Abstract
While much is known about DNA structure on the basepair level, this scale represents only a fraction of the structural levels involved in folding the genomic material. With recent advances in experimental and theoretical techniques, a variety of structures have been observed on the fiber, gene, and chromosome levels of genome organization. Here we view chromatin architecture from nucleosomes and fibers to genes and chromosomes, highlighting the rich structural diversity and fiber fluidity emerging from both experimental and theoretical techniques. In this context, we discuss our recently proposed folding mechanism, which we call “hierarchical looping”, similar to rope flaking used in mountain climbing, where 10-nm zigzag chromatin fibers are compacted laterally into self-associating loops which then stack and fold in space. We propose that hierarchical looping may act as a bridge between fibers and genes as well as provide a mechanism to relate key features of interphase and metaphase chromosome architecture to genome structural changes. This motif emerged by analysis of ultrastructural internucleosome contact data by electron microscopy-assisted nucleosome interaction capture cross-linking experiments, in combination with mesoscale modeling. We suggest that while the local folding of chromatin can be regulated at the fiber level by adjustment of internal factors such as linker-histone binding affinities, linker DNA lengths, and divalent ion levels, hierarchical looping on the gene level can additionally be controlled by posttranslational modifications and external factors such as polycomb group proteins. From a combination of 3C data and mesoscale modeling, we suggest that hierarchical looping could also play a role in epigenetic gene silencing, as stacked loops may occlude access to transcription start sites. With advances in crystallography, single-molecule in vitro biochemistry, in vivo imaging techniques, and genome-wide contact data experiments, various modeling approaches are allowing for previously unavailable structural interpretation of these data at multiple spatial and temporal scales. An unprecedented level of productivity and opportunity is on the horizon for the chromatin structure field.
Main Text
Deciphering the structure and function of the genomic material has proven to be an enduring challenge in modern science. As our appreciation for the diversity and flexibility of the double helix has deepened, its large-scale coiling around histone proteins to form the chromosomal material in eukaryotic organisms has opened new structural and mechanistic questions. These questions span from single nucleosomes (hundreds of basepairs) to chromatin fibers (thousands of basepairs, kb) to condensed chromosomes (millions of basepairs, Mb); see Fig. 1. Not only do we lack a full understanding of the structure of the chromatin fiber and chromosomal arrangements from the kb to Mb levels, we know much less about how structural transformations between these states occur. Because transitions from open, transcriptionally active to closed, transcriptionally silent epigenetic states are of crucial importance to genome function—from transcription to differentiation to cell senescence—there is great interest in deciphering these states and transitions by modeling and instrumentation.
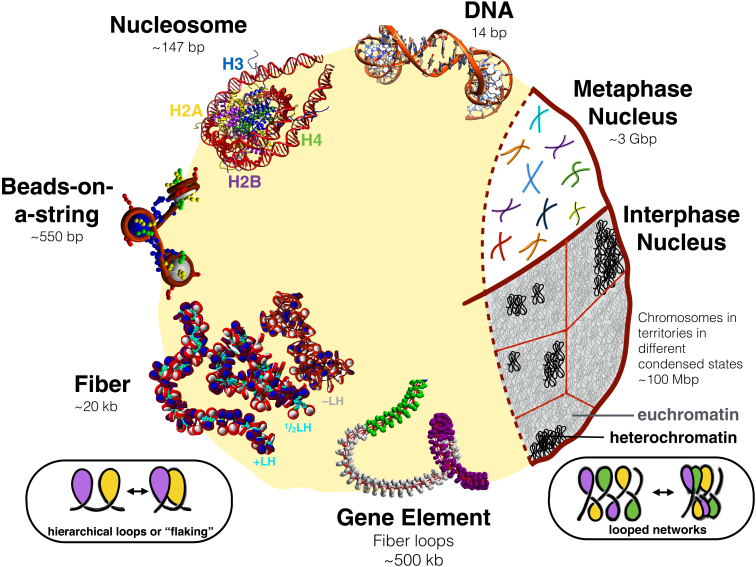
Figure 1.
A schematic view of the levels and structures involved as the chromatin fiber packs genomic DNA in the cell nucleus. Several levels of structural and functional states are known but not well understood. Double-stranded DNA binds to histone proteins to form complexes, known as nucleosomes, the building blocks of chromatin. Each nucleosome is composed of ∼147 bp of DNA coiled around eight histone proteins, two dimers each of H2A, H2B, H3, and H4. Many nucleosomes have been resolved at atomic resolution. Nucleosomes connect to one another to form the chromatin fiber, where an open state forms at low salt without linker histones called beads-on-a-string. A more condensed state forms at high salt with linker histones like H1 or H5. The long-assumed compact 30-nm fiber may adopt more variable forms in vivo, as shown for the subsaturated LH fibers (one LH per two nucleosomes, or ½LH) and LH-deficient (−LH) fibers shown in the bottom left. Relatively straight +LH fibers show little self-association, whereas ½LH fibers show moderate self-association, and −LH fibers show high levels of self-association. All three types show variable widths. Additionally, these self-associating fibers may involve hierarchical looping or flaking, as used in mountain climbing and rappelling, which is more pronounced in ½LH and −LH fibers. Flaking, in this context, refers to the lateral compaction of a long polymer due to loops of various size undergoing further folding in space, while avoiding tangles or knots. In the context of rope folding, flaking refers to packaging flexible elongated rope into neatly stacked loops (by winding the loops back and forth) so that the rope can be easily unraveled. A simple scheme for such rope stacking is shown at the bottom left. At bottom right, we show another possible variation of such compact networks of stacked loops. Active gene elements form large loose loops that bridge gene promoters to gene-transcribing regions. In cooperation with other epigenetic marks and scaffolding proteins, these gene elements organize into chromosomes. In interphase chromosomes, individual chromosomes are separated into different chromosomal territories. In the metaphase cell, individual chromosomes can be distinguished, as shown by different colors. Interphase chromosomes are also known to exist in different levels of condensation. The more loosely packed genomic state is known as “euchromatin”, while the more densely packed genomic material is denoted as “heterochromatin”. A possible internal organization for these two related states involves tight versus open networks of chromatin loops, which build upon the proposed flaking or hierarchical looping motif. To see this figure in color, go online.
Exciting recent advances are providing important anchors for addressing these puzzles. Namely, rapid developments in x-ray crystallography, cryo-electron microscopy (cryo-EM), in situ biochemistry, single-nucleosome resolution nanoscopy, and genome-wide high throughput experiments are providing a wide range of images and data across several orders of magnitude of spatial and temporal scales. In tandem, in silico techniques now make possible reliable modeling of the chromatin polymer: atomic nucleosomes, coarse-grained chromatin fibers, and segments of chromosomes. Together, a wide range of multiscale models are helping bridge experimental data with increased accuracy and insight, as recently reviewed (1, 2). An unprecedented level of opportunity and productivity for chromatin science is on the horizon.
