Abstract
Ectomycorrhizal fungi are an essential component of forest ecosystems, most of which can form edible and medical fruiting bodies. Although many studies have focused on the fructification of ectomycorrhizal fungi in phenology, the impact of environmental contamination, especially living garbage, on the formation of fruiting body is still unknown. A field investigation, combined with a high‐throughput sequencing method, was used to study the effect of living garbage pollution on the fructification and hypogeous community structure of ectomycorrhizal fungi symbiosing with cedar (Cedrus deodara (Roxb.) G. Don). The results showed that garbage significantly altered soil abiotic and biotic properties, increasing soil urease activity, decreasing the soil exchangeable metal content and phosphatase activity, and ultimately inhibiting the formation of fruiting bodies. The pollution of garbage also changed the community structure of hypogeous ectomycorrhizal fungi where ectomycorrhizal ascomycetes dominated. In unpolluted sites, the relative abundance of ectomycorrhizal ascomycetes and basidiomycetes were almost equal. Although no fruiting bodies were observed in that soil polluted by living garbage, the sequencing result showed that various ectomycorrhizal fungi were present underground, suggesting that these taxonomic fungi had the potential to cope with adverse conditions. This study not only provided a deeper understanding of the relationship between ectomycorrhizal fungal communities and prevailing environmental conditions, but provided a new pathway for the excavation and utilization of the resource of antistress ectomycorrhizal fungi.
Keywords: Cedrus deodara, community structure, ectomycorrhizal fungi, environmental contamination, fruiting body
1. Introduction
Ectomycorrhizal (ECM) fungi are one of the most important participants in nutrient cycling in terrestrial ecosystems, especially for nitrogen, phosphorus, and potassium (Behie & Bidochka, 2014; Cairney, 2011; Nasholm, Kielland, & Ganeteg, 2009; Szuba, 2015). ECM fungi can acquire soil‐insoluble elements by biological weathering and transfer them to their host plants in exchange for plant‐derived carbohydrates. ECM fungi can form symbiotic associations with less than 5% of terrestrial plant species (Landeweert, Hoffland, Finlay, Kuyper, & Van Breemen, 2001), and 28–60% of those plants' photosynthetic products are transferred to the fungi (Cairney, 2012; Fogel & Hunt, 1983; Leake, Donnelly, Saunders, Boddy, & Read, 2001) to form a major source of soil organic carbon. Hence, a stable, and appropriate, ECM fungal community structure can not only accelerate the biogeochemical cycle of soil mineral elements, but maintain the healthy growth of plants and stabilization of its surrounding ecosystem. Moreover, many ECM fungi can form conspicuous mushrooms (Henkel et al., 2011; Natarajan, Senthilarasu, Kumaresan, & Riviere, 2005; Tedersoo, May, & Smith, 2010), with important economic and medicinal value. For instance, truffles, chanterelles, and boletes are delicious wild edible mushrooms that cannot be completely artificial cultivated (Hall, Yun, & Amicucci, 2003); modified amanitin extracted from Amanita fruiting bodies has potential medicinal value (Hechler, Kulke, Mueller, Pahl, & Anderl, 2014; Liu et al., 2015). Therefore, the study of artificial cultivation of fruiting bodies, and the ecological function, of ECM fungi has been a focus of attention for ecologists and mycologists.
The continued excessive exploitation of natural resources and the consequent anthropogenic pollution disturbs ecosystem equilibria; however, knowledge of the effect of these human factors on the formation of fruiting bodies and community structure of ECM fungi remains deficient. Thus, it is necessary to study the fructification and community structure of ECM fungi under different environmental conditions, which contributes to the understanding of the effect of environmental disruption on these two aspects.
Cedar (Cedrus deodara (Roxb.) G. Don), a widespread greening tree species in Nanjing, can form symbiotic associations with many ECM fungi (Itoo & Reshi, 2014; Jabeen, Ashraf, & Khalid, 2015; Singh & Lakhanpal, 2004). Many cedar trees have been planted on Nanjing Normal University's campus. Many ECM fungi can be observed in the summer and autumn seasons beneath such cedar trees, however, few are seen near the campus avenues adjacent to living garbage dumps, mainly consist of plastics, waste paper and clothes, building wastes, few food residues, etc. So whether ECM fungal mycelia still exist in the rhizosphere of cedar and their community composition changes compared to those rhizospheres unpolluted by living garbage requires further research. Answering these questions will help us to understand fruiting body formation conditions, and it may even contribute to planting activities in polluted areas. Furthermore, investigating the community structure in polluted conditions will provide a feasible approach to restoration in contaminated environments by plants coupled with microbes. We therefore carried out this study on fruiting body formation and community structure of ECM fungi in areas polluted by living garbage and unpolluted environments.
2. Materials and Methods
2.1. Sample collection
Three cedar trees with an accumulation of garbage in the immediate vicinity were randomly chosen as polluted sites (PS) on 11 September, 2015. Three cedar trees, without garbage, were randomly chosen to represent unpolluted sites (UPS). The trees were at least 200 m apart to ensure statistical independence between sites. At each site, three soil samples were collected from the rhizosphere beneath each cedar tree (about 0.5 m separated each soil sample, samples measured 150 mm × 150 mm to 100 mm depth). A total 18 soil samples were collected and labeled. Then, 18 soil samples were placed in sterile polyethylene bags and brought to the laboratory. All soil samples were sieved (passing a 2 mm square aperture nylon mesh), and some soil samples were air‐dried for the determination of pH, total organic carbon, total nitrogen, and exchangeable element concentration. The remainder soil samples were stored at 4°C for determination of enzyme activity, or at −80°C for ECM fungal DNA extraction.
