Abstract
Autophagy influences how cancer cells respond to nutrient deprivation and hypoxic stress, two hallmarks of the tumor microenvironment (TME). In this study, we explored the impact of autophagy on the pathophysiology of breast cancer cells, using a novel hypoxia-dependent, reversible dominant negative strategy to regulate autophagy at the cellular level within the TME. Suppression of autophagy via hypoxia-induced expression of the kinase-dead unc-51 like autophagy activating kinase (ULK1) mutant K46N increased lung metastases in MDA-MB-231 xenograft mouse models. Consistent with this effect, expressing a dominant-negative mutant of ULK1 or ATG4b or a ULK1-targeting shRNA facilitated cell migration in vitro. Functional proteomic and transcriptome analysis revealed that loss of hypoxia-regulated autophagy promotes metastasis via induction of the fibronectin integrin signaling axis. Indeed, loss of ULK1 function increased fibronectin deposition in the hypoxic TME. Together, our results indicated that hypoxia-regulated autophagy suppresses metastasis in breast cancer by preventing tumor fibrosis. These results also suggest cautions in the development of autophagy-based strategies for cancer treatment.
Keywords: Autophagy, Hypoxia, Tumor Microenvironment, Metastasis, Breast Cancer
INTRODUCTION
The tumor microenvironment (TME) is frequently subject to fluctuating oxygen concentrations due to varying access to vasculature. Tumor cells within the hypoxic TME respond to oxygen-deficiency through global gene expression alterations, mediated primarily by the transcription factor, hypoxia-inducible factor-1 (HIF1). The effects of hypoxia, and the corresponding increase in HIF1α expression, on cancer progression have been well studied(1). In a clinical context, hypoxia within the TME is associated with poor prognosis due to increased angiogenesis, chemo-resistance, and metastatic potential (2,3).
Similarly, it has become clear that autophagy plays paradoxical roles in tumorigenesis. In general, autophagy provides a survival mechanism that gives cancer cells metabolic flexibility, allowing for their survival in nutrient and oxygen poor TMEs (4,5). Here, autophagy catabolizes malfunctioning proteins and organelles, yielding nutrients that can be recycled and reallocated to essential cellular pathways, allowing for tumor progression. Conversely, autophagy also removes reactive oxygen species, which prevents genomic instability and cancer driving mutations(6). Indeed, autophagy has been shown to be activated in response to hypoxia and ischemia, and may be a driving mechanism behind HIF1-associated survival and chemo-resistance (7,8).
A growing body of research has begun exploring the effects of hypoxia-regulated autophagy on tumor progression in efforts to better understand the biological dynamics associated with the hypoxic TME. Several of these studies have suggested hypoxia induced autophagy increases tumor cell resistance to chemotherapy and promotes cell survival (9–12). However, these investigations are dependent on in vitro model systems, in which cells undergo an “all-or-none” hypoxia treatment that does not accurately represent the dynamic and heterogeneous nature of the hypoxic TME. Similarly, due to the context-dependent nature of autophagy, traditional autophagy inhibitory strategies that do not incorporate TME dynamics may not be ideal for examining the role of autophagy within the TME. Taken together, a more physiologically accurate assessment which incorporates the contextual nature of hypoxia-regulated autophagy on tumor biology has remained elusive.
Accordingly, we investigated the role of autophagy within the hypoxic TME of a triple negative breast cancer model using a novel hypoxia-dependent reversible dominant-negative ULK1 (dnULK1) gene expression strategy to physiologically modulate autophagy at single cell levels. ULK1 (Unc-51 like kinase 1) is the only autophagy gene to encode a serine/threonine kinase, and forms a complex with multiple regulatory subunits, comprising of ATG13 and FIP200. Importantly, ULK1 has been well studied in the context of nutrient and oxygen deprivation, in which the mechanistic target of rapamycin complex 1 (mTORC1) negatively regulates ULK1 through hyper-phosphorylation in nutrient-rich environments; while AMP-activated protein kinase (AMPK) binds and phosphorylates ULK1 to activate autophagy under hypoxic and nutrient deprivation (13,14). Thus, ULK1 plays a critical up-stream role in regulating autophagy in response to hypoxic conditions, and provides an excellent target for investigating hypoxia-regulated autophagy. Here, we demonstrate that the inhibition of autophagy selectively in hypoxic tumor cells through HIF1α dependent expression of dnULK1 increases breast cancer metastasis to the lung, and provide a comprehensive overview of the underlying mechanisms driving metastasis within the hypoxic TME.
MATERIALS AND METHODS
Cell Lines
Human cancer cell lines MDA-MB-231 and HT-1080 were purchased from ATCC (Rockville, MD), and ULK1−/− mouse embryonic fibroblasts (MEFs) were a kind gift from Dr. Mondira Kundu (St. Jude Children’s Research Hospital, Memphis, TN). All cell lines were passaged in our laboratory for fewer than 6 months before use and periodically authenticated by morphologic inspection, growth curve analysis, and mycoplasma testing.
Orthotopic Xenograft mouse model of breast cancer using MDA-MB-231 cells
All animal studies were performed according to the guidelines established by the Institutional Animal Care and Use Committee (IACUC) at the Penn State College of Medicine. An orthotopic xenograft breast cancer model was generated by injecting 2.0×106 MDA-MB-231 cells into the inguinal mammary fat pad of female NOD SCID Gamma (NSG) mice, aged 6–8 weeks. Cells were injected in a 50:50 mixture of PBS and matrigel basement membrane matrix (Fisher Scientific; cat#CB-40234). Mice were imaged for luciferase expression on a weekly basis for 7 weeks via a Xenogen IVIS bioluminescent imager. Mice were injected with 5ul/gram body weight of 30mg/ml Luciferin-D (Gold Biotechnology, cat# LUCK-1G) in PBS, 5 minutes prior to imaging. Photon flux was calculated using region of interest (ROI) measurements of either the primary tumor site, or the ventral thoracic area for lung metastasis. Tumor volume was also measured using calipers and calculated as πLW2/6. At the experiment end point, mice were euthanized and tumor, lung, and liver tissue were harvested for ex vivo analysis and subsequent histology.
In vivo tail-vein metastasis assay
Lung seeding assays were performed by injecting 5.0×105 MDA-MB-231 cells into the tail vein of 6 to 8 week old female NSG mice. Mice were imaged on the ventral side for luciferase expression on a weekly basis for 7 weeks via Xenogen IVIS imaging, the same way described above. At the experiment end point, mice were euthanized and tumor, lung, and liver tissue were harvested for ex vivo analysis and subsequent histological analysis.
Immunoblotting
Cells were cultured in normal culture medium under normoxia (21% O2) or hypoxia (1% O2) conditions for the indicated times. Cells were subsequently lysed in RIPA lysis buffer containing protease inhibitors and subjected to immunoblotting with primary antibodies: Flag (sigma, cat#F1804_200UG); p62 (American Research Products, cat#03-GP62-C); HIF1 (Cell Signaling, cat#3716S); β-actin (Sigma-Aldrich, cat#A5441); mCherry (Abcam, cat#ab125096); Fibronectin (Abcam, cat#ab2413); Tom20 (Santa Cruz, cat#sc-11415); LOX (abcam, cat#ab174316); HSP60 (Cell signaling, cat#12165P) and detection with a Li-Cor Odyssey CLx Imager.
Immunohistochemistry
Tissues from NSG mice were subjected to immunohistochemical analysis as described previously(15). For detection of hypoxic areas, mice were treated with an intraperitoneal injection of hypoxyprobe-1 (60mg/kg Hypoxyprobe, cat# HP1-100), one hour prior to euthanasia. Hypoxyprobe was subsequently detected using the antibody specific to pimonidazole adducts (Hypoxyprobe, cat# HP1-100).
qPCR quantification
Peripheral blood from orthotopic xenograft mouse models was taken biweekly. Circulating DNA was isolated from 100µL of each plasma sample using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Cat. 55114.). The amount of human derived DNA in each sample was quantified via real time real-time PCR (QuantStudio 12K Flex) using a human specific FOXP2 gene primer/probe set (Life Technologies, cat#4400291). ULK1 gene expression was assessed in MDA-MB-231 cells using ULK1 gene primer/probe set (Life Technologies, cat#4331182)
Fluorescent microscopy and phalloidin-mediated actin labeling
After fixing in 4% paraformaldehyde for 1 min, followed by permeabilization with 0.1% Triton X-100 for 5 minutes, cells were stained with primary antibody or Alexa Flour 488 Phalloidin (Life Technologies, cat# A12379) in PBS and 1% bovine serum albumin (BSA) for 20 minutes at room temperature. Cells were mounted with ProLong Gold antifade reagent with DAPI (Invitrogen, cat#P36936) and visualized on an Olympus IX81 microscope. Lamellipodia were quantified by counting the number of cells displaying lamellipodia within a field at 200× magnification, and averaging 15 randomly selected fields.