As always, however, new advances raise new questions. While a key goal over the past decade has been identification of the assumed structure of a compact 30-nm chromatin fiber—whether zigzag, solenoid, or other—an intensely debated new topic concerns the existence of this ordered fiber itself in the nucleus of living cells. Many in vivo experiments and models are now suggesting that less ordered, polymorphic forms of chromatin fibers may be more functionally relevant (3, 4, 5, 6, 7, 8, 9). Additionally, although it is becoming apparent that chromatin structure depends on many internal and external variables such as the DNA linker length, linker histone density, and ion concentration (10, 11, 12, 13, 14), there is still uncertainty about how these parameters regulate chromatin structure across cell stages (15). Clearly, various discrete states are now being recognized at different cell cycle stages and differentiation states, and new insights into the structure of interphase and metaphase chromatin are emerging (15, 16, 17). Identifying these various states, interpreting their transitions and functions, and relating these functions to global or local epigenetic marks for manipulation of cell differentiation states, now form intense and exciting areas of research (12, 18, 19).
In this perspective, we discuss several of these different structural arrangements on multiple length scales—namely the nucleosome, fiber (kb), and gene levels (∼100–800 kb), and relate our emerging folding motif, hierarchical looping or flaking (see Fig. 1) to other models for interphase and metaphase chromatin and to various levels in chromatin structure and function. We suggest that the combined field advances mark a turning point in our understanding of the inner workings of genomic DNA, as we rapidly move from a static, discrete structure of the chromatin fiber to self-organizing functional systems composed of fluid, interconverting fibers.
From nucleosomes to chromosomes
Chromatin foundations
The building block of eukaryotic DNA in the nucleus is the nucleosome, a nucleoprotein complex where two copies of each histone protein H2A, H2B, H3, and H4 form an octamer around which ∼147 bp of DNA are wrapped tightly in 1.75 left-handed supercoil turns; see Fig. 1 (20). This stable structure forms spontaneously in solution under native conditions, and can be stabilized by certain conserved sequence motifs (21). Each core histone protein also features a polypeptide tail, or a terminal domain that projects away from the nucleosome into the solvent. These tails are generally disordered and have been implicated in a variety of functional roles, including nucleosome stability (22), cell signaling (23), large-scale chromatin compaction (24), and genetic activation/silencing (23). Due to their distinct positions on the nucleosome, the various histone tails have unique roles in stabilizing chromatin architecture. The H2A and H2B tails have been implicated in maintenance of nucleosome stability by point mutation studies in combination with Förster resonance energy transfer (FRET) experiments (22). Tail deletion studies show that H2A and H2B tails maintain cross-nucleosome interactions (25). The H4 tails play dual roles in interacting with an acidic patch on the surface of the nucleosome and forming contacts with nonparent nucleosomes (26). The H4 tail is also a common target for numerous chemical modifications during or after translation (termed “posttranslational modifications” or PTMs) such as acetylation, phosphorylation, or methylation (23). EM imaging and biochemical assays have shown that these alterations induce global structural changes on chromatin (27). While these four core histones define canonical nucleosomes, many variants exist for each histone, although the specific functions of most variants are not well understood. Among the well-known variants, H2A.Z alters the stability of the resulting nucleosome and is associated with transcription start sites (TSSs), while H3.3 helps establish the CENP-A nucleosome found in centromeric regions (12).
Additionally, recent modeling work has further illuminated the roles of tails in stabilizing higher-order chromatin structure. For example, mesoscale modeling has corroborated that the H4 tail bridges internucleosome contacts via the acidic patch; the H3 tail screens the DNA linkers and stabilizes stems when linker histones are present; the H2A and H2B tails establish cross-nucleosome contacts (28); and the H4 and H3 tails play significant roles in magnesium dependent compaction of fibers (29). Recent multiscale studies also suggest that the mechanisms by which histone tails mediate higher-order chromatin structure are complex. For example, atomic resolution modeling coupled with coarse-grained models have suggested a stepwise mechanism by which the H3 tail may stabilize nucleosomal DNA unwrapping by binding to partially unwrapped nucleosomal DNA (30), and that lysine acetylation may regulate global chromatin architecture not through direct charge modulation per se, but by inhibiting crucial long-range internucleosome contacts due to decreased tail flexibility (31).
Another crucial protein implicated in chromatin architecture is the linker histone (LH), which associates with the linker DNA near the entry/exit point to/from the nucleosome (32). The linker histone has a crucial role in chromatin condensation by forming stems with the linker DNA (33). H1 and H5 linker histones are common, and variants for different tissues exist, which compete in the cell nucleus for binding time (34).
The linker histone is composed of three domains—an N-terminal domain; an unstructured C-terminal domain; and a small globular head, which is highly positive and binds tightly to the entering/exiting DNA of the nucleosome. The C-terminal domain, which is larger than 100 amino acids, stretches out along the linker DNA and condenses in response to increasing cellular salt concentrations (35). Binding of linker histones dramatically alters global chromatin fiber properties. Namely, saturated LH chromatin (i.e., one LH per nucleosome) encourages condensation of chromatin fibers by forming tight stems with the DNA (36, 37). Although saturated LH densities are noted in major cell lines (15), the affinities of nucleosomes to linker histones are highly dynamic (38). This binding affinity is also affected by PTMs such as phosphorylation and citrullination (39); citrullination involves converting an arginine residue into a noncoding amino acid citrulline, in which the terminal region of the side chain has been neutralized (39). Such PTMs have also been implicated in cell differentiation states (39) and cell cycle maintenance (38).
The length of linker DNA between nucleosomes plays an important role in chromatin fiber structure and architecture (11, 32). Average reported linker lengths vary widely depending on the tissue, organism, cell cycle, and differentiation state (40, 41). It has been observed experimentally for yeast (42) and mouse (43) cell lines that chromatin fiber linker lengths tend to periodic values of 10n or 10n + 5 bp (where n = 1, 2, 3, 4, 5 for yeast and n = 1, 2, 3, …, 10 for mice). Such effects were explained by theoretical modeling (44, 45). How this effect extends to systems with linker histone, however, remains unknown.
The linker length is often described by the nucleosome repeat length (NRL), or the sum of DNA linker basepairs plus 147 bp of the DNA wrapped around each nucleosome (14). For example, on average, human genes have ∼44 bp separating each nucleosome, with NRL = 191 bp (40). NRLs as short as 167 bp are found in fission yeast and neuronal cells, whereas the longest linkers with an NRL of ∼240 bp exist in echinoderm sperm (14).
While early quantification of NRLs in various cells was performed by a micrococcal nuclease assays (14), EM provided the first images demonstrating fiber structure dependence on the NRL (32). Such patterns have been corroborated by mesoscale modeling, where short NRLs of 173 or 182 bp (26 or 35 bp linker) favor stiff compact fibers with strict repeating structures; medium NRLs of 191 or 200 bp (44 or 53 bp linker) show polymorphic bent fibers; and long NRLs such as 227 bp (80 bp linker) favor highly bent globular states where individual fibers can be difficult to define (46, 47, 48). Recent force-extension studies also showed the different mechanical response for fibers of different linker lengths (49).
When fibers are saturated with linker histone, the most compact fibers emerge at moderate values of NRL (50). These patterns occur because compact stems form optimally when the length of the LH is approximately the same as the length of the linker DNA. When the DNA is much longer, it necessarily bends, encouraging less compact fibers (48).
A linear relationship between the NRL and LH density has long been observed experimentally (14). Recently, Luque et al. (50) have interpreted this relationship by formation of compact zigzag fibers at a critical LH density that is NRL-dependent. Importantly, at this compact zigzag state, different fibers have similar tail interaction patterns, suggesting how cells can sense these patterns to adjust the NRL and LH density as needed for cell function at specific cell stages. The LH content of chromatin fibers is thus not only important for regulating the local chromatin structure; the effects may translate into global features, as we discuss below.
