Abstract
p38 MAP kinase (MAPK) isoforms α, β, and γ, are expressed in the heart. p38α appears pro-apoptotic whereas p38β is pro-hypertrophic. The mechanisms mediating these divergent effects are unknown; hence elucidating the downstream signaling of p38 should further our understanding. Downstream effectors include MAPK-activated protein kinase (MK)-3, which is expressed in many tissues including skeletal muscles and heart. We cloned full-length MK3 (MK3.1, 384 aa) and a novel splice variant (MK3.2, 266 aa) from murine heart. For MK3.2, skipping of exons 8 and 9 resulted in a frame-shift in translation of the first 85 base pairs of exon 10 followed by an in-frame stop codon. Of 3 putative phosphorylation sites for p38 MAPK, only Thr-203 remained functional in MK3.2. In addition, MK3.2 lacked nuclear localization and export signals. Quantitative real-time PCR confirmed the presence of these mRNA species in heart and skeletal muscle; however, the relative abundance of MK3.2 differed. Furthermore, whereas total MK3 mRNA was increased, the relative abundance of MK3.2 mRNA decreased in MK2−/− mice. Immunoblotting revealed 2 bands of MK3 immunoreactivity in ventricular lysates. Ectopically expressed MK3.1 localized to the nucleus whereas MK3.2 was distributed throughout the cell; however, whereas MK3.1 translocated to the cytoplasm in response to osmotic stress, MK3.2 was degraded. The p38α/β inhibitor SB203580 prevented the degradation of MK3.2. Furthermore, replacing Thr-203 with alanine prevented the loss of MK3.2 following osmotic stress, as did pretreatment with the proteosome inhibitor MG132. In vitro, GST-MK3.1 was strongly phosphorylated by p38α and p38β, but a poor substrate for p38δ and p38γ. GST-MK3.2 was poorly phosphorylated by p38α and p38β and not phosphorylated by p38δ and p38γ. Hence, differential regulation of MKs may, in part, explain diverse downstream effects mediated by p38 signaling.
Keywords: p38 MAPK, MK3, Post-transcriptional regulation, Alternative splicing
1. Introduction
Myocardial hypertrophy occurs when the heart is exposed to a chronic increase in peripheral resistance and/or neurohormonal factors. These agents activate several targets including the MAP kinase (MAPK) pathways. The MAPK signaling pathways consist of protein kinase cascades linking extracellular stimuli to various targets in the cytoplasm, cytoskeleton, membrane, and nucleus and are involved in awide variety of cellular processes such as proliferation, differentiation, transcription regulation and development [1,2]. There are at least three distinct MAPK signaling pathways in mammals including the extracellular signal-regulated kinases (ERKs), the c-Jun N-terminal kinases (JNKs) and the p38 MAPKs [3,4]. These kinases are activated in cascades by phosphorylation on both threonine and tyrosine residues in the regulatory T–X–Y loop present in all typical MAPKs [5]. p38 (also known as CSBP,mHOG1, RK, and SAPK2) is the archetypal member of the second MAPK-related pathway in mammalian cells [6,7]. There are four known p38 isoforms (α, β, δ and γ) (reviewed in[8]). p38α, β, and γ [9], are expressed in the heart. In neonatal cardiac ventricular myocytes, p38α induces phenotypic changes resembling apoptosis whereas p38β is pro-hypertrophic [10]. The mechanisms by which p38 MAPKs have such different effects remain to be elucidated; however, a better understanding of the downstream targets of p38 should answer these questions. The MAPK-activated protein kinases (MKs) -2, -3 and -5 are directly activated by p38 MAPKs [11] although activation of MK5 by p38 in vivo has become controversial (see [12]). Hence, the divergent effects of the p38 cascade may be mediated by differential regulation of the MKs.
MK3 (chromosome 3P kinase, 3pK) is a serine/threonine protein kinase that is expressed in many tissues including skeletal muscles and heart [13]. Its physiological substrates include the small heat shock protein hsp27/hsp25 [14,15], 5-lipoxygenase [16], polycomb group proteins [17], and the transcription factor E47 [18]. In vitro studies demonstrated that ERK, p38 MAPK and Jun N-terminal kinase were all able to phosphorylate and activate this kinase, which suggested the role of this kinase as an integrative element of signaling in both mitogen and stress responses [19]. Although MK2 and MK3 show extensive similarities in terms of structure, regulation and substrate specificity [1,13,19–21], the phenotype of MK3−/− mice differs from the inflammation-impaired phenotype of MK2−/− mice [22,23]. Specificity may result from cell-type specific or developmental differences in the expression of MK2 versus MK3 [24]. Alternatively, as arsenite induces higher levels of MK3 activation than anisomycin, sorbitol or interleukin-1, the nature of the activating stimulus may also influence the relative extent of MK2 and MK3 activation [20]. However, as the expression of MK2 is generally much greater than that of MK3 [20,23], the differences in phenotype of the MK2−/− versus the MK3−/− mice may result from differences in the level of expression. Consistent with this, ectopic expression of either MK2 or MK3 can rescue MK2−/− mouse embryonic fibroblasts [23]. Furthermore, relative to MK2−/− mice, MK2−/−:MK3−/− mice show further destabilization of p38α and reduction in both LPS-induced tristetraprolin and LPS-induced TNF-α expression [23]. Hence, experiments to date suggest that MK2 and MK3 overlap in their physiological function. Alternatively, although both MK2 and MK3 form a stabilizing complex with p38 in the heart [23], and MK2 is generally expressed at much higher levels than MK3, transgenically expressed catalytically inactive mutant FLAG-p38α precipitates MK3, but not MK2, from heart lysates [25], suggesting that p38α/β may preferentially signal via MK3 in the myocardium.
The aim of this study was to clone and characterize the full-length form of MK3 and a novel splice variant, referred to herein as MK3.2, from murine cardiac ventricular total RNA to further improve our understanding on heart-specific MAPK signaling pathways. Our data suggest that both MK3.1 and MK3.2 are translated in the heart but differ in their subcellular localization. Upon activation of p38 MAPK by hyperosmotic stress, MK3.1 rapidly translocated to the cytoplasm whereas MK3.2 was rapidly degraded and this degradation was inhibited by SB203580, MG132, or replacement of Thr-203 with a non-phorphorylatable residue. Finally, although only MK3.1 shows binding to p38α and p38β, both MK3.1 and MK3.2 are substrates for p38α and β in vitro, and hence, may play distinct roles in mediating downstream effects of the p38 MAPK pathway.
2. Materials and methods
2.1. Materials
[γ-32P]ATP was from Amersham Pharmacia Biotech. Membrane grade (reduced) Triton X-100 (TX-100), leupeptin, and PMSF were from Roche Molecular Biochemicals. SDS-polyacrylamide gel electrophoresis reagents, nitrocellulose, and Bradford protein assay reagent were from Bio-Rad. The cAMP-dependent protein kinase inhibitor peptide (PKI, amino acid sequence TTYADFIASGRTGRRNAIHD) was from the University of Calgary Peptide Synthesis Core Facility. Canine hsp27, cloned into the pET24a expression vector [26], was from Dr. William Gerthoffer (Reno, NV). Antibodies to MK2 (sc-6621), MK3 (sc-1973, sc-28782), andMK5 (sc-8253)were from Santa Cruz Biotechnology Inc. Anti-GFP and -V5were from BD Biosciences and Invitrogen, respectively. HRP-conjugated secondary antibodies were from Jackson Laboratories. Myelin basic protein (MBP) was purified as described previously [27]. Anti-humanMK2 phospho-threonine-222 (3044), plus phosphorylated, active, recombinant ERK1, ERK2, p38α, β2, δ, and γ were from Upstate Cell Signaling Solutions. All other reagents were of analytical grade or best grade available. pGEX-2T murine MK5 and pGEX-2T murine MK2 were cloned by PCR. A plasmid containing human p38γ was a gift from Dr. Jiahuai Han. cDNA for human p38α and β was from OPEN Biosystems and p38δ was from ATCC. Each p38 isoform was subcloned into pGEX-6p-2 (Amersham) modified to include a hexahistidine tag 3′ to the Precision protease cleavage sequence to create tandem affinity-tagged recombinant proteins. All constructs were confirmed by sequencing. Plasmids were introduced into E. coli strain BL21 and expression induced by the addition of 1 mM isopropyl-β-D-thiogalactosidase. GST-fusion proteins were purified by affinity chromatography on glutathione Sepharose.
2.2. Immunoblotting
SDS-PAGE, transfer and visualization were performed as described previously [28].
2.3. Protein kinase assay
Purified GST-MK3.1 or GST-MK3.2 (5 μg) were incubated with phosphorylated, activated forms of ERK1, ERK2 or each p38 MAPK isoform (5 mU) in 30 μl of a medium comprising 20 mM Tris-Cl (pH 7.5), 10 mM MgCl2, 1 mM EGTA, 1 μM PKI, 10 mM DTT, 10 μg/ml leupeptin, 1 mM Na3VO4, 5 μg hsp27 and 10 μM [γ-32P]ATP. MBP was employed to demonstrate that reactions contained similar levels of p38 or ERK activity. After 1 h at 30 °C, reactions were stopped by the addition of 10 μl 4× SDS-PAGE sample buffer, heated at 70 °C for 90 s, and the phosphorylated products analyzed by SDS-PAGE and autoradiography.
