an important reason for the metastasis and recurrence of human cancer [23], we explored whether miR-489 could modulate the migration and invasion of HCC cells. Transfection of miR-489 mimic obviously up-regulated the level of miR-489 in HCCLM3 cells (P < .05, Figure 2A). The wound healing assays showed that miR-489 overexpression notably reduced cell migration in HCCLM3 cells (P < .05, Figure 2B). And Transwell assays explored that ectopic expression of miR-489 significantly reduced the numbers of migrated and invaded HCCLM3 cells (P < .05, Figure 2C). In a pulmonary metastasis model, miR-489 overexpression remarkably reduced the number of lung metastases in nude mice (P < .05, Figure 2D). In turn, miR-489 inhibitor significantly decreased the level of miR-489 in HepG2 cells (P < .05, Figure 3A). Subsequently, miR-489 silencing notably facilitated HepG2 cell migration and invasion (P < .05, respectively, Figure 3, B and C). Furthermore, overexpressing and underexpressing miR-489 were performed in MHCC97H and MHCC97L cells, respectively. miR-489 showed similar effects on migration and invasion of MHCC97H and MHCC97L cells (P < .05, respectively, Supplementary Figure 1). Notably, CCK-8 assays indicated that miR-489 alteration did not significantly influenced HCC cell growth (Supplementary Figure 2). Thus, miR-489 exerts a anti-metastatic role in HCC cells.
Figure 2.
miR-489 overexpression inhibits the mobility of HCCLM3 cells. (A) HCCLM3 cells that were transduced with miRNA scrambled control clones (NC) or miR-489 mimics were confirmed by qRT-PCR. n = 3 repeats with similar results, *P < .05 by t test. (B) Wound healing assays indicated that miR-489 overexpression inhibited the migration of HCCLM3 cells. (C) Transwell assays confirmed that miR-489 overexpression restrained HCCLM3 cell migration and invasion. n = 3 repeats with similar results, *P < .05 by t test. (D) HCCLM3 cells that were transfected miR-489 mimic and miRNA scrambled control clones (NC) were intravenously injected into nude mice (n = 8). HE staining revealed that miR-489 overexpression significantly reduced lung metastases of HCCLM3 cells in vivo. *P < .05 by t test.
Figure 3.
miR-489 knockdown facilitates the metastasis of HepG2 cells. (A) HepG2 cells that were transduced with miRNA inhibitor control clones (anti-NC) or miR-489 inhibitors were confirmed by qRT-PCR. n = 3 repeats with similar results, *P < .05 by t test. (B) miR-489 knockdown notably facilitated the migration of HepG2 cells. (C) miR-489 knockdown prominently increased HepG2 cell migration and invasion. n = 3 repeats with similar results, *P < .05 by t test.
Supplementary Figure 1.
miR-489 regulates migration and invasion in HCC cells. (A) MHCC97H cells were transduced with miRNA scrambled control clones (NC) or miR-489 mimics. Transwell assays confirmed that miR-489 overexpression prohibited cell migration and invasion. n = 3 repeats with similar results, *P < .05 by t test. (B) MHCC97L cells were transduced with miRNA inhibitor control clones (anti-NC) or miR-489 inhibitors. miR-489 knockdown prominently facilitated cell migration and invasion. n = 3 repeats with similar results, *P < .05 by t test.
Supplementary Figure 2.
miR-489 alteration shows no obvious effect on HCC cell growth. HCCLM3 and HepG2 cells that were transfected with miR-489 mimic and inhibitor, respectively, were subjected to CCK-8 proliferation assays. Both miR-489 overexpression and knockdown did not significantly influenced HCC cell growth.