Experimental background on nucleosomes, fibers, and chromosomes
Mononucleosomes and short oligonucleosome fibers have been investigated using x-ray crystallography, cryo-EM imaging, small angle x-ray scattering techniques, and single molecule FRET experiments, to name a few (15). Cryo-EM imaging recently detailed a compact tetranucleosome fiber (with short DNA linkers) at high resolution (51), while x-ray crystallography has revealed configurations of histone tails (52). Single molecule FRET experiments have mapped conformational intermediates of wrapped and unwrapped DNA around the nucleosome, suggesting several states involved in spontaneous unfolding (53), as well as characterizing the binding configurations and salt-dependent compaction of the LH C-terminal domain (54).
At low salt levels, a short chain of nucleosomes appears like “beads on a string”, or uncondensed 10 nm chromatin fibers, as observed on the electron microscope (Fig. 1). This level of organization constitutes chromatin secondary structure. With recent advances in genome sequencing, however, some researchers have characterized this level of organization in terms of nucleosome occupancy, where the binding sites of nucleosomes are plotted against genomic sequence. Nucleosome occupancy profiles have been characterized in both somatic and embryonic cells, often revealing local fluctuations in nucleosome binding tendencies near TSSs (55).
At higher levels of salt concentration, nucleosomes associate to form chromatin fibers (up to ∼200 nucleosomes). These fibers are generally studied by contact probability profiles, where cross-linking experiments in vitro and in vivo determine nucleosome contact tendencies. Short-range contacts, ranging from 1 to 25 nucleosomes, can be determined by the EM-assisted nucleosome interaction capture (EMANIC) technique, which cross links nucleosomes and then relaxes the contacts, followed by EM visualization (9, 56). These contact probability profiles serve as a fingerprint for chromatin fiber type, and can distinguish ordered from nonordered fibers, as well as highlighting fiber characteristics. Zigzag fibers, for example, feature a next-neighbor dominance, or i ± 2 peaks in local contacts, whereas solenoid fibers show strong near-neighbor contacts, or i ± 1 peaks in the local contact probability profile. Ideal solenoids also show lower peaks in the i ± 6 nucleosome range. Further contacts for idealized zigzag or solenoid fibers are not expected, due to heteromorphicity in natural cellular conditions. Other imaging techniques have also been used to study chromatin fibers at this level, including atomic force microscopy and optical tweezers used for force-pulling, although the resolution obtained is lower compared to that for small oligonucleosome fibers. See Schlick et al. (57) for a review.
Gene loci and functional gene elements that span from 100 to 800 kb (∼400–4000 nucleosomes) can be examined by chromatin conformation capture techniques (3C), where chromatin contacts are fixed via formaldehyde titration and then sequencing technology is used to determine the distance from specified sequences to cross-linked regions. These contact profiles have been used to investigate the structure of active (58) and inactive (59) gene loci, roughly corresponding to closed heterochromatin and open euchromatin regions (see Fig. 1). These studies suggest that active euchromatin may be composed of 2–3 large loops on the order of 100–500 kb each, which act to bring TSSs within close proximity to form “active chromatin hubs” (58). Genes associated with low levels of transcription in healthy cells, like the GATA-4 locus, feature smaller loops (8–30 kb); their transcription start site is near the center of the connection hub and is associated with trimethylations of Lys27 on the H3 tail (H3K27Me3) and recruitment of polycomb group (PcG) proteins (60). Together, these data suggest that loop size in the fiber and gene levels is important for distinguishing between open and closed chromatin states (59). Superresolution imaging techniques like STORM have also investigated the structure of active and inactive chromatin at the gene level, providing information about volume and surface area of gene loci (61). Details of the three-dimensional structures of these contacts, however, have not yet been observed directly.
Contact probabilities that span millions of basepairs are also emerging for different cell lines and species via genomewide chromosome experiments like Hi-C, an extension of 3C techniques developed for high-throughput implementation (62). Contact maps show that chromosomes self-associate with marked division in the nucleus (as sketched in Fig. 1), corroborating data by in situ fluorescence techniques (62). Contacts on the Mb length scales can be further decomposed from chromosomes to topologically associating domains (TADs) (63), which vary in size. The CTCF protein is associated with long-term maintenance of these TADs (64), and a recently proposed loop extrusion mechanism suggests that cohesin proteins work cooperatively with CTCF proteins to recognize specific DNA sequences and establish the loops (65).
Contact probabilities at this scale, however, are difficult to understand directly, and require polymer or other models for their interpretation. Traditionally, polymer models neglect details at the kb length scale, as well as other variables known to influence chromatin structure, such as discussed above (i.e., NRL, linker histone density, divalent ion concentration, and PTMs) (66). The recent energy landscape approach (67, 68) offers another interesting way to interpret genomic data on this level.
General computational approaches
All atom, mesoscale, and polymer models
Computational techniques that can systematically explore chromatin variables and features are helping interpret and provide key insights into choromatin structure and function not available by experiments alone. New modeling approaches have been successful in determining the structure, energy, dynamics, and effects of biochemical modifications for a wide variety of chromatin-related structures and processes (1, 2). Computational techniques can be classified as atomistic, general coarse-grained, mesoscale, polymer, and continuum approaches.
While powerful, atomic resolution computational approaches are generally limited to a few nucleosomes due to high computational requirements. These models, which incorporate results from well-parameterized protein and nucleic acid force fields, are applied to a variety of interesting questions. These include the dynamics of histone tails and linker DNA in solution (69), effects of PTMs on global chromatin architecture (31), dynamics of DNA unwrapping (70), and the roles of histone tails in stabilizing the nucleosome core particle (30). See Ozer et al. (1), Dans et al. (2), and Schlick et al. (57) for recent reviews.
Coarse-grained approaches are necessary for the study of larger systems of chromatin. For example, recent investigations explored the free energy of wrapping/unwrapping of the nucleosome (68), tail-driven chromatin aggregation, and interactions with explicit ions (71), force spectroscopy curves (72), tail dynamics (30), and the interaction of tails with regulatory proteins under ionic conditions (73).
Mesoscale approaches often treat the nucleosome as a basic subunit of modeling, while using other theories to treat linker DNA and flexible protein segments. For example, in our model (Fig. 2), we treat the nucleosome, without the protruding tails, as a rigid electrostatic object parameterized by Debye-Hückel pseudo-surface charges that reproduce the electrostatic environment of an atomistic nucleosome (74). The linker DNA is treated with a modified wormlike chain (48), and the flexible histone tails and linker histones are coarse-grained by Brownian dynamics to approximate atomic dynamics in united-atom protein representations (75). By combining these various representations into a single mesoscale potential, we have developed a robust and scalable model that treats systems from 1 to 500 nucleosomes (9, 76). The model incorporates varying internal parameters such as linker DNA length (46, 47, 48), histone tails and their PTMs (28, 31, 75), dynamic LH binding (50, 77, 78), and divalent ion concentrations (56).
