Abstract
The N-terminal tail of histone H4 is an indispensable mediator for inter-nucleosome interaction, which is required for chromatin fiber condensation. H4K16 acetylation (H4K16Ac) activates gene transcription by influencing both chromatin structure and interplay with nonhistone proteins. To understand the influence of H4K16Ac on inter-nucleosome interaction, we performed a simulation study for the H4 tail in the context of two nucleosomes in neighboring unit cells in the crystal structure. The binding conformation of H4 tail with/without K16Ac was sampled by replica exchange with solute tempering, and the free energy landscape was explored by metadynamics. The results indicate two important features of H4K16: 1) it is the first button to anchor the H4 tail on the adjacent nucleosome; and 2) it is the only acetylation site interacting with the acidic patch. H4K16Ac disrupts the electrostatic interactions of K16, weakens H4 tail-acidic patch binding, and significantly increases H4 tail conformation diversity. Our study suggests that H4K16Ac directly reduces the inter-nucleosome interaction mediated by the H4 tail, which might further encourage the binding of nonhistone proteins on the acidic patch.
Introduction
In the eukaryotic nucleus, the compactness of chromatin determines the accessibility of DNA and fundamentally relates to gene transcription. The hierarchical folding of higher-order chromatin starts from the basic building block, the nucleosome. The nucleosome is formed by ∼150 bp DNA wrapping around a positively charged protein core, consisting of two copies each of histones H2A, H2B, H3, and H4. Each histone protein contains a well folded helical core and disordered terminals, the histone tails. The flexible and positively charged histone tails are essential mediators of intra-/inter-nucleosome interaction and ideal binding partners for nonhistone proteins, thus a key to chromatin folding and compaction.
The specific function of different histone tails in chromatin folding has been intensively investigated (1, 2, 3, 4). All histone tails can influence chromatin compaction and accessibility, depending on salt concentration, construction of the nucleosome arrays, and the type of assembly process; however, the H4 tail probably plays the most important role in inter-nucleosome interaction, because its deletion significantly reduces the compaction of nucleosome arrays (5). Inter-nucleosome interactions mediated by the H4 tail may involve either DNA or histone from the adjacent nucleosome, depending on the compactness of the chromatin fiber. Several nucleosome structures have been described, some of which show an interaction of the H4 tail with the H2A/H2B acidic patch of the adjacent particle (6, 7), whereas in the condensed tetra-nucleosomes or stacked nucleosome, this is sterically disfavored (8). In self-associated nucleosome arrays, cross-linking experiments captured inter-nucleosome interaction between the H4 tail and H2A (9), but rarely between the H4 tail and DNA (10). The mediating role of the H4 tail is not only induced via general electrostatic effect, but also via specific interactions involving certain residues on the H4 tail and the H2A/H2B dimer (11, 12).
The middle part of the H4 tail, the KRHRK segment (residues 16–20), contains strongly positive charges and forms a “basic patch”. On the H2A-H2B dimer, the glutamic acid and aspartic acid residues (H2A E56, E61, E64, D90, E91, E92, and H2B E102 and E110) build up a negatively charged area, called the “acidic patch”. Due to the spatial proximity and the electrostatic attraction, stable salt bridges can be formed between these two parts from neighboring nucleosomes (6). The acidic patch is also an important binding site for many nucleosome binding proteins. These proteins compete with the H4 tail or among themselves for binding to the acidic patch, thus modulate chromatin remodeling (13).
The flexible and basic histone tails are ideal to bind the substrate pocket of enzymes, thus undergo a variety of posttranslational modifications. Posttranslational modifications regulate gene transcription by altering chromatin structure or (and) nonhistone protein interaction. The acetylation on H4 lysine 16 (H4K16Ac) is a prevalent posttranslational modification, associated with gene activation and DNA damage repair (14, 15). It plays a prominent role in the histone modification cascade that leads to gene activation and silencing, e.g., the MOF protein responsible for dosage compensation in Drosophila is a H4K16 acetylase (16). Loss of H4K16Ac, on the other hand, was globally found in cancer cells (17). The transcriptional function of H4K16Ac is independent from the cumulative effect induced by other H4 lysine acetylations (18). Remarkably, at the molecular level, H4K16Ac influences both chromatin structure and protein interplay (19, 20). On the structural level, H4K16Ac significantly reduces nucleosome-nucleosome stacking and chromatin folding, standing out from H4K16Q or other H4 tail acetylations. It indicates that H4K16Ac influences chromatin structure via a specific mechanism rather than a conventional electrostatic effect (21, 22, 23).
To elucidate the special role of H4K16Ac in regulating chromatin higher-order structure on a molecular level, multiscale simulation studies have been performed. Potoyan and Papoian (24) proposed that K16Ac enhances the binding between H4 tail and its own nucleosomal DNA, and therefore reduces inter-nucleosome interactions. Another effect of lysine acetylations is to increase the secondary structure of the H4 tail (25). Ordered histone tails are less available to the adjacent nucleosome, and thus induce unfolding of chromatin fiber (26). However, an opposite effect of H4K16Ac, which decreases the α-helix in the isolated H4 tail, has also been reported by a previous replica exchange molecular dynamics (REMD) simulation study (24, 27). Moreover, in the context of the nucleosome, the intratail hydrogen bonds are disrupted by interaction between H4 tail and DNA, which drastically disorder the secondary structure of isolated histone tails (28).
Despite the intensive discussion on the structural effect of H4K16Ac, a direct investigation of its influence on the inter-nucleosome H4 tail-acidic patch interaction is still missing. In this work, we explored, for the first time to our knowledge, the binding conformation of the H4 tail and the influence of H4K16Ac in the context of two adjacent nucleosomes, in explicit solvent using replica exchange with solute tempering (REST). The free energy landscape of H4 tail was explored by metadynamics. The results indicate that two segments of H4 tail are separately responsible for the intranucleosome interaction with DNA and the inter-nucleosome interaction with the acidic patch. Interestingly, K16 is the first residue and the only acetylation site located in the acidic patch binding part of the H4 tail. In contrast to wild-type K16, the acetylated K16 loses the stable contact to the acidic patch. Acetylation of K16 significantly weakens the H4 tail-acidic patch interaction and increases the diversity of H4 tail binding conformations. Further analysis suggested that the particular position of K16 in the H4 tail enhances the effect of K16Ac on the inter-nucleosome interaction.