miR-489 Post-Transcriptionally Regulates MMP7 Expression
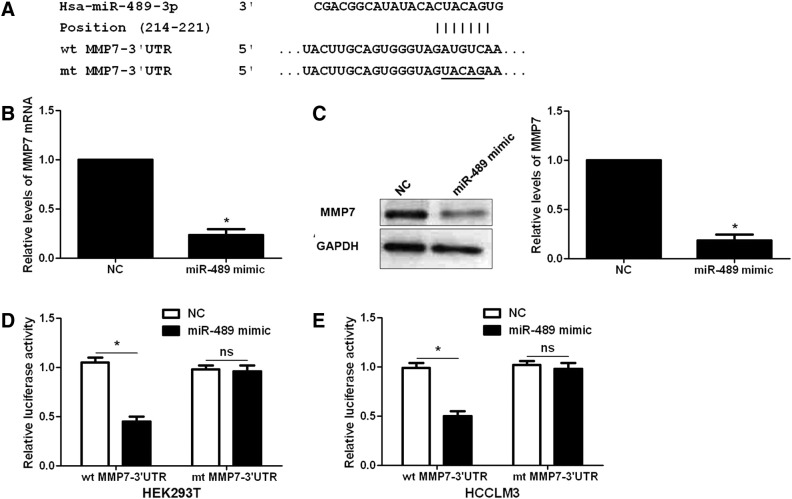
To disclose the potential molecular mechanisms involved in the role of miR-489 in HCC cells, we searched for candidate target genes of miR-489 using publicly available databases, including TargetScan, miRanda and PicTar. MMP7, a pro-metastatic molecule in HCC [24], was considered as one of the candidates with which miR-489 could bind directly (Figure 4A). Further experiments were performed to confirm the above hypothesis. Interestingly, miR-489 overexpression reduced the expressions of MMP7 mRNA and protein (P < .05, respectively, Figure 4, B and C). We further explored whether MMP7 was a downstream target molecule of miR-489. Then, our data indicated that miR-489 overexpression decreased the luciferase activity of wt MMP7 3′-UTR (P < .05, respectively, Figure 4, D and E), while alteration of miR-489 did not have any influence on the luciferase activity of mt MMP7 3′-UTR in HEK293T and HCCLM3 cells (Figure 4, D and E). Therefore, these data indicate miR-489 can regulate the expression of MMP7 by directly interacting with its 3′-UTR in HCC.
Figure 4.
MMP7 is a downstream molecule of miR-489. (A) The potential miR-489 binding site in wild type (wt) 3′-UTR sequence of MMP7. The underlined part is the mutant site designed for mutant (mt) 3′-UTR sequence of MMP7. (B) and (C) HCCLM3 cells that were transduced with NC or miR-489 mimics were confirmed by qRT-PCR and immunoblotting for MMP7 mRNA and protein expression. n = 3 repeats with similar results, *P < .05 by t test, respectively. (D) and (E) Overexpression of miR-489 decreased the luciferase activity of wild type (wt) MMP7 3′-UTR in both HEK293T and HCCLM3 cells. While alteration of miR-489 showed non-effect on the luciferase activity of mutant (mt) MMP7 3′-UTR. n = 3 repeats with similar results, *P < .05 by t test, respectively.
miR-489 Inversely Correlates with MMP7 in HCC Specimens
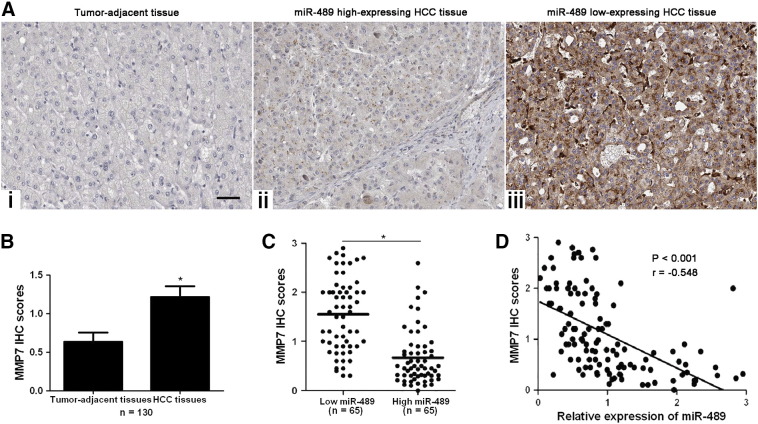
Next, IHC was performed to detect MMP7 in HCC and noncancerous tissues. IHC scores evaluation indicated that MMP7 was overexpressed in HCC tissues (P < .05, Figure 5, A and B). Notably, miR-489 high expressing tumors showed weak staining of MMP7 (Figure 5A–ii), while miR-489 low expressing tumors showed strong staining of MMP7 (Figure 5A–iii). Quantification data suggested that the expressions of MMP7 in miR-489 low expressing tumors were notably higher than those in miR-489 high expressing cases (P < .05, Figure 5C). Spearman's correlation analysis disclosed that miR-489 was negatively correlated with MMP7 in HCC specimens (r = −0.548, P < .001, Figure 5D). These data showed a negative correlation between miR-489 and MMP7 in HCC.
Figure 5.
