Summary
Neural circuits in the cerebral cortex consist of excitatory pyramidal cells and inhibitory interneurons. These two main classes of cortical neurons follow largely different genetic programs, yet they assemble into highly specialized circuits during development following a very precise choreography. Previous studies have shown that signals produced by pyramidal cells influence the migration of cortical interneurons, but the molecular nature of these factors has remained elusive. Here, we identified Neuregulin 3 (Nrg3) as a chemoattractive factor expressed by developing pyramidal cells that guides the allocation of cortical interneurons in the developing cortical plate. Gain- and loss-of-function approaches reveal that Nrg3 modulates the migration of interneurons into the cortical plate in a process that is dependent on the tyrosine kinase receptor ErbB4. Perturbation of Nrg3 signaling in conditional mutants leads to abnormal lamination of cortical interneurons. Nrg3 is therefore a critical mediator in the assembly of cortical inhibitory circuits.
Keywords: cerebral cortex, interneuron, migration, neuregulins, lamination, Erbb4, GABA, inhibition, cortical plate, pyramidal cell
Graphical Abstract
Highlights
-
•
Nrg3 acts a short-range chemoattractive molecule for cortical interneurons
-
•
Nrg3 functions through ErbB4 to attract interneurons into the cortical plate
-
•
Interneurons prefer Cxcl12 over Nrg3 during tangential migration
-
•
Disruption of Nrg3 signaling causes abnormal interneuron lamination in the cortex
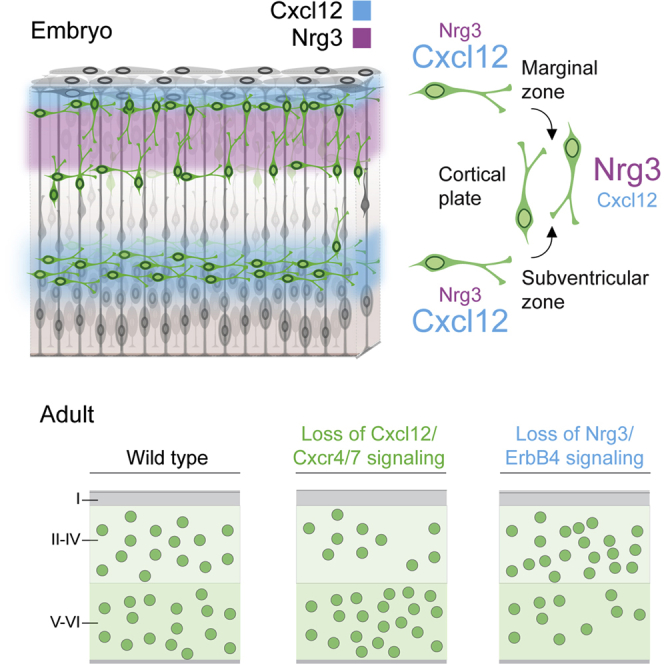
The integration of interneurons in the developing cerebral cortex depends on signals generated by pyramidal cells. In this study, Bartolini et al. identify Neuregulin 3 as a developmentally regulated factor that controls the migration of interneurons into the cortical plate and influences their final laminar distribution.
Introduction
The balance of excitation and inhibition is crucial for the normal functioning of the cerebral cortex (Vogels and Abbott, 2009, Xue et al., 2014, Yizhar et al., 2011), yet the mechanisms controlling the assembly of excitatory pyramidal cells and inhibitory gamma-aminobutyric acid-containing (GABAergic) interneurons into common circuits are poorly understood. Although most neural circuits consist of different classes of neurons, the cerebral cortex is relatively unique in that excitatory and inhibitory neurons are born in distinct progenitor regions of the embryonic telencephalon (Marín and Rubenstein, 2001, Wonders and Anderson, 2006), and therefore their assembly requires the precise coordination of two largely different migration programs (Marín and Rubenstein, 2003).
Cortical GABAergic interneurons are highly heterogeneous and comprise distinct functional classes with unique morphological, electrophysiological, and molecular features (Tremblay et al., 2016). Recent transcriptomic analyses in the mouse suggest that the neocortex contains over 20 molecularly distinct classes of interneurons (Tasic et al., 2016, Zeisel et al., 2015), a number comparable to those identified in the hippocampus through morphological and electrophysiological studies (Klausberger and Somogyi, 2008). Nevertheless, neocortical interneurons can be classified into several cardinal groups based on their developmental origin and the expression of key molecular markers (Tremblay et al., 2016). The large majority of cortical interneurons derives from the embryonic medial ganglionic eminence (MGE) and comprises two major classes: parvalbumin-expressing (PV+) fast-spiking interneurons such as chandelier cells and basket cells, and somatostatin-expressing (SST+) interneurons comprising mostly Martinotti cells.