Materials and Methods
Model system setup
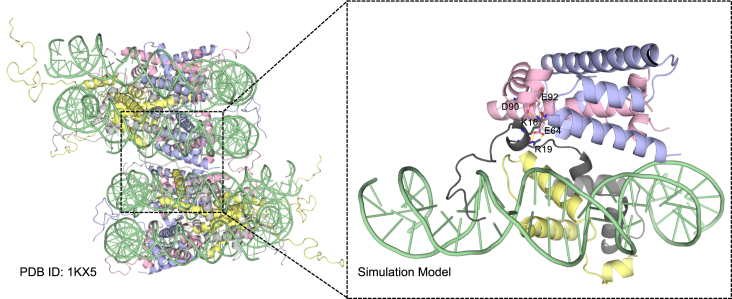
The model system under investigation is based on the nucleosome crystal structure (PDB: 1KX5), where the H4 tail is attached to the neighboring acidic patch in the crystal packing (7). To reduce the system size, only the N-terminal part of H4 (chain F, residues 1–45) and the surrounding residues within 1.0 nm from the H4 tail (residues 1–32) were selected for simulation, including segments of DNA, H3 from the same nucleosome, and H2A, H2B of the neighboring nucleosome (see Fig. 1). With the experimental evidence that H4 residues 14–19 are crucial for chromatin fiber compaction, there is a strong possibility that these residues are participating in inter-nucleosome interaction. However, in this structure with a complete H4 tail (PDB: 1KX5), residues K16, R17, and R19 are all pointing toward DNA. In another nucleosome structure (PDB: 1AOI), in which H4 residues 1–15 are invisible, K16 and R19 are attached to the acidic patch (6). Therefore, 30 ns preliminary REMD simulation with implicit solvent (29, 30, 31) was performed to generate possible conformations of the H4 tail. The parameters for the implicit solvent model are the same as our previous work (28). A conformation with K16 and R19 attached to the acidic patch, which is similar to the H4 tail in the crystal structure (PDB: 1AOI), was sampled. Afterwards 90 ns implicit solvent REMD was performed with this new conformation to verify it. This structure was used as the initial structure for the production simulation with explicit solvent (Fig. 1).
Figure 1.
Initial structure. Structure of two nucleosomes from crystal packing (left) and the model structure used in simulation (right). (Green) DNA; (yellow) H3; (gray) H4; (pink) H2A; and (blue) H2B. To see this figure in color, go online.
To investigate the influence of K16 acetylation on the H4 tail binding conformation, we prepared a system with H4K16 acetylated. The point charge for the acetylated lysine was calculated following the protocol used in the development of the AMBER force field (32). We optimized the conformation of acetylated lysine by the density functional theory with a B3LYP scheme and a 6-31G∗ basis, then calculated the electrostatic potential surfaces by HF/6-31G∗. All quantum chemistry calculations were done with Gaussian 09 (33). The restrained electrostatic potential method within Antechamber was applied to fit the electrostatic potential surfaces to point charges (34, 35).
REST simulation
REMD simulations are widely used for sampling of protein conformations. Several replicas of the system were run in parallel at different temperatures, and in a given time interval the coordinates and velocity of the neighboring two replicas were exchanged if the Metropolis criterion is fulfilled. Thus the system is heated up and cooled down, to overcome the energy barrier of conformational transitions (29). The potential difference between two replicas has to be chosen to achieve a specific acceptance ratio of replica exchange. Therefore, the number of needed replicas increases with (where N is the degrees of freedom in the system) (36).
Because we are interested in the conformation of the solute, we want to keep the temperature of solvent unchanged to reduce the potential difference between two replicas. In this case we applied so-called REST (37), a particular form of Hamiltonian replica exchange, to modulate the temperature of the solute. The potential energy of the system can be written as sum of three parts: solvent-solvent, solvent-solute, and solute-solute. We rescaled the last two potentials according to a temperature factor λ = T0/Ti (T0 is the lowest temperature, Ti is the temperature for the ith replica). In molecular dynamics, the potential consists of Coulomb, Lennard-Jones, and bonded potential (bond stretching, angle bending, torsion, etc.). For the Coulomb interaction, we rescaled the partial charges in the solute . For the Lennard-Jones potential, we rescaled the depth of the potential well: (38). For the bonded potential, we rescaled the dihedral term: .
To perform REST simulations we used GROMACS 4.6 (39, 40, 41) patched with Plumed2 (42, 43), in which the Hamiltonian REMD scheme is implemented (44). For both the wild-type (WT) and K16Ac (AC) H4 tail system, 28 replicas are used to achieve the exchange acceptance ratio of 0.2–0.3. The temperatures used to rescale the topology files are: 300.0, 304.5, 309.1, 313.7, 323.2, 328.0, 333.0, 337.9, 343.0, 348.2, 353.4, 358.7, 364.1, 369.5, 375.1, 380.7, 386.4, 392.2, 398.1, 404.1, 410.1, 416.3, 422.5, 428.8, 435.3, 441.8, and 448.4 K. The system energy was minimized by the conjugate gradient method. After 200 ps NVT and NPT equilibration with restraint on solute, 100 ns REST simulation was performed under NVT condition for both systems. To maintain the stability of the system and to reduce the number of degrees of freedom, position restraints were applied to atoms except for the H4 tail (H4 residues 1–32) and side-chain atoms of H4-tail-attached residues (H4 residues 55, 58, and 59; H3 residues 62, 63, 69, 70, 73, 76, 77, and 80; H2A residues 56, 60, 61, 64, 65, 68, 71, and 90–92; and H2B residues 44, 45, 102, and 110), and the force constants on x/y/z vectors were all set to 1000 kJ/mol/nm2 (45). To justify this approach, a comparison of the fully flexible two-nucleosome system and the truncated restrained system can be found in Fig. S11 in the Supporting Material. Solute atoms were parameterized with the Amber99SB force field (32, 46), and water molecules with the TIP3P model (47). Sodium ions were added to neutralize the charges. The distance between solute atoms and the box edges was at least 1.0 nm. The total system contains 132,039 atoms. The short-range cutoff for nonbond interaction was 1.2 nm, and long-range electrostatic interaction was treated with the PME method (48). The LINCS algorithm was applied to constrain all bonds of hydrogen atoms (49), and the time step for integration was set to 2 fs.