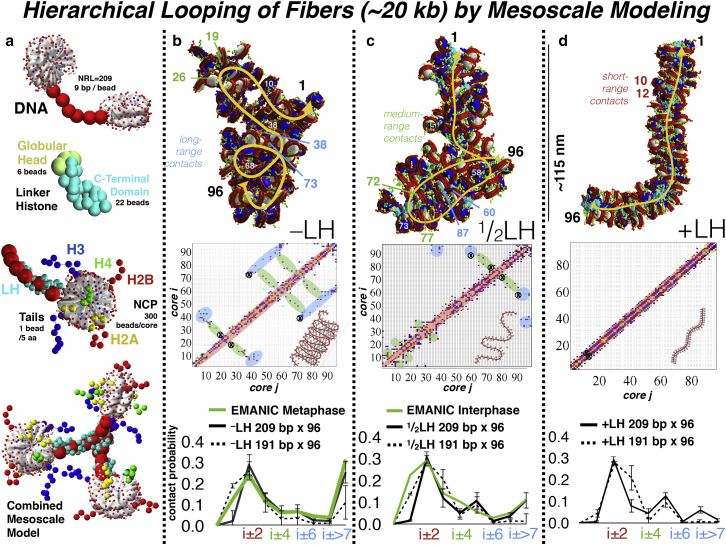
Figure 2.
Hierarchical looping at the fiber level. (a) Our mesoscale model treats the various chromatin constituents separately. DNA is modeled as a modified worm-like chain, with 9 bp per bead. LHs are coarse-grained from all-atom molecular dynamics simulations so that six beads represent the globular head and 22 beads represent the C-terminal domain. The histone tails are coarse-grained from united-atom representations similarly, so that 5 amino acids define a tail bead. Nucleosome cores, without tails, are treated as rigid bodies with 300 Debye-Hückel pseudo-charges placed on the irregular nucleosome surface using our program DiSCO (92), so that the electrostatic field is reasonably reproduced as a function of the monovalent salt concentration. These DNA, LH, and histone tail beads are combined with the nucleosome to form our mesoscale chromatin model. (b–d) Sample 96-nucleosome configurations are shown for LH-deficient (−LH) (b), partially LH-saturated (one LH per two nucleosomes, or ½LH) (c), and LH-saturated (one LH per nucleosome, or +LH) (d) fibers. Internucleosome contact matrices are calculated from each mesoscale configuration by measuring the distance between any two core, tail, or linker DNA beads belonging to separate nucleosomes, where any distance measured <2 Å is considered a contact. In these matrices, local contacts are highlighted in red, whereas medium-range contacts are highlighted in green, and long-range contacts are colored in blue. To help visualize the three types of internucleosome contacts, we mark specific nucleosome interactions in the contact matrices with an x symbol, and indicate the corresponding pair of nucleosomes in the configurations above. We also sketch, at each bottom-right corner of the matrix, a simple polymer view to illustrate the evolution of flaking. At bottom, EMANIC contact probabilities for metaphase and interphase chromatin were derived by cross linking native HeLa cell contacts in vivo, before relaxing the structures and imaging via EM (9). When compared to contact probabilities derived from mesoscale models (black and dashed lines) of varying LH concentrations, the patterns of −LH fibers show good agreement with metaphase chromatin, whereas ½LH fibers correspond well to interphase chromatin. LHs can modulate both local and global compaction: more compact fibers with saturated LH content have less self-association and thus are globally less compact, whereas ½LH or −LH fibers are locally less compact, but globally more compact due to enhanced long-range (i ± ≥7) contacts. To see this figure in color, go online.
Kb-Mb-sized loops were first modeled in an attempt to interpret fluorescence in situ hybridization (FISH) data of a section of Chromosome 4 (79). In models based on these data, the chromatin fiber was treated as an ideal polymer and 120 kb-sized loops were proposed to be arranged in a rosette-like structure in space (80), predicting compartmentalization on the Mb level (81). More recent polymer models (82) are similarly helping interpret Hi-C contacts in the Mb range. These synergistic approaches have successfully established the global behavior of chromosomal organization in the nucleus (83), highlighting the organization of TADs (65, 80), and describing formation of metaphase chromatin (84). The physical nature of the polymer model subunit, however, is still an active area of debate. For example, proposed models include rosette-like multiloop structures, loops with confined volume, and polymer melts (82). The lack of detailed resolution regarding gene- and fiber-sized contacts (kb to sub-Mb) is a drawback. The notable absence of connections between the fiber and chromosomal scales between experiment and theory naturally suggests the development of multiscale models that can bridge these gaps (1, 2, 3, 31). The multiscale model of Collepardo-Guevara et al. (31) combines the nucleosome and fiber levels by a three-tiered computation (microsecond atomistic molecular dynamics for tails and dinucleosomes, with coarse-grained mesoscale fiber sampling), but more general frameworks are also required to reach chromosomal levels.
Insights from mesoscale modeling
Kb fibers are structurally diverse
The mesoscale model described above has reproduced experimentally observed properties of chromatin for both local structural features (internucleosome distances, entry/exit angles) and global features. For example, calculated sedimentation coefficients reproduce the salt dependence of in vitro studies of fibers with 60 bp linker DNAs (or NRL = 207 bp) and of purified rat chromatin (56, 78). Similarly, local properties such as the entry/exit angles, triplet angles, and packing ratios have been validated as a function of salt for chicken erythrocyte chromatin that are summarized and cited in Luque et al. (78). Force-pulling experiments identify similar plateau values in force-extension curves for short and long linker lengths and LH orientations (85), and structural studies show good agreement for fibers of short, medium, and long linker lengths (46, 48) compared to solved structures (51, 52). See Arya and Schlick (29), Luque et al. (78), and Collepardo-Guevara and Schlick (85) for a summary of experimental validations.
We have also described the variability of viable secondary structures (48, 56), highlighting the rich structural diversity of oligonucleosome fibers (57). Simulations have suggested that interconversions between structures are tuned by internal parameters such as NRL (46), divalent ion concentration (56), LH densities (50), and LH dynamic binding/unbinding (85), all of which vary throughout cell cycle and differentiation state (15). Thus, nuclear organization and architecture may be best understood as an ensemble of chromatin states, tied together by common folding motifs and compaction states, rather than chemical species or static structure (57).
The long-standing controversy between zigzag and solenoid forms of chromatin can be reconciled by considering that the addition of magnesium ions into solution encourages a small degree of linker DNA bending so that zigzag and solenoid features combine into one compact fiber (56). Namely, computational contact profiles compared with EMANIC contact profiles reveal that most compact fibers blend as mostly zigzag with 20–30% solenoid features optimally, to produce a more compact fiber than a pure zigzag chain. These polymorphic features likely encourage interconversions in the cell, as needed to fit evolving biological roles.
Modeling has suggested that by varying the NRL value along 24 nucleosome fibers, three different classes of fiber conformations form, termed bent-ladders, canonical, and polymorphic, each with distinct structural properties. Bent ladders, which incorporate alternating short or medium linker lengths (with alternating NRL values of 173/182, 173/209, and 173/227 bp), favor short loops, hairpins, or bends ∼10–20 nucleosomes long. Canonical fibers, composed of alternating medium and long linkers with medium average linker length (NRLs of 182/191, 182/200, 191/200, 191/200, 200/209, 209/218, and 218/227 bp) form irregular/heteromorphic zigzag fibers similar to those seen with uniform medium linker lengths. Polymorphic fibers, composed of alternating medium and long linker lengths with long linkers on average (NRLs of 191/218, 200/218, 191/227, and 200/227 bp), tend to form highly bent or looped structures (48). In vivo chromatin is likely to exhibit natural variations from the above ideal models of NRLs. Because the diameters of compact 30-nm chromatin fibers are similar to the dimension of side-by-side 10-nm fibers or self-associating fibers (see Fig. 1), either one is a viable constituent for larger chromatin architectures.