Our understanding of the mechanisms controlling the development of cortical interneurons has increased exponentially since Anderson et al. (1997) discovered the origin of cortical interneurons in the embryonic subpallium. However, although the mechanisms controlling the tangential migration of interneurons from the ganglionic eminences toward the cortex have been widely explored (Baudoin et al., 2012, Flames et al., 2004, Hernández-Miranda et al., 2011, Marín, 2013, Marín et al., 2001, Nóbrega-Pereira et al., 2008, Polleux et al., 2002, Pozas and Ibáñez, 2005, Rakić et al., 2015, van den Berghe et al., 2013), the cellular and molecular events governing the final allocation of interneurons in their corresponding cortical layers remain largely unknown. Interneurons follow two main routes of migration as they disperse tangentially throughout the cortex, the marginal zone (MZ) and the subventricular zone (SVZ) (Lavdas et al., 1999, Marín and Rubenstein, 2001, Miyoshi and Fishell, 2011, Tanaka et al., 2009). Interneurons initially avoid migrating into the cortical plate (CP), where pyramidal cells differentiate and form the different cortical layers (López-Bendito et al., 2008). Chemokines are responsible for maintaining migrating interneurons in their migratory streams (Tiveron et al., 2006), and deficits in this signaling system lead to laminar and regional defects in the distribution of cortical interneurons (Li et al., 2008, López-Bendito et al., 2008, Sánchez-Alcañiz et al., 2011, Wang et al., 2011). These results revealed that the timing of CP invasion is crucial for the final allocation of interneurons in cortical layers, but the molecular factors controlling this process have not yet been identified.
Neuregulins comprise a large family of widely expressed epidermal growth factor (EGF)-like proteins that regulates multiple aspects of neurodevelopment, including neuronal migration, axon guidance, myelination, and synapse formation (Mei and Nave, 2014, Mei and Xiong, 2008). Neuregulins are EGF domain-containing proteins encoded by six individual genes that activate receptor tyrosine kinases of the ERBB family, and some of the members of this signaling network have been shown to play crucial roles in the assembly of GABAergic circuits (Rico and Marín, 2011). In particular, ErbB4 is highly expressed by interneurons migrating from the MGE (Rakić et al., 2015, Villar-Cerviño et al., 2015, Yau et al., 2003), most notably among PV+ interneurons (Fazzari et al., 2010, Vullhorst et al., 2009). ErbB4 is required for the tangential migration of GABAergic interneurons from the subpallium to the developing cortex, and this effect is mediated by different isoforms of Nrg1 (Flames et al., 2004, Rakić et al., 2015). Here, we have identified Nrg3 as a chemoattractive factor regulating the final allocation of GABAergic interneurons in the cortex. Our results demonstrate that ErbB4-mediated Nrg3 signaling controls CP entry by GABAergic interneurons and influences their final laminar allocation. Together with previous results, our observations unravel a hierarchical system of chemoattractive signals that regulate the distribution of interneurons in the cerebral cortex.
Results
A Candidate Gene Approach to Identify Factors Regulating Cortical Plate Invasion by Interneurons
Different studies have shown that the chemokine Cxcl12 is responsible for maintaining the migration of interneurons within the MZ and the SVZ as they disperse tangentially throughout the cortex (Li et al., 2008, López-Bendito et al., 2008, Sánchez-Alcañiz et al., 2011, Stumm et al., 2007, Tiveron et al., 2006, Wang et al., 2011). Genetic deletion of chemokine receptors (Abe et al., 2014, Sánchez-Alcañiz et al., 2011, Tiveron et al., 2006, Vogt et al., 2014, Wang et al., 2011) or acute inhibition of chemokine signaling (Lysko et al., 2014) (Figure S1; Movies S1 and S2) lead to the rapid accumulation of interneurons in the CP, which suggests the existence of chemoattractive signals in this region regulating the final allocation of interneurons.