2.2. Determination of soil physical and chemical properties and enzymatic activity
Soil pH was determined in water (ratio 1:2.5 w/v, stirring thoroughly, and settling for 30 min before measurement) (Islam & Weil, 2000). Soil total nitrogen was determined by vario EL III Element Analyzer (Elementar, Germany). Soil organic carbon was measured by TOC Analyser SSM‐5000A (Shimadzu, Japan). Soil exchangeable cations and phosphorus were extracted by the ammonium bicarbonate‐diethylenetriaminepentaacetic acid (AB‐DTPA) multi‐extractant method (Soltanpour, 1985), and the concentration of exchangeable cations were determined by IRIS Intrepid II XSP (Thermo Electron, USA). Soil exchangeable phosphorus was determined by molybdenum blue colorimetry (Murphy & Riley, 1962). The method of Eivazi and Tabatabai (1977) was used to assess soil phosphatase activity. The determination of urease activity was based on the colorimetric determination of the ammonia released after incubation of soil samples with urea solution for 24 hr at 37°C (Alef & Nannipieri, 1995).
2.3. DNA extraction and sequencing of ECM fungi
Soil DNA was extracted using a FastDNA® Spin Kit for soil (MP Biomedicals, USA) according to the manufacturer's instructions. According to its concentration, the DNA solution was diluted to 1 ng/μl with sterile deionized water. The primers ITS1‐F(5′‐CTTGGTCATTTAGAGGAAGTAA‐3′) (Gardes & Bruns, 1993) and ITS2(5′‐GCTGCGTTCTTCATCGATGC‐3′) (White, Bruns, Lee, & Taylor, 1990) were used to amplify the internal transcribed spacer 1 (ITS1) of ECM fungal rDNA. To distinguish different samples, barcodes were added to forward and reverse primers to attribute sequences to each sample. All PCR reactions were carried out using a Phusion® High‐Fidelity PCR Master Mix (New England Biolabs, USA). PCR products were electrophoresed on 2% agarose gel. According to the concentration, PCR products were mixed at an equimolar ratio and re‐electrophoresed on 2% agarose gel. Then the target DNA strips were extracted using a QIAquick® Gel Extraction Kit (Qiagen, Germany). Sequencing libraries were generated using TruSeq® DNA PCR‐Free Sample Preparation Kit (Illumina, USA) following manufacturer's recommendations and index codes were added. The library quality was assessed on the Qubit® 2.0 Fluorometer (Thermo Scientific, USA) and Agilent Bioanalyzer 2100 system (Agilent Technologies, USA). Finally, the library was sequenced on an Illumina HiSeq2500 platform by Novogene (Beijing, China, http://www.novogene.cn/) and 250 bp paired‐end reads were generated.
2.4. Data processing
Sequenced raw data generated by Illumina were separated by sample according to barcode sequences. Then barcode and primer sequences were removed from each sample. Due to the Illumina Hiseq2500 platform adopting a paired‐end sequencing strategy, reads were merged into a complete sequence as supposed from valid tags. Raw valid tags were produced in FLASH (Magoč & Salzberg, 2011). Merged raw valid tags were filtered in QIIME referring to the default parameters (Bokulich et al., 2013; Caporaso et al., 2010) and high‐quality clean tags were obtained. Chimeric sequences in clean tags were picked out and removed using UCHIME (Edgar, Haas, Clemente, Quince, & Knight, 2011) according to the Unite database (https://unite.ut.ee/) and effective tags were obtained. Effective tags for all samples were clustered into operational taxonomic units (OTU) at 97% similarity: a representative tag with the highest abundance in each OTU was selected for taxonomic assignment by the Uparse algorithm (Edgar, 2013). Taxonomic annotation of representative tags was performed in QIIME by use of the BLAST method (Altschul, Gish, Miller, Myers, & Lipman, 1990), again, with reference to the Unite database (Kõljalg et al., 2013). Finally, to ensure a fair comparison between samples, all samples were normalized to the minimum number of effective tags. Subsequent analyses were based on the normalized data.
2.5. Statistical analysis
Rarefaction curves and the α‐diversity indices of Chao1, Shannon, and Simpson were produced in QIIME. For the β‐diversity index, an OTU level‐based dissimilarity Binary‐Ochiai metric (Boyce & Ellison, 2001) was used to measure the pair‐wise community similarity between samples, and principal coordinate analysis (PCoA) was used to visualize the distance matrix of all 18 samples in QIIME. Adonis (Fierer et al., 2010) performed in QIIME was used to test whether, or not, differences among groups of samples were significant. Canonical correspondence analysis (CCA) was performed in R using the Vegan package. t test was performed in SPSS 20 (IBM, USA). Heatmaps were generated using HemI1.0 (Deng, Wang, Liu, Cheng, & Xue, 2014).
3. Results
3.1. Soil physical and chemical properties and enzymatic activities
Soil pH and the content of exchangeable metals and phosphorus are as listed in Table 1. The soil pH at UPS was significantly lower than that at PS (t = −2.275, p = .023). The soil exchangeable calcium, exchangeable iron, exchangeable manganese, exchangeable magnesium, exchangeable nickel, and exchangeable zinc contents at UPS were noticeably higher than those in polluted areas (t = 10.994, p < .001; t = 2.827, p = .012; t = 3.650, p = .003; t = 2.907, p = .010; t = 2.900, p = .014; t = 2.305, p = .035, respectively). The content of soil total organic carbon at UPS was lower than that at PS. The assessment of soil enzymatic activity showed that phosphatase activity at UPS was noticeably higher than that at PS, however, urease activity was lower than that at PS (Fig. 1).
Table 1.
