Abstract
Despite increasing use of nanotechnology in neuroscience, the characterization of interactions between magnetic nanoparticles (MNPs) and primary cortical neural networks remains underdeveloped. In particular, how the age of primary neural networks affects MNP uptake and endocytosis is critical when considering MNP-based therapies for age-related diseases. In this study, primary cortical neural networks are cultured up to 4 weeks and with CCL11/eotaxin, an age-inducing chemokine, to create aged neural networks. As the neural networks aged, their association with membrane-bound starch-coated ferromagnetic nanoparticles (fMNPs) increased while their endocytic mechanisms are impaired, resulting in reduced internalization of chitosan-coated fMNPs. The age of the neurons also negates the neuro-protective effects of chitosan coatings on fMNPs, attributing to decreased intracellular trafficking and increased co-localization of MNPs with lysosomes. These findings demonstrates the importance of age and developmental stage of primary neural cells when developing in vitro models for fMNP therapeutics targeting age-related diseases.
Keywords: CCL11/eotaxin, nanoparticles, age, cortical neurons, conditioned media
1. Introduction
With progress in nanotechnology, magnetic nanoparticles (MNPs) have found utility in applications such as drug delivery and neural stimulation.[1, 2] However, the interactions between MNPs and primary neural cells have been under-characterized. Most studies on MNP uptake in neurons made use of differentiated neurons[3] and neuron-like cells such as PC12[4, 5] whose interactions with MNPs differ from primary neurons[6, 7] (Table S1).
To motivate greater nanotechnology innovation in neuroscience, it is paramount to first understand the interactions such as membrane-association and endocytic mechanisms between MNPs and primary neural cultures.[1] As such interactions are likely to be specific[8, 9] depending on the properties of MNPs (coating, size etc.) and neural cell type (neurons, astrocytes etc.) in different brain regions, it is important to minimize variables of study and comprehensively investigate one type of interaction at a time.
In this paper, we characterized the interactions between MNPs and primary cortical neural networks of different developmental stages[10] (immature: day 0–6, developing: day 6–19, mature: day 20 onwards) and molecular aging. Cortical neural networks are chosen as: (1) the cortices are implicated in memory formation,[11] (2) with increments in human lifespan globally, more people are expected to suffer from aged-cortex-related neurodegenerative diseases,[12, 13] (3) the level of characterization of aged cortical neural networks has been minor in comparison to those of hippocampal origins despite its importance for diverse cognitive functions (Table S2).
One other motivation for this research is to better guide the diverse applications of nanotechnology to tackle developmental- and age-related neurodegenerative diseases where the ultimate location and uptake of MNPs often affect the efficacy of treatments. To stimulate neural networks, Tay et al. (100–300 nm in conditioned media), Chen et al. (200 nm in media) and Huang et al. (10 nm in PBS) made use of MNPs to open mechano/heat-sensitive ion channels.[2, 14, 15] In these applications, ion influxes were maximized when MNPs associated with cell membranes and were not internalized. There are also groups introducing, with the use of MNPs, siRNA and drugs into neural networks.[16, 17] In these applications, the objective is to maximize internalization and transport of MNPs to specific organelles. As these applications are targeted to understand or reduce the impact of developmental- and age-related neurodegenerative diseases, the characterization of interactions between MNPs and primary neural networks of different developmental stages and ages will be valuable for the design of MNPs.
Ideally, to create cortical neural networks of different ages, the neural cells would be cultured for varying periods of time. However, it is a challenge to maintain in vitro primary cortical neural cultures for more than 4 weeks healthily (i.e. maintaining ~1:1 astrocytes/glia: neuron ratio[18] and MNP-induced cytotoxicity).
Porter et al. found that neurons up to 4 weeks of culture exhibited increase in L-type calcium channels similar to aging,[19] suggesting that neural cultures are suitable to study different phases of neuronal development. Recently, it was also found that the level of a chemokine called CCL11 (eotaxin) in the cerebral spinal fluid (CSF) increases with age.[20] In the central nervous system (CNS), CCL11 is mainly produced by neurons[21, 22] which also express the main CCL11 receptor i.e. CCR3.[21, 23–26] Villeda et al. reported that increasing peripheral levels of CCL11 decreased neurogenesis in mice model.[20] Additionally, CCL11 has been shown to be implicated in aging[27–30] and is a potential drug to induce aging in neural networks. From these recent findings, we thus rationalized that the effects of developmental stage and molecular aging on MNP-neural networks interactions should be captured with culturing cortical neural networks for varying weeks and with CCL11 (see Table S3 for comparison with other models).
Here, we show through extensive characterizations that CCL11 can induce aging-like morphologies and cellular behaviors, and that neural networks of different ages interacted differently with NPs. Finally, we show that neuroprotection conferred by chitosan coatings on MNPs is age-dependent due to differences in lysosomal co-localization and intracellular trafficking of the NPs.
2. Results and Discussions
2.1 CCL11-treated and aged neural networks shared similar morphologies
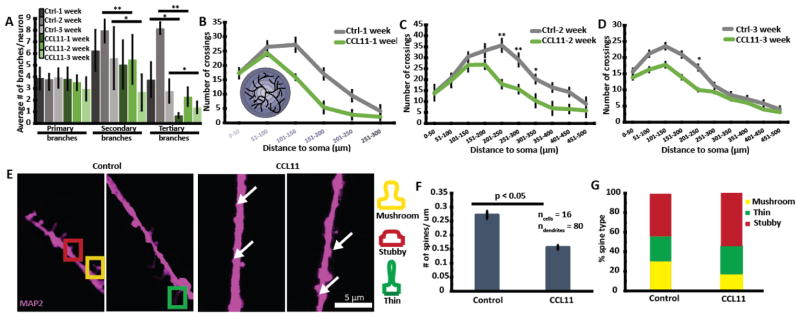
Extensive studies have confirmed that with age, there is no regression of dendritic length[31] but there is reduction in dendritic branching in the cortices of rats[32] and humans.[33] Sholl’s analyses revealed that CCL11 treated neurons exhibited reduced distal dendritic branching compared to control neurons (week 2 and 3) and consistent with literature findings[32–34] (Figure 1A–D).