2.4. Kinetic assays
Different concentrations of purified GST-MK3.1, GST-MK3.2, GST-MK2 or GST-MK5 fusion proteins were incubated with phosphorylated, activated forms of each p38MAPK isoformin a buffer containing 20 mM Tris-Cl (pH 7.5), 10mM MgCl2, 1mM EGTA, 1μM PKI, 10 mM DTT, 10 μg/ml leupeptin, 1 mM Na3VO4, 100 μM [γ-32P]ATP (3.5 Ci/mmol), and 167 mU/ml of p38 MAPK activity. The reaction volume was 450 μl. The activity of each lot of activated p38MAPKwas verified upon receipt using MBP as the phosphate acceptor: one unit of p38 MAPK activity is defined as 1 nmol/min of phosphate incorporated into 0.5 mg/ml MBP at 30 °C in the presence of 100 μM ATP. This ensured that lot-to-lot differences in the purity of activated p38 did not result in variability in the concentration of activated p38 employed in the reaction medium. To determine the reaction velocity (V) at each substrate concentration ([S]), reactions were allowed to proceed for various incubation times (0–60 min at 5 min intervals) at 30 °C, then stopped by transfer of 30 μl of reaction mix to a microcentrifuge tube containing 10 μl of 4× SDS-PAGE sample buffer, heating to 70 °C for 90 s and resolved on 10–20% acrylamide-gradient SDS-PAGE. Gels were stained using Coomassie Brilliant Blue R-250, dried, bands corresponding to the phosphorylated substrate protein (i.e., MBP, MK2, MK3, MK5) excised and 32P incorporation determined by liquid scintillation counting. The specific activity of the 100 μM [γ-32P]ATP was also determined by scintillation counting following appropriate dilution with water. The slope of the linear region of a plot of phosphate incorporation versus time, determined by linear regression, was taken as V for each substrate concentration and p38 isoform. The kinetic parameters Km and Vmax were calculated from [S] and the experimentally determined values for V by a non-linear regression curve fit of a parabola to the Michaelis–Menten formula y=Vmax(x(Km+x)−1) using Graphpad Prism™ software (Version 4.0c for Mac). For kinetic assays, the protein concentration for each recombinant MK was determined by resolving both the MK and a standard curve of BSA on SDS-PAGE, staining with Coomassie Brilliant Blue R-250, and quantifying the intensity of the appropriate bands using a gel imaging system and Bio-Rad Quantity One® software (Version 4.5.2 for Mac).
2.5. Binding of MK3-V5 to GST-p38
HEK cells transfected with either V5 epitope-tagged MK3.1 or MK3.2 were rinsed with cold TBS and lysed in ice-cold lysis buffer (50 mM Tris–HCl, pH 7.5 at 5 °C, 20 mM β-glycerophosphate, 20 mM NaF, 5 mM EDTA, 10 mM EGTA, 1 mM Na3VO4, 1 μM microcystin LR, 10 mM benzamidine, 0.5 mM PMSF, 10 μg/ml leupeptin, 5 mM DTT, and 1% TX-100). Lysates (1 mg) were incubated with 1 μg of GST-p38 (α, β, γ, or δ) or GST for 1 h at 5 °C in 200 μl of binding buffer containing 50 mM Tris–HCl (pH 7.5 at 5 °C), 270 mM sucrose, 50 mM NaF, 1 mM EDTA, 1 mM EGTA, 1 mM Na3VO4, 1mM sodium pyrophosphate, and 1% TX-100. Glutathione-Sepharose (30 μl of a 50% suspension) was added and the lysates incubated at 5 °C for an additional 30 min. The beads were washed 3 times with binding buffer, twice with TBST, and then resuspended in 50 μl of 1× SDS-sample buffer. Co-precipitated MK3.1-V5 or MK3.2-V5 was detected by SDS-PAGE and then immunoblotting with V5-specific antiserum.
2.6. Confocal microscopy
The intracellular localization of the MK3.1-V5 and MK3.2-V5 proteins were studied using a scanning confocal fluorescence microscope (LSM 510 Carl Zeiss, Oberkochen, Germany) as described previously [28,29]. HEK cells expressing V5 epitope-tagged proteins were plated on laminin-coated coverslips overnight (37 °C, 95% O2–5% CO2), starved for 16 h, and then incubated for 15 min in the presence or absence of 0.3 Msorbitol. Cells were fixed for 20 min in 2% paraformaldehyde solution, rinsed three times in PBS, blocked in a solution containing 2% donkey serum and 0.2% TX-100 and incubated overnight at 4 °C in PBS containing 1% donkey serum, 0.05% TX-100 and V5 antibody (1:200). The coverslips were then rinsed with PBS, drained, incubated for 1 h at room temperature with the appropriate secondary antibody, washed with PBS, drained, and mounted onto glass slides using a drop of 0.1% DABCO/glycerol medium.
2.7. Preparation of murine cardiac lysates
Hearts were rapidly removed, trimmed, rinsed in ice-cold TBS, snap-frozen in liquid nitrogen and pulverized under liquid nitrogen using a mortar and pestle. The powdered tissue was resuspended, using a 2 ml Potter–Elvehjem tissue grinder (15 passes), in 1.25 ml of ice-cold lysis buffer (see above). Homogenates were then cleared of insoluble cellular debris by centrifugation for 30 min at 100000×g (48000 rpm) and 4 °C in a Beckman TLA-100.3 rotor. Finally, supernatants were collected, aliquoted, snap-frozen using liquid nitrogen, and stored at −80 °C.
2.8. Miscellaneous methods
Protein concentrations were determined by the method of Bradford [30] using γ-globulin as standard.
2.9. Nucleotide sequence accession number
The sequence for murine MK3.2 has been deposited in GenBank (accession number DQ899946).
3. Results
3.1. Cloning and sequencing of MK3 cDNAs
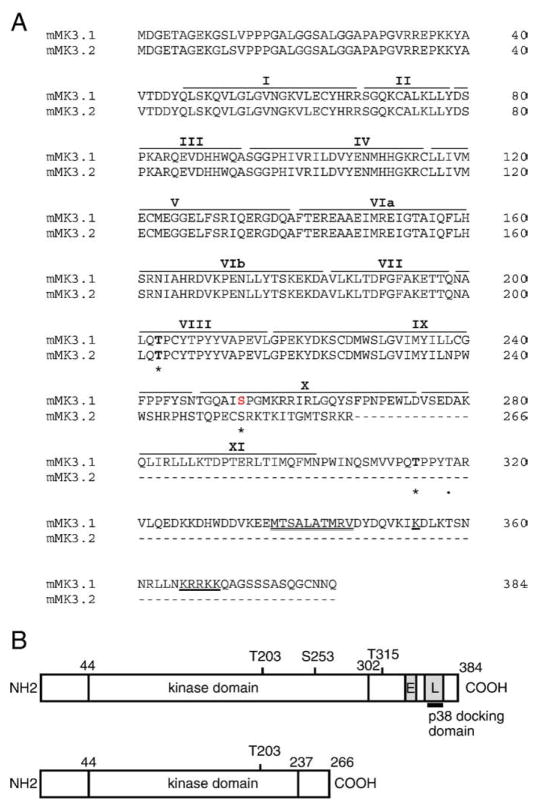
MK3 cDNA was cloned from total RNA isolated from murine ventricular myocardium. Initial attempts to amplify full-length MK3 were unsuccessful and hence overlapping 5′ and 3′ segments were amplified separately. The forward and reverse primers employed for PCR amplification of MK3 were designed according to the mouse MK3 cDNA sequence on NCBI (XM_204309) and are shown in Supplemental Table 1. PCR using the 5′ primer set always resulted in a single product. Upon ligation into pCR2.1 and amplification, cDNA from 3 clones was purified and sequenced. All other clones were confirmed by restriction mapping. The sequences of all 5′ clones were identical to the known sequence for the corresponding region of murine MK3. In contrast, upon amplifying the 3′-terminal fragment two products, differing in size on agarose gels, were obtained. Cloning and sequencing of the 3′ fragments revealed the larger of the two to be identical to the 3′ sequence for MK3 already in the database. Both 3′ fragments were ligated to the 5′ fragment as described in Materials and methods. Based upon their relative size, we will refer to the form encoding full-length MK3 as MK3.1 and the novel shorter form as MK3.2. Alignment of MK3.1 and MK3.2 with the full-length mRNA encoding MK3 (NM_178907.1) and the murine genomic sequence (gene ID 102626) indicated that MK3.2 (GenBank accession DQ899946) is a splice variant resulting from the exclusion of exons 8 and 9. Skipping exons 8 and 9 produced a shift in reading frame in exon 10, resulting in an mRNA wherein translation continues for an additional 29 amino acids followed by an in-frame stop codon. Hence, MK3.2, with an open reading frame of 798 bp, putatively encodes a 266 aa protein, whereas MK3.1 has an open reading frame of 1152 bp and encodes a 384 aa protein. The predicted amino acid sequence for the C-terminal of MK3.2, starting at Leu-237, differs from that of MK3.1 (Fig. 1A,B), and similar sequence was not found in any other protein in the Swissprot database. MK3.2 is missing (i) part of the conserved protein kinase subdomain IX and entire subdomains X and XI, (ii) two (Ser-253 and Thr-315) of the three potential proline-directed phosphorylation sites for p38 (Thr-203, Ser-253, and Thr-315), (iii) the nuclear localization signal (NLS) [19] overlapping with (iv) the putative p38α/β-docking site, and finally (v) also the nuclear export signal (NES) [21] (Fig. 1B). MK3.1 and MK3.2 were also obtained using RNA isolated from adult mouse ventricular myocytes (not shown).