Soil properties of unpolluted and polluted sites
| Sites | pH | TOC | TN | Ca | Cu | Fe | K | Mg | Mn | Ni | P | Zn |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mg/g | mg/kg | |||||||||||
| UPS | 6.98 ± 0.67 | 14.65 ± 4.79 | 1.27 ± 0.45 | 516.78 ± 0.13 | 3.11 ± 0.95 | 63.19 ± 18.42 | 42.52 ± 13.38 | 102.28 ± 29.64 | 41.96 ± 17.24 | 0.97 ± 0.38 | 0.42 ± 0.42 | 6.67 ± 1.75 |
| PS | 7.55 ± 0.56 | 16.36 ± 2.30 | 0.74 ± 0.12 | 362.56 ± 30.56 | 2.73 ± 0.24 | 43.80 ± 9.17 | 51.72 ± 16.16 | 61.75 ± 15.19 | 23.67 ± 7.69 | 0.57 ± 0.17 | 0.48 ± 0.38 | 4.90 ± 1.49 |
TOC, total organic carbon; TN, total nitrogen; Ca, exchangeable calcium; Cu, exchangeable copper; Fe, exchangeable iron; K, exchangeable potassium; Mg, exchangeable magnesium; Mn, exchangeable manganese; Ni, exchangeable nickel; P, exchangeable phosphorus; Zn, exchangeable zinc. Values are mean ± standard deviation. UPS, unpolluted sites; PS, polluted sites.
Figure 1.
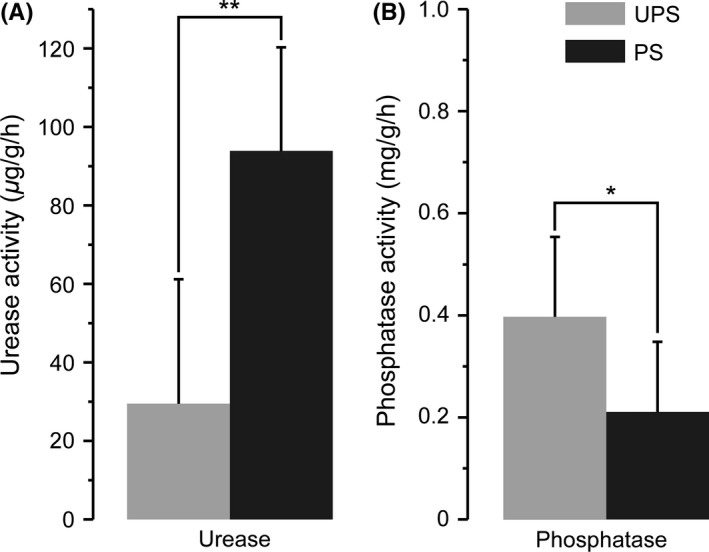
Activities of soil urease and phosphatase. (A) Urease activity is expressed as μg of NH 3‐N produced through hydrolysis of urea per hr per g of soil at 37°C. (B) Phosphatase activity is expressed as mg of phenol produced per hour per g of soil at 37°C. Values are mean± SD. Asterisks show significant differences identified by t test at p < .05. UPS, unpolluted sites; PS, polluted sites. *p < .05, **p < .01
3.2. Data analysis
More than 30,000 effective tags of the ITS1 region were obtained for each sample after merging and quality control. The average length of the effective tags was 260.19 bp. Rarefaction curves of all samples are shown in Figure 2, indicating that sequencing depth can reveal soil fungal community composition. OTUs belonging to the ECM fungal taxonomy (EOTUs) were picked out according to Tedersoo et al. (2010) and the Unite database. Subsequent analyses (α‐ and β‐diversity) were based on selected EOTUs. The α‐diversity indices were calculated as shown in Table 2.
Figure 2.
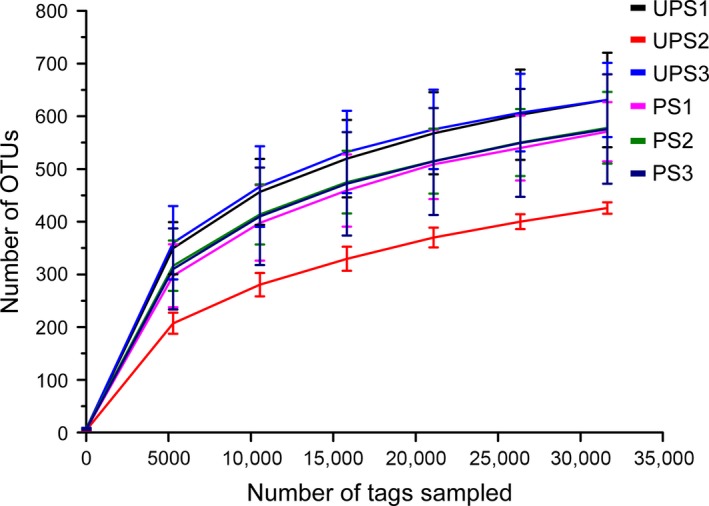
Rarefaction curves of the OTU number at 97% similarity for each sample. Average value of three replicates and error bar are shown. UPS, unpolluted sites; PS, polluted sites.
Table 2.
α‐Diversity indices: all sites
| Site | α‐Diversity indices | |||
|---|---|---|---|---|
| EOTUs | Chao1 | Shannon | Simpson | |
| UPS | 32.0 ± 6.9 | 37.4 ± 9.8 | 4.5 ± 1.1 | 0.84 ± 0.09 |
| PS | 30.0 ± 7.6 | 34.6 ± 7.7 | 4.7 ± 0.9 | 0.85 ± 0.09 |
Values are mean ± standard deviation. UPS, unpolluted sites; PS, polluted sites.