The difference of MMP7 expression between HCC and non-tumor tissues. (A) Representative IHC staining of MMP7 in HCC and tumor-adjacent tissues. miR-489 high expressing tumors showed weak staining of MMP7 (ii), while miR-489 low expressing tumors showed strong staining of MMP7 (iii). (B) The expression difference of MMP7 between HCC and adjacent non-tumor tissues. n = 130, *P < .05 by t test. (C) The expression difference of MMP7 between miR-489 low and high expressing HCC specimens. n = 65, *P < .05 by t test. (D) An negative correlation between miR-489 and MMP7 was confirmed in HCC tissues. n = 130, P < .05 by Spearman's rank correlation analysis.
Restoration of MMP7 Abrogates the Anti-Metastatic Role of miR-489
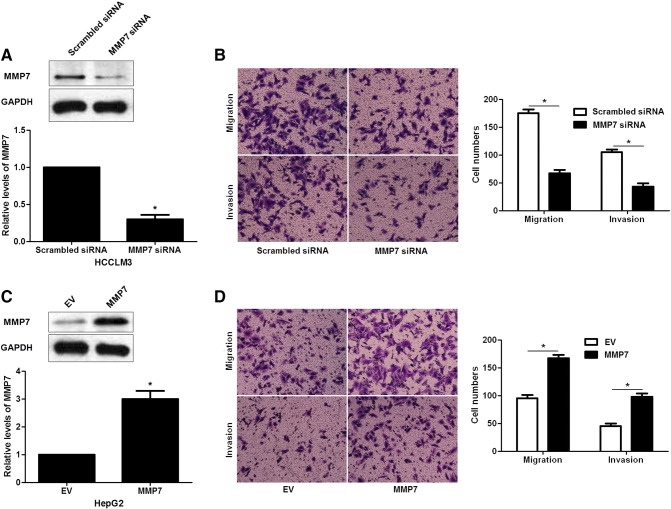
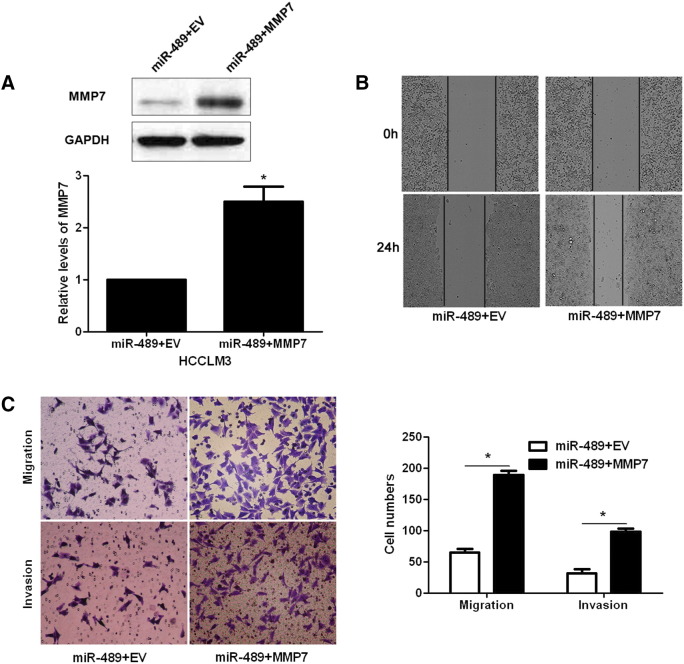
The role of MMP7 was further confirmed in our study. MMP7 knockdown by a specific siRNA showed a significant reduction of migrated and invaded cells in HCCLM3 cells (P < .05, respectively, Figure 6, A and B). In contrast, MMP7 overexpression that was assessed by immunoblotting promoted cell migration and invasion in HepG2 cells (P < .05, respectively, Figure 6, C and D). Since we confirmed MMP7 was a target molecule of miR-489, MMP7 retroviruses were employed to disclose whether MMP7 restoration could abolish the anti-metastatic role of miR-489 in HCC cells. As shown in Figure 7A, MMP7 retroviruses infection significantly increased the level of MMP7 in miR-489 overexpressing HCCLM3 cells (P < .05). Consequently, restoration of MMP7 promoted the metastatic behavior of miR-489 overexpressing HCCLM3 cells with enhanced cell migration and invasion (P < .05, respectively, Figure 7, B and C). These rescue experiments suggest that miR-489 inhibits the migration and invasion of HCC cells possibly by targeting MMP7.
Figure 6.