We took a candidate gene approach to identify molecules regulating the migration of interneurons into the CP. We hypothesized that factors relevant for this process might be upregulated in pyramidal cells at the time interneurons invade the CP. To analyze gene expression in equivalent cohorts of pyramidal cells, we performed in utero electroporation experiments with a plasmid encoding GFP targeting the dorsal pallium of E14.5 mice (Figure 1A). We then used fluorescence-activated cell sorting (FACS) to isolate GFP+ pyramidal cells from these experiments at two different stages: embryonic day 17.5 (E17.5), when pyramidal cells are still migrating toward the CP, and postnatal day 4 (P4), when pyramidal cells have already reached their final position (Figures 1B–1D). To examine the differential gene expression at these two stages in the development of pyramidal cells, we customized a TaqMan array with ∼100 genes known to be involved in neuronal migration, adhesion, and axon guidance during corticogenesis (Table S1), including members of the eph, ephrin, semaphorin, plexin, cadherin, protocadherin, and neuregulin families. We also included several genes known to encode proteins expressed in pyramidal cells, such as Cux1 and Tbr1, as positive controls, and genes that encode proteins exclusively expressed in interneurons, such as Cxcr7, Cxcr4, Sst, Lhx6, and Htr3a, as negative controls.
Figure 1.
Identification of Factors Regulating the Invasion of the Cortical Plate by Interneurons
(A) Schematic of the experimental design. A plasmid encoding Gfp was electroporated in the neocortex of E14.5 mouse embryos. The electroporated region was then isolated at E17.5 or P4, and GFP+ cells were dissociated and recovered via FACS.
(B and C) Distribution of GFP+ pyramidal cells at E17.5 (B) and P4 (C), prior to isolation.
(D) FACS of GFP+ cells.
(E) Among the pool of candidate genes (Table S1), we found 42 that were differentially expressed between the two stages. The corresponding p values from t-tests are shown in Table S2. Histograms show average ± SEM. Scale bar represents 250 μm.
We identified 42 genes that are differentially expressed between the two stages (Figure 1E; Table S2). We focused our attention on 19 genes that are significantly more expressed by pyramidal cells at P4 compared to E17.5, because these are more likely to be involved in the chemoattraction of interneurons into the CP. To examine the pattern of expression of these genes, we performed in situ hybridization at E17.5 and P4 for this later list of candidate genes (Cdh6, Cdh20, Epha6, Epha10, Ephb2, Ephb3, Ephb6, Efna3, Efna5, Efnb3, Plxndc2, Plxna1, Plxnd1, Pcdh9, Pcdh15, Pcdh20, Nrg3, and Sema7a). Analysis of the expression of candidate genes revealed different patterns. For example, some genes were preferentially expressed in superficial layers of the cortex, and their expression increased during early postnatal stages (Figures S2A–S2F). In other cases, candidate genes were expressed throughout all layers of the neocortex (Figures S2G and S2H).
Nrg3 Is Expressed in the Developing Cortical Plate
We noticed that one of the genes that is expressed throughout the CP and that is significantly more expressed by pyramidal cells at later stages is Neuregulin 3 (Nrg3), a member of the neuregulin family. In vitro experiments have shown that Nrg3 binds preferentially to ErbB4 receptors (Zhang et al., 1997), which are highly enriched in migrating cortical interneurons and excluded from pyramidal cells (Fazzari et al., 2010, Flames et al., 2004, Vullhorst et al., 2009, Yau et al., 2003). Based on this evidence, we hypothesized that Nrg3 might regulate the intracortical migration of GABAergic interneurons and focused our subsequent work on this molecule.
We investigated the pattern of expression of Nrg3 in the developing cortex from mid-embryonic until early postnatal stages using in situ hybridization (Figure 2). We observed that Nrg3 is highly expressed in the developing CP since its inception, and that Nrg3 expression is maintained in pyramidal cells as they mature and start forming differentiated layers. Pyramidal cells therefore express Nrg3 as soon as they reach the CP, and its expression is subsequently maintained throughout all layers of the neocortex, including the subplate. Nrg3 is, however, largely absent from the MZ and the SVZ at all stages examined (Figure 2). These results support the hypothesis that Nrg3 might be involved in the regulation of interneuron migration into the CP.
Figure 2.
Expression of Nrg3 mRNA in the Developing Mouse Cortex
(A–E′) Coronal sections through the telencephalon of E13.5 (A and A′), E15.5 (B and B′), E18.5 (C and C′), P2 (D and D′), and P4 (E and E′) embryos and neonates showing mRNA expression for Nrg3. Ac, anterior commissure; CP, cortical plate; H, Hippocampus; ic, internal capsule; LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence; MZ, marginal zone; NCx, neocortex; PCx, piriform cortex; Str, striatum; SVZ, subventricular zone; Th, thalamus; VZ, ventricular zone; I–VI, cortical layers I–VI. Scale bars represent 250 μm.