Wound Healing Assay
Confluent cells were scratched with a sterile P1000 pipette tip to generate a defined scratch wound. Detached cells were removed by washing with PBS prior to incubation in compete medium in either normoxia or hypoxia. Images were taken using a digital camera mounted on an Olympus CKX41 microscope every 24 hours at fixed locations on the plate and compared to the starting position. The total distance was quantified using imageJ software.
Boyden Chamber Invasion Assay
Boyden Chambers (Corning BioCoat Matrigel Invasion Chambers, Cat#354480) were prepared according to the manufacturer’s instructions. MDA-MB-231 Cells were placed in either Normoxia (21% O2) or Hypoxia (1% O2) for 48 hours and subsequently fixed in 10% formaldehyde and stained with 0.05% Crystal Violet. The amount of invaded cells was quantified using imageJ software.
Functional proteomic analysis via reverse phase protein array (RPPA)
The expression levels of 298 proteins were assessed by Reverse phase protein array (RPPA) through the MD-Anderson cancer center RPPA core facility. Proteins significantly (p<0.05) enriched in the HRE-dnULK1 tumors compared with control tumors were submitted to Ingenuity pathway analysis (IPA) software (IPA, QIAGEN Redwood City, www.qiagen.com/injenuity) to generate network-module gene set analysis to visualize previously described relationships among the proteins.
RNA Isolation, Library Preparation and Sequencing
Total RNA from cells was isolated with mirVana™ miRNA isolation lysis buffer (Ambion, cat#8540G21). RNA integration number (RIN) was measured using an Agilent 2100 Bioanalyzer RNA 6000 Nano kit to confirm RIN above 7. cDNA libraries were prepared using the NEXTflexTK Rapid RNA Sequencing kit (BioO Scientific) as per the manufacturer’s instructions. The denatured libraries were diluted to 10 pM by pre-chilled hybridization buffer and loaded onto a TruSEQ Rapid flow cell on an Illumina HiSeq 2500 and run for 50 cycles using a single-read recipe according to the manufacturer’s instructions.
mRNA Sequencing Data Processing and Pathway Analysis
De-multiplexed sequencing reads passing the default purify filtering of Illumina CASAVA pipeline (released version 1.8) were quality trimmed/filtered using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit) by having a quality score cutoff of 20. Filtered reads were aligned to human reference genome (hg38) using Tophate (v2.0.9) by allowing up to 2 mismatches(16). FPKM values were calculated using Cufflinks v2.0.2 provided with the Ensemble gene annotation (release 78)(17). Ingenuity Pathway Analysis (IPA, QIAGEN Redwood City, www.qiagen.com/ingenuity) was used to identify functionally relevant signaling pathways in differentially expressed genes and proteins.
Statistics
Data were analyzed using Graph Pad Prism and statistical software SAS version 9.4 (SAS Institute, Cary, NC, USA). Student t-test (two-tailed) was used for single comparisons. Group differences were evaluated using ANOVA or repeated-measure ANOVA models. Data were considered statistically significant when p<0.05.
RESULTS
HRE-dnULK1 provides hypoxia-dependent suppression of autophagy
In order to investigate the role of hypoxia-regulated autophagy within the TME, we generated a plasmid vector utilizing the hypoxia response element (HRE) promoter to regulate expression of a dominant-negative ULK1(K46N) gene (dnULK1), to allow for hypoxia-dependent inhibition of ULK1 (Figure 1a). A construct containing the HRE promoter, but lacking the dnULK1 gene, was also included as a negative control. With this, naturally occurring hypoxic regions generated by tumor growth will activate the HRE-dnULK1 construct, allowing for physiological inhibition of the essential autophagy kinase, ULK1. Likewise, if a hypoxic tumor cell regains access to oxygen, the construct will be turned off, allowing endogenous ULK1 to reactivate to restore autophagic function (Supplementary figure 1a). To validate this, upon transduction of the HRE-dnULK1 construct into a triple negative breast cancer cell line, MDA-MB-231; the expression of dnULK1 and mCherry were verified to be dependent upon hypoxic exposure, and correlate with HIF1α expression (Figure 1a–b, Supplementary figure 1b). Hypoxia-induced autophagy was monitored through several established autophagy markers, namely LC3 lipidation and p62 accumulation (18). The HRE-dnULK1 construct was verified to inhibit autophagy under hypoxic conditions, as evident by a significant level of p62 protein accumulation under hypoxic conditions, as compared to control cells (p<0.05) (Figure 1c–e). While LC3 lipidation was not blocked by HRE-dnULK1, an increase in both LC3-I and LC3-II suggest reduced autophagy flux. Furthermore, degradation of two mitochondrial markers, HSP60 and Tom20, were found to be blocked in HRE-dnULK1 expressing cells in hypoxic conditions, indicating a suppression of mitophagy (Figure 1c). Together, these results indicate that HRE-dnULK1 provides a system that allows for hypoxia inducible suppression of autophagy.
Figure 1. Hypoxia induced expression of dnULK1 suppresses autophagy in MDA-MB-231 cells, in vitro.
a, Schematic representation of plasmid vectors expressing the hypoxia-inducible dominant negative ULK1 gene (HRE-dnULK1), and the hypoxia inducible blank control (HRE-Empty). Each vector contains hypoxia dependent mCherry expression via an internal ribosomal entry site (IRES). b, Immunoblot assessing hypoxia inducible expression of dnULK1 and mCherry. c, Immunoblot of autophagy markers in HRE-dnULK1 in in MDA-MB-231 cells compared to HRE-Empty control at normoxia (N), Hypoxia (1% O2) for 48 hours (H), and reoxygenation for 24 hours after hypoxia (H/R). d, Quantification of p62 accumulation from three independent experiments using Image Studio Digits Software (*p<0.05, t-test). e, Immunofluorescent staining of p62 in MDA-MB-231 cells expressing HRE-Empty or HRE-dnULK1 in Normoxia or Hypoxia (1% O2) for 48 hours.