Figure 1.
CCL11 treatment decreased distal dendritic branching and dendritic spine density. (A) CCL11 treatment significantly reduced the extent of secondary and tertiary dendritic branching. (B–D) Bottom left hand corner of (B) shows the schematic of Sholl’s analysis. Briefly, circles of different radii are drawn around the soma/cell body. Each intersection between the dendrites and the circumferences of the circles was counted and tabulated for graph plotting. The circumferences of the circles are also color coded to match their radii on the horizontal axis. CCL11 treatment reduced distal dendritic branching as shown with Sholl’s analyses. * represents p < 0.05 and ** represents p < 0.001 using Student’s t-test. (E) Representative images of spines with and without CCL11 treatment. White arrows indicated the presence of small spines that were present in larger numbers in CCL11 treated neurons. (F) CCL11 treatment reduced spine density especially (G) % of mushroom-shaped spines.
Dendritic spines undergo active re-modelling during the growth (day 1–6), maintenance (day 7–19), and decline (beyond day 19) phases.[35] Reduction in dendritic spine density, especially mushroom-shaped spines associated with plasticity, is observed in older animals.[36] Figure 1E–F shows that CCL11 treatment decreased spine density in day 18 neurons. On average, there were 0.27 spines/10 μm dendritic length in the control neurons compared to 0.15 in CCL11 treated neurons. There was also greater number of highly small dendritic spines found in CCL11 treated neurons (white arrows in Figure 1E) that had been. reported previously by Ricardo et al.[37] We also observed a reduction in % mushroom-shaped spines in CCL11 treated neurons (Figure 1G).
Collectively, our results showed that CCL11 treatment reduced dendritic branching and spine density, and disrupted calcium homeostasis (Figure S3–4) which are typically observed in aged neurons. These observations suggest that CCL11 can be used to create phenotypes resembling aged neural networks which provide us, now with a model to investigate the effects of developmental stages and age of neural networks and their interactions with NPs.
2.2 Characterization of magnetic nanoparticles
We next investigated whether the interactions between starch- and chitosan-coated ferromagnetic nanoparticles (fMNPs) and cortical neural networks are affected by developmental stages and molecular aging with CCL11. Chitosan and starch coated iron oxide fMNPs were chosen as model particles for a variety of reasons: (1) they are commercially available and fluorescently labeled for easy visualization, (2) the properties of the fMNPs used here have been well-characterized (chitosan[38–41] and starch[42, 43]) for drug delivery and phagocytosis, and (3) chitosan and starch coated MNPs are one of the most widely used biopolymer coatings employed in brain-related biomedical interventions. Chitosan-coated MNPs were found to reduce secondary oxidative stress in neurons.[5] They can also improve specificity of brain targeting for estradiol stimulation[44] and nucleic acid delivery.[45] Starch-coated fMNPs have also been approved by the US Food and Drug Administration for clinical use as magnetic resonance contrast agents with reportedly low cytotoxicity.[46] Thus, this study using starch- and chitosan-coated fMNPs has great research and translational value.
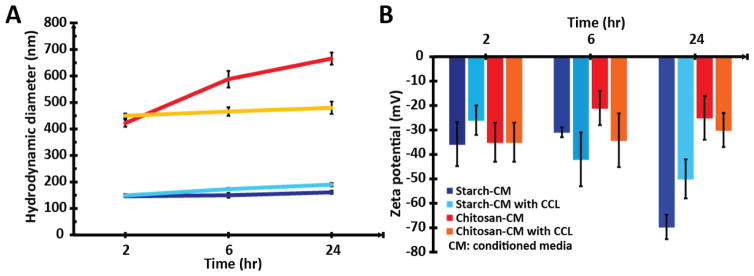
The hydrodynamic diameters (Dh) and zeta potential (ζ) of fMNPs in PBS, serum-free Neurobasal media and conditioned Neurobasal media (media grown with cells for a week) was measured (Table S4). To the best of our knowledge, no group has yet investigated the effects of conditioned media on nanoparticle properties. This latter characterization was performed as neural cells can modify their environment with active depletion/secretion of factors, hence affecting protein corona formation.[47] Furthermore, nanoparticle incubation with primary neural networks has to be performed in conditioned media due to the high sensitivity of neural cells to environmental changes. Hence the properties of the fMNPs in conditioned media are more accurate for assessing their interactions with neural cells. The Dh of the fMNPs in conditioned media grew as incubation time increased, indicative of protein corona formation (Figure 2A). However, as time increased, the ζ of starch-coated fMNPs became more negative while that of chitosan-coated fMNPs became less negative (Figure 2B). The time-dependence properties can affect the interactions between the fMNPs and neural cells. For instance, as the cell membrane is negatively charged, the interactions of positively charged NPs with the cell membrane are favored and might increase uptake of chitosan-coated fMNPs.
Figure 2.
Hydrodynamic diameter (Dh) and zeta potential (ζ) of starch- and chitosan-coated fMNPs as a function of time in conditioned media with and without CCL11. (A) As time progressed, the Dh of the fMNPs increased due to protein corona formation. (B) As time progressed, the ζ of starch-coated fMNPs became more negative while that for chitosan-coated fMNPs became more positive. The time-dependence impact of conditioned media on the properties of fMNPs can affect their interactions with neural cells.