Fig. 1.
cDNA and amino acid alignment of MK3 splice variants. (A) Predicted amino acid sequence of MK3.1 and MK3.2. Putative sites of phosphorylation by p38 MAPK (Thr-203, Ser-253, and Thr-315) are indicated by an asterisk and the auto-phosphorylation site (Thr-319) is indicated by a dot. Roman numerals above refer to the subdomains (I–IX) conserved in all protein kinases. The consensus bipartite nuclear localization signal is underlined whereas the putative nuclear export sequence is indicated by a double underline. Dashes represent gaps. (B) Schematic representation of MK3.1 and MK3.2. ‘E’ nuclear export signal; ‘L’, nuclear localization signal.
3.2. In vivo expression of MK3.1 and MK3.2
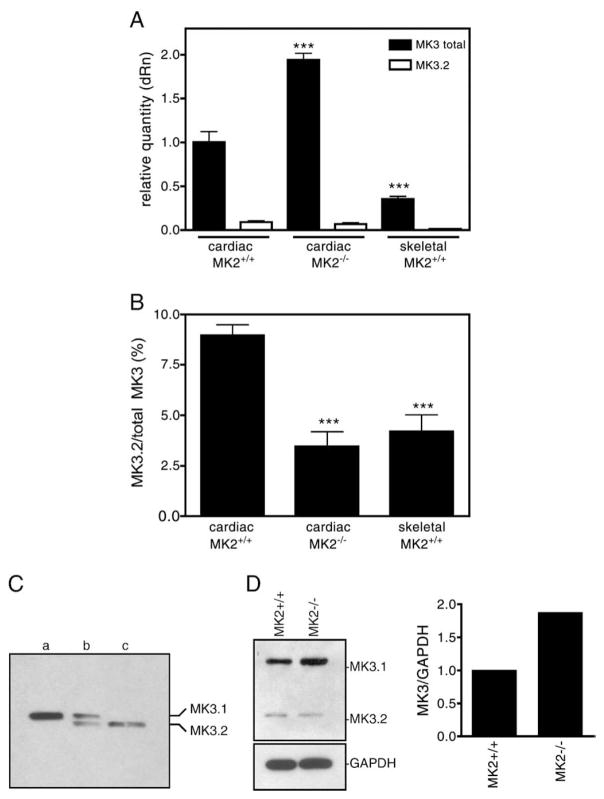
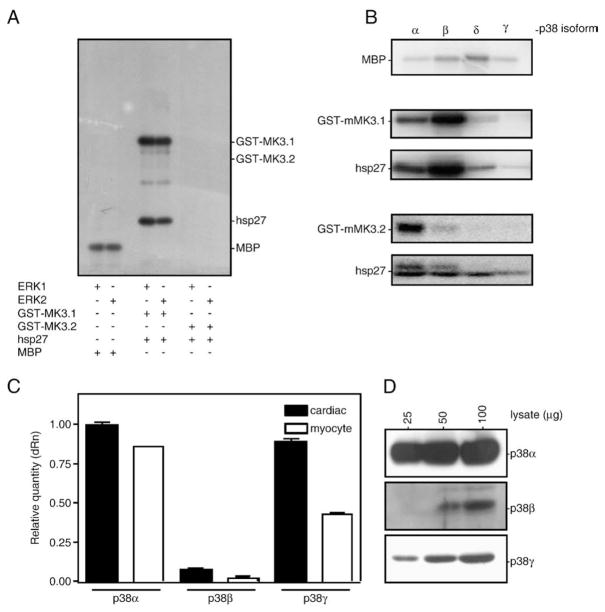
To confirm the existence of MK3.2 mRNA in vivo, a real-time quantitative PCR (qPCR) approach was employed. Primers were designed according to the mouse MK3 cDNA sequence in the NCBI database (XM_204309), using Primer Express (Version 2, ABI). Primers for total MK3 flanked the boundary between exons 4 and 5, which is common to both MK3.1 and MK3.2. To selectively detect MK3.2, the forward primer was specific to a sequence in exon 5 whereas the reverse primer was designed to span the exon 7/10 junction, a sequence only present if exons 8 and 9 were absent. To detect MK3.1, primers that flanked the boundary between exons 8 and 9 were chosen (not shown). Expression levels of the splice variants in both cardiac and skeletal muscle were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH, stable expression in all samples) expression (Fig. 2A). Based upon melting-curve analysis, MK3.2-selective primers amplified a single product from total mouse heart RNA, which sequencing confirmed to be the appropriate target sequence. Both cardiac and skeletal muscles express high levels of MK3 mRNA relative to other tissues [13]: the amount of total MK3 mRNA detected in the heart was 2.7-fold greater than that in skeletal muscle (Fig. 2A). Analysis of the abundance of MK3.2 mRNA revealed that the ratio of MK3.2/total MK3 mRNA was 2-fold greater in RNA isolated from the ventricular myocardium than RNA from skeletal muscle (Fig. 2B). Interestingly, mRNA levels of MK3.2 did not vary with pressure-overload induced hypertrophy nor through the different stages of post-natal development (data not shown).
Fig. 2.
Detection of the MK3.1 and MK3.2 in murine ventricular myocardium. (A) The relative expression levels of total MK3 and MK3.2 mRNA were measured in RNA isolated from murine ventricular myocardium (filled bars) or skeletal muscle (open bars) of wild-type (MK2+/+) or MK2−/− mice by qPCR. Values are normalized to the amount of total MK3 transcript in hearts from MK2+/+ litter mates. (B) Abundance of MK3.2 mRNA expressed as percentage of total MK3 mRNA. (C) MK3.1 and MK3.2 protein are detected in mouse heart by immunoblotting. Recombinant GST-MK3.1 (lane a) and GST-MK3.2 (lane c), treated with thrombin to remove the GST moiety, were electrophoresed alongside 100 μg of lysate prepared from mouse ventricular myocardium (lane b). (D) Detection of MK3 protein in lysates from MK2−/− hearts and MK2+/+ litter mates by immunoblotting. Shown on the right, immunoreactive bands were quantified and normalized to GAPDH content. qPCR data are the mean and S.E. (n=8–10). ***, p<0.001 vs. total MK3 in MK2+/+ hearts (A) or MK3.2/total MK3 in MK2+/+ hearts (B); one-way ANOVA with Newman–Keuls post-hoc analysis.
Human MK3 and MK2 show sequence identity of 75% at the amino acid level [13,19] and results obtained employing MK2−/− and MK3−/− mice indicate that MK3 and MK2 may serve similar physiological function [23]. In light of this similarity, we examined the possibility that the absence of functional MK2 may result in compensatory changes in the expression or splicing of MK3. Analysis of MK3 mRNAs in RNA isolated from hearts of MK2−/− mice [22] by real-time PCR revealed a 2-fold increase in total MK3 mRNA (Fig. 2A) and a 3-fold reduction in the quantity of MK3.2 relative to total MK3 mRNA, compared with wild-type (Fig. 2B). Hence, the absence of functional MK2 results in compensatory changes in both the amount of MK3 mRNA and a reduction in exon skipping that results in formation of MK3.2 mRNA.
The expression of MK3.1 and MK3.2 at the protein level was examined in the heart by immunoblotting using antisera directed against the N-terminus of MK3. Two immunoreactive bands were revealed in lysates prepared from intact murine ventricular myocardium and the upper and lower bands displayed mobilities on SDS-PAGE similar to recombinant MK3.1 and MK3.2 (see below), respectively, once the GST moiety was removed with thrombin (Fig. 2C). Furthermore, in the MK2-deficient hearts, immunoblotting indicated that MK3 protein levels were about twice that detected in wild-type litter mates (Fig. 2D).