3.3. ECM fungal community structures: all sites
All EOTUs were classified into Ascomycota, Basidiomycota, and Zygomycota, due to the negligible number of sequences obtained for Zygomycota, this part of the EOTUs was not analyzed here. At site UPS1, the top five genera were: Trichophaea, Pisolithus, Scleroderma, Cortinarius, and Inocybe. At site UPS2, the top five genera were as follows: Trichophaea, Russula, Astraeus, Scleroderma, and Cortinarius. At site UPS3, the top five genera were as follows: Trichophaea, Tomentella, Scleroderma, Wilcoxina, and Cortinarius. At UPS, most ECM genera were similar, while they were highly dissimilar at PS. At site PS1, the top five genera were as follows: Tomentella, Trichophaea, Wilcoxina, Scleroderma, and Cortinarius. At site PS2, the top five genera were as follows: Trichophaea, Wilcoxina, Scleroderma, Russula, and Tuber. At site PS3, the top five genera were as follows: Tuber, Scleroderma, Hydnobolites, Trichophaea, and Pseudaleuria. A graphical representation, at genus level, is shown in Figure 3.
Figure 3.
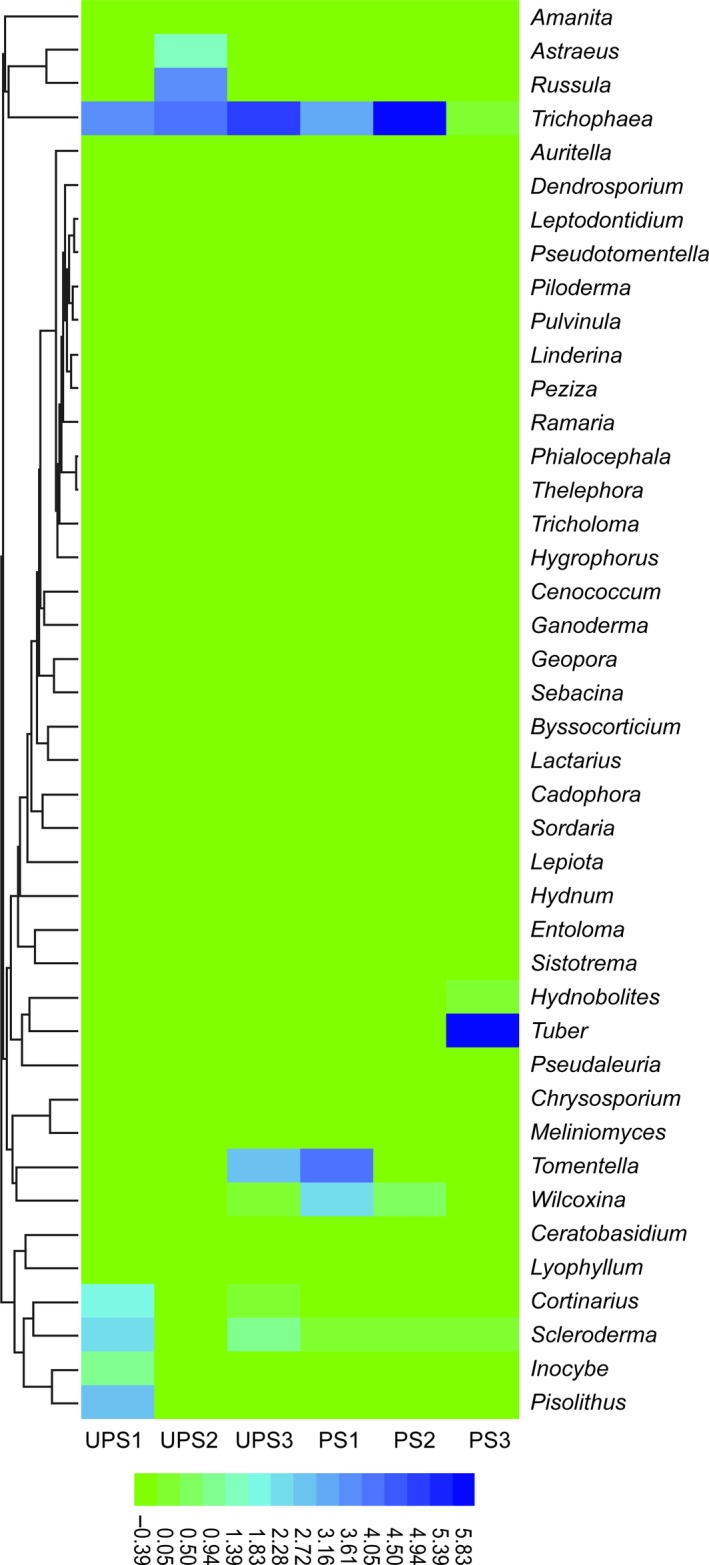
Heatmap of the Ectomycorrhizal (ECM) fungal community structure of all sites at genus level. The highest and lowest abundance of genera are clustered, respectively: the color gradient and similarity reflect the similarity and dissimilarity of ECM fungal community structure in polluted and unpolluted sites. The horizontal axis shows sample information and the vertical axis shows information pertaining to species annotation
PCoA were performed to represent the differences in ECM fungal community composition (Fig. 4). The ECM fungal community structures of UPS can be separated from those at PS on PC1 (Adonis p = 1 × 10−4, R 2 = 0.142, number of permutations = 9999). In the polluted group, ECM fungal community structures of different sites were also completely dissimilar (Adonis p = .003, R 2 = 0.376, number of permutations = 9999).
Figure 4.