2.3 Starch-coated fMNPs primarily associated with membrane while chitosan-coated fMNPs were internalized
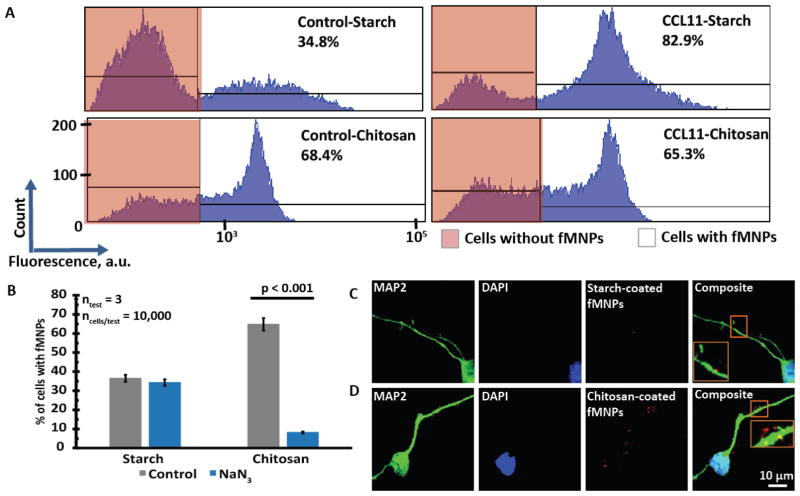
We first made use of flow cytometry to investigate the interactions between starch/chitosan-coated fMNPs and untreated/CCL-treated 1-week-old neural networks. The results showed that the untreated neural networks interacted more with chitosan-coated fMNPs and CCL11 treatment increased the interaction between the neural networks and starch-coated fMNPs (Figure 3A).
Figure 3.
Starch-coated fMNPs were associated with the cell membrane while chitosan-coated fMNPs were internalized by the neural networks. (A) Flow cytometry results showed that chitosan-coated fMNPs interacted more with the neural networks. CCL11 treatment increased the interactions of starch-coated fMNPs but it decreased the interactions of chitosan-coated fMNPs with 1-week-old neural networks. (B) Flow cytometry results showed that the uptake of chitosan-coated fMNPs and not starch-coated fMNPs was affected by addition of sodium azide, an energy depletion agent that inhibited endocytosis. This indicated that starch-coated fMNPs were mostly localized at the cell membrane while the chitosan-coated fMNPs were internalized. (C) Confocal images showing that starch-coated fMNPs were localized at the membranes while (D) chitosan-coated fMNPs were internalized.
However, one limitation of flow cytometry is that it is unable to differentiate between cells with internalized fMNPs or cells with fMNPs associated with the membrane. We thus inhibited endocytosis by depleting the energy of the neural networks with sodium azide. Figure 3B shows that while uptake of chitosan-coated fMNPs were significantly reduced (suggesting endocytosis), the interactions between starch-coated fMNPs and the neural networks were not affected, suggesting that most of the starch-coated fMNPs were associated with the cell membranes rather than being internalized after 24 hr incubation. This is also supported by confocal images which show membrane-bound starch-coated fMNPs (Figure 3C and S5A) and internalized chitosan-coated fMNPs (Figure 3D and S5B). We hypothesize that although starch-coated fMNPs have smaller Dh (Table S4), their more negative ζ may have disfavored their interactions with negatively charged cell membranes,[48] resulting in association rather than internalization, suggesting the greater role of ζ in NP uptake by cortical neural cells. Starch- and chitosan-coated fMNPs can thus function as our model systems to evaluate the effects of age on membrane localization and internalization of NPs respectively.
Lastly, we also found that unlike other cell types such as cancer cell lines that interacted with fMNPs in the minute scale,[49] cortical neural networks needed at least 24 incubation before there were substantial interactions with the fMNPs (Figure S9). This result highlight the differences in behaviors between primary neural cultures and neuron-like cell lines such as PC12 and that a timescale in the hr range should be considered for applications such as drug delivery.
2.4 The age of cortical neural network affected their interaction with fMNPs
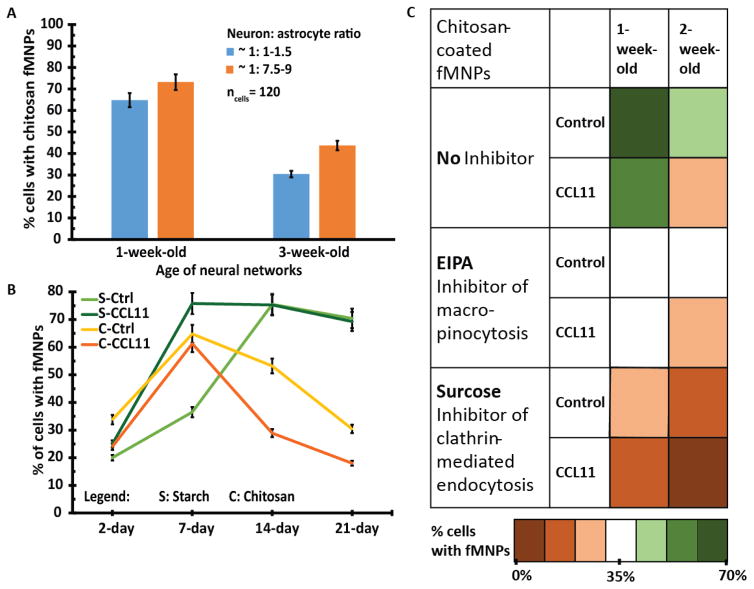
As the cortical neural networks consisted of approximately equal number of neurons to astrocytes, we decided to investigate whether the uptake of chitosan-coated fMNPs were due to one particular cell type. This is important as there is a large number of publications using chitosan-coated NPs to ameliorate brain diseases due to dysfunctional neuronal activities (Table S5) but none of them have evaluated whether their NPs were targeting neurons or other cell types present in the culture. By changing the composition of the media, we were able to generate neural networks composing of 1:1.5 and 1:7.5–9 neuron: astrocytes ratio. We did not observe significant differences in the uptake of chitosan-coated fMNPs by same age neural networks with different cellular compositions (Figure 4A). This result favors the use of chitosan-coated MNPs in neuroscience applications as this class of NPs can be significantly internalized by neurons.
Figure 4.