MMP7 regulates the metastatic behaviors of HCC cells. (A) HCCLM3 cells that were transduced with MMP7 siRNA or scrambled siRNA were subjected to western blotting for MMP7 expression. n = 3 repeats with similar results, *P < .05 by t test. (B) Quantitative data indicated that MMP7 knockdown inhibited the migration and invasion of HCCLM3 cells. n = 3 repeats with similar results, *P < .05 by t test. (C) HepG2 cells that were transduced with empty vector (EV) or MMP7 retroviruses were subjected to immunoblotting for MMP7 expression. n = 3 repeats with similar results, *P < .05 by t test. (D) Transwell assays demonstrated that MMP7 overexpression facilitated the metastatic behaviors of HepG2 cells with increased cell migration and invasion. n = 3 repeats with similar results, *P < .05 by t test.
Figure 7.
MMP7 restoration reverses the effects of miR-489. (A) miR-489 overexpressing HCCLM3 cells that were infected with empty vector (EV) or MMP7 retroviruses were confirmed by western blotting for MMP7 expression. n = 3 repeats with similar results, *P < .05 by t test. (B) MMP7 restoration significantly promoted the migration of miR-489 overexpressing HCCLM3 cells. (C) MMP7 restoration evidently facilitated cell migration and invasion in miR-489 overexpressing HCCLM3 cells. n = 3 repeats with similar results, *P < .05 by t test.
Discussion
Emerging evidences have confirmed that miRNAs are actively involved in the pathogenic process of HCC [25]. And miRNAs have been reported to be important mediator of the metastasis and epithelial mesenchymal transition of HCC cells [26]. According to the important function of miRNAs in HCC, miRNAs have been considered as potential diagnostic biomarkers and drug-targets of HCC [27]. In this study, miR-489 was found to be significantly under-expressed in HCC. HepG2 and MHCC97L were low invasive cell lines, while MHCC97H and HCCLM3 were high metastatic cell lines. Thus, the relative levels of miR-489 in MHCC97H and HCCLM3 cells were lower than those in HepG2 and MHCC97L cells. And low expression of miR-489 conferred adverse clinical parameters of HCC patients including more tumor nodes, venous invasion and advanced clinical stage. More importantly, decreased expression of miR-489 correlated with shortened 5-year overall and recurrence-free survival. Therefore, miR-489 played tumor suppressive role in HCC and could potentially serve as a promising biological tag for the prognosis of patients.
Systemic metastasis is the important reason for the unsatisfactory prognosis of HCC patients [28]. Increased migratory and invasive ability of cancer cells underlies the systemic metastasis of HCC. Thus, it is fundamental to disclose the underlying mechanisms for the metastasis of HCC cells. Here, we explored that miR-489 inhibited the migration and invasion of HCC cells in vitro and in vivo. These data confirmed that miR-489 exerted tumor suppressive role in HCC by inhibiting metastatic behaviors of HCC cells. MMP7, a secreted protein, exerts a role in the destruction of extracellular matrix substrates in human cancers [24]. Otherwise, MMP7 was found to function as a pro-metastatic factor by promoting the migratory and invasive ability of cancer cells [29]. Recent studies reported that overexpression of MMP7 was found in HCC specimens and cells [30], [31]. Moreover, MMP7 was considered as an crucial target molecule of dickkopf1 (DKK1) and mediated DKK1-induced HCC cell migration and invasion [24]. Here, we also revealed that MMP7 promoted HCC cell migration and invasion. Notably, we disclosed that miR-489 suppressed the expression of MMP7 in HCC cells. And the levels of MMP7 in HCC tissues were negatively correlated with the expressions of miR-489. Moreover, we found that miR-489 could directly interact with the 3′-UTR of MMP7. These experiments suggest that MMP7 is a downstream molecule of miR-489. Furthermore, we found that restoration of MMP7 could abrogate the anti-metastatic effects of miR-489 on HCC cells migration and invasion. These suggest that miR-489 inhibits the migration and invasion of HCC cells possibly by targeting MMP7.
Altogether, our study demonstrates that miR-489 expression is significantly decreased in HCC. The low level of miR-489 correlates with adverse clinical parameters of HCC patients and shortened survival. And miR-489 inhibits the metastasis of HCC cells. Furthermore, MMP7 is a direct target of miR-489 in HCC. Altogether, miR-489 exerts its inhibitory effects on HCC metastasis, at least in part, by targeting MMP7.