Suppression of ULK1 in the hypoxic TME increases metastasis of MDA-MB-231 cells to the lungs in xenograft mouse models
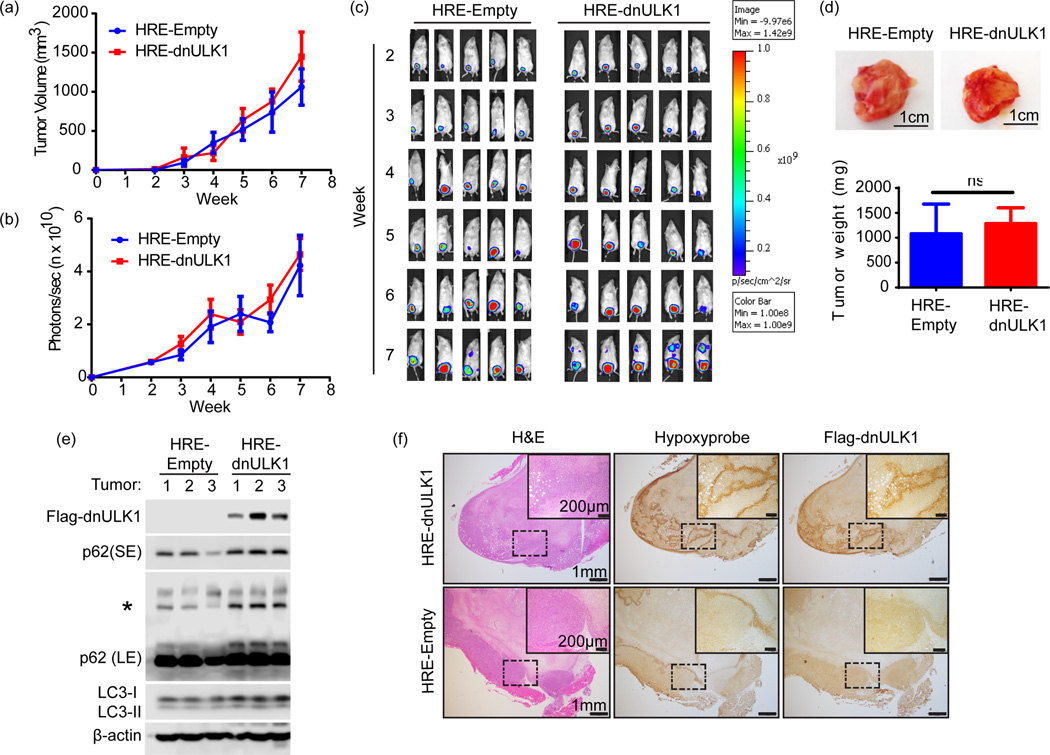
To assess the effects of hypoxia-regulated autophagy in a physiological setting, MDA-MB-231 breast cancer cells harboring either the HRE-Empty or HRE-dnULK1 construct were injected into the inguinal mammary fat pad of NSG mice, generating an orthotopic xenograft mouse model of triple negative breast cancer. A luciferase reporter gene was also introduced into each cell type to allow for noninvasive monitoring of tumor growth and metastasis. Measurements of growth of the primary tumor site revealed no significant difference between the HRE-dnULK1 and HRE-Empty groups (Figure 2a–c). Ex vivo analysis of xenograft tissues confirmed that the primary tumor size remained similar between experimental groups (Figure 2d). Importantly, primary tumors expressing the HRE-dnULK1 construct contained higher levels of p62 accumulation as well as an increase in LC3-I, indicating inhibition of autophagy in vivo (Figure 2e). Furthermore, the expression of dnULK1 within tissues strongly correlated to hypoxic areas within the primary tumors, demonstrating dnULK1 expression is indeed hypoxia-dependent in vivo (Figure 2f).
Figure 2. Loss of ULK1 function in the hypoxic tumor cells does not affect primary tumor growth.
a, Tumor volume of primary xenograft tumors over 7 weeks (mean ± s.d, n=15 mice per group). b, Quantification of primary tumor luciferase expression photon flux (photons/sec) in HRE-dnULK1 and HRE-Empty xenografts (mean ±SEM, n=15 mice per group). c, Representative images of luciferase expression from primary xenograft tumors over a 7 week time period. d, Representative images of tumors from HRE-dnULK1 and HRE-Empty orthotopic xenograft mice and quantification of final weight of excised primary tumors (mean ± s.d, n=15, ns=no significance, t-test). e, Immunoblot analysis of dnULK1 and p62 expression from 3 primary tumors from HRE-empty and HRE-dnULK1 xenografts (SE= short exposure, LE = Long Exposure, Asterisks (*)= p62 aggregate). f, Immunohistochemistry staining of primary tumors and lung serial sections from HRE-dnULK1 xenografts at week 7. Hypoxyprobe™ detects hypoxic areas within each tissue, which correlates to dnULK1 expression.
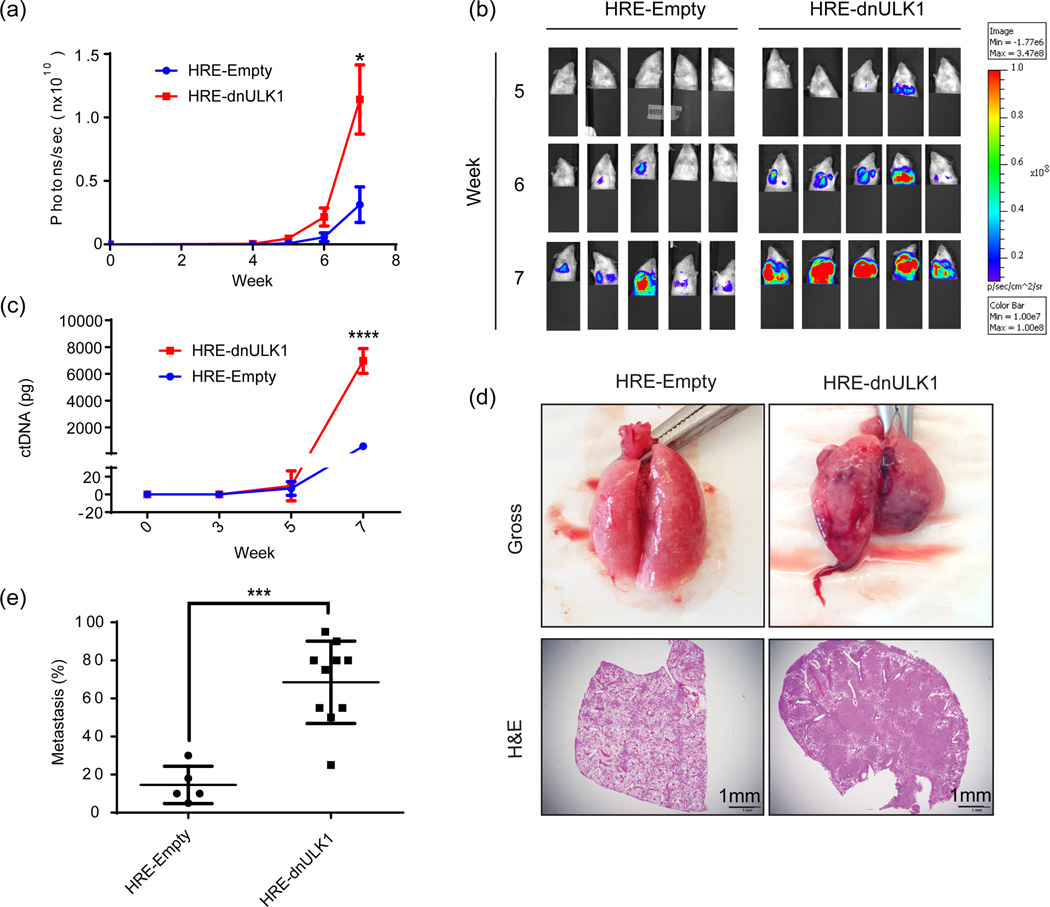
Although primary tumor growth remained similar between the HRE-Empty and HRE-dnULK1 groups, unexpectedly, metastasis from the primary tumor site to lung tissue was significantly higher in mice containing HRE-dnULK1 expressing tumors at the week 7 time point (p<0.05) (Figure 3a–b). A marked increase in circulating tumor DNA (ctDNA) levels was also detected at this point in HRE-dnULK1 mice (p<0.0001)(Figure 3c), indicating higher levels of intravasation of tumor cells and/or circulating tumor cells in the HRE-dnULK1 group. Gross tissue and histological analysis confirmed an increase in metastasis to the lungs in the HRE-dnULK1 group (Figure 3d–e). Interestingly, tumor growth within the lung generated a hypoxic TME; which in turn, activated the HRE-dnULK1 construct within the lung tissue (Supplementary figure 2a). A trend of increased metastasis of HRE-dnULK1 tumors to the liver was also observed; however, quantification of liver metastasis through histological analysis did not reach statistical significance between the groups (Supplementary figure 2b).
Figure 3. Loss of ULK1 function in the hypoxic TME increases metastases of MDA-MB-231 xenografts to the lungs.
a, Quantification of metastasis of MDA-MB-231 xenografts to the lungs via luciferase expression (photons/sec) in the thoracic cavity (mean ± SEM, n=15, *p<0.05, t-test). b, Representative images of luciferase expression in MDA-MB-231 xenograft mice within the thoracic cavity in weeks 5 through 7, of a 7 week time course. Primary tumor signal was shielded to allow luciferase signal in the lung cavity to be measured. c, Quantitative PCR analysis of circulating MDA-MB-231 xenograft DNA (mean ± s.d, n=3, ****p<0.0001, t-test). d, Representative gross lung tissue and H&E stains from HRE-Empty and HRE-dnULK1 MDA-MB-231 xenografts at week 7 illustrating the degree of metastasis to the lung. f, Quantification of lung metastasis as measured by the percent of tumor tissue within a lung section (mean ± s.d, n=5 (HRE-Empty);10 (HRE-dnULK1), ***p<0.001, t-test).