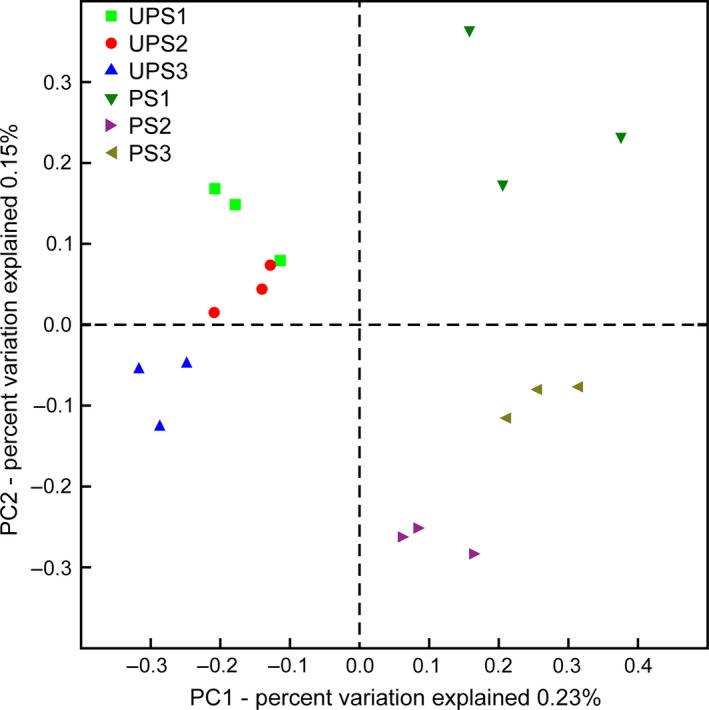
PCoA of community composition at all sites at EOTU level based on richness and relative abundance. The distance between points reflects the differences in Ectomycorrhizal (ECM) fungal community structure in the samples
3.4. Relationship between environmental factors and soil ECM fungal communities
A subset of environmental parameters (exchangeable K+, exchangeable Mg2+, and urease) was selected by the BioEnv function of the Vegan package in R: this had the highest Pearson correlation with ECM fungal communities. The selected environmental parameters were used to perform CCA analysis (Fig. 5). The Mantel test indicated a highly significant correlation between environmental variables (exchangeable K+ and exchangeable Mg2+, and urease activity) and ECM fungal community (r = .33, p = .004; r = .40, p = .003; r = 0.25, p = .017, respectively).
Figure 5.
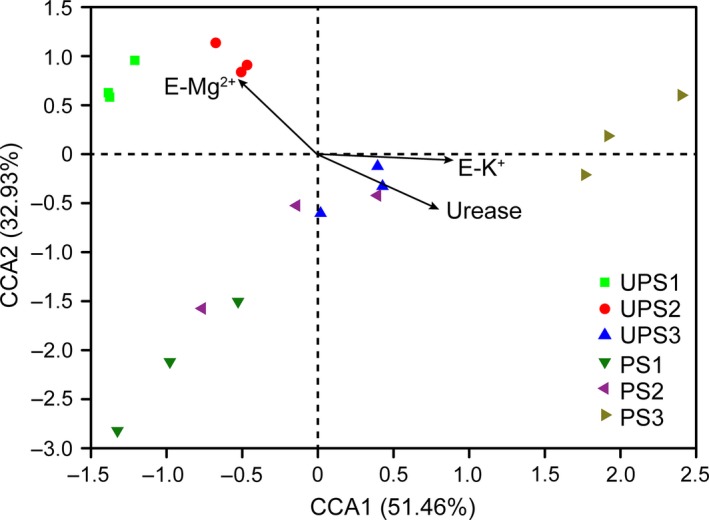
Canonical correspondence analysis (CCA) analysis shows the relationships between Ectomycorrhizal (ECM) fungal community and environmental factors across six sites. The arrows indicate environmental factors, the length of each arrow represents the strength of the relationship between the environmental variable and the distribution of ECM fungi. E‐Mg2+, exchangeable Mg2+; E‐K+, exchangeable K+
4. Discussion
During 3 years of investigation, we observed fruiting bodies of ECM fungi Calvatia, Cortinarius, Pisolithus, Russula, and Scleroderma at unpolluted sites, although the types of fruiting bodies observed were different each year due to the effects of phenological factors (Barroetaveña, La Manna, & Alonso, 2008; Gange, Gange, Sparks, & Boddy, 2007; Jang & Kim, 2015; Kauserud et al., 2008; Lilleskov, Bruns, Dawson, & Camacho, 2009). However, ECM and even saprophytic mushrooms cannot be observed in garbage‐polluted sites, indicating that living garbage inhibited the epigeous fructification of ECM fungi. Although living garbage inhibited the formation of fruiting bodies, sequencing results still showed many kinds, and at high relative abundance, of ECM fungal mycelia in the rhizosphere of cedar (Fig. 3), suggesting that these taxonomic fungi can survive in contaminated environments but demand more suitable conditions to enable fruiting. In garbage‐polluted sites, ECM fungal species were dominated by ascomycetes (Fig. 6), but ECM basidiomycetes showed fruiting bodies in unpolluted sites albeit at a very low relative abundance of hypogeous mycelia. The result may indicate that living garbage pollution inhibit the formation of ECM fungal fruiting bodies by (1) reducing ECM fungal mycelia biomass which are needed in the early stage of fruiting body formation and (2) altering soil abiotic and biotic factors to inhibit fruiting even although significant numbers of mycelia are present.
Figure 6.