Interaction between fMNPs and cortical neural networks of different ages. (A) Neural networks composed of approximately equal numbers of neurons to astrocytes (1: 1–1.5) or mainly of astrocytes (1: 7.5–9) did not exhibit significant differences in their uptake of chitosan-coated fMNPs, although slightly higher uptake was observed in astrocytes. (B) Young neural networks (day 2) exhibited less interactions than older networks probably due to less extensive surface area. 2- and 3-week-old cortical neural networks internalized less chitosan-coated fMNPs but their association with starch-coated fMNPs remained stable. CCL11 treatment consistently decreased the internalization of chitosan-coated fMNPs probably due to poorer metabolism in aged neural networks. (C) Heat map shows that neural networks most likely uptake chitosan-coated fMNPs via micropinocytosis and clathrin-mediated endocytosis. As the neural networks aged, macro-pinocytosis played a lesser role in uptake of chitosan-coated fMNPs.
Next, we investigated the effects of age of neural networks and its impact with fMNPs interactions. We first found that the association/internalization of starch/chitosan-coated fMNPs increased from 2-day-old to 1-week-old neural networks (Figure 4B). This can be attributed largely to a significant increase in surface area as the networks grew. We also discovered that the uptake of chitosan-coated fMNPs by the neural networks decreased from week 1–3 (Figure 4B) and that CCL11 treated neural networks always internalized less chitosan-coated fMNPs than the controls. This could be attributed to poorer metabolism in aged neural networks as endocytosis requires energy.[47] However, the association of starch-coated fMNPs with older neural networks increased from week 1 to 2 and stabilized till week 3 (Figure 4B). This finding could be attributed to the membrane-associating property of starch-coated fMNPs which did not require energy.
Taking into consideration the prevalence of clathrin coats in neurons[50] and absence of caveolae in most neurons,[51] we investigated the impact of age on macro-pinocytosis (size limit: 0.5–5 μm) and clathrin-mediated endocytosis (size limit: 100–200 nm).[52] We found that blocking clathrin-mediated endocytosis macro-pinocytosis decreased uptake of chitosan-coated fMNPs significantly. Figure 4C shows that both mechanisms contributed almost equally to internalization of chitosan-coated fMNPs in 1-week-old neural networks. However, as the neural network ages (either with CCL11 treatment or at week 2), the efficiency of macro-pinocytosis was more greatly reduced. This finding has implications for the efficacy of MNP designed to target specific endocytic pathways.[53]
2.5 Neuro-protective effects of chitosan-coated magnetic nanoparticles are age-dependent
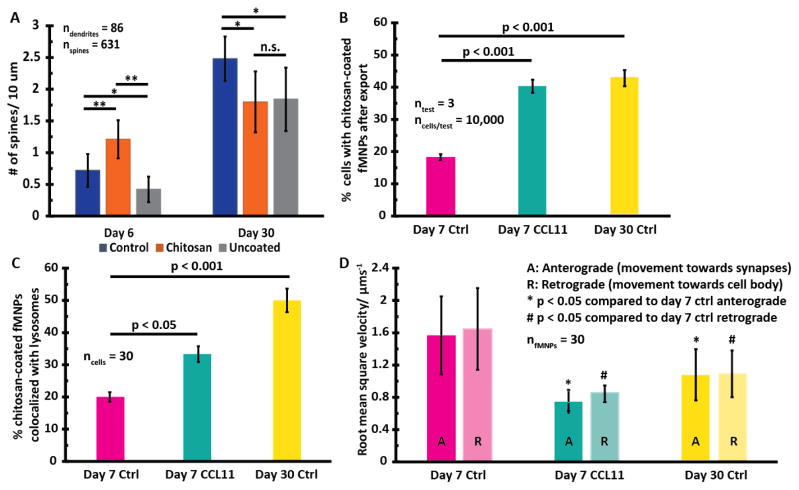
Although chitosan-based MNPs are known to confer neuro-protective benefits in multiple studies (Table S5), no study, to the best of our knowledge, has investigated the impact of the age of neural networks on neuro-protection. We incubated chitosan-coated and uncoated iron oxide-based fMNPs with 6-(immature) and 30-day-old (mature) neural networks for 4 days (the age of the neural networks was intentionally chosen as dendritic spine development in neurons typically matures and stabilizes after 22 days of culture[37]) and found that chitosan coatings conferred neuro-protection to 6-day-old neurons but not 30-day-old neurons (Figure 5A, S9).
Figure 5.
Increased co-localization of chitosan-coated fMNPs with lysosomes and reduced intracellular trafficking might contribute to age-dependent neuro-protective effects of chitosan-coatings on fMNPs. (A) Chitosan-coatings increased dendritic spine density in immature (6-day-old) neurons but decreased dendritic spine density in mature (30-day-old) neurons. (B) There was greater net retention of NPs and (C) co-localization of NPs with lysosomes in older neurons (either induced by CCL11 treatment or natural aging). (D) The root mean square velocity of NPs (both retrograde (* p < 0.05) and anterograde (# p < 0.05) transport) in older neurons was significantly less than that in younger neurons.
2.6 The age of neurons affects co-localization with lysosomes and intracellular trafficking
The export of MNPs out of neural networks plays an important role for MNP-based-therapeutics as it influences decisions such as dosage and time gaps between doses.[52] The time MNPs are retained in the cells can also impact their degradation and cytotoxicity.[54] We found that neural networks can export MNPs, consistent with findings from Wong et al.[3] This behavior is different from cell types such as A549 that does not export MNPs after internalization.[55, 56] As the neurons aged (either induced by CCL11 treatment or naturally), the degree of export decreased,[50] possibly due to poorer metabolism. In older neurons (CCL11 treated or natural aging), we found greater net retention (Figure 5B) and co-localization of chitosan-coated fMNPs with lysosomes (Figure 5C).