3.3. Subcellular localization of MK3 splice variants
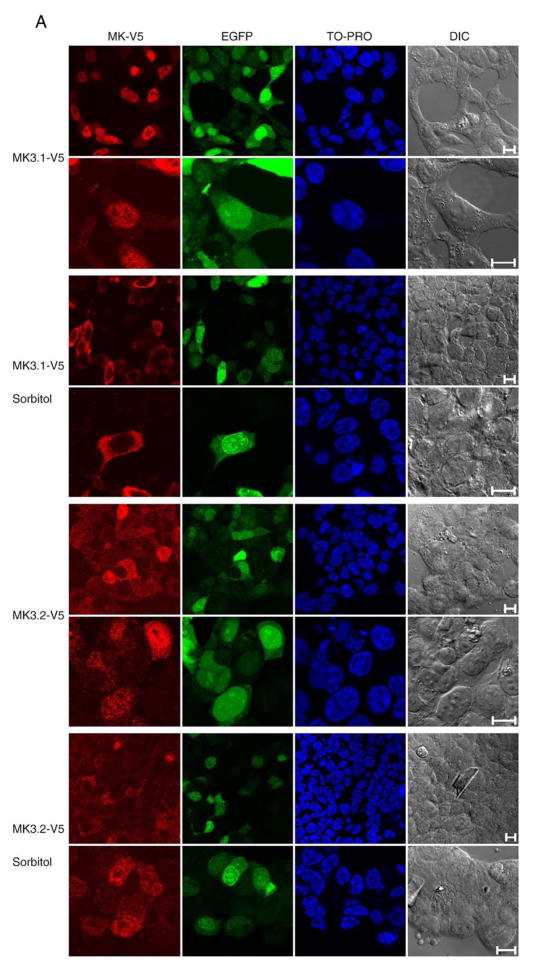
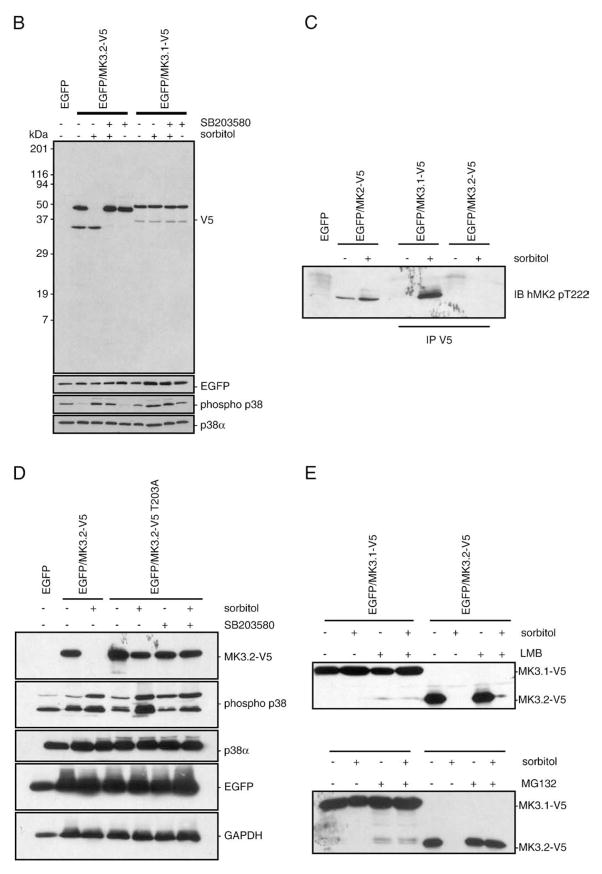
Inactive MK3 is sequestered in the nucleus [18,31]; however, the C-terminal of MK3.2 does not include the NLS and NES found in MK3.1 (Fig. 1C). To compare the subcellular localization ofMK3.1 and MK3.2, constructs were engineered in a pIRES-EGFP vector wherein MK3.1 orMK3.2 were modified to include a C-terminal V5 epitope tag and inserted upstream of the internal ribosomal entry site. pIRES is a bicistronic vector that allows 2 separate proteins (e.g., MK3 and EGFP) to be simultaneously expressed from the same mRNA. Immunoblotting for EGFP or the V5 epitope tag in lysates of HEK293t cells, transfected with either pIRES-MK3.1-V5-EGFP or pIRES-MK3.2-V5-EGFP, demonstrated that both forms of MK3 were expressed (Fig. 3). To determine the subcellular localization of MK3.1 and MK3.2, and the effect of activation upon this localization, HEK293t cells, transfected with either pIRES-MK3.1-V5-EGFP or pIRES-MK3.2-V5-EGFP, were serum-starved and then incubated in the presence of 0.3 Msorbitol for 15 min to stimulate p38 MAPK [19]. The intracellular localization of MK3.1-V5 and MK3.2-V5 was revealed by decorating fixed cells with anti-V5 antisera and imaging using a Zeiss LSM-510 confocal fluorescence microscope. MK3.1-V5 localized to the nucleus in non-stimulated HEK293t cells whereas MK3.2-V5 showed a more uniform subcellular distribution (Fig. 3A). However, in response to hyperosmotic stress MK3.1-V5 translocated to the cytosol, whereas the total cellular level of MK3.2-V5 immunoreactivity appeared to be reduced. Subsequent analysis of MK3.1-V5 and MK3.2-V5 by SDS-PAGE and immunoblotting in lysates from sorbitol-treated cells confirmed the loss of MK3.2-V5 immunoreactivity (Fig. 3B). Furthermore, pretreatment with the p38α/β inhibitor SB203580 (10 μM) prevented the sorbitol-induced breakdown of MK3.2. In contrast, phorbol 12-myristate 13-acetate (0.1 μM, 15 min), a potent activator of ERK1/2, did not induce the breakdown of MK3.2 (data not shown). To confirm that sorbitol was indeed inducing the phosphorylation of MK3 at p38-specific sites, an antibody against phosphorylated Thr-222 of human MK2 was employed. This site is conserved in mouse MK2 (Thr-208) and MK3 (Thr-203). Phosphorylation of MK3.1-V5 was observed following exposure to osmotic stress; however, no phosphorylated MK3.2-V5 was detected in V5 immunoprecipitates (Fig. 3C). One explanation for these data is that MK3.2 becomes unstable and is rapidly degraded upon phosphorylation by p38. To further examine this possibility, Thr-203, the only remaining site for phosphorylation by p38 in MK3.2, was mutated to alanine. MK3.2 T203A was no longer degraded in response to osmotic stress (Fig. 3D). Blocking nuclear export of MK3.2 with leptomycin B did not alter the stability of MK3.2-V5 whereas the preincubation with the proteosome inhibitor MG132 prevented the sorbitol-induced breakdown of MK3.2 (Fig. 3E). Taken together, these results suggest that phosphorylation of exogenously expressed MK3.2-V5 at T203 resulted in its rapid degradation by the proteosome.
Fig. 3.
Characterization of recombinant, expressed MK3.1-V5 and MK3.2-V5. (A) Subcellular localization of MK3.1-V5 or MK3.2-V5 in response to osmotic stress. HEK293t cells were transfected with pIRES-MK3.1-V5-EGFP or pIRES-MK3.2-V5-EGFP, starved, stimulated for 15 min with 0.3 M of sorbitol, fixed, labeled with V5 antisera and visualized by confocal fluorescence microscopy. Scale bar is 10 μM. (B) Osmotic stress induces a rapid breakdown of MK3.2-V5. HEK293t cells were transfected with pIRES-EGFP, pIRES-MK3.1-V5-EGFP or pIRES-MK3.2-V5-EGFP and, where indicated, were starved, stimulated for 15 min with 0.3Mof sorbitol, rinsed with cold TBS, lysed, and V5 immunoreactivity revealed following SDS-PAGE and immunoblotting. Where indicated, cells were pretreated with SB203580 (10 μM) for 10 min and then maintained for an additional 15 min in the presence of SB203580 and in the presence or absence of 0.3M sorbitol. Membranes were then stripped using 0.2 M NaOH (2×10 min) and reprobed using EGFP-specific antisera. In separate immunoblots, employing the same samples, filters were probed for phosphorylated p38 MAPK, stripped, and reprobed for total p38α immunoreactivity. Numbers at the left indicate the positions of the molecular mass marker proteins (in kDa). (C) Osmotic stress induces the phosphorylation of MK3. HEK293t cells were transfected with pIRES-EGFP, pIRES-MK2-V5-EGFP, pIRES-MK3.1-V5-EGFP or pIRES-MK3.2-V5-EGFP and, where indicated, were starved, stimulated for 15 min with 0.3 Mof sorbitol, rinsed with cold TBS, lysed, and probed with an antibody against human MK2phospho-threonine-222 following SDS-PAGE and immunoblotting. MK3.1-V5 and MK3.2-V5 were immunoprecipitated from lysates (1 mg) using an anti-V5 antibody. (D) MK3.2-V5T203A does not degrade in response to osmotic stress. HEK293t cells were transfected with pIRES-EGFP, pIRES-MK3.2-V5-EGFP or pIRES-MK3.2-V5 T203A-EGFP and, where indicated, were treated as described for panel B. (E) Effect of leptomycin B and MG132 upon the stability of MK3.2 during osmotic stress. HEK293t cells were transfected with pIRES-MK3.1-V5-EGFP or pIRES-MK3.2-V5-EGFP and, where indicated, were starved, stimulated for 15 min with 0.3 M of sorbitol, rinsed with cold TBS, lysed, and V5 immunoreactivity revealed following SDS-PAGE and immunoblotting. Where indicated, cells were pretreated with leptomycin B (LMB; 20 nM, 30 min) [21] or MG132 (50 μM, 4 h) [59] and then maintained for an additional 15 min in the presence of LMB or MG132 and in the presence or absence of 0.3 M sorbitol. Cells were then rinsed with cold TBS, lysed, and V5 immunoreactivity revealed following SDS-PAGE and immunoblotting.