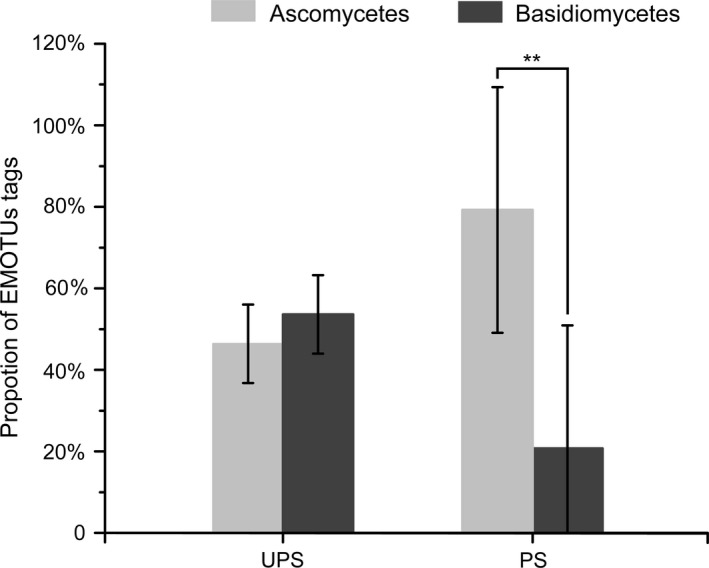
The relative abundance of Ectomycorrhizal (ECM) Ascomycota and Basidiomycota between unpolluted and polluted sites. Asterisks show significant differences identified by t test at p < .05. **p < .01
Living garbage not only inhibits the formation of ectomycorrhizal mushrooms, but affects the hypogeous ECM fungal community. In living garbage‐polluted sites, the community composition of ECM fungi was dissimilar to that at unpolluted sites and that differed between groups: this may have been caused by the type of garbage (Fig. 4). From sequencing data, it was found that the proportion of ECM Ascomycota and Basidiomycota were similar at unpolluted sites, but ECM Ascomycota predominated at polluted sites (Fig. 6), which indicated ECM basidiomycetes required preferable environmental conditions and ECM ascomycetes can perhaps adapt to various environments in accordance with studies by Grogan et al. (2000), Torres and Honrubia (1997), Visser (1995), and Vralstad, Myhre, and Schumacher (2002) showing that ECM ascomycetes can survive in stressful environments.
The accumulation of living garbage in soil changes soil abiotic and biotic properties. In this study, the soil exchangeable calcium, iron, magnesium, manganese, nickel, and zinc contents and total nitrogen at unpolluted sites were higher than those in polluted sites (Table 1). The decrease in these elements essential to ECM fungal life and activity may be a key abiotic factor affecting ECM fungal fructification; many of these elements can also affect ECM fungal communities. CCA analysis showed that soil exchangeable Mg2+ did affect the ECM fungal communities between unpolluted and polluted sites (Fig. 5). These abiotic factors such as metal ions concentration, nitrogen availability, and alkalinity‐induced stresses not only alter ECM fungal community, but affect the formation of fruiting body. In addition, urease activity was not only a key biotic factor inducing change in ECM fungal communities (Fig. 5), but it was also related to the formation of fruiting bodies. The urease activity at polluted sites exceeded that at unpolluted sites (Fig. 1A). Due to soil total nitrogen was lower at polluted sites (Table 1), high urease activity was for accelerating the nitrogen availability in the soil. Furthermore, the uptake of ammonium by EM fungi came at a high carbon cost, which led to less carbon availability for mycelial growth and fruiting body formation (Bidartondo, Ek, Wallander, & Söderström, 2001; Wallander, 1995). Although ECM fungi can exchange nutrients for more plant‐derived carbohydrates (Wallander et al., 2013), when the soil nutrients are insufficient to maintain their own demand, they will supply less nutrients to their hosts and thus obtain less carbohydrates from their host plants so that their biomass declines. However, the hydrolysis of urea would also increase soil pH and the formation of metal carbonates leading to exchangeable metal contents declining (Fujita, Ferris, Lawson, Colwell, & Smith, 2000; Garau, Castaldi, Santona, Deiana, & Melis, 2007; Stocks‐Fischer, Galinat, & Bang, 1999), consequently, reducing mycelial growth and fruiting body formation. Additionally, soil phosphatase activity decreased at polluted sites (Fig. 1B), which mainly because of declined ECM mycelial biomass and that led to lower phosphorus. Phosphorus is required for ECM fungal growth, so a decrease in the amount thereof affected the formation of fruiting bodies. Soil enzyme activities related to soil mineral element mobilization could affect ECM fungi with regard to their mineral element utilization and mycelial growth, thus affecting fruiting and ECM fungal community structure. In summary, soil ECM fungal community structure is not only affected by pH, edaphic enzyme activities, and effective metal content, but is affected by other factors, such as hypogeous saprophytes and host plants (Koide, Fernandez, & Petprakob, 2011).
5. Conclusion
The formation of ECM fungal fruiting bodies requires suitable environmental conditions. The pollution of living garbage changes soil abiotic and biotic properties in the rhizosphere of cedar, decreasing mycelial biomass, inhibiting the formation of ECM fungal fruiting bodies, and altering the community structure of ECM fungi. That many ECM fungi detected in polluted sites promoted understanding of ECM fungal fruiting body formation conditions and their environmental adaptability. This research provided a new way of screening and utilizing the resource of antiadversity fungi, which can resist the deficiency of nutrients, even organic and inorganic compounds pollution.
Funding Information
This work was jointly supported by the National Natural Science Foundation of China (Grant no. 41373078) and a project funded by the Priority Academic Programme Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of Interest
None declared.
Acknowledgments
This work was jointly supported by the National Natural Science Foundation of China (Grant no. 41373078) and a project funded by the Priority Academic Programme Development of Jiangsu Higher Education Institutions (PAPD).