Nrg3 Is a Short-Range Chemoattractant for MGE-Derived Interneurons
We have previously shown that different isoforms of Nrg1 act both as short- and long-range chemoattractive molecules for tangentially migrating interneurons (Flames et al., 2004). To examine whether Nrg3 may exert a similar effect on cortical interneurons, we performed confrontation assays in three-dimensional matrices in which we cultured MGE explants obtained from E13.5 GFP-expressing embryos together with aggregates of COS cells transfected with control or Nrg3 encoding plasmids (Figure 3A1). In parallel experiments, we carried out co-cultures in which COS cells were transfected with Ig-Nrg1, which encodes for a diffusible form of Nrg1. As described before (Flames et al., 2004), we observed that Ig-Nrg1 exerts a prominent chemoattractive response in MGE-derived cells (Figures 3B, 3C, and 3H). In contrast, we found no significant differences in the response of MGE-derived cells to Nrg3 compared to controls (Figures 3B, 3D, and 3H). Thus, Nrg3 does not seem to function as a long-range chemoattractant for MGE-derived interneurons.
Figure 3.
Nrg3 Functions as a Short-Range Chemoattractant for MGE-Derived Interneurons and Requires ErbB4 Receptors
(A) Schematic of the experimental design. MGE explants were confronted to transfected COS cells located at relative long (A1) or short (A2) distances.
(B–G) Migration of MGE-derived interneurons in response to mock- (B and E), Ig-Nrg1- (C), CRD-Nrg1- (D), or Nrg3-transfected (D and G) COS cells located at relatively long (B–D) and short (E–G) range. Dotted lines indicate the limits of the explants and COS cell aggregates.
(H) Quantification of long-distance confrontation assays. Control: MGE versus mock, n = 19; MGE versus Ig-Nrg1, n = 24; one-way ANOVA, ∗∗p < 0.01. MGE versus Nrg3, n = 20; one-way ANOVA, p > 0.05.
(I) Quantification of short-distance confrontation assays. Control: MGE versus mock, n = 29; MGE versus CRD-Nrg1, n = 24, MGE versus Nrg3, n = 27; one-way ANOVA, ∗∗p < 0.01.
(J) Schematic of the experimental design. MGE explants were confronted to control and Nrg3-transfected COS cells located at relative short distances.
(K, L, N, and O) Migration of MGE-derived cells derived from Erbb4+/+;HER4heart (K and L) and Erbb4−/−;HER4heart (N and O) mice in response to mock- (K and N) or Nrg3-transfected (L and O) COS cell aggregates cultured in matrigel matrices for 48 hr.
(M) Quantification of confrontation assays. Erbb4+/+;HER4heart versus mock, n = 33; Erbb4+/+;HER4heart versus Nrg3, n = 25; Erbb4−/−;HER4heart versus mock, n = 15; Erbb4−/−;HER4heart versus Nrg3, n = 14; one-way ANOVA, ∗∗∗p < 0.001. Histograms show average ± SEM. NCx, neocortex; MGE, medial ganglionic eminence. Scale bar represents 200 μm.
To investigate whether Nrg3 may function as a short-range chemoattractant for migrating interneurons, we carried out a new set of co-culture experiments in which COS cell aggregates, transfected with control or Nrg3-encoding plasmids, were placed at a relatively short distance from MGE explants (Figure 3A2). In this new set of experiments, we used COS cells transfected with CRD-Nrg1 as a positive control, because this membrane-bound form of Nrg1 has been shown to induce short-range chemoattraction during the migration of cortical interneurons through the subpallium (Flames et al., 2004). We found that both CRD-Nrg1 and Nrg3 evoke a potent chemoattractive effect on migrating interneurons, which can be visualized by the great abundance of cells accumulating around the proximal side of COS cell aggregates (Figures 3E–3G and 3I). Thus, Nrg3 induces a potent short-range chemoattractive effect on MGE-derived interneurons.