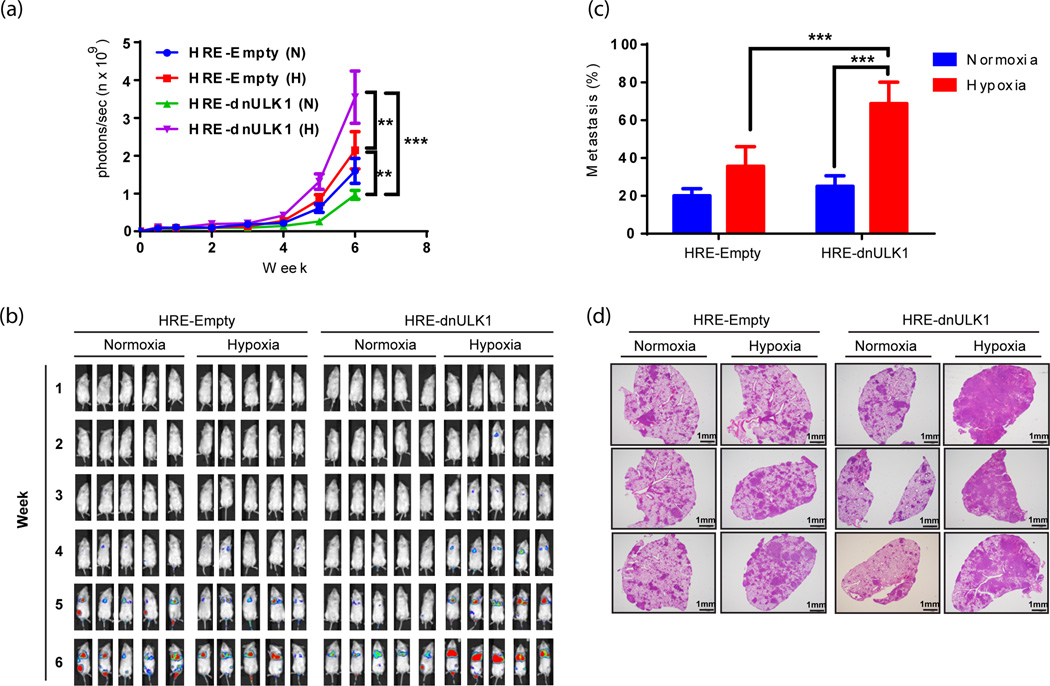
Metastasis is a multistep process that involves: (1) intravasation of tumor cells into the blood stream or lymphatic circulation, (2) survival of tumor cells in circulation, (3) extravasation out of the blood stream, and (4) engraftment and growth at the secondary site. To further assess how hypoxia-regulated autophagy influences metastasis, we performed an in vivo lung seeding assay which allows the intravasation step of metastasis to be bypassed. Thus, the effects of hypoxia-regulated autophagy on survival in the blood stream and engraftment into the lungs can be assessed. Here, HRE-Empty and HRE-dnULK1 MDA-MB-231 cells cultured in normoxic conditions prior to tail-vein injection yielded similar levels of lung metastasis, as detect by luciferase expression in the thoracic cavity (Figure 4a–b). However, pre-incubation of the two respective cell lines in hypoxic conditions for 48hrs prior to injection resulted in a significant increase in lung engraftment in the HRE-dnULK1 expressing group (p<0.01)(Figure 4a–b). This verifies that the hypoxia activated dnULK1 gene is indeed causing the metastatic phenotype, and that the loss of autophagy under hypoxic conditions increases engraftment into the lungs. Histological analysis of lung tissue from these groups verified this finding, as the hypoxia-incubated HRE-dnULK1 group exhibited higher levels of tumor engraftment at significant levels (p<0.001) (Figure 4c–d).
Figure 4. Loss of hypoxia-regulated autophagy increases engraftment of MDA-MB-231 cells into the lung.
MDA-MB-231 cells expressing HRE-Empty or HRE-dnULK1 were pre-incubated in normoxia (N) or hypoxia (H) (1% O2) for 48 hours and subjected to tail vain injection into NSG mice (10 mice per group). a, Quantification of metastasis of MDA-MB-231 xenografts to the lungs via luciferase expression (photons/sec) (mean ± SEM, n=10, **p<0.01, ***p<0.001, repeated-measure ANOVA). b, Representative images of luciferase expression from MDA-MB-231 HRE-dnULK1 and HRE-Empty cell lines within the lungs of NSG mice over a 6 week time period. c, Quantification of lung metastasis as measured by the percent of tumor tissue within a lung section (mean ± s.d, n=8 per group (4 for HRE-dnULK1 at 1% 02), ***p<0.001, t-test). d, Representative images of hematoxylin and eosin (H&E) stained lung tissues.
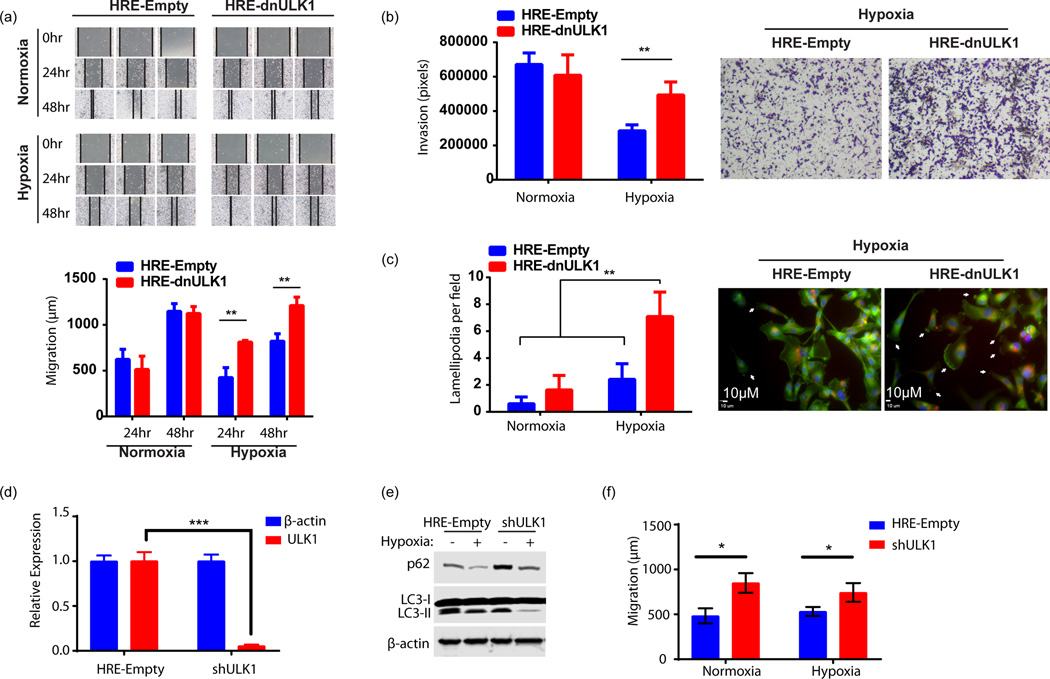
Inhibition of hypoxia-mediated autophagy increases migration and invasion in vitro
To corroborate the increased metastasis exhibited by MDA-MB-231 cells expressing the HRE-dnULK1 construct in vivo, we aimed to replicate this phenotype in vitro. Accordingly, migration and invasion of MDA-MB-231 cells were assessed using wound healing and invasion assays, respectively. In both cases, incubation of HRE-dnULK1 expressing MDA-MB-231 cells in hypoxia increased migration and invasion compared to the HRE-Empty counterpart (Figures 5a–b). This indicates that the loss of autophagy can increase the motility of MDA-MB-231 cells, as well as their ability to invade, which may facilitate both intravasation and extravasation. Furthermore, there was a significant increase (p<0.01) in lamellapodia formation observed in HRE-dnULK1 expressing MDA-MB-231 cells in hypoxia, indicating these cells exhibit a morphology conducive to migration and characteristic of mesenchymal migration and metastasis (Figure 5c)(19). As an alternative approach, shRNA targeting ULK1 (shULK1) was used to confirm that the loss of ULK1 function indeed results in an increase in migration in both hypoxic and normoxic conditions (Figure 5d–f). Furthermore, we targeted an additional autophagy gene, ATG4b, to determine whether the increase in migration is a ULK1-depedent phenotype. The protease ATG4b lies downstream of ULK1 and is required for processing pro-LC3 prior to lipidation, a key step in autophagosome biogenesis. Here, expression of a dominant negative ATG4b gene (mStrawberry-ATG4b(C74A)), was verified to suppress autophagic flux as evident by increased p62 levels (Supplementary figure 3a). Indeed, expression of dnATG4b increased migration of MDA-MB-231 cells compared to empty control cells (p<0.05; Supplementary figure 3b). Together, these results indicate the loss of autophagy in hypoxia promotes migration and invasion of MDA-MB-231 cells through mesenchymal associated mobility.