Trajectory analysis
Clustering analysis and calculation of RMSD, minimum distance, distance, contact, and covariance matrices were done using GROMACS modules (40). The trajectory was clustered with respect to the RMSD of the H4 tail nonhydrogen atoms, performing the following steps. The trajectory was first clustered using the GROMOS method (45, 50), with the cutoff set to 0.25 nm. The generated clusters were merged if the RMSD of their average structure was less than the cutoff. We repeated this step until the result converged. Last, the small clusters with <10 members were compared to any member in another cluster, and merged into the big cluster if the RMSD is below cutoff. The covariance matrices of WT and AC H4 tail were calculated from the RMSD of Cα atoms. The covariance of AC (Cov(AC)orig) was normalized to the relative root mean square fluctuation (RMSF) of Cα atoms:
Metadynamics
Metadynamics (51, 52) is applied to explore the free energy landscape of the system and verify the results of REST simulation. Metadynamics is an enhanced sampling technique aimed at reconstructing free energy along defined dimensions, by adding an external potential to fill up the wells in the energy landscape. The external potential is a sum of Gaussians deposited along the space of selected degree of freedom—collective variables (CVs). Principal component analysis (PCA) was performed on the REST trajectory. The first principal component (PC) of Cα of H4 residue 2, 9, 12, 15, 18, and 25 was used as the first CV, according to their RMSD along the eigenvector (Fig. S5). The second CV is the minimum distance (Mindist) between the side chains of H4 (Ac)K16 and H2A E61/D90. A 350 ns well-tempered metadynamics simulation (53) was performed for the WT and AC systems. Every 1 ps, a Gaussian with a height of 1.0 kJ/mol is deposited along the space of CVs. The bias factor for well-tempered metadynamics is set to 15. The width of the Gaussian is 0.1 nm in the dimension of PC and 0.05 nm for Mindist. Fig. S7 shows the sampling of CVs. The sampling of the PC for AC is not as efficient as for WT, because the second principal component in AC is also an important motion (Fig. S5 A); nevertheless, a wide range of the PC space was visited for both systems. Figs. S8 and S9 show the convergence of the metadynamics.
Results
Overview of REST sampling
Efficient replica exchange was achieved during 100 ns REST simulation, with the acceptance ratio ranging from 0.21 to 0.28. The probability distributions of the potential energies overlap well with each other (Fig. S1), and each of the 28 replicas has explored the whole temperature space (Fig. S2). The evolution of the structure clusters shows the convergence of the sampling (Fig. S3). In the following, we analyzed only the 300 K trajectory.
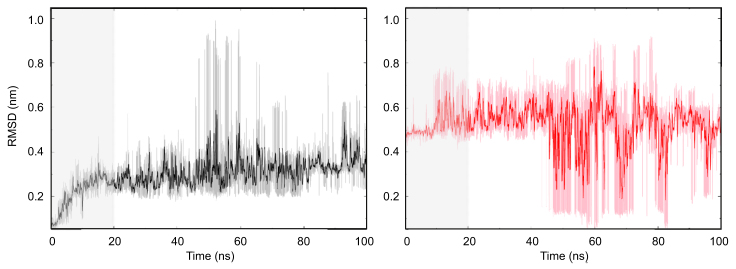
To get a preliminary view of the H4 tail conformational evolution, we calculated the RMSD of the H4 tail backbone atoms compared to the initial structure (Fig. 2). It is obvious that the AC tail exhibits larger conformational fluctuations than the WT tail. In the initial phase (0–20 ns), the RMSD of WT rises gradually from 0 to ∼0.25 nm, whereas that of AC jumped to 0.5 nm at the beginning of the simulation. It indicates that the initial structure is strongly disfavored by the AC tail. For both systems, the drastic fluctuation after 50 ns corresponds to efficient crossing of energy transition barriers and the occurrence of a new conformation. In the last 30 ns, convergence is roughly reached for the sampling of the WT system, as indicated by the steady RMSD, but not for the AC system. It could be due to the lack of distinguishable minimum in the AC H4 tail conformation space, which will be discussed in the following sections. For the following parameter calculation and conformation analysis, we used the trajectory starting at 20 ns.
Figure 2.
RMSD of H4 tail. RMSD of the backbone atoms of the WT (left) and the AC (right) H4 tail was calculated every 10 ps (lines with light color), with the initial structure as reference. Running average of every 200 ps was plotted with deep color. To see this figure in color, go online.
Acetylation weakens the stable interaction between H4K16 and the H2A-H2B dimer from the neighboring nucleosome
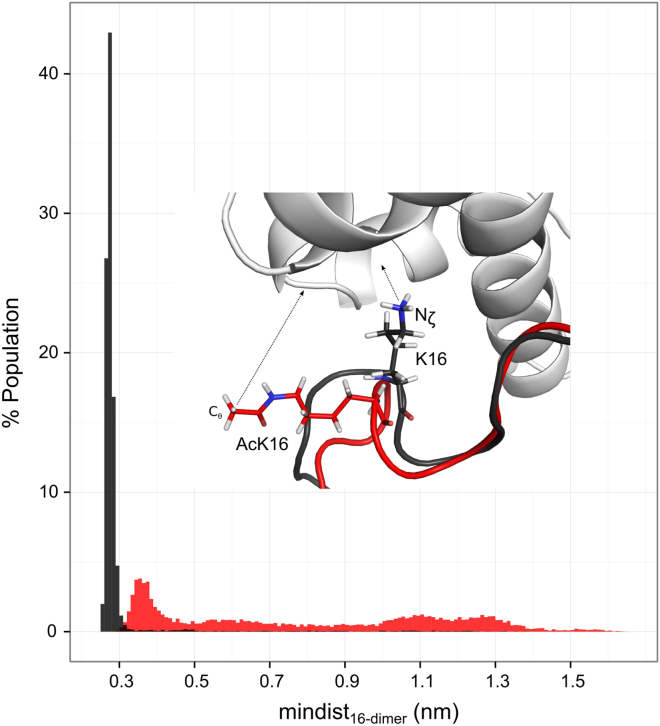
In the crystal structure (PDB: 1AOI), K16 is in contact with the acidic patch, forming salt bridges with H2A E61, D90, and E92. To study whether these contacts survive during the replica exchange simulation, we measured the minimum distance between the H4K16(Ac) side chain and the center of mass of the H2A-H2B fragment (mindist16-dimer). The position of H4K16 in WT is quite stable, as demonstrated by the sharp distribution of mindist16-dimer (Fig. 3). The mindist16-dimer of WT is always (population > 99%) under 0.35 nm, which we defined as the cutoff for contacts. This result confirms the strong interaction between K16 and the neighboring H2A-H2B dimer. In contrast, the mindist16-dimer of AC has an extremely diffuse distribution, reflecting the uncertainty of the K16Ac position. In only 10% of the sampled structures, K16Ac contacts the dimer (mindist16-dimer < 0.35 nm), whereas 43% of the K16Ac positions are completely detached from the neighboring nucleosome (mindist16-dimer > 0.90 nm). Acetylation neutralizes the positive charge on K16 and disrupts its electrostatic interaction with the acidic patch. Although the acetyl group brings an extra hydrogen bond acceptor and additional atoms for van der Waals interactions, AcK16 still loses its stable configuration.