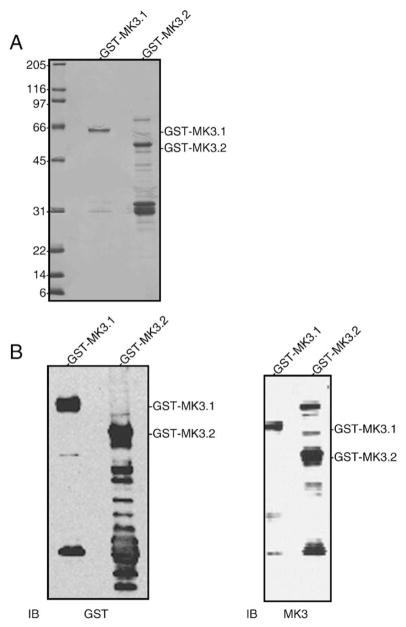
3.4. Expression and purification of recombinant MK3.1 and MK3.2
Full-length MK3.1 and MK3.2 cDNAs were cloned into bacterial expression vector pGEX-2T to produce fusion proteins with GST at the N-terminus, thus permitting purification of the MK3 splice variants as GST-fusion proteins by affinity chromatography on glutathione Sepharose. Induction with IPTG followed by purification on glutathione Sepharose and SDS-PAGE revealed GST-MK3.1 and GST-MK3.2 fusion proteins of approximately 65-kDa and 50-kDa, respectively, were expressed (Fig. 4A). The identity of these bands was confirmed by immunoblotting using antisera against both GST and the N-terminus of MK3 (Fig. 4B). Interestingly, although affinity purification resulted in relatively pure preparations of GST-MK3.1 (Fig. 4A), preparations of GST-MK3.2 were characterized by numerous other bands. Immunoblotting with antisera to GST or MK3 also revealed many bands in GSTMK3.2 preparations following affinity purification (Fig. 4B). As these bands were generally absent in preparations of GST-MK3.1, GST-MK3.2 may be prone to degradation.
Fig. 4.
Expression of recombinant MK3 splice variants. (A) Aliquots of purified GST-MK3.1 and GST-MK3.2 were resolved by 10–20% acrylamide-gradient SDS-PAGE then stained with Coomassie blue. (B) Purified GST-MK3.1 and GST-MK3.2 were separated by 10–20% acrylamide-gradient SDS-PAGE, transferred to nitrocellulose and probed with antisera raised against GST (left) or MK3 (right). Numbers at the left indicate the positions of the molecular mass marker proteins (in kDa).
3.5. Activation of the MK3.1 and MK3.2 by ERK1/2 and p38 MAPKs
To compare the phosphorylation and activation of MK3.1 and MK3.2 by ERK1/2 and p38 MAPKs, in vitro kinase assays were performed. Based upon a sequence alignment of MK3 with MK2, there are 3 putative proline-directed sites in human MK3 for phosphorylation by p38 MAPK: Thr-201, Ser-251 and Thr-313 [19], which correspond to Thr-203, Ser-253 and Thr-315 in murine MK3. ERK1/2 phosphorylates human MK3 at Thr-201 and Thr-313 [1] whereas p38 may phosphorylate all 3 sites. The predicted amino acid sequence for MK3.2 indicates the loss of Thr-315 and the replacement of Pro-254 with an arginine. As a result of this latter change, Ser-253 no longer lies within a p38 consensus phosphorylation sequence. Thus, 2 out of 3 potential sites for phosphorylation by p38 are absent in MK3.2. To determine if both MK3.1 and MK3.2 are phosphorylated and activated by ERK1/2 or p38 MAPK, purified recombinant GST-MK3.1 or GST-MK3.2 was incubated with hsp27, an in vitro substrate of MK3 [1], together with the phosphorylated, activated forms of ERK1, ERK2 (Fig. 5A) or each p38 MAPK isoform (Fig. 5B) in kinase buffer containing [γ-32P]ATP and Mg2+. As p38α, β, and γ mRNAs were detected in RNA from both heart and ventricular myocytes (Fig. 5C) and p38α, β, and γ were detected at the protein level (Fig. 5D), all 4 isoforms of p38 were examined in order to assess potential differences in the ability of GST-MK3.1 and/or GST-MK3.2 to act downstream in the p38 cascade. Low levels of p38δ mRNA were also detected (not shown). To ensure that similar levels of activity were employed in each case, the specific activity of ERK1/2 and each p38 isoform was determined using MBP as a substrate. Although Thr-203 remained unaffected by splicing, GST-MK3.1, but not GST-MK3.2, was phosphorylated by ERKs 1 and 2. When phosphorylated by ERK1/2, GST-MK3.1 was activated and phosphorylated hsp27. Both GST-MK3.1 and GST-MK3.2 were phosphorylated by p38α and p38β (Fig. 5B); however, GST-MK3.2 was phosphorylated to a much lesser extent than GST-MK3.1. Phosphorylation of GST-MK3.1 and GSTMK3.2 by p38α/β resulted in increased catalytic activity towards hsp27. In contrast, both MK3 variants were poor substrates for p38δ and p38γ (Fig. 5B).
Fig. 5.
Phosphorylation of MK3 splice variants by ERK1/2 and p38 MAPK isoforms. Purified proteins GST-MK3.1 and GST-MK3.2 were incubated for 1 h at 30 °C with phosphorylated, activated forms of (A) ERK1, ERK2 or (B) each p38 MAPK isoform (5 mU). The specific activity of each MAPK was determined using myelin basic protein (5 μg of MBP). Following phosphorylation, samples were solubilized using SDS-PAGE sample buffer and separated using SDS-PAGE. Gels were dried and 32P incorporation determined by autoradiography. (C) The relative expression levels of p38α, p38β, and p38γ mRNA were measured in RNA isolated from murine cardiac ventricular myocardium (filled bars) or cardiac ventricular myocytes (open bars) of wild-type mice by qPCR using the primers listed in Supplemental Table 2. Quantitation by qPCR was performed in triplicate: the results shown are the means (±S.E.) of 3 (heart) or 2 (myocyte) biological replicates. (D) The expression of p38α, p38β, and p38γ proteins were examined in the indicated amounts of murine ventricular lysate by immunoblotting using isoform-specific antisera.
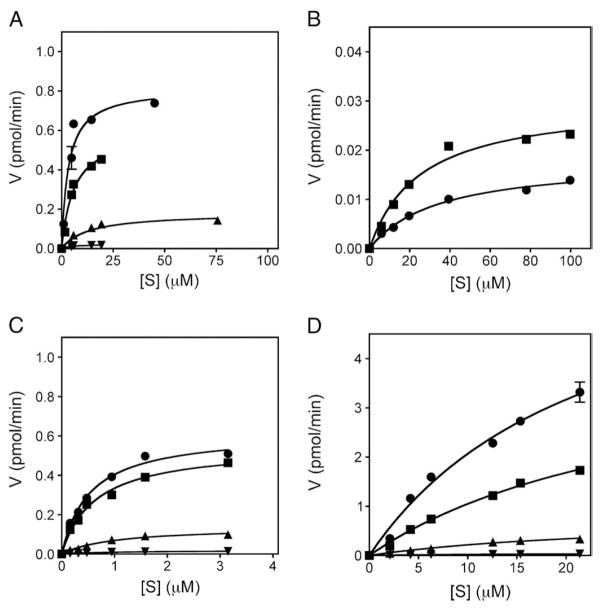
To further characterize the ability of GST-MK3.1 and GST-MK3.2 to serve as substrates for p38 MAPKs, and thus their role in p38 signaling, kinetic assays were performed using the four p38 MAPK isoforms and MK3.1 and MK3.2 as well as MK2 and MK5 as controls (see Materials and methods). As shown previously [32], MK3.1 was phosphorylated by p38α and β, and to a lesser extent δ and γ (Fig. 6A, Table 1). MK3.2 was only poorly phosphorylated by p38α and β (Fig. 6B, Table 1), this being characterized both by a higher Km and by Vmax values that were over 20-fold lower than observed for MK3.1. Due to the low levels of phosphate incorporation into MK3.2 catalyzed by p38γ and δ (Fig. 5B), further kinetic analysis was not undertaken. MK2, which is 70% identical to MK3, was phosphorylated to similar levels as MK3.1 by each isoform of p38 MAPK (Fig. 6C, Table 1). Phosphorylation of MK5 was characterized by Km values similar to those of MK2 and MK3, but 10–100-fold greater Vmax. In fact, under the in vitro conditions employed herein, MK5 was the substrate with the largest Vmax for each p38 isoform (Fig. 6D, Table 1). In each case, the observed kinetic constants indicated that the MKs were better substrates for p38α and β than for δ and γ.