Nrg3 Chemoattraction Requires ErbB4 Function
Because Nrg3 is thought to bind preferentially to ErbB4 receptors (Zhang et al., 1997), we next examined whether ErbB4 function mediates the chemoattractive responses elicited by Nrg3 in MGE-derived interneurons. To this end, we carried out a new set of co-culture experiments with MGE explants obtained from control and Erbb4 mutant embryos (Figure 3J). Because loss of ErbB4 causes early lethality due to cardiac defects, Erbb4 mutants carried a human transgene under a cardiac-specific myosin promoter (HER4heart) to circumvent this problem (Tidcombe et al., 2003). In contrast to controls, we observed that Nrg3 does not exert any effect on MGE-derived cells obtained from Erbb4 mutant embryos (Figures 3K–3O). Thus, ErbB4 is necessary for the migration of cortical interneurons in response to Nrg3. Altogether, these experiments indicate that the chemoattractive effect elicited by Nrg3 on migrating interneurons is mediated by ErbB4 function.
Tangentially Migrating MGE-Derived Interneurons Prefer Cxcl12 to Nrg3
It has been previously shown that the chemokine Cxcl12 strongly promotes the tangential migration of MGE-derived cells throughout the embryonic cortex. Cxcl12 is expressed by the meninges and in the SVZ of the pallium during embryonic development (Borrell and Marín, 2006, Stumm et al., 2003, Tiveron et al., 2006). Because Nrg3 is expressed in the CP from early stages of development (Figures 2A and 2A′), tangentially migrating interneurons encounter both cues as they reach the embryonic cortex. Both Cxcl12 and Nrg3 seem to function as chemoattractive factors for migrating MGE-derived interneurons, so we next explored whether interneurons display a preference for any of these molecules. To this end, we cultured MGE explants obtained from E13.5 embryos from GFP-expressing mice together with aggregates of COS cells placed at a short distance and transfected with either a mock plasmid, Nrg3 or Nrg3 and Cxcl12 together (Figure 4A1). As expected, we observed that Cxcl12 enhances the migration of MGE-derived interneurons (Figures 4B, 4D, and 4E). In addition, we found that Cxcl12 does not block the chemoattractive effect elicited by Nrg3 (Figures 4B–4E). These results suggested that tangentially migrating MGE-derived interneurons are equipped to respond to both Cxcl12 and Nrg3 simultaneously.
Figure 4.
MGE-Derived Interneurons Display Preferential Responses to Cxcl12 and Nrg3
(A) Schematic of the experimental designs. MGE explants were confronted to transfected COS cells located at relative short distances (A1) or placed on coated matrices in stripe choice assays (A2).
(B–D) Migration of MGE-derived interneurons in response to mock- (B), Nrg3- (C), or Nrg3- and Cxcl12-transfected (D) COS cells. Dotted lines indicate the limits of the explants and COS cell aggregates.
(E) Quantification of MGE-derived interneurons migrating away from explants. MGE versus mock, n = 18; MGE versus Nrg3, n = 15; MGE versus Nrg3 + Cxcl12, n = 27; one-way ANOVA, ∗∗∗p < 0.001.
(F) Quantification of short-distance confrontation assays. MGE versus mock, n = 18; MGE versus Nrg3, n = 15; MGE versus Nrg3 + Cxcl12, n = 27; one-way ANOVA, ∗∗∗p < 0.001, ∗p < 0.05.
(G and H) Migration of MGE-derived cells in the stripe choice assay, with control and Nrg3-coated (G) or Nrg3- and Cxcl12-coated alternating stripes (H).
(I) Quantification of stripe choice assays. Nrg3/Nrg3 (n = 11) versus Nrg3/GST (n = 20); one-way ANOVA, ∗∗∗p < 0.001. Cxl12/Cxl12 (n = 13) versus Nrg3/Cxcl12 (n = 18); one-way ANOVA, ∗∗∗p < 0.001. Histograms show average ± SEM. NCx, neocortex; MGE, medial ganglionic eminence. Scale bar represents 200 μm.
We next wondered whether MGE-derived interneurons display any preference for Cxcl12 or Nrg3. To answer this question, we performed stripe choice assays using recombinant proteins, as described previously (Walter et al., 1987). In brief, E13.5 MGE explants obtained from GFP-expressing embryos were placed on top of stripes coated with a control peptide (GST), Nrg3-GST or recombinant Cxcl12 in alternate combinations (Figure 4A2), and their lane preference was scored after 48 hr. As expected, MGE-derived cells showed no migratory preference when alternative stripes were coated with the same recombinant protein (GST/GST, Nrg3/Nrg3 or Cxcl12/Cxcl12) in control experiments (Figure 4I; data not shown). In contrast, MGE-derived interneurons displayed a strong preference for Nrg3-coated stripes compare to control lanes (Figures 4G and 4I). Moreover, MGE-derived cells exhibited a strong preference toward Cxcl12 when they were given the possibility to migrate on alternating stripes containing Nrg3 and Cxcl12 (Figures 4H and 4I). Taken together, these experiments suggest that tangentially migrating interneurons can respond simultaneously to Cxcl12 and Nrg3, but they display stronger affinity for the chemokine. These observations are consistent with the in vivo behavior of MGE-derived interneurons, which initially disperse through the cortex via Cxcl12-rich territories (MZ and SVZ) without accumulating in the CP (López-Bendito et al., 2008).