Figure 3.
Distribution of minimum distance between the H4(Ac)K16 and H2A-H2B fragments. The top atom of H4(Ac)K16 side chain (Nζ of K16, Cθ of AcK16) and the nonhydrogen atoms on H2A-H2B fragment were selected for measurement. (Black) WT; (red) AC. To see this figure in color, go online.
K16 acetylation alters completely the conformation space of the H4 tail and induces the tail away from neighboring nucleosome
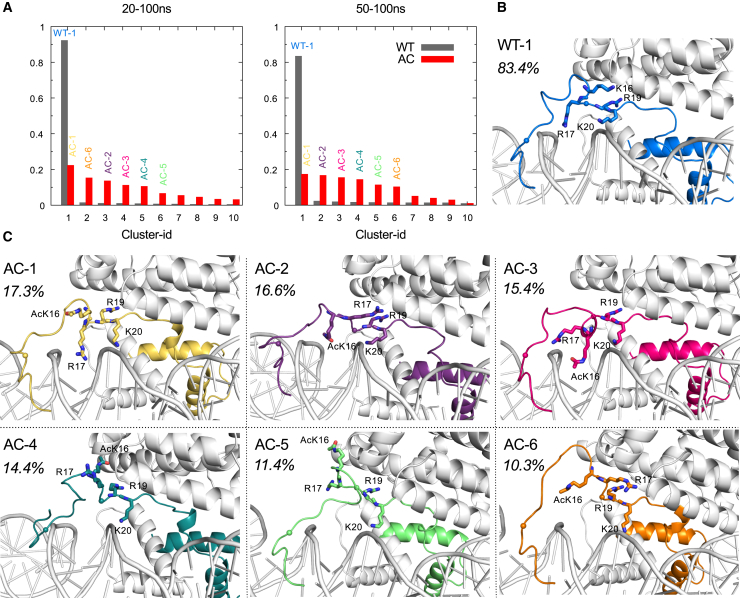
The single acetylation of K16 impacts the whole binding conformation of H4. We did a cluster analysis on the trajectory between 20 and 100 ns to show the ensemble of the whole production trajectory, and 50–100 ns to demonstrate the ensemble of the efficient exchanging phase. The occupancy of major clusters reveals that K16 acetylation drastically increases the diversity of H4 tail conformational space (Fig. 4 A). In the WT system, the dominant cluster covers 93.3% of all frames and 83.4% of the frames in the last 50 ns, indicating the existence of strong and specific interactions. In comparison, the AC ensemble is distributed quite evenly over several clusters. The largest cluster occupies only 22.3% of 20–100 ns frames and 17.3% of 50–100 ns frames. Although the AC system exhibits a convergence to the top six clusters in the last 50 ns, there is no preference for any of them.
Figure 4.
Cluster analysis. The size of the top 10 clusters (A) and the center structure of major clusters for WT (B) and AC (C) systems. (Stick) Residues 16–20; (ball) position of residues 7 and 18. The occupancy of each cluster in the last 50 ns is labeled as number next to corresponding structure. To see this figure in color, go online.
As illustrated by the representative structure of each cluster, the first residues of both WT and AC tail are always binding to DNA. The main difference between WT and AC shows up on residues 7–17, which bind either to DNA or to the acidic patch (Fig. 4, B and C). In the WT system, each of the four charged residues on the basic patch (residues 16–20) has a defined role: K16 and R19 attach to the acidic patch; R17 and K20 bind to DNA (Fig. 4 B). This conformation takes full advantage of the negatively charged neighbors, and avoids the repulsion among the positively charged side chains. In the AC system, due to charge neutralization and steric hindrance, AcK16 leaves the acidic patch and attempts to compensate this loss by (1) promoting intratail hydrogen bonds (AC-1, AC-2, and AC-6); (2) approaching DNA surface via van der Waals interaction (AC-2); (3) inserting into the DNA groove due to hydrophobic interaction between methyl group and the DNA rings system (AC-3); and (4) forming hydrogen bonds with the open and basic region of H2A-H2B dimer (AC-4 and AC-5) (Fig. 4 C). R19 and K20 still maintain the binding to the H2A-H2B dimer and DNA, respectively. The conformation of residues 7–17 varies from cluster to cluster, reflecting that the H4 tail loses stable binding conformation in the two-nucleosome interface.
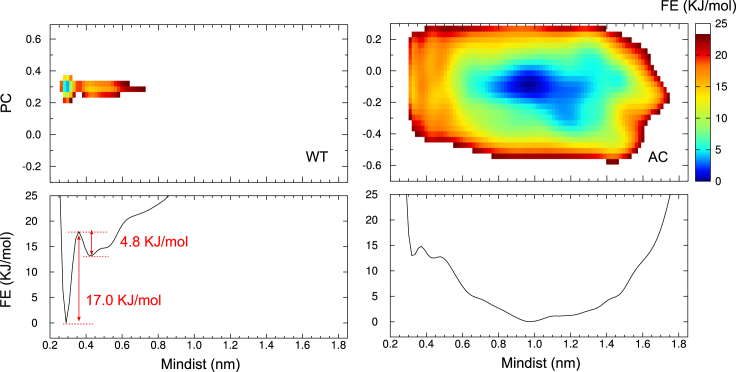
To quantify the difference between the WT and AC H4 tail, we profiled the free energy landscape (Fig. 5). In accordance with the distribution of mindist16-dimer in Fig. 3, the WT H4 tail has a distinguishable minimum with the K16 binding to the adjacent H2A E61/D90, whereas the landscape of the AC H4 tail is very flat and favors the area away from the adjacent H2A. Even though the free energy surface of the WT shows another local minimum for Mindist ≈ 0.4 nm, the most favored conformation of the system is represented by the global and distinct minimum for Mindist ≈ 0.3 nm. In contrast, the AC H4 tail prefers a conformation with the K16 away from the acidic patch, with the Mindist value ranging from 0.8 to 1.5 nm. The energy surface of the AC H4 tail has a flat and broad minimum. Fig. 5 shows the results after 350 ns metadynamics, which can be compared to the 300 ns results (Fig. S9): the free energy landscape of the WT is almost the same, whereas that of the AC shows a transition, but the minimum is still within the range of 0.8–1.5 for the Mindist dimension. Apparently, within this Mindist range, the AC H4 tail has no preferred conformation population. The significant difference between the free energy landscape of WT and AC explains the result of cluster analysis: WT H4 tail has one dominant cluster, whereas the AC H4 tail has six clusters of similar size (Fig. 4).