Sun, Q. , Liu, Y. , Yuan, H. , Lian, B. The effect of environmental contamination on the community structure and fructification of ectomycorrhizal fungi. MicrobiologyOpen. 2017;6: e00396. https://doi.org/10.1002/mbo3.396
References
- Alef, K. , & Nannipieri, P. (1995). Methods in Applied Soil Microbiology and Biochemistry. London: Academic Press. [Google Scholar]
- Altschul, S. F. , Gish, W. , Miller, W. , Myers, E. W. , & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Barroetaveña, C. , La Manna, L. , & Alonso, M. V. (2008). Variables affecting Suillus luteus fructification in ponderosa pine plantations of Patagonia (Argentina). Forest Ecology and Management, 256, 1868–1874. [Google Scholar]
- Behie, S. W. , & Bidochka, M. J. (2014). Nutrient transfer in plant‐fungal symbioses. Trends in Plant Science, 19, 734–740. [DOI] [PubMed] [Google Scholar]
- Bidartondo, M. I. , Ek, H. , Wallander, H. , & Söderström, B. (2001). Do nutrient additions alter carbon sink strength of ectomycorrhizal fungi? New Phytologist, 153, 543–550. [Google Scholar]
- Bokulich, N. A. , Subramanian, S. , Faith, J. J. , Gevers, D. , Gordon, J. I. , Knight, R. , … Caporaso, J. G. (2013). Quality‐filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nature Methods, 10, 57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce, R. L. , & Ellison, P. C. (2001). Choosing the best similarity index when performing fuzzy set ordination on binary data. Journal of Vegetation Science, 12, 711–720. [Google Scholar]
- Cairney, J. W. G. (2011). Ectomycorrhizal fungi: The symbiotic route to the root for phosphorus in forest soils. Plant and Soil, 344, 51–71. [Google Scholar]
- Cairney, J. W. G. (2012). Extramatrical mycelia of ectomycorrhizal fungi as moderators of carbon dynamics in forest soil. Soil Biology & Biochemistry, 47, 198–208. [Google Scholar]
- Caporaso, J. G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F. D. , Costello, E. K. , … Knight, R. (2010). QIIME allows analysis of high‐throughput community sequencing data. Nature Methods, 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, W. , Wang, Y. , Liu, Z. , Cheng, H. , & Xue, Y. (2014). HemI: A Toolkit for Illustrating Heatmaps. PLoS ONE, 9, e111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. C. (2013). UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nature Methods, 10, 996–998. [DOI] [PubMed] [Google Scholar]
- Edgar, R. C. , Haas, B. J. , Clemente, J. C. , Quince, C. , & Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 27, 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eivazi, F. , & Tabatabai, M. A. (1977). Phosphatases in soils. Soil Biology & Biochemistry, 9, 167–172. [Google Scholar]
- Fierer, N. , Lauber, C. L. , Zhou, N. , Mcdonald, D. , Costello, E. K. , & Knight, R. (2010). Forensic identification using skin bacterial communities. Proceedings of the National Academy of Sciences of the United States of America, 107, 6477–6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel, R. , & Hunt, G. (1983). Contribution of mycorrhizae and soil fungi to nutrient cycling in a Douglas‐fir ecosystem. Canadian Journal of Forest Research, 13, 219–232. [Google Scholar]
- Fujita, Y. , Ferris, F. G. , Lawson, R. D. , Colwell, F. S. , & Smith, R. W. (2000). Subscribed Content Calcium Carbonate Precipitation by Ureolytic Subsurface Bacteria. Geomicrobiology Journal, 17, 305–318. [Google Scholar]
- Gange, A. C. , Gange, E. G. , Sparks, T. H. , & Boddy, L. (2007). Rapid and Recent Changes in Fungal Fruiting Patterns. Science, 316, 71–71. [DOI] [PubMed] [Google Scholar]
- Garau, G. , Castaldi, P. , Santona, L. , Deiana, P. , & Melis, P. (2007). Influence of red mud, zeolite and lime on heavy metal immobilization, culturable heterotrophic microbial populations and enzyme activities in a contaminated soil. Geoderma, 142, 47–57. [Google Scholar]
- Gardes, M. , & Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes‐application to the identification of mycorrhizae and rusts. Molecular Ecology, 2, 113–118. [DOI] [PubMed] [Google Scholar]
- Grogan, P. , Baar, J. , & Bruns, T. D. (2000). Below‐ground ectomycorrhizal community structure in a recently burned bishop pine forest. Journal of Ecology, 88, 1051–1062. [Google Scholar]
- Hall, I. R. , Yun, W. , & Amicucci, A. (2003). Cultivation of edible ectomycorrhizal mushrooms. Trends in Biotechnology, 21, 433–438. [DOI] [PubMed] [Google Scholar]
- Hechler, T. , Kulke, M. , Mueller, C. , Pahl, A. , & Anderl, J. (2014). Amanitin‐based antibody‐drug conjugates targeting the prostate‐specific membrane antigen. Cancer Research, 74, 664. [Google Scholar]
- Henkel, T. W. , Aime, M. C. , Chin, M. M. L. , Miller, S. L. , Vilgalys, R. , & Smith, M. E. (2011). Ectomycorrhizal fungal sporocarp diversity and discovery of new taxa in Dicymbe monodominant forests of the Guiana Shield. Biodiversity and Conservation, 21, 2195–2220. [Google Scholar]
- Islam, K. R. , & Weil, R. R. (2000). Land use effects on soil quality in a tropical forest ecosystem of Bangladesh. Agriculture, Ecosystems & Environment, 79, 9–16. [Google Scholar]
- Itoo, Z. A. , & Reshi, Z. A. (2014). Ectomycorrhizal diversity associated with Cedrus deodara and Pinus wallichiana in the Kashmir Himalaya, India. Pakistan Journal of Biological sciences: PJBS, 17, 32–40. [DOI] [PubMed] [Google Scholar]