Nrg3 Overexpression Promotes Interneuron Invasion of the CP In Vivo
We next wondered whether unbalancing the normal levels of Nrg3 in the developing CP would interfere with the migration of cortical interneurons in vivo. To this end, we electroporated the ventricular zone of the pallium in E14.5 Nkx2-1Cre;Ai9 embryos with either Gfp-expressing plasmids or a combination of Gfp and Nrg3 (Figure 5A), and examined the distribution of MGE-derived interneurons (labeled with tdTomato) at E18.5. We observed that overexpression of Nrg3 does not seem to disrupt the migration of pyramidal cells (Figures 5B and 5E). In contrast, overexpression of Nrg3 accelerates the invasion of the CP by MGE-derived interneurons (Figures 5C, 5D, and 5F). Thus, these results suggested that Nrg3 promotes the intracortical migration of MGE-derived interneurons in vivo.
Figure 5.
Nrg3 Overexpression in Pyramidal Cells Enhances Interneuron Invasion of the CP In Vivo
(A) Schematic of the experimental design. The pallial ventricular zone of Nkx2-1Cre;Ai9 embryos was electroporated at E14.5 with Gfp-expressing or Gfp- and Nrg3-expressing plasmids, and the distribution of MGE-derived interneurons was analyzed at E18.5.
(B, C, E, and F) Distribution of MGE-derived interneurons (labeled with tdTomato) in the somatosensory cortex of E18.5 Nkx2-1Cre;Ai9 embryos following in utero electroporation of Gfp (B and C) or Gfp and Nrg3 (E and F) at E14.5.
(D) Density of MGE-derived cells in the CP; n = 6; t test, ∗∗∗p < 0.001. Histograms show average ± SEM. CP, cortical plate; MZ, marginal zone; V and VI, cortical layers V and VI, respectively. Scale bar represents 200 μm.
In Vivo Loss of Nrg3/ErbB4 Signaling Disrupts Interneuron Lamination
To examine the long-term consequences of disrupting Nrg3 signaling in vivo, we generated mice carrying conditional Nrg3 alleles (Figure S3). To delete Nrg3 specifically from developing pyramidal cells, we used Nex-Cre mice, in which exon 2 of the NeuroD6 locus has been replaced by Cre recombinase (Goebbels et al., 2006). We found that early deletion of Nrg3 in pyramidal cells does not alter their laminar position in the postnatal cortex (Figure S4). Analysis of the distribution of Erbb4-expressing neurons in the somatosensory cortex of control and conditional Nrg3 mutants at P30 revealed no differences in the total density of Erbb4+ cells (controls: 783.28 ± 48.18 cells/mm2; Nrg3 mutants: 923.69 ± 56.15 cells/mm2; n = 4; t test, p > 0.05). However, we observed that the laminar location of Erbb4-expressing neurons was significantly different between control and conditional Nrg3 mutants, with a deep to superficial layer shift in the distribution of Erbb4-expressing cells in mutants compared to controls (Figures 6A–6C). Because ErbB4 is primarily expressed by PV+ interneurons in the cortex (Fazzari et al., 2010, Neddens and Buonanno, 2010), we also analyzed their distribution at P30. Similar to the analysis of Erbb4-expressing neurons, we found no differences in the total density of PV+ interneurons (controls: 281.88 ± 25.29 cells/mm2; Nrg3 mutants: 258.68 ± 36.1 cells/mm2; n = 5; t test, p > 0.05), but a subtle shift in their laminar distribution (Figures 6D–6F). To determine whether these differences in the laminar distribution of cortical interneurons were already present in the early postnatal cortex, we examined the distribution of Lhx6-expressing neurons in the somatosensory cortex of control and conditional Nrg3 mutants at P4. We found a significant shift toward superficial layers in the distribution of Lhx6-expressing cells in conditional Nrg3 mutants compared to controls (Figures 6G–6I). Altogether, these results revealed that conditional deletion of Nrg3 from developing pyramidal cells disrupts the laminar distribution of cortical interneurons.
Figure 6.