Intracellular trafficking of proteins along neurites are highly compartmentalized in neurons[56] and trafficking of MNPs influences the efficacy of NPs for treating neuro-degenerative diseases. We found that in older neurons (either induced by CCL11 treatment or via natural aging), the root mean square (RMS) velocity of chitosan-coated fMNPs trafficking along the neurites was reduced. The retrograde transport (i.e. movement towards cell body) and anterograde transport (i.e. movement towards synapses) in CCL-treated day 7 and day 30 neurons were significantly reduced compared to day 7 control neurons (Figure 5D, S11). This is an important finding for applications using MNPs to deliver drugs to target amyloid-beta or hyper-phosphorylated tau protein in the axons in Alzheimer’s patients where age is the greatest risk factor for getting the disease.[57]
Collectively, our data suggests that with age, there is reduced intracellular trafficking of MNPs and greater net retention and co-localization of MNPs with lysosomes. This could have resulted in higher probability of chitosan coating degradation, causing the release of free, toxic metal ions from the iron oxide core[58] and the loss of neuroprotection from chitosan coatings. These findings are aligned with recent observations that nano-toxicology is age-dependent (Table S6) and likewise, we showed here that nano-therapeutic effects can also be age-dependent.
3. Conclusion
In this study, we investigated the effects of developmental stages and molecular aging of primary cortical neural networks with fMNPs. Through extensive morphological and functional characterization, we found CCL11 to be a suitable drug to create aged phenotypes in cortical neurons although future investigation in gene expression and metabolic pathways are needed to evaluate the impact of CCL11 on aging. Next, we showed that starch- and chitosan-coated fMNPs can function as model system for MNPs that localize on membrane and MNPs that are internalized respectively. Starch-coated fMNPs associated more with the membranes of older cortical neural networks while less chitosan-coated fMNPs were internalized by the neural cells as they aged. The efficacy of macro-pinocytosis was also more adversely affected by aging compared to clathrin-mediated endocytosis. We also found that the neuro-protective effects of chitosan-coating on fMNPs are age-dependent i.e. only for immature (6-day-old) and not for mature (30-day-old) neurons. This observation may be partially explained by reduced intracellular trafficking of chitosan-coated fMNPs and increased co-localization of chitosan-coated fMNPs with the lysosomes with age, resulting in increased probability of toxic iron ions being released from the core of the fMNPs. Overall, these findings imply that future work involving nanotechnology in neuroscience should consider the effects of the age of neural cells and their interactions with the MNPs for more comprehensive evaluation of the utility of designed MNPs. We note that in this study, the findings were based on in vitro experiments with starch/chitosan-coated iron oxide core fMNPs with hydrodynamic diameter between 100–500 nm. An extension of this work would be to investigate whether the age of the neural cells also affect their interactions with nanoparticles made of different materials such as polymers or other types of metal like gold of different sizes and coatings for various biomedical applications.
4. Experimental Section
Neural cell culture
Cortical hemispheres from whole brain rats (E18, Brainbits) were transferred to PBS (33 mM glucose, 1% (v/v) Penicillin-Streptomycin (PenStrep from Gibco®) and dissected. Cortical tissues were then placed in Papain/Hibernate-E (pH 7.3) and dissociated for 15 min at 37 °C. 10% horse serum (Fisher Sci) in Neurobasal (Gibco®) was then added to quench the enzymatic activity of Papain and dissociated tissues were triturated through a 1000 μL pipette tip and filtered a 40 μm cell strainer. CCL treated neurons were exposed to CCL11 (100 pg/mL) (Novus Biologicals) from day 4 and 25% media was added every 14 days. To create neural networks composing mainly of astrocytes, the cells were grown in Dulbecco’s Modified Eagle Media (DMEM) (ThermoFisher Scientific), supplemented with 10% fetal bovine serum (FBS) (ThermoFisher Scientific) and 1% PenStrep (Gibco®).
Characterization of nanoparticle properties
ZetaPALS 90Plus particle size analyzer was used to determine the hydrodynamic diameter of NPs (54,000 × 106/mL). Briefly, NPs were suspended and pipetted into a cuvette before measurement with a laser via dynamic light scattering. Following which, the ZetaPLAS zeta potential analyzer with electrode probe AQ599 were used to determine the zeta potential of the NPs
Nanoparticle incubation
1/2/3-week-old cortical neurons were incubated with starch- or chitosan-coated fMNPs (54,000 × 106/mL) for 2, 6, 24 hr in conditioned media. After which, the cells were gently washed for 3 times with Neurobasal media to remove excess fMNPs. Cortical neurons (6-and 30-day-old) were treated with uncoated and chitosan-coated fMNPs for 4 days i.e. (at day 2 and day 26 respectively). Note: all NP incubation with the cortical neural networks occurred in conditioned media to (1) maintain the health of sensitive cells like primary neurons, (2) avoid any glia activation and related toxicity and (3) capture the effects of protein corona on NPs.
Cytotoxicity assay
Colorimetric MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assays were performed in 96 well plates where different functionalized fMNPs were added to each well seeded with cortical neurons. After 24 hr, 20 μL of MTT (5 mg/mL) in PBS was incubated for 3 hr. Then the supernatant was aspirated and 200 μL of dimethyl sulfoxide (DMSO) was added. Each 200 μL sample in DMSO was spun down to remove MNPs in solution and the supernatant was read with a 96 well plate reader for absorbance at 550 nm. Live/dead assays were also performed with propidium iodide to quantify cell death due to NPs.
Calcium dye incubation
Cortical neurons were incubated with Fluo-4 Direct™ calcium assay kit (Life Technologies) according to manufacturer’s protocol. For experiment with thapsigargin, 2 μM of the chemical was incubated with the cells 30 mins prior and during calcium dye incubation. For experiment with EGTA, EGTA was administered 90 minutes earlier to chelate free Ca2+ ions followed by incubation with calcium dye.
Immuno-fluorescent labelling
Cortical neurons were washed with DPBS and fixed with paraformaldehyde (4% v/v, Santa Cruz Biotechnologies), permeabilized with 0.1% Triton-x/DPBS and 3% BSA for 10 min and blocked with 3% goat serum in 1% BSA/DPBS. Primary antibodies were incubated overnight at 4 °C in 3% goat serum, 0.5% Tween-20 in 1% BSA/DPBS and secondary antibodies were added. Finally 4′,6-diamidino-2-phenylindole (DAPI, 300 nM in DPBS) was incubated for 15 min and pro ProLong ® Gold antifade reagent (Molecular Probes) was added. Refer to Table S7 for list of antibodies (all purchased from Life Technologies).