Figure 5. Loss of autophagic function in hypoxia increases invasion and migration, in vitro.
a, Wound healing assay assessing migration of MDA-MB-231 cells expressing either HRE-Empty or HRE-dnULK1 over 48hrs under hypoxic (1% O2) and normoxic conditions. Quantification was performed by measuring the distance between the two leading edges using imageJ software (mean ± s.d., n=3, **p<0.01, t-test). b, Transwell migration assay assessing invasion of MDA-MB-231 cells expressing either HRE-Empty or HRE-dnULK1 through a matrigel matrix over 48hrs under hypoxic (1% O2) and normoxic conditions. Quantification was performed using imageJ software (mean ± s.d., n=4, **p<0.01, t-test). c, Assessment of actin cytoskeletal dynamics by Alexo flour 488 conjugated phalloidin staining of MDA-MB-231 cells expressing either HRE-Empty or HRE-dnULK1 incubated in either normoxia and hypoxia (1% O2) (200× Magnification) (Blue = DAPI, Red=mCherry, Green=Alexa flour 488). Observed lamellapodia are indicated by white arrows. Lamellapodia were quantified by counting the number of cells displaying lamellapodia within a field, and averaging 15 randomly selected fields (mean ±s.d, **p<0.01, t-test). d, qPCR Assessment of ULK1 gene knockdown by shULK1 in MDA-MB-231 cells (mean ±s.d, n=3,***p<0.001, t-test). e, Immunoblot assessing autophagy markers in shRNA MDA-MB-231 cells versus HRE-Empty control. f, Wound healing assay assessing migration of MDA-MB-231 cells expressing either HRE-Empty or shULK1 over 24hrs under normoxic and hypoxic (1% O2) conditions. Quantification was performed by measuring the distance between the two leading edges using imageJ software (mean ± s.d., n=3 (normoxia) and 6 (hypoxia), *p<0.05, ns=no significance, t-test).
To expand these findings into an additional cell line, we examined the effects of suppressing hypoxia-induced autophagy by HRE-dnULK1 on cell migration in HT-1080 cells. Expression of dnULK1 was verified to be hypoxia- and HIF1α-dependent, and provided suppression of autophagy in hypoxia as evident by p62 accumulation compared to HRE-Empty control cells (Supplementary figure 3c). Migration of HRE-dnULK1 expressing HT-1080 cells was subsequently assessed in normoxia and hypoxia using a wound healing assay. As observed in the MDA-MB-231 cell line, there was no significant difference between HRE-dnULK1 and HRE-Empty HT-1080 migration when incubated in normoxia. However, HRE-dnULK1 expressing cells migrated significantly further than HRE-Empty cells over 24 hours in hypoxia (p<0.05) (Supplementary figure 3d). Thus, the finding that suppression of hypoxia-induced autophagy by HRE-dnULK1 promotes migration is not limited to MDA-MB-231 cells.
Loss of ULK1 function in hypoxia increases fibronectin deposition and reprograms metastatic signaling networks
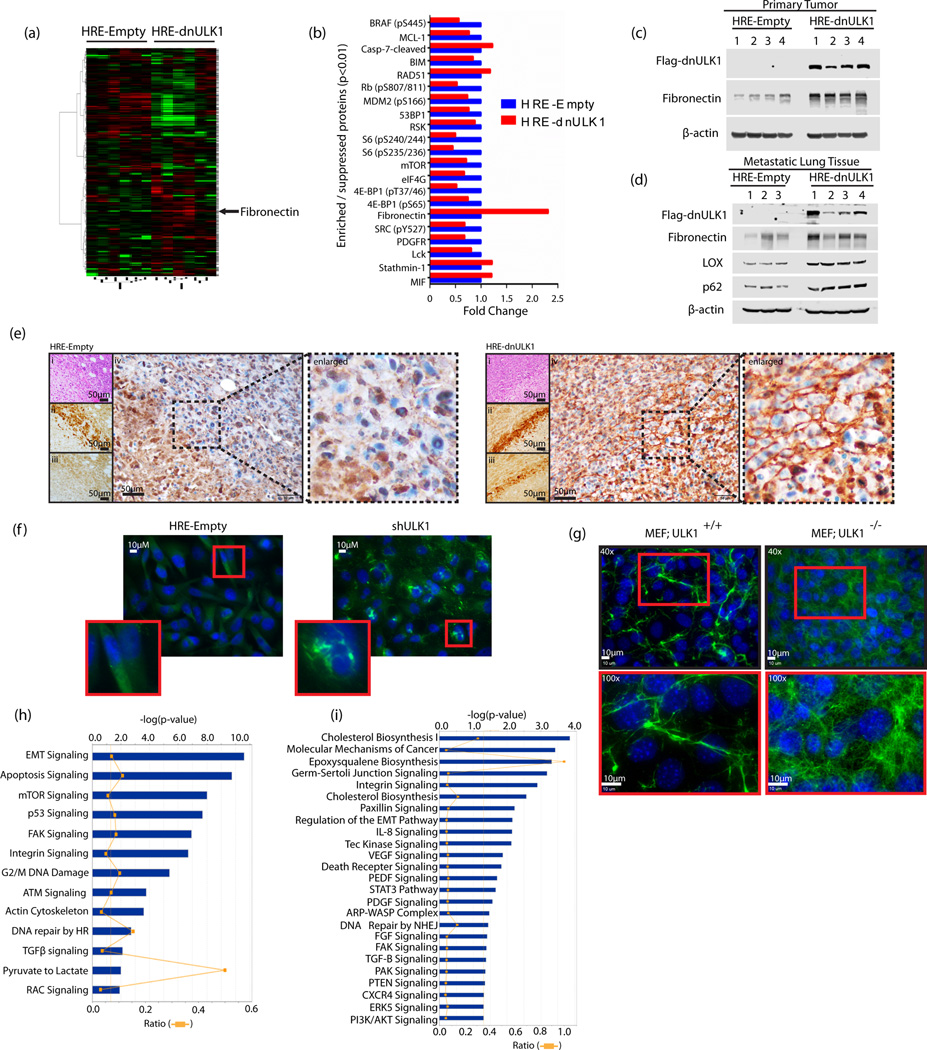
To identify the mechanisms underlying the metastatic phenotype exhibited by HRE-dnULK1 expressing cells, we performed both functional proteomic and transcriptome analysis to yield a comprehensive overview of signaling pathways involved in cancer metastasis. First, the expression of 298 proteins was assessed in primary tumors from MDA-MB-231 orthotopic xenografts using reverse phase protein array (RPPA) analysis (Figure 6a). A total of 41 proteins displayed significant enrichment or suppression (p<0.05) in HRE-dnULK1 expressing tumors compared to the HRE-Empty control (Supplementary figure 4a–e). Of these, 21 proteins reached higher significance thresholds (p<0.01), signifying a high degree of confidence for their involvement in metastasis from suppression of hypoxia-regulated autophagy (Figure 6b). In particular, fibronectin displayed the greatest enrichment with an over two-fold increase in protein expression across 6 HRE-dnULK1 tumors compared to 6 HRE-empty tumors. Enrichment in fibronectin was verified in primary tumor tissues, and was also found to be enriched in the metastatic lung tissue of HRE-dnULK1 tumors, along with p62 (figure 6c–d). Importantly, a pronounced increase in fibronectin staining was found in the extracellular matrix (ECM) of HRE-dnULK1 xenograft tumors in areas corresponding to hypoxia-induced expression of dnULK1 (Figure 6e). This increase in fibronectin in hypoxic areas was absent in HRE-Empty tumors, indicating hypoxia-induced dnULK1 promotes fibronectin deposition in vivo. A similar effect was found in vitro, where hypoxic ULK1 knockdown MDA-MB-231 cells exhibited increased fibronectin deposition (Figure 6f). Moreover, this effect was very pronounced in fibroblast cells, as MEF cells lacking ULK1 displayed dense networks of fibronectin between cells compared to wildtype control (Figure 6g). This increase in fibronectin was less dramatic in both MDA-MB-231 and MEF cells incubated at normoxia, indicating hypoxia-regulated autophagy in particular is critical in preventing tumor fibrosis (Supplementary figure 3e–f).