Abnormal Lamination of Cortical Interneurons in Conditional Nrg3 Mutants
(A and B) Distribution of Erbb4-expressing interneurons in the somatosensory cortex of control (A) and conditional Nrg3 (B) mice at P30.
(C) Quantification of the distribution of Erbb4-expressing cells; n = 4; χ2 test, ∗p < 0.05.
(D and E) Distribution of PV+ interneurons in the somatosensory cortex of control (D) and conditional Nrg3 (E) mice at P30.
(F) Quantification of the distribution of PV+ interneurons; n = 5; χ2 test, ∗p < 0.05.
(G and H) Distribution of Lhx6-expressing cells in the somatosensory cortex of control (G) and conditional Nrg3 (H) mice at P4.
(I) Quantification of the laminar distribution of Lhx6-expressing cells; n = 5; ∗p < 0.05, χ2 test. Histograms show average ± SEM. Scale bars represent 200 μm.
Because Nrg3 signaling requires ErbB4 function in cortical interneurons (Figures 3J–3O), we next wondered whether similar layering defects exist in conditional Erbb4 mutants. To this end, we bred Lhx6-Cre mice with mice carrying loxP-flanked (F) Erbb4 alleles and analyzed the distribution of PV+ interneurons in the somatosensory cortex. We have previously shown that the total number of interneurons in the cortex in Lhx6-Cre;Erbb4F/F mice is similar to controls at P30 (Del Pino et al., 2013). However, detailed analysis of the laminar distribution of PV+ interneurons revealed a clear shift in conditional Erbb4 mutants, with fewer cells in deep layers and more cells in superficial layers than in controls (Figures 7A–7F). To elucidate whether this defect reflects an overall shift in the allocation of cortical interneurons, we examined the laminar distribution of specific cohorts of PV+ cells by injecting BrdU in control and conditional Erbb4 mutants at E12.5 or E15.5 (Figure 7A). We found that interneurons born at E12.5 and E15.5 tend to occupy deep and superficial layers of the cortex, respectively, in both controls and Erbb4 mutants (Figures 7B–7E, 7G, and 7H). However, the distribution of PV+ interneurons was shifted toward progressively more superficial layers for both cohorts of cells in conditional Erbb4 mutants compared to controls (Figures 7G and 7H), a result that is consistent with the overall shift in the distribution of PV+ interneurons toward superficial layers (Figure 7F). Altogether, these experiments indicate that ErbB4 mediates the function of Nrg3 in the intracortical migration of interneurons, and that this signaling system is required for the appropriate laminar allocation for these cells.
Figure 7.
Abnormal Lamination of Cortical Interneurons in Conditional Erbb4 Mutants
(A) Schematic of the experimental design.
(B–E) Laminar distribution of PV+ interneurons (red) and specific cohorts of PV+ interneurons labeled after BrdU injections at E12.5 (B and C) and E15.5 (D and E) (red and green) in the somatosensory cortex of control (B and D) and conditional Erbb4 mutant (C and E) mice at P30.
(F) Quantification of the distribution of PV+ interneurons; n = 5; χ2 test, ∗p < 0.05.
(G and H) Quantification of the distribution of specific cohorts of PV+ interneurons born at E12.5 (G) and E15.5 (H); n = 5; χ2 test, ∗p < 0.05. Histograms show average ± SEM. Scale bar represents 200 μm.
Discussion
We have identified Nrg3 as a factor regulating the assembly of inhibitory neurons in the developing cerebral cortex. We found that Nrg3 is highly expressed by developing pyramidal cells during the entire period of interneuron allocation in the cerebral cortex. Gain- and loss-of-function experiments suggest that Nrg3 functions specifically as a chemoattractive signal regulating CP invasion by interneurons, and that this process is mediated by the tyrosine kinase receptor ErbB4. Nrg3 is therefore a critical mediator in the assembly of inhibitory circuits, controlling the orderly entry of interneurons in the CP prior to their subsequent sorting into specific cortical layers.
Nrg3 in Cortical Interneuron Development
Nrg3 was identified as a neural tissue-enriched molecule nearly 20 years ago (Zhang et al., 1997), yet very little is known about its role in brain development and function. An isoform of NRG3 that is specifically expressed in the human embryonic CNS has been shown to influence oligodendrocyte survival in vitro (Carteron et al., 2006). In addition, recent in vitro experiments revealed that MGE-derived interneurons are able to respond to Nrg3 (Li et al., 2012, Rakić et al., 2015). This is consistent with the observation that MGE-derived interneurons express ErbB4 (Flames and Marín, 2005, Yau et al., 2003), and that ErbB4 is the preferred receptor for Nrg3 (Zhang et al., 1997). Our experiments confirm and extend these observations by demonstrating a specific role for Nrg3 in the entry of GABAergic interneurons in the developing CP in vivo.