Co-localization
Lysotracker™ deep red (Life Technologies) were used to assess co-localization of fMNPs with lysosomes. Live cells were stained with Lysotracker™ deep red in Neurobasal media for 1 hr at 37 °C and 4% CO2. The media was then replaced and washed three times before imaging with a fluorescent microscope.
Flow Cytometry Analysis
1.0 × 106 neurons were seeded in individual PLL-coated 12 well-plate (Corning) and allowed to grow for 1/2/3 weeks. Fluorescently labeled MNPs (green: ex (476 nm), em (490 nm); red: ex (578 nm), em (613 nm)) were then added at a concentration of 54000 × 106/mL and incubated for 2/6/24 hr before triple washing with Neurobasal media. For experiment involving EIPA, sodium azide and sucrose, neural networks were also incubated with 100 μM EIPA, 5 mg/mL sodium azide, 450 mmol/L sucrose solution for 24 hr respectively. For experiments on MNP export, neurons were incubated with fMNPs for 24 hr following another 24 hr for export before flow cytometry analysis. Neurons were then detached from plate surface using Accutase® (Stemcell Technologies) and centrifuge at 600 g for 30 min to collect the pellet. Cell pellet was then resuspended in 500 μL DPBS and analyzed using BD LSRII flow cytometer.
Image acquisition and analysis
Wide-field fluorescent and phase contrast images were acquired an inverted fluorescent microscope (Nikon, 10×, 20×, 40× air objective) for morphological analysis, relative protein quantification and calcium imaging. For morphological analysis, ImageJ neurite tracer plug-in was used to trace the dendrites (identified with MAP2 staining). Following which, the line traces were used for Sholl’s analyses. Briefly, a threshold was set followed by selecting the soma/cell body. The starting and ending radii were then set at 5 and 500 pixels (1 pixel = 1 μm) and the number of primary branches were determined from the bright field/fluorescent images. For relative protein quantification, Cy5 for TAU (ex: 650 nm, em: 670 nm) and FITC for MAP2 (ex: 476 nm, em: 490 nm) channels were used. The parameters for each channels were kept consistent for fair comparison across samples. For calcium imaging: The relative fluorescence change ΔF/F0 of somatic fluorescence signals was acquired using ImageJ for all neurons within a particular trial. Action potential events were considered if they fulfil two criteria: (1) the fluorescence increase was at least 5 standard deviation above baseline, which was defined as first 15 ms of each trace and (2) if the event persisted more than 3 ms. Raster plots were then generated from this method. For dendritic spine imaging, confocal microscope (Leica TCS SP5) was used. The fixed and immuno-stained samples were excited with 364 nm (blue/DAPI) and 488 nm (green/MAP2) laser lines. A 63× oil objective lens were then used to capture the images of the dendritic spines. Sorting of the subtypes of dendritic spines (mushroom, thin, stubby) was performed based on the presence/absence of head and then based on the length of the protrusion. Spines with heads were classified as mushroom shaped. Spines that did not have a head and had protrusions shorter than 0.7 μm were classified as stubby shaped while spines with no head and having protrusions ≥ 0.7 μm were classified as thin shaped. For NP trafficking analysis, stacked images are first corrected for bleaching and then transformative shift using StackReg plugin in ImageJ [59]. Subsequently, NPs were tracked using TrackMate plugin [60] (DoG (difference of Gaussian) detector, 2.0 μm blob diameter, linking: 2 μm, filter: paths shorter than 60 frames were rejected, Δt = 87 s, τ = 3 s). The raw data was then exported into an excel sheet calculated for root mean square speed (RMSS) using the formula: . The definition of anterograde and retrograde transport and their analysis methods were based on [61]. Briefly, the location of the NP, cell body and neurites were determined from bright-field images and an arbitrary direction is set. Anterograde is defined as transport towards neurites (increasing coordinates) while anterograde transport is defined as transport towards cell body (decreasing coordinates).
Statistical analysis
Statistical significance was evaluated using Student’s t-test after testing for normality using either one-way ANOVA, p < 0.05 (no rejection of normality).
Acknowledgments
This work was performed with funding from US National Institutes of Health Director’s New Innovator Award (1DP2OD007113). We would also like to thank Prof Felix Schweizer for his advice on calcium-related imaging and Prof Carlos Portera-Cailliau for his advice on dendritic spine imaging. Flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility that is supported by National Institutes of Health awards P30 CA016042 and 5P30 AI028697, and by the JCCC, the UCLA AIDS Institute, the David Geffen School of Medicine at UCLA, the UCLA Chancellor’s Office, and the UCLA Vice Chancellor’s Office of Research. Confocal laser scanning microscopy was performed at the CNSI Advanced Light Microscopy/Spectroscopy Shared Resource Facility at UCLA, supported with funding from NIH-NCRR shared resources grant (CJX1-443835-WS-29646) and NSF Major Research Instrumentation grant (CHE-0722519). A.T. performed experiments and analyzed data. D.J. taught A.T. how to measure size and zeta potential of nanoparticles. A.T. wrote the manuscript and A.K., D.J., E.H and D.D. revised the manuscript.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the authors.