Figure 6. Functional proteomic and transcriptomic assessment of hypoxia-regulated autophagy.
a, Functional Proteomic analysis by reverse phase protein array (RPPA) comparing expression levels of 298 proteins in 6 HRE-dnULK1 primary xenograft tumors and 6 HRE-Empty primary xenograft tumors. b, Proteins displaying highly significant enrichment or suppression (p<0.01) in HRE-dnULk1 primary xenograft tumors compared to HRE-Empty primary xenograft tumors. c, Immunoblot analysis of primary tumor protein lysate from HRE-dnULK1 and HRE-Empty orthotopic xenografts. d, Immunoblot analysis of metastatic lung tissue protein lysate from HRE-dnULK1 and HRE-Empty orthotopic xenografts. e, Immunohistochemistry staining of primary tumors from HRE-Empty and HRE-dnULK1 xenografts at week 7 with Hematoxylin and Eosin (i), Hypoxyprobe (ii), Flag (Flag-dnULK1) (iii), and Fibronectin (iv). All images taken at 40× magnification. f, Immunofluorescent staining of fibronectin in the indicated MDA-MB-231 cells incubated in hypoxia (1% O2) for 48 hours. g, Immunofluorescent staining of fibronectin in MEF wildtype cells (MEF;ULK1+/+) and MEF ULK1 knockout cells (MEF;ULK1−/−) incubated in hypoxia (1% O2) for 48 hours. h, Canonical pathway analysis of significantly enriched or suppressed proteins (p<0.05) in RPPA analysis. i, Canonical pathway analysis of significantly enriched or suppressed (p<0.05) genes in HRE-dnULK1 MDA-MB-231 cells in hypoxia (1% O2) compared to HRE-Empty MDA-MB-231 cells in hypoxia (1% O2) as assessed through RNAseq analysis.
Subsequently, input of significantly modified proteins (p<0.05) from RPPA analysis into pathway analysis revealed enrichment in integrin signaling and metastasis related pathways (i.e. EMT signaling, FAK signaling, Actin Cytoskeleton signaling, TGF-β signaling, Rac signaling) (Figure 6h). A reduction in mTOR activity and protein synthesis was also observed, as well as an increase in DNA damage and cell cycle arrest pathways (p53 signaling, G2/M DNA damage checkpoint signaling, ATM signaling). To gain further insight, RNAseq was performed to evaluate global gene expression changes in HRE-dnULK1 MDA-MB-231 cells in hypoxia compared to HRE-Empty MDA-MB-231 cells in hypoxia. Genes that were significantly upregulated and downregulated (p<0.05) were used to analyze pathway enrichment in HRE-Empty and HRE-dnULK1 groups under hypoxia. Pathway analysis revealed HRE-dnULK1 expressing cells exhibited enrichment in many pathways known to be involved in cytoskeletal remodeling, migration, and metastasis. Specifically, we observed enrichment in integrin signaling, paxillin signaling, regulation of EMT pathways, Tec Kinase Signaling, IL8 signaling, VEGF Signaling,, ARP-WASP complex signaling, FGF signaling, FAK signaling, TGF-β signaling, PAK signaling, and CXCR4 signaling (Figure 6i). In addition, we also observed strong enrichment in cholesterol biosynthesis, which is known to promote metastasis in breast cancer, amongst others(20,21). Genes showing significant enrichment (p<0.05) that were major contributors to these pathways are presented (supplementary figures 4f–h).
Lastly, compounding candidate molecules from both proteomic and transcriptomic analyses into a single network analysis indicated fibronectin is acting as a central node connecting many of the genes found to have significant expression differences, suggesting fibronectin may be acting as a driver of metastasis through the activation of integrin signaling pathways (Supplementary figure 4i). In addition, previous reports have found the secretion of LOX/LOXL4 is strongly implicated in breast cancer metastasis and pre-metastatic niche formation (22–24). Indeed, these molecules were found to be significantly upregulated in the both the RNA expression and metastatic lung tissue in HRE-dnULK1 group, and may be key contributors to extravasation and engraftment into the pulmonary tissue (Supplementary figure 4g and figure 6d). Altogether, these data indicate that the loss of autophagy within the hypoxic TME promotes fibronectin deposition in the TME and subsequent stimulation of integrin signaling and migratory pathways.
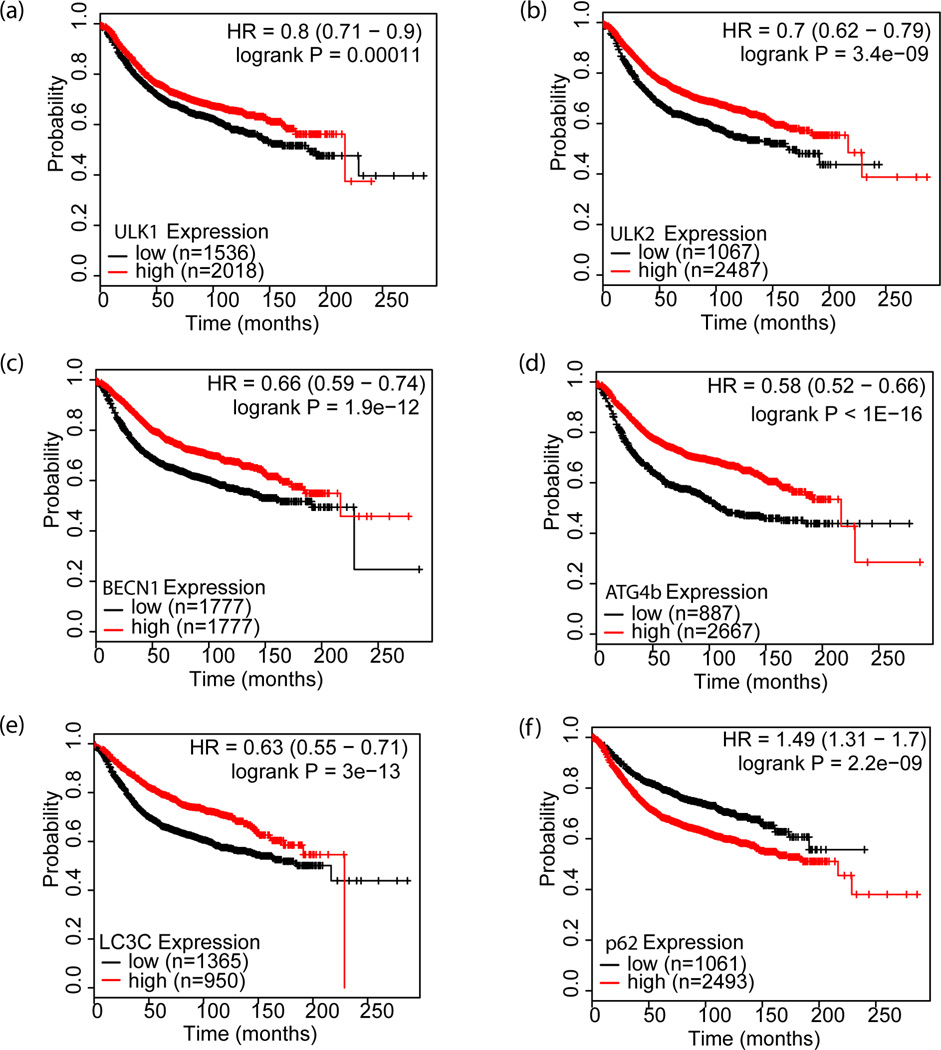
Reduced autophagy gene expression predicts poor prognosis in breast cancer
As our results indicate that the loss of autophagy in the TME may promote breast cancer progression by increasing metastasis, we investigated whether autophagy gene expression correlates with prognosis in human breast cancer patients using the Kaplan-Meier Plotter database(http://kmplot.com) which has the ability to assess the effect of over 54,000 genes on the survival and relapse free survival (RFS) of 4,142 breast cancers (25). We evaluated the potential predictive value of 5 autophagy genes (ULK1, ULK2, BECN1, ATG4b, and LC3C) involved at distinct steps of autophagosomes biogenesis in the relapse-free survival (RFS) of breast cancer patients. As shown in Figure 7a–e, the patient group with low expression of the 5 genes had significantly poorer RFS. Similarly, the patient group with high expression of the autophagy substrate, p62, also showed a significantly reduced RFS (Figure 7f). These results indicate that the reduced autophagic activity in human breast cancer predicts worse prognosis, and thus supports our findings that the loss of autophagy within the hypoxic TME promotes breast cancer progression via metastasis.