Figure 5.
2D Free energy (FE) surface (upper panels) obtained by metadynamics and the projection of free energy on the Mindist dimension (lower panels). (PC) Eigenvector from PCA analysis of REST trajectory. Mindist is the minimum distance between the side chains of (Ac)K16 and H2A E61/D90. To see this figure in color, go online.
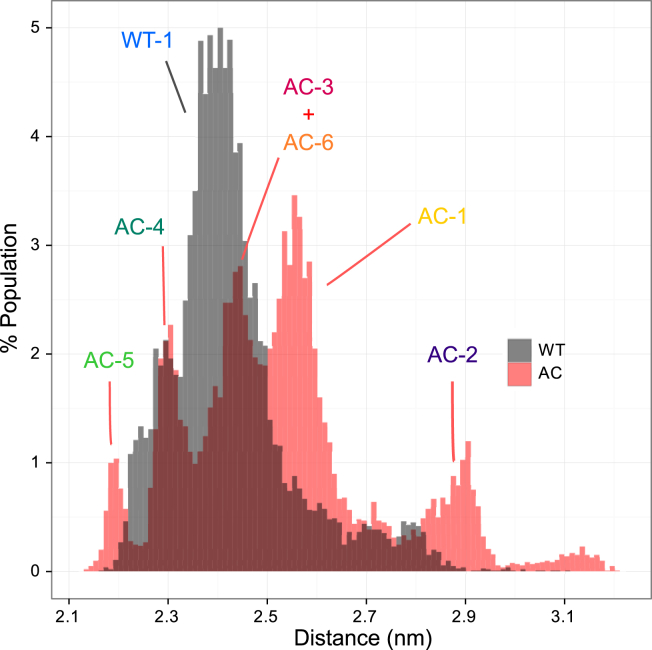
Fig. 6 shows the distribution of the distance between the flexible middle part of the tail (residues 7–17) and the H2A-H2B fragment. The major population of WT is located at ∼2.3–2.5 nm, indicating close contact between H4 tail middle part and the neighboring H2A-H2B dimer. The distribution of AC is broader, ranging from 2.2 to 3.1 nm, and the multiple peaks refer to the aforementioned diverse conformation clusters. The center of the major peak of the AC population is shifted 0.2 nm to the right of the WT center. Although both distributions, AC and WT, are overlapping significantly, the middle part of the AC H4 tail (residues 7–17) is generally further away from the adjacent H2A-H2B dimer.
Figure 6.
Distribution of the distance between the H4 tail and the neighboring nucleosome. The center of mass of the backbone atoms of H4 tail residues 7–17 and H2A-H2B dimer are used for distance measurement. The peaks can be assigned to the labeled structure clusters. To see this figure in color, go online.
K16 acetylation weakens the contacts between the H4 tail and the acidic patch, and enhances the interaction within the H4 tail residues
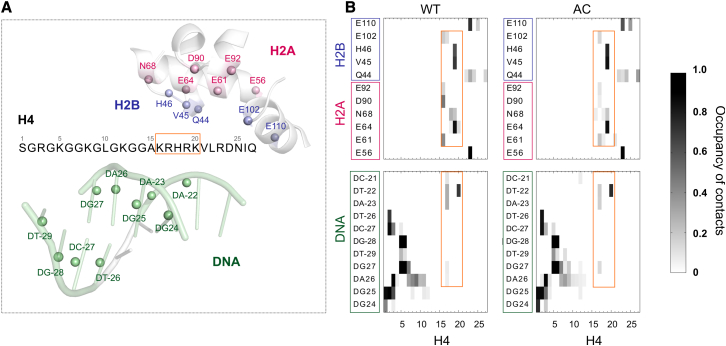
The contact maps in Fig. 7 help us to study the detailed of interactions between the H4 tail and the DNA/H2A-H2B dimer. The H4 tail is obviously divided into the “DNA binding” and the “acidic patch binding” parts. Residues 1–12 are responsible for binding with intranucleosome DNA, whereas residues 16–25 undertake inter-nucleosome interaction with the H2A-H2B dimer, especially the acidic patch. Residues 1–6 form the same contacts with DNA for the WT and the AC systems. For the acidic patch binding part, R17 and K20 are the only two residues attached to DNA. K16 in the WT system is frequently attached to H2A E61, D90 E92, and H2B E102, i.e., the acidic patch, whereas in the AC system, these interactions were completely lost, due to lack of positive charge and hindrance of large side chains. R17 tries to compensate for the loss by interacting with E61, but with a very low frequency. Some important interactions are conserved in the WT and AC systems. R19 and R23 maintain the interaction with the acidic patch (R19 with H2A E64, H2B V45 and H46; and R23 with H2A E56 and H2B E110), so that the H4 tail will not completely detach from the neighboring nucleosome. The stable contact between R23 and the acidic patch agrees with the B-factor analysis of the nucleosome crystal structure (6, 54).
Figure 7.
(A) Contact map of the H4 tail. A contact is defined as a distance between nonhydrogen atoms of <0.35 nm. The residue contact occupancy is the highest atom contact occupancy within the two residues. In the contact map (B) the x axis represents the H4 tail residues, and the y axis represents the residues from the H2A-H2B fragment and DNA (location shown in A). (Framed in orange) The basic patch (H4 residues 16–20). To see this figure in color, go online.