- Jabeen, S. , Ashraf, T. , & Khalid, A. N. (2015). Peziza succosella and its ectomycorrhiza associated with Cedrus deodara from Himalayan moist temperate forests of Pakistan. Mycotaxon, 130, 455–464. [Google Scholar]
- Jang, S. K. , & Kim, S. W. (2015). Relationship between Ectomycorrhizal Fruiting Bodies and Climatic and Environmental Factors in Naejangsan National Park. Mycobiology, 43, 122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauserud, H. , Stige, L. C. , Vik, J. O. , Økland, R. H. , Høiland, K. , & Stenseth, N. C. (2008). Mushroom Fruiting and Climate Change. Proceedings of the National Academy of Sciences of the United States of America, 105, 3811–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide, R. T. , Fernandez, C. , & Petprakob, K. (2011). General principles in the community ecology of ectomycorrhizal fungi. Annals of Forest Science, 68, 45–55. [Google Scholar]
- Kõljalg, U. , Nilsson, R. H. , Abarenkov, K. , Tedersoo, L. , Taylor, A. F. , Bahram, M. , … Larsson, K. H. (2013). Towards a unified paradigm for sequence‐based identification of fungi. Molecular Ecology, 22, 5271–5277. [DOI] [PubMed] [Google Scholar]
- Landeweert, R. , Hoffland, E. , Finlay, R. D. , Kuyper, T. W. , & Van Breemen, N. (2001). Linking plants to rocks: Ectomycorrhizal fungi mobilize nutrients from minerals. Trends in Ecology & Evolution, 16, 248–254. [DOI] [PubMed] [Google Scholar]
- Leake, J. R. , Donnelly, D. P. , Saunders, E. M. , Boddy, L. , & Read, D. J. (2001). Rates and quantities of carbon flux to ectomycorrhizal mycelium following 14C pulse labeling of Pinus sylvestris seedlings: Effects of litter patches and interaction with a wood‐decomposer fungus. Tree Physiology, 21, 71–82. [DOI] [PubMed] [Google Scholar]
- Lilleskov, E. A. , Bruns, T. D. , Dawson, T. E. , & Camacho, F. J. (2009). Water sources and controls on water‐loss rates of epigeous ectomycorrhizal fungal sporocarps during summer drought. New Phytologist, 182, 483–494. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Zhang, X. , Han, C. , Wan, G. , Huang, X. , Ivan, C. , … Liu, X. (2015). TP53 loss creates therapeutic vulnerability in colorectal cancer. Nature, 520, 697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč, T. , & Salzberg, S. L. (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics, 27, 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, J. , & Riley, J. P. (1962). A modified single solution method for the determination of phosphate in natural waters. Analytica chimica acta, 27, 31–36. [Google Scholar]
- Nasholm, T. , Kielland, K. , & Ganeteg, U. (2009). Uptake of organic nitrogen by plants. New Phytologist, 182, 31–48. [DOI] [PubMed] [Google Scholar]
- Natarajan, K. , Senthilarasu, G. , Kumaresan, V. , & Riviere, T. (2005). Diversity in ectomycorrhizal fungi of a dipterocarp forest in Western Ghats. Current Science, 88, 1893–1895. [Google Scholar]
- Singh, A. , & Lakhanpal, T. N. (2004). Seasonal distribution of active ectomycorrhizal roots under Cedrus deodara . Proceedings of the Indian National Science Academy Part B Biological Sciences, 74, 289–298. [Google Scholar]
- Soltanpour, P. N. (1985). Use of ammonium bicarbonate DTPA soil test to evaluate elemental availability and toxicity. Communications in Soil Science & Plant Analysis, 16, 323–338. [Google Scholar]
- Stocks‐Fischer, S. , Galinat, J. K. , & Bang, S. S. (1999). Microbiological precipitation of CaCO3 . Soil Biology & Biochemistry, 31, 1563–1571. [Google Scholar]
- Szuba, A. (2015). Ectomycorrhiza of Populus, Forest Ecology and Management, 347, 156–169. [Google Scholar]
- Tedersoo, L. , May, T. W. , & Smith, M. E. (2010). Ectomycorrhizal lifestyle in fungi: Global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza, 20, 217–263. [DOI] [PubMed] [Google Scholar]
- Torres, P. , & Honrubia, M. (1997). Changes and effects of a natural fire on ectomycorrhizal inoculum potential of soil in a Pinus halepensis forest. Forest Ecology and Management, 96, 189–196. [Google Scholar]
- Visser, S. (1995). Ectomycorrhizal fungal succession in jack pine stands following wildfire. New Phytologist, 129, 389–401. [Google Scholar]
- Vralstad, T. , Myhre, E. , & Schumacher, T. (2002). Molecular diversity and phylogenetic affinities of symbiotic root‐associated ascomycetes of the Helotiales in burnt and metal polluted habitats. New Phytologist, 155, 131–148. [DOI] [PubMed] [Google Scholar]
- Wallander, H. (1995). A new hypothesis to explain allocation of dry matter between mycorrhizal fungi and pine seedlings in relation to nutrient supply. Plant and Soil, 168, 243–248. [Google Scholar]
- Wallander, H. , Ekblad, A. , Godbold, D. L. , Johnson, D. , Bahr, A. , Baldrian, P. , … Rudawska, M. (2013). Evaluation of methods to estimate production, biomass and turnover of ectomycorrhizal mycelium in forests soils – A review. Soil Biology & Biochemistry, 57, 1034–1047. [Google Scholar]
- White, T. J. , Bruns, T. , Lee, S. , & Taylor, J . (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: PCR protocols: A guide to methods and applications (pp. 315–322). New York, Academic Press. [Google Scholar]