References
- 1.Silva GA. Nature reviews neuroscience. 2006;7(1):65–74. doi: 10.1038/nrn1827. [DOI] [PubMed] [Google Scholar]
- 2.Tay A, Kunze A, Murray C, Di Carlo D. ACS nano. 2016 doi: 10.1021/acsnano.5b07118. [DOI] [PubMed] [Google Scholar]
- 3.Wong Y, Markham K, Xu ZP, Chen M, Lu GQM, Bartlett PF, Cooper HM. Biomaterials. 2010;31(33):8770–8779. doi: 10.1016/j.biomaterials.2010.07.077. [DOI] [PubMed] [Google Scholar]
- 4.Hasadsri L, Kreuter J, Hattori H, Iwasaki T, George JM. Journal of Biological Chemistry. 2009;284(11):6972–6981. doi: 10.1074/jbc.M805956200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho Y, Shi R, Borgens RB. Journal of biological engineering. 2010;4(1):2. doi: 10.1186/1754-1611-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardoso A, Costa P, De Almeida L, Simoes S, Plesnila N, Culmsee C, Wagner E, de Lima MP. Journal of Controlled Release. 2010;142(3):392–403. doi: 10.1016/j.jconrel.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Kim ID, Lim CM, Kim JB, Nam HY, Nam K, Kim SW, Park JS, Lee JK. Journal of Controlled Release. 2010;142(3):422–430. doi: 10.1016/j.jconrel.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 8.Chithrani BD, Chan WC. Nano letters. 2007;7(6):1542–1550. doi: 10.1021/nl070363y. [DOI] [PubMed] [Google Scholar]
- 9.Limbach LK, Li Y, Grass RN, Brunner TJ, Hintermann MA, Muller M, Gunther D, Stark J. Environmental science & technology. 2005;39(23):9370–9376. doi: 10.1021/es051043o. [DOI] [PubMed] [Google Scholar]
- 10.Zheng K, Bard L, Reynolds JP, King C, Jensen TP, Gourine AV, Rusakov DA. Neuron. 2015;88(2):277–288. doi: 10.1016/j.neuron.2015.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berman DE, Dudai Y. Science. 2001;291(5512):2417–2419. doi: 10.1126/science.1058165. [DOI] [PubMed] [Google Scholar]
- 12.Hedden T, Gabrieli JD. Nature reviews neuroscience. 2004;5(2):87–96. doi: 10.1038/nrn1323. [DOI] [PubMed] [Google Scholar]
- 13.Mattson MP, Magnus T. Nature reviews neuroscience. 2006;7(4):278–294. doi: 10.1038/nrn1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen R, Romero G, Christiansen MG, Mohr A, Anikeeva P. Science. 2015;347(6229):1477–1480. doi: 10.1126/science.1261821. [DOI] [PubMed] [Google Scholar]
- 15.Huang H, Delikanli S, Zeng H, Ferkey DM, Pralle A. Nature nanotechnology. 2010;5(8):602–606. doi: 10.1038/nnano.2010.125. [DOI] [PubMed] [Google Scholar]
- 16.Dobson J. Gene therapy. 2006;13(4):283–287. doi: 10.1038/sj.gt.3302720. [DOI] [PubMed] [Google Scholar]
- 17.Arruebo M, Fernández-Pacheco R, Ibarra MR, Santamaría J. Nano today. 2007;2(3):22–32. [Google Scholar]
- 18.Ospina KR, Parada S, Lapuente C, Céspedes AE. Revista Colombiana de Ciencia Animal. 2015;7(1) [Google Scholar]
- 19.Porter NM, Thibault O, Thibault V, Chen KC, Landfield PW. The Journal of Neuroscience. 1997;17(14):5629–5639. doi: 10.1523/JNEUROSCI.17-14-05629.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villeda SA, Luo J, Mosher KI, Zou B, Britschgi M, Bieri G, Stan TM, Fainberg N, Ding Z, Eggel A. Nature. 2011;477(7362):90–94. doi: 10.1038/nature10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adzemovic MZ, Ockinger J, Zeitelhofer M, Hochmeister S, Beyeen AD, Paulson A, Gillett A, Thessen Hedreul M, Covacu R, Lassmann H. PLoS One. 2012;7(7):e39794–e39794. doi: 10.1371/journal.pone.0039794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fryer AD, Stein LH, Nie Z, Curtis DE, Evans CM, Hodgson ST, Jose PJ, Belmonte KE, Fitch E, Jacoby DB. Journal of Clinical Investigation. 2006;116(1):228. doi: 10.1172/JCI25423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, Wittchen ES, Jiang Y, Ambati B, Grossniklaus HE, Hartnett ME. Investigative ophthalmology & visual science. 2011;52(11):8271. doi: 10.1167/iovs.11-8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maysami S, Nguyen D, Zobel F, Heine S, Höpfner M, Stangel M. Journal of neuroimmunology. 2006;178(1):17–23. doi: 10.1016/j.jneuroim.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 25.Wainwright DA, Xin J, Mesnard NA, Beahrs TR, Politis CM, Sanders VM, Jones KJ. ASN neuro. 2009;1(5):AN20090017. doi: 10.1042/AN20090017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parajuli B, Horiuchi H, Mizuno T, Takeuchi H, Suzumura A. Glia. 2015 doi: 10.1002/glia.22892. [DOI] [PubMed] [Google Scholar]
- 27.Dumanis SB, Tesoriero JA, Babus LW, Nguyen MT, Trotter JH, Ladu MJ, Weeber EJ, Turner RS, Xu B, Rebeck GW. The Journal of Neuroscience. 2009;29(48):15317–15322. doi: 10.1523/JNEUROSCI.4026-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baloyannis S. Journal of the neurological sciences. 2009;283(1):153–157. doi: 10.1016/j.jns.2009.02.370. [DOI] [PubMed] [Google Scholar]
- 29.Perez RG, Zheng H, Van der Ploeg LH, Koo EH. The Journal of Neuroscience. 1997;17(24):9407–9414. doi: 10.1523/JNEUROSCI.17-24-09407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Llorens-Martin M, Fuster-Matanzo A, Teixeira C, Jurado-Arjona J, Ulloa F, Rábano A, Hernández F, Soriano E, Ávila J. Molecular psychiatry. 2013;18(4):451–460. doi: 10.1038/mp.2013.4. [DOI] [PubMed] [Google Scholar]