Figure 7. Kaplan-Meier survival curves relative to autophagy gene expression for breast cancer.
Kaplan-Meier relapse-free survival (RFS) curves of breast cancer patients with high and low expression of autophagy-related genes: a, ULK1; b, ULK2; c, BECN1; d, ATG4b; e, LC3C and f, p62 were analyzed with KM-plotter software (http://kmplot.com/analysis/).
DISCUSSION
The influence of hypoxia on gene expression and cancer progression has long been appreciated. Likewise, autophagy has emerged as a powerful cellular process that can greatly influence how cancer cells respond to stress and the microenvironment as a whole. Here, we assessed the role of autophagy in the hypoxic TME. Specifically, we found that the suppression of hypoxia-induced autophagy increased the metastases of MDA-MB-231 cancer cells to the lung using two separate mouse model approaches. Interestingly, the primary tumor sizes remained similar between HRE-Empty and HRE-dnULK1 xenografts, indicating that overall growth rate was not affected by the loss of hypoxia-regulated autophagy. Although autophagy has been previously found to effect cellular migration and invasion, the definitive role of autophagy in cell movement remains contentious, as autophagy can act as both a suppressor(26–30) and a promoter(31–34) of migration and metastasis. This is in agreement with a growing body of evidence displaying a context dependent nature of autophagy, and suggests the influence of autophagy on metastasis is dependent on multiple factors. Our approach that allows for physiological regulation of autophagy within the hypoxic TME provides evidence that hypoxia-induced autophagy suppresses metastasis, and thus gives unique and valuable insight into the physiologic role of autophagy within the TME.
To further understand the mechanisms of how inhibition of hypoxia-mediated autophagy promotes metastasis, an assessment of both protein and RNA expression was performed to provide a comprehensive overview of alterations in signaling pathways. Transcriptome analysis indicated that the loss of autophagy under hypoxia stimulates cellular movement pathways through expression of many cytoskeleton remodeling proteins, and activation of integrin signaling and pathways directly associated with cell migration. In vitro assessment of HRE-dnULK1 expressing MDA-MB-231 cells supported this finding, as an increase in migration and invasion was observed, as well as an increase in lamellipodia formation, illustrating cytoskeleton remodeling pathways are indeed being activated. More importantly, ex vivo RPPA analysis of xenograft tumors yielded significant enrichment in many of the same migratory pathways, indicating these pathways are activated in both in vitro and in vivo systems.
Several proteins in particular displayed marked expression changes within the primary tumors, and may be acting as drivers of metastasis. Fibronectin displayed significant enrichment within HRE-dnULK1 MDA-MB-231 primary tumors as well as in the metastatic lung tissue. Moreover, a marked increase in fibronectin deposition was found in the hypoxic TME of xenograft tumors when autophagy is suppressed. Notably, fibronectin expression was not found to be increased through RNAseq analysis, indicating the observed increase in fibronectin levels within the TME is unlikely due to increased gene expression. Thus, the source of fibronectin may be due to a reduction in autophagic degradation or an increase in secretion of fibronectin protein. Fibronectin is a key component of the extracellular matrix (ECM) that is expressed by both cancer-associated fibroblasts (CAFs) and cancer cells, and drives metastasis via mechanotransduction(35–37). Importantly, fibronectin engages integrin receptors leading to the activation of intracellular GTPases for cytoskeletal remodeling and cell motility(35,38). Supporting this, transcriptome and functional proteomic analysis indicated both integrin and several downstream pathway constituents were enriched in HRE-dnULK1 tumors and cell lines. These findings are in agreement with recent studies reporting autophagy to be a regulator of fibrogenesis and fibronectin-mediated migration(26,39–41). Furthermore, autophagy has been shown to modulate cell migration through β1-integrin membrane recycling, in which the loss of autophagy prevents integrin sequestration within autophagosomes and subsequent degradation in lysosomes (26). Thus, we hypothesize that the loss of hypoxia-inducible autophagy promotes metastasis through extracellular remodeling and a fibronectin-integrin signaling axis.
Importantly, the use of physiological regulation of autophagy provides an inherent induction of heterogeneity within the TME. That is, modulation of autophagy can only take place in areas of sufficient hypoxia to activate the suppressing gene (i.e. dnULK1), while leaving autophagy in normoxic cells intact. Thus, this generates a heterogeneous population of autophagy-competent and autophagy-deficient cells within a single tumor, which may create the opportunity for cooperation between the two populations. This may allow for the highly stressed autophagy-deficient tumor cells to be supported by autophagy-competent tumor cells during the process of migration out of a nutrient poor environment. Indeed, previous studies have shown tumor-stromal cell interactions promote tumor progression through different levels of autophagic activity(42,43). However, autophagy-mediated cell cooperativity has yet to be fully investigated between tumor cells, and may elucidate key intercellular networks within the TME. Thus, we provide a promising physiological model system that will allow for future research of tumor cell-tumor cell interactions within the hypoxic TME.
In summation, we used a novel hypoxia-inducible dominant-negative strategy to demonstrate that the suppression of autophagy in the hypoxic TME promotes metastasis both in vitro and in vivo. Gene and protein expression analysis indicate fibronectin and downstream integrin signaling are key mediators of the observed metastatic phenotype. This study provides key insight into the mechanistic interplay of hypoxia-induced autophagy and metastasis within the TME.
Supplementary Material
Acknowledgments
We thank Yuka Imamura (Penn State Hershey Genome Science Facilities) for her technical help with the RNAseq analysis, as well as Sriranga (TJ) Iyyanki and Jay Zhu for their help with large data set analysis and statistics. We are grateful to the Wang lab members who gave insight and technical help. We would like to thank JM Brown (Stanford University) and Mondira Kundu (St. Jude Children's Research Hospital) for providing us the 5×HRE promoter and ULK1−/− MEFs, respectively.