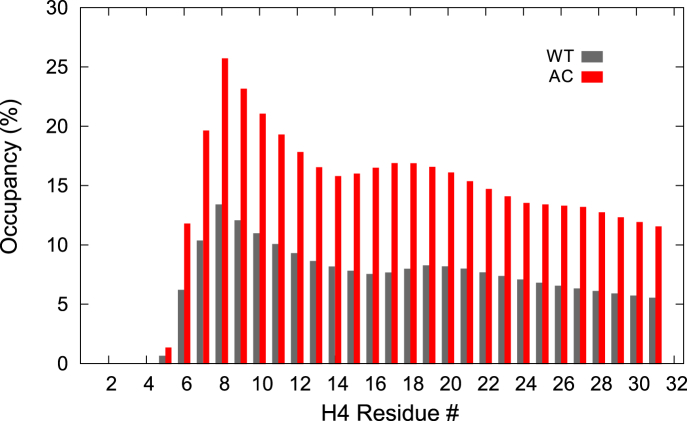
In accordance with our previous result, the H4 tail in the context of the nucleosome is basically disordered due to active electrostatic interaction with outside partners (28). Indeed, only some low-frequency 310-helix structures (formed by i+3→i hydrogen bonds) were found in the WT system (Fig. S4). Nevertheless, we observed an increasing occupancy of 310-helix structures in the AC system (Fig. 8). Evidently, with the reduction of contacts between the H4 tail and the acidic patch, the intratail interaction increases in the AC system. The relative tendency of each residue to form a 310-helix is similar in WT and AC, but the occupancy in AC is twice as that in WT. This is in agreement with the simulation study of Winogradoff et al. (55), which reported a 310-helix structure at residues 7–9 in the K16 acetylated H4 tail. It indicated that acetylations relieve the electrostatic repulsion among positively charged side chains, thus increasing the secondary structure propensities of the isolated H4 tail. The increased α-helix in the H4 tail induced by acetylations was also shown by circular dichroism experiment (25). In our case, acetylation disrupts the interaction between the H4 tail and the acidic patch, which gives the H4 tail the flexibility to form intratail hydrogen bonds. The increasing intratail interaction helps to stabilize these structures.
Figure 8.
Occupancy of 310- helix structure in the H4 tail. To see this figure in color, go online.
K16 plays a central role in stabilizing the H4 tail binding conformation
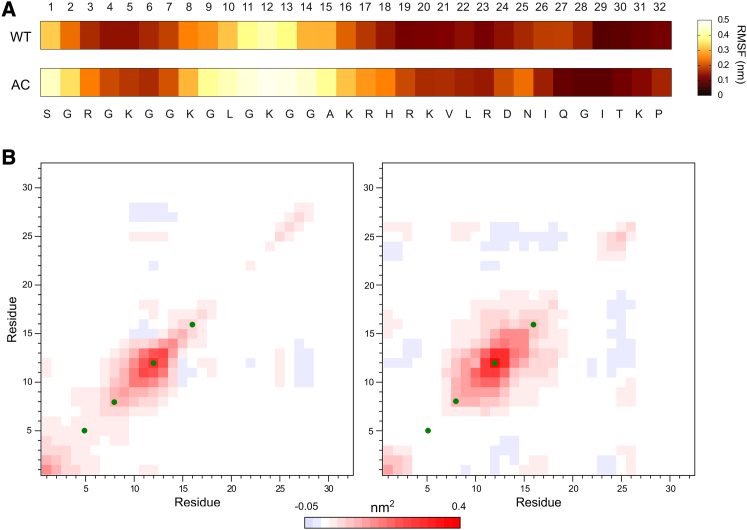
The effect of K16 acetylation on chromatin folding is more significant than other H4 tail acetylations (21, 22, 23). As mentioned before, K16 is on the border separating the DNA-binding and acidic-patch binding parts of the H4 tail (Fig. 7). This special location might relate to the distinguishable function of K16Ac. As shown by the RMSF of the H4 tail residues in Fig. 9 A, the most flexible area of both the WT and AC systems is centered at K12. This lysine forms moderate contacts with DNA, and is flanked by small amino acids like glycine and alanine, which contribute little to the interaction with either DNA or the acidic patch. In the WT H4 tail, A15 is the last flexible residue with RMSF > 0.2 nm. However, without the electrostatic interactions of K16, the area of the AC H4 tail with RMSF > 0.2 nm is extended up to H18. The covariance analysis further reveals the important role of K16 to fix H4 tail conformation. The covariance matrix of WT exhibits a narrow and weak correlation area along the diagonal, indicating that the residues are only influenced by their direct neighbors (Fig. 9 B). The most correlated area is that of residues 7–14, whereas residues after A15 (especially residues 19–21) are more independent. Here K16 plays the role of an anchor, which keeps the flexible region from fluctuating. In comparison, the red area in the AC matrix is obviously larger and deeper, corresponding to stronger correlation between residues. The correlated area extends to cover residues 7–19, showing that AcK16 is flexible enough to pass on the fluctuation.
Figure 9.
RMSF of the H4 tail backbone atoms (A) and covariance matrix of the H4 tail C-α atom RMSD (B). The RMSF of the H4 tail backbone atoms for WT and AC is shown as a color map (A), labeled with residue number and type of amino acid. The covariances of WT (B, left) and AC (B, right) were normalized (see Materials and Methods). (Red) Correlated behavior between the residue in the x and y axes; (blue) anticorrelation. The closer the value is to 0, the less correlated is the pair of residues. (Green dots) Positions of K5, K8, K12, and K16. To see this figure in color, go online.
Thus, K16 acts as the first button to fix the H4 tail on the acidic patch. The K16 acetylation diminishes this function, destabilizes the middle part of the H4 tail, and weakens the interaction with the neighboring nucleosome. Remarkably, all the other acetylation sites of H4 tail (K5, K8, and K12) are located on DNA binding part (Fig. 7), and therefore have less influence on inter-nucleosome interactions.
Discussion
A previous simulation study of the isolated H4 tail pointed out that K16Ac reduces the heterogeneity of H4 tail conformation space, by promoting the hydrogen bond between the H4 tail residues (55). However, in the context of two nucleosomes in neighboring unit cells in the crystal structure, we found increased conformation diversity of the H4 tail with K16Ac. These results reveal the dual effects of lysine acetylation. In isolated H4 tails, the positively charged side chains of lysine and arginine repel each other. Lysine acetylation releases this tension, facilitates intratail folding, and thus stabilizes the conformation. However, in the presence of negatively charged DNA and the acidic patch, the system is dominated by intermolecule electrostatic interaction, such as salt bridges between the H4 tail and the acidic patch. After acetylation and the loss of the charge, the increase of intratail interaction is not enough to compensate the reduction of intermolecule interaction. Therefore, the AC H4 tail becomes very unstable, as demonstrated by the more diverse conformation space.