Kb fibers form flaked networks or hierarchical loops
Self-associating fibers, expected in the crowded milieu of the cell nucleus, may explain the absence of ordered 30 nm fibers. EMANIC data from chromatin in vivo revealed medium-range internucleosome contacts for both interphase and metaphase chromatin, and long-range interactions for metaphase chromatin (9). Our corroborating modeling of 96 nucleosome fibers at various levels of linker histone concentrations suggests a regular looping network with a prevalence of stacked, hierarchical loops as illustrated in Figs. 1 and 2. The term “hierarchical looping”, or “flaking”, refers to a polymer that has been compacted laterally without knotting, with loops of various size undergoing further folding in space. We find that linker histone-deficient (−LH) fibers show significantly greater long-range contacts and self-association, whereas fibers with one LH per two nucleosomes (½LH) show medium-range and fewer long-range contacts. In contrast, LH saturated (+LH) fibers show mostly short-range interactions (Fig. 2). Moreover, the average contact probability profiles of −LH fibers computed from mesoscale modeling show excellent agreement with local contact probability profiles in bulk metaphase chromatin as determined by EMANIC. Similarly, contact profiles for simulated ½LH fibers agree with local contact probability profiles derived from bulk interphase chromatin by EMANIC (Fig. 2, bottom row). Experimentally, it is known that LH binding affinities are lower in metaphase chromatin (38). Together, these findings suggest that stacks of such hierarchical loops may represent local chromatin contacts in vivo for both interphase and metaphase chromatin, similar to rope flaking used by mountain climbers and rapelers. This folding motif features zigzag-dominant chains with the added benefit of avoiding global tangles or knots (9), and is consistent with an earlier mapping of nucleosome proximities in living cells (86).
The self-associating fibers define highly dynamic networks. Despite binding with high affinity, linker histones are known to exchange spontaneously in solution (38). Indeed, in our separate modeling work of dynamic LH binding in the context of fiber stretching due to applied forces, we found that dynamic LH binding leads to significant softening of the fiber during force extension and reproduces experimental force extension curves better. Thus, both the binding rate and local binding affinity play an important role in structural characteristics of the chromatin fiber, highlighting its fluid nature (77).
Interphase and metaphase chromatin transitions are mediated by LH binding, NRL, and salt concentration
While some characteristics of both interphase and metaphase chromatin have been elucidated experimentally, these characteristics largely rely on long-lived contacts bridged by external proteins such as CTCF and cohesin proteins on the Mb scale. Candidates for dynamic, local folding motifs on the fiber scale (kb), however, are lacking. Hierarchical looping could help suggest how transitions between interphase and metaphase forms could be mediated through dynamic LH binding on both local and global structural scales. That is, the higher levels of LH densities associated with interphase chromatin yield more locally condensed and stiff fibers (Fig. 2), whereas lower levels of LH associated with metaphase chromatin promote more condensed global chromatin architectures due to increased midrange contacts and enhanced self-association and nucleosome intradigitation (Fig. 2). Thus, LH density likely plays a pivotal structural role in the formation of condensed chromosomes.
Indeed, although LH binding affinity can vary throughout the cell cycle, it is notably at a minimum during metaphase, corresponding to the most globally condensed chromatin state (38, 87). Healthy cells actively regulate the binding affinity of LHs to nucleosomes throughout the cell cycle in many ways, including LH phosphorylation (14) and citrullination (39). The sensitive relationship between NRL and LH binding affinity is also associated with histone tail patterns and cellular constraint aspects (50). Specifically, when the length of the linker DNA corresponds to the optimal length of LH’s C-terminal domain condensation, a stable LH stem forms in a salt-dependent manner (50, 78). Taken together, these findings suggest that chromatin internal characteristics are important components that regulate interphase and metaphase chromatin transitions, and that local contacts formed by hierarchical loops, or flaking networks, can connect different condensation states fluidly. These dynamic internal parameters that evolve frequently are in contrast to external proteins such as chromatin remodeling factors that act on longer timescales and result in more stable structures.
Gene level folding is mediated by external proteins and tail epigenetic marks
Because loop architectures are important features of gene elements, hierarchical looping may play a role on the sub-Mb level as well, in tandem with external protein elements and epigenetic modifications. For example, the GATA-4 gene locus, which resides on the short arm of chromosome 8, has been implicated in epigenetic silencing via recruitment of long-lived chromatin remodeling factors at nucleosomes associated with trimethylation of the H3 tail at Lys27. Specifically, PcG proteins, which bind tightly to H3K27Me3-enriched nucleosomes and methylated DNA, establish important bridges via loops on the sub-Mb scale to regulate transcription. To examine these structures, we have incorporated 3C contact data of a 5-loop hub for the GATA-4 system as harmonic restraints in our mesoscale model. Analysis suggests that hierarchical loops operate on this scale (∼100 kb) (76) to condense the chromatin and form a 5-loop stacked globule (see Fig. 3). Specifically, these stacked loops accommodate the GATA-4 connections seen in epigenetically silenced cells, and envelop the TSS, which lies between the third and fourth loop (in the direction of transcription; see Fig. 3). Thus, hierarchical looping may play a structural and functional role by helping guide the 5-loop gene element into the arrangement associated with gene silencing, in cooperation with external proteins and PTMs of the H3 tail.
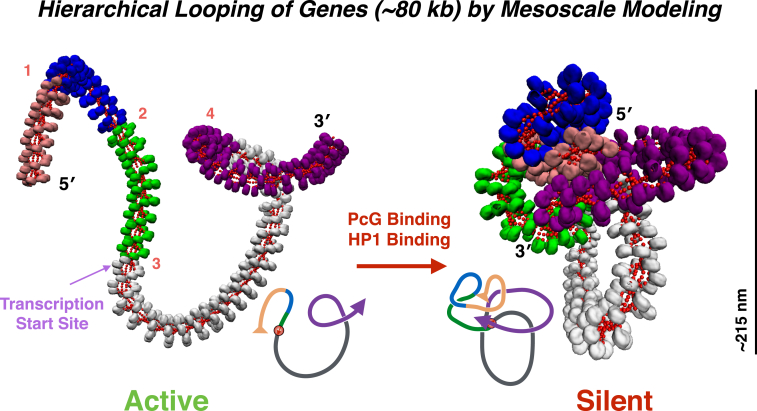
Figure 3.
Hierarchical looping at the gene level. Structural model of the GATA-4 gene locus in an open, active state (left) versus an epigenetically silent state (right) as suggested by a combination of 3C contact data and mesoscale modeling (76). The GATA-4 gene has been shown to exhibit five distinct chromatin loops of ∼43, 61, 57, 157, and 109 nucleosomes each (colored in tan, blue, green, white, and purple, respectively); each loop is associated with enriched trimethylation of Lys27 of histone tail H3 (H3K27Me3) and PcG protein binding. The TSS, which resides between loop 3 and 4 (from 5′ to 3′), is enveloped by the chromatin loops when these contacts are enforced in our mesoscale model, suggesting a structural mechanism for epigenetic silencing of the GATA-4 gene. To see this figure in color, go online.