Grant Support: This work was supported by National Institutes of Health Grant CA171501, and by the Lois High Berstler Endowment Fund and the Four Diamonds Fund of the Pennsylvania State University College of Medicine
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
REFERENCES
- 1.Semenza GL. The hypoxic tumor microenvironment: A driving force for breast cancer progression. Biochim Biophys Acta. 2015 doi: 10.1016/j.bbamcr.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Z-J, Semenza GL, Zhang H-F. Hypoxia-inducible factor 1 and breast cancer metastasis. J Zhejiang Univ Sci B. 2015;16:32–43. doi: 10.1631/jzus.B1400221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cosse J-P, Michiels C. Tumour hypoxia affects the responsiveness of cancer cells to chemotherapy and promotes cancer progression. Anticancer Agents Med Chem. 2008;8:790–797. doi: 10.2174/187152008785914798. [DOI] [PubMed] [Google Scholar]
- 4.Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 5.Sato K, Tsuchihara K, Fujii S, Sugiyama M, Goya T, Atomi Y, et al. Autophagy is activated in colorectal cancer cells and contributes to the tolerance to nutrient deprivation. Cancer Res. 2007;67:9677–9684. doi: 10.1158/0008-5472.CAN-07-1462. [DOI] [PubMed] [Google Scholar]
- 6.Kongara S, Karantza V. The interplay between autophagy and ROS in tumorigenesis. Front Oncol. 2012;2:171. doi: 10.3389/fonc.2012.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, et al. Mitochondrial Autophagy Is an HIF-1-dependent Adaptive Metabolic Response to Hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Mazure NM, Pouysségur J. Hypoxia-induced autophagy: cell death or cell survival? Curr Opin Cell Biol. 2010;22:177–180. doi: 10.1016/j.ceb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 9.He W-S, Dai X-F, Jin M, Liu C-W, Rent J-H. Hypoxia-induced autophagy confers resistance of breast cancer cells to ionizing radiation. Oncol Res. 2012;20:251–258. doi: 10.3727/096504013x13589503483012. [DOI] [PubMed] [Google Scholar]
- 10.Hu Y-L, DeLay M, Jahangiri A, Molinaro AM, Rose SD, Carbonell WS, et al. Hypoxia-induced autophagy promotes tumor cell survival and adaptation to antiangiogenic treatment in glioblastoma. Cancer Res. 2012;72:1773–1783. doi: 10.1158/0008-5472.CAN-11-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noman MZ, Janji B, Kaminska B, Van Moer K, Pierson S, Przanowski P, et al. Blocking hypoxia-induced autophagy in tumors restores cytotoxic T-cell activity and promotes regression. Cancer Res. 2011;71:5976–5986. doi: 10.1158/0008-5472.CAN-11-1094. [DOI] [PubMed] [Google Scholar]
- 12.Peng X, Gong F, Chen Y, Jiang Y, Liu J, Yu M, et al. Autophagy promotes paclitaxel resistance of cervical cancer cells: involvement of Warburg effect activated hypoxia-induced factor 1-α-mediated signaling. Cell Death Dis. 2014;5:e1367. doi: 10.1038/cddis.2014.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egan DF, Kim J, Shaw RJ, Guan K-L. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy. 2011;7:645–646. doi: 10.4161/auto.7.6.15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JW, Park S, Takahashi Y, Wang H-G. The association of AMPK with ULK1 regulates autophagy. PloS One. 2010;5:e15394. doi: 10.1371/journal.pone.0015394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi Y, Hori T, Cooper TK, Liao J, Desai N, Serfass JM, et al. Bif-1 haploinsufficiency promotes chromosomal instability and accelerates Myc-driven lymphomagenesis via suppression of mitophagy. Blood. 2013;121:1622–1632. doi: 10.1182/blood-2012-10-459826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinforma Oxf Engl. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 20.Antalis CJ, Uchida A, Buhman KK, Siddiqui RA. Migration of MDA-MB-231 breast cancer cells depends on the availability of exogenous lipids and cholesterol esterification. Clin Exp Metastasis. 2011;28:733–741. doi: 10.1007/s10585-011-9405-9. [DOI] [PubMed] [Google Scholar]
- 21.Nelson ER, Chang C, McDonnell DP. Cholesterol and breast cancer pathophysiology. Trends Endocrinol Metab TEM. 2014;25:649–655. doi: 10.1016/j.tem.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erler JT, Bennewith KL, Nicolau M, Dornhöfer N, Kong C, Le Q-T, et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 23.Erler JT, Bennewith KL, Cox TR, Lang G, Bird D, Koong A, et al. Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell. 2009;15:35–44. doi: 10.1016/j.ccr.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox TR, Bird D, Baker A-M, Barker HE, Ho MW-Y, Lang G, et al. LOX-mediated collagen crosslinking is responsible for fibrosis-enhanced metastasis. Cancer Res. 2013;73:1721–1732. doi: 10.1158/0008-5472.CAN-12-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PloS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tuloup-Minguez V, Hamaï A, Greffard A, Nicolas V, Codogno P, Botti J. Autophagy modulates cell migration and β1 integrin membrane recycling. Cell Cycle Georget Tex. 2013;12:3317–3328. doi: 10.4161/cc.26298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Catalano M, D’Alessandro G, Lepore F, Corazzari M, Caldarola S, Valacca C, et al. Autophagy induction impairs migration and invasion by reversing EMT in glioblastoma cells. Mol Oncol. 2015;9:1612–1625. doi: 10.1016/j.molonc.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chourasia AH, Tracy K, Frankenberger C, Boland ML, Sharifi MN, Drake LE, et al. Mitophagy defects arising from BNip3 loss promote mammary tumor progression to metastasis. EMBO Rep. 2015;16:1145–1163. doi: 10.15252/embr.201540759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Indelicato M, Pucci B, Schito L, Reali V, Aventaggiato M, Mazzarino MC, et al. Role of hypoxia and autophagy in MDA-MB-231 invasiveness. J Cell Physiol. 2010;223:359–368. doi: 10.1002/jcp.22041. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida T, Tsujioka M, Honda S, Tanaka M, Shimizu S. Autophagy suppresses cell migration by degrading GEF-H1, a RhoA GEF. Oncotarget. 2016 doi: 10.18632/oncotarget.8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharifi MN, Mowers EE, Drake LE, Collier C, Chen H, Zamora M, et al. Autophagy Promotes Focal Adhesion Disassembly and Cell Motility of Metastatic Tumor Cells through the Direct Interaction of Paxillin with LC3. Cell Rep. 2016;15:1660–1672. doi: 10.1016/j.celrep.2016.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nitta T, Sato Y, Ren XS, Harada K, Sasaki M, Hirano S, et al. Autophagy may promote carcinoma cell invasion and correlate with poor prognosis in cholangiocarcinoma. Int J Clin Exp Pathol. 2014;7:4913–4921. [PMC free article] [PubMed] [Google Scholar]
- 33.Macintosh RL, Timpson P, Thorburn J, Anderson KI, Thorburn A, Ryan KM. Inhibition of autophagy impairs tumor cell invasion in an organotypic model. Cell Cycle Georget Tex. 2012;11:2022–2029. doi: 10.4161/cc.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lock R, Kenific CM, Leidal AM, Salas E, Debnath J. Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov. 2014;4:466–479. doi: 10.1158/2159-8290.CD-13-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Topalovski M, Brekken RA. Matrix control of pancreatic cancer: New insights into fibronectin signaling. Cancer Lett. 2015 doi: 10.1016/j.canlet.2015.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park J, Schwarzbauer JE. Mammary epithelial cell interactions with fibronectin stimulate epithelial-mesenchymal transition. Oncogene. 2014;33:1649–1657. doi: 10.1038/onc.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Au A, Vasel M, Kraft S, Sens C, Hackl N, Marx A, et al. Circulating fibronectin controls tumor growth. Neoplasia N Y N. 2013;15:925–938. doi: 10.1593/neo.13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh KK, Lovren F, Pan Y, Quan A, Ramadan A, Matkar PN, et al. The essential autophagy gene ATG7 modulates organ fibrosis via regulation of endothelial-to-mesenchymal transition. J Biol Chem. 2015;290:2547–2559. doi: 10.1074/jbc.M114.604603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghavami S, Cunnington RH, Gupta S, Yeganeh B, Filomeno KL, Freed DH, et al. Autophagy is a regulator of TGF-β1-induced fibrogenesis in primary human atrial myofibroblasts. Cell Death Dis. 2015;6:e1696. doi: 10.1038/cddis.2015.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W, Zou J, Yue F, Song K, Chen Q, McKeehan WL, et al. Defects in MAP1S-mediated autophagy cause reduction in mouse lifespans especially when fibronectin is overexpressed. Aging Cell. 2016 doi: 10.1111/acel.12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez-Outschoorn UE, Pavlides S, Howell A, Pestell RG, Tanowitz HB, Sotgia F, et al. Stromal–epithelial metabolic coupling in cancer: Integrating autophagy and metabolism in the tumor microenvironment. Int J Biochem Cell Biol. 2011;43:1045–1051. doi: 10.1016/j.biocel.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez-Outschoorn U, Sotgia F, Lisanti MP. Tumor microenvironment and metabolic synergy in breast cancers: critical importance of mitochondrial fuels and function. Semin Oncol. 2014;41:195–216. doi: 10.1053/j.seminoncol.2014.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.