In this work, we explored the influence of H4K16 acetylation on the inter-nucleosome interaction mediated by the H4 tail. The model represents the situation of a less condensed nucleosome array, where the H4 tail is attached to the acidic patch from the adjacent nucleosome. As mentioned before, H4K16Ac weakens but never abolishes the inter-nucleosome interaction. H4R17, R19, and R23 stay attached to the acidic patch on the neighboring nucleosome. This observation might be partly due to technical reasons. In our setup, constraints were applied to all atoms except the H4 tail and side chains of acidic patch. The effect of H4K16Ac might be magnified in a more complete and flexible environment, where larger-scale conformation changes with respect to nucleosome stability and relative position of two nucleosomes are allowed (see Fig. S11). Besides, only a very few ions were added in the solvent to neutralize the system. Ions impair the electrostatic interaction between H4 tail and DNA or the acidic patch by screening the charges. Therefore, higher salt concentration might increase the effect of H4K16Ac. Despite of the technical limitation, it might be a fact that the direct structure alteration caused by H4K16Ac is moderate. Wakamori et al. (54) suggested that the effect of H4 tail acetylations on inter-nucleosome histone-histone interaction is negligible. The B-factors of the H4 tail and DNA were increasing in the nucleosome with tetra-acetylated H4 tails, whereas those of the acidic patch and H4R23 were not affected. In accordance, we also observed stable interaction between H4-R23 and H2A-E56/H2B-E110. Nevertheless, the interaction between the H4 tail residues 7–17 and the acidic patch is significantly weakened by K16Ac. In this case, other acidic patch binding proteins, which compete with the H4 tail, might win the chance to bind to the nucleosome and cause downstream transcriptional effects.
It is roughly a consensus that K16Ac activates transcription by influencing both nucleosome structure and interaction with external proteins. The reduction of inter-nucleosome interaction induced by H4K16Ac could be due to 1) increase of the interaction between H4 tail and its own DNA (24); 2) decrease of the conformation flexibility of H4 tail (24, 26, 55); and 3) elongated H4 tail conformation and exposure of the H4 tail for interaction with nonhistone protein (55). In this work, we sampled the conformation of the H4 tail in the context of two nucleosomes in neighboring unit cells in the crystal structure and showed that the H4K16Ac destabilizes the binding conformation of the H4 tail on the neighboring acidic patch. We further proposed that the unique influence of H4K16Ac might be due to its special location in the H4 tail, which makes it the first anchor on the acidic patch, as well as the only acetylation site involved in acidic patch binding. Although the K16Ac H4 tail is not completely detached from the neighboring nucleosome, the probability increases for other nucleosome-binding proteins to approach the acidic patch. However, from molecule to cell, the context of discussion (elements involved in the interaction network, timescale, etc.) can completely change. Experiment in vivo indicated that H4K16Ac does not change the chromatin structure in embryonic stem cells, but marks active enhancers (56). Future studies are expected to fill the gap between microscopic molecular investigations and experiments in a larger and more complicated environment.
Author Contributions
R.Z. designed the research, performed the simulation, analyzed the data, and wrote the article; J.E. analyzed the data and assisted in writing the article; and J.L. designed the research and assisted in writing the article.
Acknowledgments
All simulations were performed on the HELICS computer cluster at the Institute for Scientific Computing at the University of Heidelberg. We thank Professor Andreas Dreuw for providing software and advice for our quantum chemistry calculations. We are also grateful to Professor Markus Lill for valuable discussions.
R.Z. was funded by the Heidelberg Graduate School of Mathematical and Computational Methods for the Sciences (HGS MathComp), and by Deutsche Forschungsgemeinschaft (DFG) grant No. GSC 220 from the German Universities Excellence Initiative. The visit of R.Z. to Professor Markus Lill’s group was supported by the Boehringer Ingelheim Fonds.
Editor: Tamar Schlick.
Footnotes
Eleven figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)31043-8.
Supporting Material
References
- 1.Zheng C., Hayes J.J. Intra- and inter-nucleosomal protein-DNA interactions of the core histone tail domains in a model system. J. Biol. Chem. 2003;278:24217–24224. doi: 10.1074/jbc.M302817200. [DOI] [PubMed] [Google Scholar]
- 2.Pepenella S., Murphy K.J., Hayes J.J. Intra- and inter-nucleosome interactions of the core histone tail domains in higher-order chromatin structure. Chromosoma. 2014;123:3–13. doi: 10.1007/s00412-013-0435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arya G., Schlick T. A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments. J. Phys. Chem. A. 2009;113:4045–4059. doi: 10.1021/jp810375d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saurabh S., Glaser M.A., Maiti P.K. Atomistic simulation of stacked nucleosome core particles: tail bridging, the H4 tail, and effect of hydrophobic forces. J. Phys. Chem. B. 2016;120:3048–3060. doi: 10.1021/acs.jpcb.5b11863. [DOI] [PubMed] [Google Scholar]
- 5.Dorigo B., Schalch T., Richmond T.J. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 6.Luger K., Mäder A.W., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 7.Davey C.A., Sargent D.F., Richmond T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 8.Schalch T., Duda S., Richmond T.J. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 9.Sinha D., Shogren-Knaak M.A. Role of direct interactions between the histone H4 tail and the H2A core in long range nucleosome contacts. J. Biol. Chem. 2010;285:16572–16581. doi: 10.1074/jbc.M109.091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pepenella S., Murphy K.J., Hayes J.J. A distinct switch in interactions of the histone H4 tail domain upon salt-dependent folding of nucleosome arrays. J. Biol. Chem. 2014;289:27342–27351. doi: 10.1074/jbc.M114.595140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howell S.C., Andresen K., Qiu X. Elucidating internucleosome interactions and the roles of histone tails. Biophys. J. 2013;105:194–199. doi: 10.1016/j.bpj.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorigo B., Schalch T., Richmond T.J. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 13.Kalashnikova A.A., Porter-Goff M.E., Hansen J.C. The role of the nucleosome acidic patch in modulating higher order chromatin structure. J. R. Soc. Interface. 2013;10:20121022. doi: 10.1098/rsif.2012.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verdone L., Caserta M., Di Mauro E. Role of histone acetylation in the control of gene expression. Biochem. Cell Biol. 2005;83:344–353. doi: 10.1139/o05-041. [DOI] [PubMed] [Google Scholar]