Fig. 6.
Kinetic analysis of p38 MAPK-mediated phosphorylation of MK2, MK3 and MK5. Different amounts of purified GST-MK3.1 (A), GST-MK3.2 (B), GST-MK2 (C), or GST-MK5 (D) were incubated with the phosphorylated, activated forms of p38α (■), p38β (●), p38δ (▲), or p38γ (▼). A time course of 32P incorporation was performed at 30 °C for each indicated [S] as described in Materials and methods. Following phosphorylation, samples were solubilized using SDS-PAGE sample buffer and separated using SDS-PAGE. Gels were stained with Coomassie Brilliant Blue R-250, dried, the substrate band excised, and 32P incorporation determined by liquid scintillation counting. The specific activity of the [γ32P]ATP was determined for each experiment.
Table 1.
Kinetic parameters describing p38 MAPK isoform-mediated phosphorylation of MK2, MK3, and MK5.
| p38α | p38β | p38δ | p38γ | |
|---|---|---|---|---|
| Km (nM) | ||||
| MK2 | 15.2 | 12.7 | 23.7 | 22.7 |
| MK3.1 | 6.01 | 5.54 | 13.9 | 3.40 |
| MK3.2 | 19.0 | 25.6 | nd | nd |
| MK5 | 17.5 | 14.8 | 15.8 | 8.91 |
| Vmax (pmol/min) | ||||
| MK2 | 0.546 | 0.618 | 0.138 | 0.0185 |
| MK3.1 | 0.537 | 0.883 | 0.208 | 0.0214 |
| MK3.2 | 0.0296 | 0.0177 | nd | nd |
| MK5 | 2.59 | 4.62 | 0.503 | 0.0402 |
| Vmax/Km | ||||
| MK2 | 36 | 49 | 5.8 | 0.82 |
| MK3.1 | 89 | 160 | 8.4 | 2.4 |
| MK3.2 | 1.6 | 0.69 | nd | nd |
| MK5 | 150 | 310 | 32 | 4.5 |
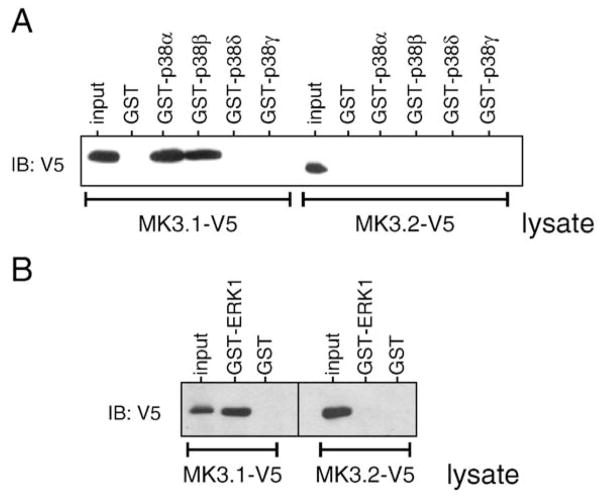
As discussed above, MK3.2 lacks two of the three sites for phosphorylation by p38 and this may underlie its reduced capacity to serve as a substrate for p38; however, p38α and p38β displayed high Vmax for phosphorylation of MK5, which also possesses only a single site for p38. Alternatively, MK3.1 interacts with p38α/β via a docking domain located in its C-terminal and this domain is absent in MK3.2. To study further the interaction between MK3.2 and p38, GST pull-down assays were employed using lysates from cells transfected with either MK3.1-V5 or MK3.2-V5 plus purified recombinant p38 MAPK isoforms (Fig. 7A). MK3.1-V5 bound to p38α and p38β but not p38δ or p38γ whereas MK3.2-V5 was not precipitated by any of the p38 isoforms. In conclusion, MK3.2 was only poorly phosphorylated by p38 and this likely resulted from the absence of a p38α/β-docking domain and, hence, an inability to form high affinity interactions with p38. Similarly, MK3.1-V5, but not MK3.2-V5, was pulled down by GST-ERK1 (Fig. 7B), and this inability to form a high affinity interaction with ERK1 may underlie the ability of ERK1 and ERK2 to phosphorylate MK3.1-V5 but not MK3.2-V5 (Fig. 5A).
Fig. 7.
MK3.2 does not bind to p38 MAPK or ERK1. To determine if there are direct interactions between MK3.2 and p38 (A) or ERK1 (B), lysates from HEK cells transfected with either MK3.1-V5 or MK3.2-V5 (1 mg) were incubated with recombinant GSTp38α, GST-p38β, GST-p38δ, GST-p38γ, GST-ERK1 (1 μg) or GST. Co-precipitated MK3.1-V5 or MK3.2-V5 was detected by immunoblotting using anti-V5 antisera.
4. Discussion
We have demonstrated the presence of a novel splice variant of MK3, which we have termed MK3.2, in murine striated muscle. MK3.2, with an open reading frame of 798 bp, putatively encodes a 266 aa protein, whereas MK3.1 has an open reading frame of 1152 bp and encodes a 384 aa protein. The predicted amino acid sequence for the C-terminal of MK3.2, starting at Leu-237, is unique and differs from that of MK3.1. As a result, MK3.2 is lacking part of conserved protein kinase subdomain IX and all of subdomains X and XI. Furthermore, two (Ser-253 and Thr-315) of three potential phosphorylation sites for p38 (Thr-203, Ser-253, and Thr-315), both nuclear localization [19] and nuclear export [21] signals, and a putative p38-docking domain are also absent. Northern analysis of MK3 expression revealed a major mRNA of 3.5-kb as well as a minor mRNA of 2-kb that was absent in brain and showed highest abundance in cardiac and skeletal muscle [19]. Quantitative realtime PCR analysis revealed a predominance of mRNA for the MK3.1 variant with respect to that of the MK3.2 variant in mouse heart. MK3.2 was also detected in skeletal muscle and, interestingly, the abundance of MK3.2 mRNA, relative to the level of total MK3 transcript, was less in skeletal muscle than in the heart. MK3 and MK2 show sequence identity of about 75% [13,19] and may serve similar physiological functions [23]; hence, we examined the possibility of compensatory changes in the expression or splicing of MK3 in the absence of functional MK2. Although MK3 expression at the protein level was reportedly unaltered in MK2−/− mice [23], the abundance of total MK3 mRNA was 2-fold greater in MK2-deficient hearts (Fig. 2A) and the relative abundance of MK3.2 mRNA was 3-fold lower (Fig. 2B). A comparable increase in MK3.1 protein was also observed (Fig. 3D). Hence, in response to a deficiency of MK2, there was a compensatory increase in MK3 mRNA as well as altered splicing of theMK3 transcript; however, it is not currently known if the increase in mRNA resulted from alterations in promoter activity versus mRNA stability.
MK2 and MK3 contain both functional NLS [13,19,33,34] and NES [21] in their C-termini. The NLS is accessible in both the inactive and active forms of MK2 whereas access to the NES is prevented by intramolecular interactions when MK2 is inactive. Phosphorylation at Thr-317 by p38 MAPK induces a conformational change, activating MK2 and freeing the NES. As a result, MK2 is rapidly exported from the nucleus. Hence, these motifs function together to modulate the subcellular distribution of MK2 in a phosphorylation-dependent manner [21]. Regulation of MK3 is thought to follow the same pattern. MK3.2 lacks both NLS and NES, and ectopically expressed MK3.2-V5 displayed uniform distribution in unstimulated HEK cells. In contrast, a fusion protein comprising GFP plus MK2 C-terminal amino acids 315–383, containing both the NLS and the NES, localized exclusively to the cytosol [21]. Unexpectedly, whereas MK3.1 translocated to the cytosol in response to osmotic stress, MK3.2 was degraded and this breakdown required the activation of an SB203580-sensitive p38 MAPK, a phosphorylatable residue at amino acid position 203, and an active proteosome.