While external remodeling factors such as PcG proteins and HP1 may induce long-lived hierarchical looping on the gene level, short-lived local contacts on the fiber level can be mediated by internal parameters such as LHs (78), NRLs (48, 50), H4 tail modification states (31), and divalent ion concentration (56). These internal parameters act primarily by modulating linker DNA flexibility, LH binding affinities, and nucleosome-nucleosome contacts, whereas larger sub-Mb contacts are likely guided into their arrangement by external proteins and epigenetic marks such as H3 tail modifications.
Hierarchical looping is consistent with experiment
A general paradigm of hierarchical chromatin folding has long been assumed for DNA compaction (88). However, a specific mechanism of folding has rarely been suggested beyond coiling as implied by 30-nm structures. Recent Hi-C data in conjunction with polymer models of metaphase chromatin do not suggest hierarchical coiling, but instead condensation, which proceeds first along the main axis and then orthogonal to the main axis, where orthogonal compaction occurs with linear arrays of regular loops on the order of hundreds of kb (84). Hi-C-based computational models of interphase chromatin are more heterogenous in terms of compaction states, but the data underscore the role of large loops anchored by CTCF proteins. Early FISH experiments have suggested models of 120 kb-sized loops arranged into rosettelike subcompartments as early as 1992 (79, 81), predicting loop networks for TADs, which were later supported by Hi-C data (63).
The zigzag aspects of hierarchical looping are also supported by recent experiments. Single nucleosome resolution experiments such as superresolution nanoscopy can quantify local contacts in bulk conditions. For example, superresolution imaging of mouse embryonic stem cells recently showed a prevalence of 4-nucleosome clusters (19), fitting with similar clusters found in high-resolution Micro-C data obtained from budding yeast genomes (89). Such clusters emerged naturally in mesoscale simulations that employed forced unraveling of zigzag fibers, suggesting that small nucleosome clusters are a feature of highly fluid chromatin chains under tension (90). The pliant zigzag chains present in flaked networks that do not form 30-nm fibers are an important feature of hierarchical looping (see Figs. 1 and 2). They also naturally lead to small clusters of nucleosomes corresponding to medium-range interactions.
Imaging of condensed metaphase chromosomes, in conjunction with in situ fluorescence assays like FISH, has shown that metaphase chromatin is structurally condensed in bands that lie perpendicular to the longitudinal axis of the chromosome. These results have motivated the thin-plate model for metaphase chromatin, where nucleosomes are organized in single layers of nucleosomes, resulting in bands with a width of ∼6 nm (17). Hierarchical looping shares extensive nucleosomal self-association with this model (Figs. 1 and 2), but also makes connections with the more disordered notion of a polymer melt (4), which arose from cryo-EM imaging of metaphase HeLa cells that lack 30-nm fibers. Hierarchical looping helps reconcile these models by combining structural zigzag layers with fluid networks whose geometries can be controlled by internal parameters and external factors (Fig. 1).
Looking ahead
Considering the wide variety of fluid secondary and tertiary chromatin structures now coming to light, it is clear that highly dynamic and polymorphic structures, which readily interconvert, likely describe the genome material in live eukaryotic cells, from the kb to the sub-Mb level, and possibly further. Common folding motifs, such as hierarchical looping or flaking, may help bridge the architecture of fibers (kb) to genes (sub-Mb) and possibly chromosome territories (Mb) as proposed in Fig. 1. Hierarchical loops bridge available experimental data, suggesting ordered zigzag fibers on one level, but also loose, self-associating fibers in living cells whose structure can be regulated by internal and external factors. Specifically, the binding affinities of linker histones to nucleosomes may be modulated dynamically throughout cell cycle or differentiation state (91). These variable LH densities may in turn affect the extent of fiber self-association, and hence chromatin compaction throughout cell cycle stage or differentiation state. Further protein binding and epigenetic marks work in tandem to regulate fiber accessibility and condensation across multiple scales. The precise mechanistic details about how transitions between these states occur, remain to be uncovered.
New opportunities for modelers are now available as never before, particularly as various experimental approaches such as EMANIC and high-resolution Hi-C-like technologies such as Micro-C, and superresolution imaging technologies begin to overlap. Initial results of these rapid developments are highlighting the fluid exchange of chromatin secondary structure on the local level, along with global nuclear features specific to cellular differentiation state and stage (56, 89). Despite the great progress made, however, important challenges lie ahead. It will be important to fully characterize the various levels of folding hierarchies, transitions between various states, and the associated dynamics by a combination of experiment and theory. Bridging these scales as suggested in Ozer et al. (1) will undoubtedly offer translational ramifications for understanding genetic diseases and developing novel epigenetic-based treatments (59).
Acknowledgments
We thank Dr. Sergei Grigoryev for comments on a draft of this article.
This work was supported by National Institutes of Health grant No. R01-055164 to T.S., and by Phillip-Morris USA and Phillip Morris International to T.S.
Editor: Wilma Olson.