- 15.Krishnan V., Chow M.Z.Y., Zhou Z. Histone H4 lysine 16 hypoacetylation is associated with defective DNA repair and premature senescence in Zmpste24-deficient mice. Proc. Natl. Acad. Sci. USA. 2011;108:12325–12330. doi: 10.1073/pnas.1102789108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akhtar A., Becker P.B. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell. 2000;5:367–375. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- 17.Fraga M.F., Ballestar E., Esteller M. Loss of acetylation at Lys16 and trimethylation at Lys-20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 18.Dion M.F., Altschuler S.J., Rando O.J. Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl. Acad. Sci. USA. 2005;102:5501–5506. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shogren-Knaak M., Ishii H., Peterson C.L. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 20.Oppikofer M., Kueng S., Gasser S.M. A dual role of H4K16 acetylation in the establishment of yeast silent chromatin. EMBO J. 2011;30:2610–2621. doi: 10.1038/emboj.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allahverdi A., Yang R., Nordenskiöld L. The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 2011;39:1680–1691. doi: 10.1093/nar/gkq900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y., Lu C., Nordenskiöld L. Influence of histone tails and H4 tail acetylations on nucleosome-nucleosome interactions. J. Mol. Biol. 2011;414:749–764. doi: 10.1016/j.jmb.2011.10.031. [DOI] [PubMed] [Google Scholar]
- 23.Robinson P.J.J., An W., Rhodes D. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J. Mol. Biol. 2008;381:816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Potoyan D.A., Papoian G.A. Regulation of the H4 tail binding and folding landscapes via Lys-16 acetylation. Proc. Natl. Acad. Sci. USA. 2012;109:17857–17862. doi: 10.1073/pnas.1201805109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., Moore S.C., Ausió J. Acetylation increases the α-helical content of the histone tails of the nucleosome. J. Biol. Chem. 2000;275:35013–35020. doi: 10.1074/jbc.M004998200. [DOI] [PubMed] [Google Scholar]
- 26.Collepardo-Guevara R., Portella G., Orozco M. Chromatin unfolding by epigenetic modifications explained by dramatic impairment of internucleosome interactions: a multiscale computational study. J. Am. Chem. Soc. 2015;137:10205–10215. doi: 10.1021/jacs.5b04086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang D., Arya G. Structure and binding of the H4 histone tail and the effects of Lysine 16 acetylation. Phys. Chem. Chem. Phys. 2011;13:2911–2921. doi: 10.1039/c0cp01487g. [DOI] [PubMed] [Google Scholar]
- 28.Erler J., Zhang R., Langowski J. The role of histone tails in the nucleosome: a computational study. Biophys. J. 2014;107:2911–2922. doi: 10.1016/j.bpj.2014.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugita Y., Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999;314:141–151. [Google Scholar]
- 30.Tsui V., Case D.A. Molecular dynamics simulations of nucleic acids with a generalized Born solvation model. J. Am. Chem. Soc. 2000;122:2489–2498. [Google Scholar]
- 31.Onufriev A., Bashford D., Case D.A. Exploring protein native states and large-scale conformational changes with a modified generalized Born model. Proteins. 2004;55:383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 32.Cornell W.D., Cieplak P., Kollman P.A. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 33.Frisch M.J., Trucks G.W., Fox D.J. Gaussian; Wallingford, CT: 2009. Gaussian 09. [Google Scholar]
- 34.Bayly C.I., Cieplak P., Kollman P.A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges—the RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- 35.Wang J., Wang W., Case D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 36.Fukunishi H., Watanabe O., Takada S. On the Hamiltonian replica exchange method for efficient sampling of biomolecular systems: application to protein structure prediction. J. Chem. Phys. 2002;116:9058–9067. [Google Scholar]
- 37.Liu P., Kim B., Berne B.J. Replica exchange with solute tempering: a method for sampling biological systems in explicit water. Proc. Natl. Acad. Sci. USA. 2005;102:13749–13754. doi: 10.1073/pnas.0506346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terakawa T., Kameda T., Takada S. On easy implementation of a variant of the replica exchange with solute tempering in GROMACS. J. Comput. Chem. 2011;32:1228–1234. doi: 10.1002/jcc.21703. [DOI] [PubMed] [Google Scholar]
- 39.van der Spoel D., Lindahl E., Berendsen H.J.C. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 40.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 41.Pronk S., Páll S., Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonomi M., Branduardi D., Parrinello M. PLUMED: a portable plugin for free-energy calculations with molecular dynamics. Comput. Phys. Commun. 2009;180:1961–1972. [Google Scholar]
- 43.Tribello G.A., Bonomi M., Bussi G. PLUMED 2: new feathers for an old bird. Comput. Phys. Commun. 2014;185:604–613. [Google Scholar]
- 44.Bussi G. Hamiltonian replica exchange in GROMACS: a flexible implementation. Mol. Phys. 2014;112:379–384. [Google Scholar]
- 45.van der Spoel, D., B. H. E. Lindahl, and the GROMACS Development Team. 2013. GROMACS User Manual version 4.6.3, www.gromacs.org.
- 46.Hornak V., Abel R., Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 48.Darden T., York D., Pedersen L. Particle mesh Ewald—an N.Log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 49.Hess B., Bekker H., Fraaije J. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 50.Daura X., Gademann K., Mark A.E. Peptide folding: when simulation meets experiment. Angew. Chem. Int. 1999;38:236–240. [Google Scholar]
- 51.Laio A., Parrinello M. Escaping free-energy minima. Proc. Natl. Acad. Sci. USA. 2002;99:12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laio A., Gervasio F.L. Metadynamics: a method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science. Rep. Prog. Phys. 2008;71:126601. [Google Scholar]
- 53.Barducci A., Bussi G., Parrinello M. Well-tempered metadynamics: a smoothly converging and tunable free-energy method. Phys. Rev. Lett. 2008;100:020603. doi: 10.1103/PhysRevLett.100.020603. [DOI] [PubMed] [Google Scholar]
- 54.Wakamori M., Fujii Y., Yokoyama S. Intra- and inter-nucleosomal interactions of the histone H4 tail revealed with a human nucleosome core particle with genetically incorporated H4 tetra-acetylation. Sci. Rep. 2015;5:17204. doi: 10.1038/srep17204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winogradoff D., Echeverria I., Papoian G.A. The acetylation landscape of the H4 histone tail: disentangling the interplay between the specific and cumulative effects. J. Am. Chem. Soc. 2015;137:6245–6253. doi: 10.1021/jacs.5b00235. [DOI] [PubMed] [Google Scholar]
- 56.Taylor G.C.A., Eskeland R., Bickmore W.A. H4K16 acetylation marks active genes and enhancers of embryonic stem cells, but does not alter chromatin compaction. Genome Res. 2013;23:2053–2065. doi: 10.1101/gr.155028.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.