Differences in substrate selectivity indicate that signaling through the p38 cascade diverges following activation of the p38 MAPKs [35–37]. Activation of MK2 and MK3 in vivo is mediated by SB203580-sensitive p38 MAPK isoforms α and β [1,19,20]. In vitro, MK3 and MK2 are phosphorylated by p38α and p38β [1,19,35] and p38α phosphorylates MK2 and MK3 with similar substrate specificity constants (kcat/Km) [38]. Phosphorylation of MK2 and MK3 by p38γ has been controversial; however, when observed, the kinetics of phosphorylation do not correlate with those of activation, suggesting that phosphorylation of MK2 and MK3 by p38γ is at sites unrelated to activation [35,39–41]. In the present study, p38α and p38β phosphorylated MK3.2 but to a lesser extent than MK3.1. A splice variant of MK2, MK2b, which lacks a C-terminal p38-docking domain [34,42,43] fails to co-precipitate with p38, suggesting the p38-docking domain is required for MK2 to form a high affinity complex with p38 [34]. Furthermore, disruption or prevention of the formation of this complex using a synthetic peptide corresponding to the p38-docking domain (MK2 residues 370–400) inhibits phosphorylation of MK2 by p38α [44]. Similarly, MK3.1-V5, but not MK3.2-V5, co-immunoprecipitated with p38α or β, suggesting that the lower levels of phosphorylation of MK3.2, relative to MK3.1, observed in vitro were a result of the lack of high affinity binding between MK3.2 and either p38α or p38β. This may also underlie the inability of ERK1/2 to phosphorylate MK3.2, as MK3.1 was pulled down by ERK1 whereas MK3.2 was not. All MKs were poor substrates for p38δ and p38γ. Although missing conserved protein kinase subdomains X and XI, including two of the predicted sites for phosphorylation by p38 MAPK, MK3.2 was catalytically active, albeit weakly, upon phosphorylation. Reduced catalytic activity may result from the absence of components within the catalytic domain; however for full activation, MK3 must also be phosphorylated at two of the three potential regulatory sites [1] and only one of these sites is functional in MK3.2. The relevance of MK3.2 activation is unclear at present as ectopically expressed MK3.2 was degraded upon activation of the p38α/β by osmotic stress.
Within the heart, the p38 cascade is activated by pro-hypertrophic stimuli including endothelin-1, phenylephrine, angiotensin II, mechanical stretch, and pressure overload [45–48]. The targets for p38 MAPK signaling in the heart remain largely unknown. Overexpression of constitutively active MKK3 together with wild-type p38α controls the expression of genes related to cell division, inflammation, cell signaling, cell adhesion and transcription [49]. Forced activation of p38 with arsenite or by transgenic overexpression of constitutively active mutant MKK3bE or MKK6bE is associated with increased fibrosis, negative inotropy, reduced cardiac function, and premature death due to cardiac failure [25,50–52]. Alternatively, left ventricular remodeling secondary to coronary artery ligation is unaffected by ablation of MKK3 [53] suggesting potential redundancy or functional overlap in signaling upstream of p38. Furthermore, the scaffold protein TAB-1 activates p38α in an MKK3-independent manner and p38α activated in this manner is retained within the cytosol, thus targeting a spectrum of substrates distinct from those downstream of MKK3-p38α signaling [54,55]. The role of p38β has not been extensively studied in the heart. Although the results from the in vitro experiments presented herein suggest that p38α and β may play similar roles, others have shown that the functional roles of p38α and β in the myocyte are markedly different [10]. Kinetic analysis revealed that, among the MKs, MK5 was a better substrate for all the p38 isoforms in vitro (Table 1), although its role as a physiological substrate for p38 in vivo is controversial [56]. Furthermore, as shown previously, p38α and β were better activators of the MKs than p38δ and γ. In addition to p38α and β, heart expresses p38γ at both the mRNA and protein levels (Fig. 5C,D; [9]). p38γ and p38α exert opposing effects upon activating protein 1-mediated transcription [37]. Furthermore, the depressed contractile function associated with activation of p38 is not reversed upon pharmacologic inhibition of p38 or expression of a dominant negative mutant p38α [25,51,57] implying the involvement of p38γ. Although p38α and β have been more thoroughly studied due to the availability of selective inhibitors for these isoforms [58], the role of p38γ in regulating cardiac structure and/or function remains unknown.
5. Conclusions
We have identified a novel splice variant of MK3, MK3.2. mRNA encoding MK3.2 was detected in both skeletal and cardiac muscles and the ratio of MK3.2 to total MK3 mRNA differed between these two tissues, implying tissue-specific regulation of maturation of the MK3 transcript. MK3.2, which lacked a p38-docking domain, did not bind p38, was only poorly activated by p38α/β but displayed detectable catalytic activity towards hsp27 in vitro. Interestingly, in resting HEK cells, MK3.1-V5 localized to the nucleus whereas MK3.2-V5 immunoreactivity was both nuclear and cytosolic; however, MK3.1-V5 translocated to the cytosol upon hyperosmotic stress, whereas MK3.2-V5 was degraded. One possible explanation for this would be that it allows for a rapid, selective inactivation of MK3.2-V5 without the requirement for a phosphatase. Future work aimed at identifying such a substrate(s) is critical for understanding the role of MK3 in the heart. Finally, the biological consequence of MK3.2 activation merits further investigation as it may play a distinct role in mediating the cellular response to stress.
Supplementary Material
Acknowledgments
Supported by grants from the Canadian Institutes of Health Research (MOP-77791). BGA was a New Investigator of the Heart and Stroke Foundation of Canada and a Senior Scholar of the Fondation de la Recherche en Santé du Québec (FRSQ).
Abbreviations
- DMSO
dimethylsulfoxide
- DTT
dithiothreitol
- ERK
extracellular signal-related kinase
- FPLC
fast protein liquid chromatography
- GST
glutathione S-transferase
- MAPK
mitogen-activated protein kinase
- MK2
MAPK-activated protein kinase-2
- MK3
MAPK-activated protein kinase-3
- MK5
MAPK-activated protein kinase-5
- MBP
myelin basic protein
- NES
nuclear export signal
- NLS
nuclear localization signal
- PKI
cyclic AMP-dependent protein kinase inhibitory peptide
- PAGE
polyacrylamide gel electrophoresis
- PMSF
phenylmethylsulfonyl fluoride
- TX-100
Triton X-100
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.cellsig.2010.05.019.
References
- 1.Ludwig S, Engel K, Hoffmeyer A, Sithanandam G, Neufeld B, Palm D, Gaestel M, Rapp UR. Mol Cell Biol. 1996;16(12):6687. doi: 10.1128/mcb.16.12.6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tibbles LA, Woodgett JR. Cell Mol Life Sci. 1999;55(10):1230. doi: 10.1007/s000180050369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ammerer G. Curr Opin Genet Dev. 1994;4(1):90. doi: 10.1016/0959-437x(94)90096-5. [DOI] [PubMed] [Google Scholar]
- 4.Cobb MH, Goldsmith EJ. J Biol Chem. 1995;270(25):14843. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- 5.Hanks SK, Quinn AM, Hunter T. Science. 1988;241:42. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 6.Han J, Lee J-D, Bibbs L, Ulevitch RJ. Science. 1994;265:808. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 7.Lee JC, Laydon JT, McDonnell PC, Gallagher TF, Kumar S, Green D, McNulty D, Blumenthal MJ, Heys JR, Landvatter SW, Strickler JE, McLaughlin MM, Siemens IR, Fischer SM, Livi GP, White JR, Adams JL, Young PR. Nature. 1994;372:739. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 8.Kyriakis JM, Avruch J. Physiol Rev. 2001;81(2):807. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 9.Court NW, dos Remedios CG, Cordell J, Bogoyevitch MA. J Mol Cell Cardiol. 2002;3:413. doi: 10.1006/jmcc.2001.1523. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Huang S, Sah VP, Ross J, Jr, Brown JH, Han J, Chien KR. J Biol Chem. 1998;273:2161. doi: 10.1074/jbc.273.4.2161. [DOI] [PubMed] [Google Scholar]