References
- 1.Ozer G., Luque A., Schlick T. The chromatin fiber: multiscale problems and approaches. Curr. Opin. Struct. Biol. 2015;31:124–139. doi: 10.1016/j.sbi.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dans P.D., Walther J., Orozco M. Multiscale simulation of DNA. Curr. Opin. Struct. Biol. 2016;37:29–45. doi: 10.1016/j.sbi.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 3.Eltsov M., MacLellan K.M., Dubochet J. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc. Natl. Acad. Sci. USA. 2008;105:19732–19737. doi: 10.1073/pnas.0810057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishino Y., Eltsov M., Maeshima K. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J. 2012;31:1644–1653. doi: 10.1038/emboj.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maeshima K., Hihara S., Eltsov M. Chromatin structure: does the 30-nm fibre exist in vivo? Curr. Opin. Cell Biol. 2010;22:291–297. doi: 10.1016/j.ceb.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Fussner E., Ching R.W., Bazett-Jones D.P. Living without 30-nm chromatin fibers. Trends Biochem. Sci. 2011;36:1–6. doi: 10.1016/j.tibs.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Joti Y., Hikima T., Maeshima K. Chromosomes without a 30-nm chromatin fiber. Nucleus. 2012;3:404–410. doi: 10.4161/nucl.21222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maeshima K., Imai R., Nozaki T. Chromatin as dynamic 10-nm fibers. Chromosoma. 2014;123:225–237. doi: 10.1007/s00412-014-0460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grigoryev S.A., Bascom G., Schlick T. Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes. Proc. Natl. Acad. Sci. USA. 2016;113:1238–1243. doi: 10.1073/pnas.1518280113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Worcel A., Strogatz S., Riley D. Structure of chromatin and the linking number of DNA. Proc. Natl. Acad. Sci. USA. 1981;78:1461–1465. doi: 10.1073/pnas.78.3.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Correll S.J., Schubert M.H., Grigoryev S.A. Short nucleosome repeats impose rotational modulations on chromatin fibre folding. EMBO J. 2012;31:2416–2426. doi: 10.1038/emboj.2012.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venkatesh S., Workman J.L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015;16:178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 13.Strick R., Strissel P.L., Levi-Setti R. Cation-chromatin binding as shown by ion microscopy is essential for the structural integrity of chromosomes. J. Cell Biol. 2001;155:899–910. doi: 10.1083/jcb.200105026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodcock C.L., Skoultchi A.I., Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 15.Woodcock C.L., Ghosh R.P. Chromatin higher-order structure and dynamics. Cold Spring Harb. Perspect. Biol. 2010;2:a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eagen K.P., Hartl T.A., Kornberg R.D. Stable chromosome condensation revealed by chromosome conformation capture. Cell. 2015;163:934–946. doi: 10.1016/j.cell.2015.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daban J.-R. Stacked thin layers of metaphase chromatin explain the geometry of chromosome rearrangements and banding. Sci. Rep. 2015;5:14891. doi: 10.1038/srep14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egger G., Liang G., Jones P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 19.Ricci M.A., Manzo C., Cosma M.P. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell. 2015;160:1145–1158. doi: 10.1016/j.cell.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 20.Luger K., Mäder A.W., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 21.Widom J. Role of DNA sequence in nucleosome stability and dynamics. Q. Rev. Biophys. 2001;34:269–324. doi: 10.1017/s0033583501003699. [DOI] [PubMed] [Google Scholar]
- 22.Gautier T., Abbott D.W., Dimitrov S. Histone variant H2ABbd confers lower stability to the nucleosome. EMBO Rep. 2004;5:715–720. doi: 10.1038/sj.embor.7400182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Shogren-Knaak M., Ishii H., Peterson C.L. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 25.Tse C., Hansen J.C. Hybrid trypsinized nucleosomal arrays: identification of multiple functional roles of the H2A/H2B and H3/H4 N-termini in chromatin fiber compaction. Biochemistry. 1997;36:11381–11388. doi: 10.1021/bi970801n. [DOI] [PubMed] [Google Scholar]
- 26.Kalashnikova A.A., Porter-Goff M.E., Hansen J.C. The role of the nucleosome acidic patch in modulating higher order chromatin structure. J. R. Soc. Interface. 2013;10:20121022. doi: 10.1098/rsif.2012.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li G., Reinberg D. Chromatin higher-order structures and gene regulation. Curr. Opin. Genet. Dev. 2011;21:175–186. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arya G., Schlick T. Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model. Proc. Natl. Acad. Sci. USA. 2006;103:16236–16241. doi: 10.1073/pnas.0604817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arya G., Schlick T. A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments. J. Phys. Chem. A. 2009;113:4045–4059. doi: 10.1021/jp810375d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voltz K., Trylska J., Langowski J. Unwrapping of nucleosomal DNA ends: a multiscale molecular dynamics study. Biophys. J. 2012;102:849–858. doi: 10.1016/j.bpj.2011.11.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collepardo-Guevara R., Portella G., Orozco M. Chromatin unfolding by epigenetic modifications explained by dramatic impairment of internucleosome interactions: a multiscale computational study. J. Am. Chem. Soc. 2015;137:10205–10215. doi: 10.1021/jacs.5b04086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Routh A., Sandin S., Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. USA. 2008;105:8872–8877. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bednar J., Horowitz R.A., Woodcock C.L. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl. Acad. Sci. USA. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bustin M., Catez F., Lim J.-H. The dynamics of histone H1 function in chromatin. Mol. Cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 35.Caterino T.L., Hayes J.J. Structure of the H1 C-terminal domain and function in chromatin condensation. Biochem. Cell Biol. 2011;89:35–44. doi: 10.1139/O10-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Syed S.H., Goutte-Gattat D., Dimitrov S. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc. Natl. Acad. Sci. USA. 2010;107:9620–9625. doi: 10.1073/pnas.1000309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou B.-R., Feng H., Bai Y. Structural insights into the histone H1-nucleosome complex. Proc. Natl. Acad. Sci. USA. 2013;110:19390–19395. doi: 10.1073/pnas.1314905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen D., Dundr M., Huang S. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J. Cell Biol. 2005;168:41–54. doi: 10.1083/jcb.200407182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christophorou M.A., Castelo-Branco G., Kouzarides T. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. 2014;507:104–108. doi: 10.1038/nature12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Compton J.L., Bellard M., Chambon P. Biochemical evidence of variability in the DNA repeat length in the chromatin of higher eukaryotes. Proc. Natl. Acad. Sci. USA. 1976;73:4382–4386. doi: 10.1073/pnas.73.12.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Dieuleveult M., Yen K., Gérard M. Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature. 2016;530:113–116. doi: 10.1038/nature16505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brogaard K., Xi L., Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486:496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voong L.N., Xi L., Wang X. Insights into nucleosome organization in mouse embryonic stem cells through chemical mapping. Cell. 2016;167:1555–1570.e15. doi: 10.1016/j.cell.2016.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Norouzi D., Zhurkin V.B. Topological polymorphism of the two-start chromatin fiber. Biophys. J. 2015;108:2591–2600. doi: 10.1016/j.bpj.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clauvelin N., Lo P., Olson W.K. Nucleosome positioning and composition modulate in silico chromatin flexibility. J. Phys. Condens. Matter. 2015;27:064112. doi: 10.1088/0953-8984/27/6/064112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perišić O., Collepardo-Guevara R., Schlick T. Modeling studies of chromatin fiber structure as a function of DNA linker length. J. Mol. Biol. 2010;403:777–802. doi: 10.1016/j.jmb.