- 31.Burke SN, Barnes CA. Nature reviews neuroscience. 2006;7(1):30–40. doi: 10.1038/nrn1809. [DOI] [PubMed] [Google Scholar]
- 32.Grill JD, Riddle DR. Brain research. 2002;937(1):8–21. doi: 10.1016/s0006-8993(02)02457-5. [DOI] [PubMed] [Google Scholar]
- 33.De Brabander J, Kramers R, Uylings H. The European journal of neuroscience. 1998;10(4):1261–1269. doi: 10.1046/j.1460-9568.1998.00137.x. [DOI] [PubMed] [Google Scholar]
- 34.Uylings HB, De Brabander J. Brain and cognition. 2002;49(3):268–276. doi: 10.1006/brcg.2001.1500. [DOI] [PubMed] [Google Scholar]
- 35.Alvarez VA, Sabatini BL. Annu Rev Neurosci. 2007;30:79–97. doi: 10.1146/annurev.neuro.30.051606.094222. [DOI] [PubMed] [Google Scholar]
- 36.Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Nature neuroscience. 2011;14(3):285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mostany R, Anstey JE, Crump KL, Maco B, Knott G, Portera-Cailliau C. The Journal of Neuroscience. 2013;33(9):4094–4104. doi: 10.1523/JNEUROSCI.4825-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kunze A, Tseng P, Godzich C, Murray C, Caputo A, Schweizer FE, Di Carlo D. ACS nano. 2015;9(4):3664–3676. doi: 10.1021/nn505330w. [DOI] [PubMed] [Google Scholar]
- 39.Linemann T, Thomsen LB, Jardin KGd, Laursen JC, Jensen JB, Lichota J, Moos T. Pharmaceutics. 2013;5(2):246–260. doi: 10.3390/pharmaceutics5020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Obermeier A, Kuchler S, Matl F, Pirzer T, Stemberger A, Mykhaylyk O, Friess W, Burgkart R. Journal of Biomaterials Science, Polymer Edition. 2012;23(18):2321–2336. doi: 10.1163/156856211X616183. [DOI] [PubMed] [Google Scholar]
- 41.Pala A, Liberatore M, D’Elia P, Nepi F, Megna V, Mastantuono M, Al-Nahhas A, Rubello D, Barteri M. Molecular Imaging and Biology. 2012;14(5):593–598. doi: 10.1007/s11307-011-0526-y. [DOI] [PubMed] [Google Scholar]
- 42.Ruge CA, Schaefer UF, Herrmann J, Kirch J, Canadas O, Echaide M, Pérez-Gil J, Casals C, Müller R, Lehr C-M. 2012 doi: 10.1371/journal.pone.0040775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tseng P, Di Carlo D, Judy JW. Nano letters. 2009;9(8):3053–3059. doi: 10.1021/nl901535m. [DOI] [PubMed] [Google Scholar]
- 44.Wang X, Chi N, Tang X. European Journal of Pharmaceutics and Biopharmaceutics. 2008;70(3):735–740. doi: 10.1016/j.ejpb.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Malhotra M, Tomaro-Duchesneau C, Prakash S. Biomaterials. 2013;34(4):1270–1280. doi: 10.1016/j.biomaterials.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 46.Kim DK, Mikhaylova M, Wang FH, Kehr J, Bjelke B, Zhang Y, Tsakalakos T, Muhammed M. Chemistry of Materials. 2003;15(23):4343–4351. [Google Scholar]
- 47.Lesniak A, Fenaroli F, Monopoli MP, Åberg C, Dawson KA, Salvati A. ACS nano. 2012;6(7):5845–5857. doi: 10.1021/nn300223w. [DOI] [PubMed] [Google Scholar]
- 48.He C, Hu Y, Yin L, Tang C, Yin C. Biomaterials. 2010;31(13):3657–3666. doi: 10.1016/j.biomaterials.2010.01.065. [DOI] [PubMed] [Google Scholar]
- 49.Dos Santos T, Varela J, Lynch I, Salvati A, Dawson KA. Small. 2011;7(23):3341–3349. doi: 10.1002/smll.201101076. [DOI] [PubMed] [Google Scholar]
- 50.Blanpied TA, Scott DB, Ehlers MD. Neuron. 2002;36(3):435–449. doi: 10.1016/s0896-6273(02)00979-0. [DOI] [PubMed] [Google Scholar]
- 51.Allen JA, Halverson-Tamboli RA, Rasenick MM. Nature reviews neuroscience. 2007;8(2):128–140. doi: 10.1038/nrn2059. [DOI] [PubMed] [Google Scholar]
- 52.Bareford LM, Swaan PW. Advanced drug delivery reviews. 2007;59(8):748–758. doi: 10.1016/j.addr.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dannhauser P, Platen M, Böning H, Schaap I. Nature nanotechnology. 2015 doi: 10.1038/nnano.2015.206. [DOI] [PubMed] [Google Scholar]
- 54.Harush-Frenkel O, Rozentur E, Benita S, Altschuler Y. Biomacromolecules. 2008;9(2):435–443. doi: 10.1021/bm700535p. [DOI] [PubMed] [Google Scholar]
- 55.Shapero K, Fenaroli F, Lynch I, Cottell DC, Salvati A, Dawson KA. Molecular Biosystems. 2011;7(2):371–378. doi: 10.1039/c0mb00109k. [DOI] [PubMed] [Google Scholar]
- 56.Salvati A, Åberg C, dos Santos T, Varela J, Pinto P, Lynch I, Dawson KA. Nanomedicine: Nanotechnology, Biology and Medicine. 2011;7(6):818–826. doi: 10.1016/j.nano.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 57.Gunawardena S. Pharmaceutical research. 2013;30(10):2459–2474. doi: 10.1007/s11095-013-1037-0. [DOI] [PubMed] [Google Scholar]
- 58.Pisanic TR, Blackwell JD, Shubayev VI, Fiñones RR, Jin S. Biomaterials. 2007;28(16):2572–2581. doi: 10.1016/j.biomaterials.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 59.Thevenaz P, Ruttimann UE, Unser M. Image Processing, IEEE Transactions on. 1998;7(1):27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 60.Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G. Nature methods. 2008;5(8):695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reddy PH, Shirendeb UP. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2012;1822(2):101–110. doi: 10.1016/j.bbadis.2011.10.016. http://dx.doi.org/10.1016/j.bbadis.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]