Conclusions
To conclude, we recognize miR-489 down-regulation as a biomarker for poor prognosis prediction. The underexpression of miR-489 creates a milieu of metastasis facilitation that plays a role in HCC progression. A mechanism by which underexpressed miR-489 promotes the metastasis by targeting MMP7 plays an important role in this process. This finding will improve understanding of cancer progression mechanism and provide novel targets for the molecular treatment of HCC.
The following are the supplementary data related to this article.
Conflicts of Interest
The authors declare that they have no competing interests.
Acknowledgements
This study was supported by a grant from National Natural Scientific Foundation of Guangdong Province (No. 2016A030313555).
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11(4):441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Zhang W, Dahlberg JE, Tam W. MicroRNAs in tumorigenesis: a primer. Am J Pathol. 2007;171(3):728–738. doi: 10.2353/ajpath.2007.070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 7.Tu K, Liu Z, Yao B, Han S, Yang W. MicroRNA-519a promotes tumor growth by targeting PTEN/PI3K/AKT signaling in hepatocellular carcinoma. Int J Oncol. 2016;48(3):965–974. doi: 10.3892/ijo.2015.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang W, Dou C, Wang Y, Jia Y, Li C, Zheng X, Tu K. MicroRNA-92a contributes to tumor growth of human hepatocellular carcinoma by targeting FBXW7. Oncol Rep. 2015;34(5):2576–2584. doi: 10.3892/or.2015.4210. [DOI] [PubMed] [Google Scholar]
- 9.Dou C, Wang Y, Li C, Liu Z, Jia Y, Li Q, Yang W, Yao Y, Liu Q, Tu K. MicroRNA-212 suppresses tumor growth of human hepatocellular carcinoma by targeting FOXA1. Oncotarget. 2015;6(15):13216–13228. doi: 10.18632/oncotarget.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283(44):29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang L, He D, Yang D, Chen Z, Pan Q, Mao A, Cai Y, Li X, Xing H, Shi M. MiR-489 regulates chemoresistance in breast cancer via epithelial mesenchymal transition pathway. FEBS Lett. 2014;588(11):2009–2015. doi: 10.1016/j.febslet.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Wang YW, Xing AY, Xiang S, Shi DB, Liu L, Li YX, Gao P. Suppression of SPIN1-mediated PI3K-Akt pathway by miR-489 increases chemosensitivity in breast cancer. J Pathol. 2016;239(4):459–472. doi: 10.1002/path.4743. [DOI] [PubMed] [Google Scholar]
- 13.Chai P, Tian J, Zhao D, Zhang H, Cui J, Ding K, Liu B. GSE1 negative regulation by miR-489-5p promotes breast cancer cell proliferation and invasion. Biochem Biophys Res Commun. 2016;471(1):123–128. doi: 10.1016/j.bbrc.2016.01.168. [DOI] [PubMed] [Google Scholar]
- 14.Patel Y, Shah N, Lee JS, Markoutsa E, Jie C, Liu S, Botbyl R, Reisman D, Xu P, Chen H. A novel double-negative feedback loop between miR-489 and the HER2-SHP2-MAPK signaling axis regulates breast cancer cell proliferation and tumor growth. Oncotarget. 2016;7(14):18295–18308. doi: 10.18632/oncotarget.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kikkawa N, Hanazawa T, Fujimura L, Nohata N, Suzuki H, Chazono H, Sakurai D, Horiguchi S, Okamoto Y, Seki N. miR-489 is a tumour-suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC) Br J Cancer. 2010;103(6):877–884. doi: 10.1038/sj.bjc.6605811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H, Xiao Z, Zhang H, Wang K, Liu W, Hao Q. MiR-489 modulates cisplatin resistance in human ovarian cancer cells by targeting Akt3. Anti-Cancer Drugs. 2014;25(7):799–809. doi: 10.1097/CAD.0000000000000107. [DOI] [PubMed] [Google Scholar]
- 17.Xie Z, Cai L, Li R, Zheng J, Wu H, Yang X, Li H, Wang Z. Down-regulation of miR-489 contributes into NSCLC cell invasion through targeting SUZ12. Tumour Biol. 2015;36(8):6497–6505. doi: 10.1007/s13277-015-3340-3. [DOI] [PubMed] [Google Scholar]