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlick T., Perišić O. Mesoscale simulations of two nucleosome-repeat length oligonucleosomes. Phys. Chem. Chem. Phys. 2009;11:10729–10737. doi: 10.1039/b918629h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collepardo-Guevara R., Schlick T. Chromatin fiber polymorphism triggered by variations of DNA linker lengths. Proc. Natl. Acad. Sci. USA. 2014;111:8061–8066. doi: 10.1073/pnas.1315872111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meng H., Andresen K., van Noort J. Quantitative analysis of single-molecule force spectroscopy on folded chromatin fibers. Nucleic Acids Res. 2015;43:3578–3590. doi: 10.1093/nar/gkv215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luque A., Ozer G., Schlick T. Correlation among DNA linker length, linker histone concentration, and histone tails in chromatin. Biophys. J. 2016;110:2309–2319. doi: 10.1016/j.bpj.2016.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song F., Chen P., Li G. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376–380. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- 52.Davey C.A., Sargent D.F., Richmond T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 53.Gansen A., Valeri A., Seidel C.A. Nucleosome disassembly intermediates characterized by single-molecule FRET. Proc. Natl. Acad. Sci. USA. 2009;106:15308–15313. doi: 10.1073/pnas.0903005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fang H., Wei S., Hayes J.J. Chromatin structure-dependent conformations of the H1 CTD. Nucleic Acids Res. 2016;44:9131–9141. doi: 10.1093/nar/gkw586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Segal E., Fondufe-Mittendorf Y., Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grigoryev S.A., Arya G., Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc. Natl. Acad. Sci. USA. 2009;106:13317–13322. doi: 10.1073/pnas.0903280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schlick T., Hayes J., Grigoryev S. Toward convergence of experimental studies and theoretical modeling of the chromatin fiber. J. Biol. Chem. 2012;287:5183–5191. doi: 10.1074/jbc.R111.305763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pombo A., Dillon N. Three-dimensional genome architecture: players and mechanisms. Nat. Rev. Mol. Cell Biol. 2015;16:245–257. doi: 10.1038/nrm3965. [DOI] [PubMed] [Google Scholar]
- 59.Tiwari V.K., McGarvey K.M., Baylin S.B. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 2008;6:2911–2927. doi: 10.1371/journal.pbio.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boettiger A.N., Bintu B., Zhuang X. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature. 2016;529:418–422. doi: 10.1038/nature16496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van de Corput M.P., de Boer E., Grosveld F.G. Super-resolution imaging reveals three-dimensional folding dynamics of the β-globin locus upon gene activation. J. Cell Sci. 2012;125:4630–4639. doi: 10.1242/jcs.108522. [DOI] [PubMed] [Google Scholar]
- 62.Fraser J., Williamson I., Dostie J. An overview of genome organization and how we got there: from FISH to Hi-C. Microbiol. Mol. Biol. Rev. 2015;79:347–372. doi: 10.1128/MMBR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mifsud B., Tavares-Cadete F., Osborne C.S. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 64.Phillips J.E., Corces V.G. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanborn A.L., Rao S.S., Aiden E.L. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. USA. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yaffe E., Tanay A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat. Genet. 2011;43:1059–1065. doi: 10.1038/ng.947. [DOI] [PubMed] [Google Scholar]
- 67.Zhang B., Wolynes P.G. Topology, structures, and energy landscapes of human chromosomes. Proc. Natl. Acad. Sci. USA. 2015;112:6062–6067. doi: 10.1073/pnas.1506257112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang B., Zheng W., Wolynes P.G. Exploring the free energy landscape of nucleosomes. J. Am. Chem. Soc. 2016;138:8126–8133. doi: 10.1021/jacs.6b02893. [DOI] [PubMed] [Google Scholar]
- 69.Bishop T.C. Molecular dynamics simulations of a nucleosome and free DNA. J. Biomol. Struct. Dyn. 2005;22:673–686. doi: 10.1080/07391102.2005.10507034. [DOI] [PubMed] [Google Scholar]
- 70.Ettig R., Kepper N., Rippe K. Dissecting DNA-histone interactions in the nucleosome by molecular dynamics simulations of DNA unwrapping. Biophys. J. 2011;101:1999–2008. doi: 10.1016/j.bpj.2011.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Korolev N., Fan Y., Nordenskiöld L. Modelling chromatin structure and dynamics: status and prospects. Curr. Opin. Struct. Biol. 2012;22:151–159. doi: 10.1016/j.sbi.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 72.Kepper N., Ettig R., Rippe K. Force spectroscopy of chromatin fibers: extracting energetics and structural information from Monte Carlo simulations. Biopolymers. 2011;95:435–447. doi: 10.1002/bip.21598. [DOI] [PubMed] [Google Scholar]
- 73.Olson W.K., Clauvelin N., Zheng G. Insights into gene expression and packaging from computer simulations. Biophys. Rev. 2012;4:171–178. doi: 10.1007/s12551-012-0093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Q., Beard D.A., Schlick T. Constructing irregular surfaces to enclose macromolecular complexes for mesoscale modeling using the discrete surface charge optimization (DISCO) algorithm. J. Comput. Chem. 2003;24:2063–2074. doi: 10.1002/jcc.10337. [DOI] [PubMed] [Google Scholar]
- 75.Arya G., Zhang Q., Schlick T. Flexible histone tails in a new mesoscopic oligonucleosome model. Biophys. J. 2006;91:133–150. doi: 10.1529/biophysj.106.083006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bascom G.D., Sanbonmatsu K.Y., Schlick T. Mesoscale modeling reveals hierarchical looping of chromatin fibers near gene regulatory elements. J. Phys. Chem. B. 2016;120:8642–8653. doi: 10.1021/acs.jpcb.6b03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Collepardo-Guevara R., Schlick T. Crucial role of dynamic linker histone binding and divalent ions for DNA accessibility and gene regulation revealed by mesoscale modeling of oligonucleosomes. Nucleic Acids Res. 2012;40:8803–8817. doi: 10.1093/nar/gks600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Luque A., Collepardo-Guevara R., Schlick T. Dynamic condensation of linker histone C-terminal domain regulates chromatin structure. Nucleic Acids Res. 2014;42:7553–7560. doi: 10.1093/nar/gku491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van den Engh G., Sachs R., Trask B.J. Estimating genomic distance from DNA sequence location in cell nuclei by a random walk model. Science. 1992;257:1410–1412. doi: 10.1126/science.1388286. [DOI] [PubMed] [Google Scholar]
- 80.Sachs R.K., van den Engh G., Hearst J.E. A random-walk/giant-loop model for interphase chromosomes. Proc. Natl. Acad. Sci. USA. 1995;92:2710–2714. doi: 10.1073/pnas.92.7.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Münkel C., Eils R., Langowski J. Compartmentalization of interphase chromosomes observed in simulation and experiment. J. Mol. Biol. 1999;285:1053–1065. doi: 10.1006/jmbi.1998.2361. [DOI] [PubMed] [Google Scholar]
- 82.Tark-Dame M., van Driel R., Heermann D.W. Chromatin folding—from biology to polymer models and back. J. Cell Sci. 2011;124:839–845. doi: 10.1242/jcs.077628. [DOI] [PubMed] [Google Scholar]
- 83.Mirny L.A. The fractal globule as a model of chromatin architecture in the cell. Chromosome Res. 2011;19:37–51. doi: 10.1007/s10577-010-9177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Naumova N., Imakaev M., Dekker J. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Collepardo-Guevara R., Schlick T. The effect of linker histone’s nucleosome binding affinity on chromatin unfolding mechanisms. Biophys. J. 2011;101:1670–1680. doi: 10.1016/j.bpj.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rydberg B., Holley W.R., Chatterjee A. Chromatin conformation in living cells: support for a zig-zag model of the 30 nm chromatin fiber. J. Mol. Biol. 1998;284:71–84. doi: 10.1006/jmbi.1998.2150. [DOI] [PubMed] [Google Scholar]
- 87.Hashimoto H., Takami Y., Shinkai Y. Histone H1 null vertebrate cells exhibit altered nucleosome architecture. Nucleic Acids Res. 2010;38:3533–3545. doi: 10.1093/nar/gkq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nokkala S., Nokkala C. Coiled internal structure of chromonema within chromosomes suggesting hierarchical coil model for chromosome structure. Hereditas. 1986;104:29–40. [Google Scholar]
- 89.Hsieh T.-H.S., Weiner A., Rando O.J. Mapping nucleosome resolution chromosome folding in yeast by Micro-C. Cell. 2015;162:108–119. doi: 10.1016/j.cell.2015.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ozer G., Collepardo-Guevara R., Schlick T. Forced unraveling of chromatin fibers with nonuniform linker DNA lengths. J. Phys. Condens. Matter. 2015;27:064113. doi: 10.1088/0953-8984/27/6/064113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Misteli T., Gunjan A., Brown D.T. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 92.Beard D.A., Schlick T. Modeling salt-mediated electrostatics of macromolecules: the discrete surface charge optimization algorithm and its application to the nucleosome. Biopolymers. 2001;58:106–115. doi: 10.1002/1097-0282(200101)58:1<106::AID-BIP100>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]