- 11.Angenstein F, Greenough WT, Weiler IJ. Proc Natl Acad Sci U S A. 1998;95(25):15078. doi: 10.1073/pnas.95.25.15078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaestel M. Nat Rev Mol Cell Biol. 2006;7(2):120. doi: 10.1038/nrm1834. [DOI] [PubMed] [Google Scholar]
- 13.Sithanandam G, Latif F, Duh FM, Bernal R, Smola U, Li H, Kuzmin I, Wixler V, Geil L, Shrestha S, Lloyd PA, Bader S, Sekido Y, Tartof KD, Kashuba VI, Zabarovsky ER, Dean M, Klein G, Lerman MI, Minna JD, Rapp UR, Allikmets R. Mol Cell Biol. 1996;16(3):868. doi: 10.1128/mcb.16.3.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landry J, Huot J. Biochem Cell Biol. 1995;73(9–10):703. doi: 10.1139/o95-078. [DOI] [PubMed] [Google Scholar]
- 15.Welsh MJ, Gaestel M. Ann N Y Acad Sci. 1998;851:28. doi: 10.1111/j.1749-6632.1998.tb08973.x. [DOI] [PubMed] [Google Scholar]
- 16.Werz O, Klemm J, Samuelsson B, Radmark O. Proc Natl Acad Sci U S A. 2000;97(10):5261. doi: 10.1073/pnas.050588997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voncken JW, Niessen H, Neufeld B, Rennefahrt U, Dahlmans V, Kubben N, Holzer B, Ludwig S, Rapp UR. J Biol Chem. 2005;280(7):5178. doi: 10.1074/jbc.M407155200. [DOI] [PubMed] [Google Scholar]
- 18.Neufeld B, Grosse-Wilde A, Hoffmeyer A, Jordan BW, Chen P, Dinev D, Ludwig S, Rapp UR. J Biol Chem. 2000;275(27):20239. doi: 10.1074/jbc.C901040199. [DOI] [PubMed] [Google Scholar]
- 19.McLaughlin MM, Kumar S, McDonnell PC, Van Horn S, Lee JC, Livi GP, Young PR. J Biol Chem. 1996;271(14):8488. doi: 10.1074/jbc.271.14.8488. [DOI] [PubMed] [Google Scholar]
- 20.Clifton AD, Young PR, Cohen P. FEBS Lett. 1996;302:209. doi: 10.1016/0014-5793(96)00816-2. [DOI] [PubMed] [Google Scholar]
- 21.Engel K, Kotlyarov A, Gaestel M. EMBO J. 1998;17(12):3363. doi: 10.1093/emboj/17.12.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotlyarov A, Neininger A, Schubert C, Eckert R, Birchmeier C, Volk H-D, Gaestel M. Nat Cell Biol. 1999;1:94. doi: 10.1038/10061. [DOI] [PubMed] [Google Scholar]
- 23.Ronkina N, Kotlyarov A, Dittrich-Breiholz O, Kracht M, Hitti E, Milarski K, Askew R, Marusic S, Lin LL, Gaestel M, Telliez JB. Mol Cell Biol. 2007;27(1):170. doi: 10.1128/MCB.01456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maizels ET, Mukherjee A, Sithanandam G, Peters CA, Cottom J, Mayo KE, Hunzicker-Dunn M. Mol Endocrinol. 2001;15(5):716. doi: 10.1210/mend.15.5.0634. [DOI] [PubMed] [Google Scholar]
- 25.Vahebi S, Ota A, Li M, Warren CM, de Tombe PP, Wang Y, Solaro RJ. Circ Res. 2007;100:408. doi: 10.1161/01.RES.0000258116.60404.ad. [DOI] [PubMed] [Google Scholar]
- 26.Larsen JK, Gerthoffer WT, Hickey E, Weber LA. Gene. 1995;161(2):305. doi: 10.1016/0378-1119(95)00290-m. [DOI] [PubMed] [Google Scholar]
- 27.Chevalier D, Allen BG. Prot Exp Purific. 2000;18:229. doi: 10.1006/prep.1999.1183. [DOI] [PubMed] [Google Scholar]
- 28.Boivin B, Chevalier D, Villeneuve LR, Rousseau E, Allen BG. J Biol Chem. 2003;278:29153. doi: 10.1074/jbc.M301738200. [DOI] [PubMed] [Google Scholar]
- 29.Boivin B, Lavoie C, Vaniotis G, Baragli A, Villeneuve LR, Ethier N, Trieu P, Allen BG, Hébert TE. Cardiovasc Res. 2006;71(1):69. doi: 10.1016/j.cardiores.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 30.Bradford MM. Anal Biochem. 1976;72:248. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 31.Zakowski V, Keramas G, Kilian K, Rapp UR, Ludwig S. Exp Cell Res. 2004;299(1):101. doi: 10.1016/j.yexcr.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 32.Goedert M, Cuenda A, Craxton M, Jakes R, Cohen P. EMBO J. 1997;16(12):3563. doi: 10.1093/emboj/16.12.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engel K, Plath K, Gaestel M. FEBS Lett. 1993;336(1):143. doi: 10.1016/0014-5793(93)81628-d. [DOI] [PubMed] [Google Scholar]
- 34.Stokoe D, Caudwell B, Cohen PTW, Cohen P. Biochem J. 1993;296:843. doi: 10.1042/bj2960843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S, McDonnell PC, Gum RJ, Hand AT, Lee JC, Young PR. Biochem Biophys Res Commun. 1997;235:533. doi: 10.1006/bbrc.1997.6849. [DOI] [PubMed] [Google Scholar]
- 36.Pramanik R, Qi X, Borowicz S, Choubey D, Schultz RM, Han J, Chen G. J Biol Chem. 2003;278(7):4831. doi: 10.1074/jbc.M207732200. [DOI] [PubMed] [Google Scholar]
- 37.Askari N, Diskin R, Avitzour M, Capone R, Livnah O, Engelberg D. J Biol Chem. 2007;282(1):91. doi: 10.1074/jbc.M608012200. [DOI] [PubMed] [Google Scholar]
- 38.Hawkins J, Zheng S, Frantz B, LoGrasso P. Arch Biochem Biophys. 2000;382(2):310. doi: 10.1006/abbi.2000.2005. [DOI] [PubMed] [Google Scholar]
- 39.Li Z, Jian Y, Ulevitch RJ, Han J. Biochem Biophys Res Commun. 1996;228:334. doi: 10.1006/bbrc.1996.1662. [DOI] [PubMed] [Google Scholar]
- 40.Cuenda A, Cohen P, Buee-Scherrer V, Goedert M. EMBO J. 1997;16(2):295. doi: 10.1093/emboj/16.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gum RJ, Young PR. Biochem Biophys Res Commun. 1999;266:284. doi: 10.1006/bbrc.1999.1787. [DOI] [PubMed] [Google Scholar]
- 42.Zu YL, Ai Y, Huang CK. J Biol Chem. 1995;270(1):202. doi: 10.1074/jbc.270.1.202. [DOI] [PubMed] [Google Scholar]
- 43.Zu YL, Wu F, Gilchrist A, Ai Y, Labadia ME, Huang C-K. Biochem Biophys Res Commun. 1994;200:1118. doi: 10.1006/bbrc.1994.1566. [DOI] [PubMed] [Google Scholar]
- 44.Lukas SM, Kroe RR, Wildeson J, Peet GW, Frego L, Davidson W, Ingraham RH, Pargellis CA, Labadia ME, Werneburg BG. Biochemistry. 2004;43(31):9950. doi: 10.1021/bi049508v. [DOI] [PubMed] [Google Scholar]
- 45.Clerk A, Michael A, Sugden PH. J Cell Biol. 1998;142:523. doi: 10.1083/jcb.142.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clerk A, Fuller SJ, Michael A, Sugden PH. J Biol Chem. 1998;723:7228. doi: 10.1074/jbc.273.13.7228. [DOI] [PubMed] [Google Scholar]
- 47.Liang F, Gardner DG. J Clin Invest. 1999;104(11):1603. doi: 10.1172/JCI7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischer TA, Ludwig S, Flory E, Gambaryan S, Singh K, Finn P, Pfeffer MA, Kelly RA, Pfeffer JM. Hypertension. 2001;37(5):1222. doi: 10.1161/01.hyp.37.5.1222. [DOI] [PubMed] [Google Scholar]
- 49.Tenhunen O, Rysa J, Ilves M, Soini Y, Ruskoaho H, Leskinen H. Circ Res. 2006;99(5):485. doi: 10.1161/01.RES.0000238387.85144.92. [DOI] [PubMed] [Google Scholar]
- 50.Liao P, Georgakopoulos D, Kovacs A, Zheng M, Lerner D, Pu H, Saffitz J, Chien K, Ziao R-P, Wang Y. Proc Natl Acad Sci U S A. 2001;98(21):12283. doi: 10.1073/pnas.211086598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liao P, Wang S-Q, Wang S, Zheng M, Zheng M, Zhang S-J, Cheng H, Wang Y, Xiao R-P. Circ Res. 2002;90:190. doi: 10.1161/hh0202.104220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y, Rajashree R, Liu Q, Hofmann P. Am J Physiol. 2003;283:H2578. doi: 10.1152/ajpheart.00365.2003. [DOI] [PubMed] [Google Scholar]
- 53.Clark JE, Flavell RA, Faircloth ME, Davis RJ, Heads RJ, Marber MS. Cardiovasc Res. 2007;74(3):466. doi: 10.1016/j.cardiores.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 54.Fiedler B, Feil R, Hofmann F, Willenbockel C, Drexler H, Smolenski A, Lohmann SM, Wollert KC. J Biol Chem. 2006;281(43):32831. doi: 10.1074/jbc.M603416200. [DOI] [PubMed] [Google Scholar]
- 55.Lu G, Kang YJ, Han J, Herschman HR, Stefani E, Wang Y. J Biol Chem. 2006;281(9):6087. doi: 10.1074/jbc.M507610200. [DOI] [PubMed] [Google Scholar]
- 56.Shi Y, Kotlyarov A, Laaβ K, Gruber AD, Butt E, Marcus K, Meyer HE, Friedrich A, Volk HD, Gaestel M. Mol Cell Biol. 2003;23(21):7732. doi: 10.1128/MCB.23.21.7732-7741.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li M, Georgakopoulos D, Lu G, Hester L, Kass DA, Hasday J, Wang Y. Circulation. 2005;111:2492. doi: 10.1161/01.CIR.0000165117.71483.0C. [DOI] [PubMed] [Google Scholar]
- 58.Zhang J, Shen B, Lin A. Trends Pharmacol Sci. 2007;28:286. doi: 10.1016/j.tips.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 59.Mawji IA, Robb GB, Tai SC, Marsden PA. J Biol Chem. 2004;279(10):8655. doi: 10.1074/jbc.M312190200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.