- 18.Scapoli L, Palmieri A, Lo Muzio L, Pezzetti F, Rubini C, Girardi A, Farinella F, Mazzotta M, Carinci F. MicroRNA expression profiling of oral carcinoma identifies new markers of tumor progression. Int J Immunopathol Pharmacol. 2010;23(4):1229–1234. doi: 10.1177/039463201002300427. [DOI] [PubMed] [Google Scholar]
- 19.Zaravinos A, Lambrou GI, Mourmouras N, Katafygiotis P, Papagregoriou G, Giannikou K, Delakas D, Deltas C. New miRNA profiles accurately distinguish renal cell carcinomas and upper tract urothelial carcinomas from the normal kidney. PLoS One. 2014;9(3) doi: 10.1371/journal.pone.0091646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munari E, Marchionni L, Chitre A, Hayashi M, Martignoni G, Brunelli M, Gobbo S, Argani P, Allaf M, Hoque MO. Clear cell papillary renal cell carcinoma: micro-RNA expression profiling and comparison with clear cell renal cell carcinoma and papillary renal cell carcinoma. Hum Pathol. 2014;45(6):1130–1138. doi: 10.1016/j.humpath.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z, Miao R, Li G, Wu Y, Robson SC, Yang X, Zhao Y, Zhao H, Zhong Y. Identification of recurrence related microRNAs in hepatocellular carcinoma after surgical resection. Int J Mol Sci. 2013;14(1):1105–1118. doi: 10.3390/ijms14011105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tu K, Yang W, Li C, Zheng X, Lu Z, Guo C, Yao Y, Liu Q. Fbxw7 is an independent prognostic marker and induces apoptosis and growth arrest by regulating YAP abundance in hepatocellular carcinoma. Mol Cancer. 2014;13:110. doi: 10.1186/1476-4598-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arlt F, Stein U. Colon cancer metastasis: MACC1 and Met as metastatic pacemakers. Int J Biochem Cell Biol. 2009;41(12):2356–2359. doi: 10.1016/j.biocel.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Chen L, Li M, Li Q, Wang CJ, Xie SQ. DKK1 promotes hepatocellular carcinoma cell migration and invasion through beta-catenin/MMP7 signaling pathway. Mol Cancer. 2013;12:157. doi: 10.1186/1476-4598-12-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarocchi M, Polvani S, Marroncini G, Galli A. Molecular mechanism of hepatitis B virus-induced hepatocarcinogenesis. World J Gastroenterol. 2014;20(33):11630–11640. doi: 10.3748/wjg.v20.i33.11630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z, Tu K, Liu Q. Effects of microRNA-30a on migration, invasion and prognosis of hepatocellular carcinoma. FEBS Lett. 2014;588(17):3089–3097. doi: 10.1016/j.febslet.2014.06.037. [DOI] [PubMed] [Google Scholar]
- 27.Anwar SL, Lehmann U. MicroRNAs: Emerging Novel Clinical Biomarkers for Hepatocellular Carcinomas. J Clin Med. 2015;4(8):1631–1650. doi: 10.3390/jcm4081631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung HK, Lee DS, Yoon SS, Kim HJ. Systemic metastasis of hepatocellular carcinoma responsive to multidisciplinary treatment including debulking surgery. Ann Surg Treat Res. 2014;86(2):100–104. doi: 10.4174/astr.2014.86.2.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oshima T, Akaike M, Yoshihara K, Shiozawa M, Yamamoto N, Sato T, Yamada R, Fujii S, Rino Y, Kunisaki C. Clinicopathological significance of the gene expression of matrix metalloproteinase-7, insulin-like growth factor-1, insulin-like growth factor-2 and insulin-like growth factor-1 receptor in patients with colorectal cancer: insulin-like growth factor-1 receptor gene expression is a useful predictor of liver metastasis from colorectal cancer. Oncol Rep. 2008;20(2):359–364. [PubMed] [Google Scholar]
- 30.Gao Q, Wang XY, Qiu SJ, Zhou J, Shi YH, Zhang BH, Fan J. Tumor stroma reaction-related gene signature predicts clinical outcome in human hepatocellular carcinoma. Cancer Sci. 2011;102(8):1522–1531. doi: 10.1111/j.1349-7006.2011.01981.x. [DOI] [PubMed] [Google Scholar]
- 31.Tomimaru Y, Koga H, Yano H, de la Monte S, Wands JR, Kim M. Upregulation of T-cell factor-4 isoform-responsive target genes in hepatocellular carcinoma. Liver Int. 2013;33(7):1100–1112. doi: 10.1111/liv.12188. [DOI] [PMC free article] [PubMed] [Google Scholar]