Our results suggest that Nrg3 functions as a short-range chemoattractive factor for cortical interneurons. Analysis of the amino acid sequence of human Nrg3 reveals important homologies with Nrg1, but contrary to many Nrg1 family members the extracellular domain of Nrg3 lacks Ig-like domains (Zhang et al., 1997). Although the structure of Nrg3 resembles more closely that of type I/II Nrg1, our experiments indicate that Nrg3 functions largely as a short-range chemoattractant, most similar to membrane-bound forms of Nrg1. Even if Nrg3 is released in vivo through post-translational proteolysis, our experiments suggest that its diffusion is likely restricted to near the CP. In this context, it is worth noting that Nrg3 contains a mucin-like Ser/Thr-rich region containing abundant sites for O-linked glycosylation (Zhang et al., 1997), which may function to limit its diffusion.
Constitutive Nrg3 mutant mice have decreased impulsivity, increased activity in a novel open field, and deficient prepulse inhibition of the acoustic startle response and fear conditioning (Hayes et al., 2016, Loos et al., 2014). Interestingly, these defects are not rescued by overexpression of Nrg3 in the adult medial prefrontal cortex—the region responsible for impulsive responding (Loos et al., 2014). In contrast, systemic overexpression of Nrg3 during early postnatal development via subcutaneous injections leads to alterations in anxiety and social behaviors in adulthood (Paterson and Law, 2014). It is conceivable that the behavioral deficits reported in these mice are due to defects in the distribution of cortical interneurons, reinforcing the notion that Nrg3 function during development is required for normal brain function.
Neuregulins have been also implicated in other steps in the development of MGE-derived cortical interneurons (Rico and Marín, 2011). In particular, Nrg1 has been shown to regulate the wiring of PV+ interneurons in the postnatal cortex (Fazzari et al., 2010, Ting et al., 2011). Because Nrg3 expression is maintained in pyramidal cells into adulthood, it remains to be explored whether this neuregulin may contribute to the regulation of the connectivity of PV+ interneurons. Nrg3 is also expressed in cortical interneurons in the postnatal brain (Loos et al., 2014), which suggests that its function in cortical circuit assembly might be more complex than previously anticipated.
A Hierarchical Organization of Guidance Cues for Cortical Interneurons
Our experiments suggest that the transition from tangential to radial migration and the allocation of interneurons inside the CP depends on a delicate balance between two chemoattractive factors, Ngr3 and Cxcl12, which seems to be hierarchically organized. Our experiments indicate that tangentially migrating interneurons show a prominent preference for Cxcl12 during their tangential dispersion through the cortex, and it is only when they lose their responsiveness to this molecule that they steer toward the source of Nrg3 in the CP. One caveat of the in vitro experiments is that the tested MGE interneurons are younger than they would normally be when they encounter both molecules in the cortex in vivo. Our in vivo analyses, however, strongly suggest that interneurons respond to both of these cues during the migration through the cortex.
Cxcl12 is strongly expressed by the meninges and by intermediate progenitor cells transitorily present in the SVZ (Stumm et al., 2003, Tham et al., 2001, Tiveron et al., 2006) and is also expressed by cells in the SP (Stumm et al., 2007). Cxcl12 is a potent long-range chemoattractant for MGE-derived interneurons in vitro (Li et al., 2008, López-Bendito et al., 2008), but its limited diffusion in vivo would explain the relative confinement of interneurons to the migratory streams found in the cortex. Consistent with this idea, mouse mutants with altered expression of Cxcl12 in the meninges or in the SVZ have defects in the intracortical migration of interneurons that are specific to the affected migratory route (Abe et al., 2015, Sessa et al., 2010, Tiveron et al., 2006, Zarbalis et al., 2012).
The molecular mechanisms regulating the tangential to radial switch in the migration of cortical interneurons and the subsequent CP invasion remains unknown. The exit of interneurons from the migratory streams is coordinated with the loss of responsiveness to Cxcl12 (Li et al., 2008), and this is ultimately linked with the entry of interneurons into the CP. The analysis of Cxcr4 and Cxcr7 mutants revealed many interneurons inside CP at relatively early embryonic stages (Li et al., 2008, López-Bendito et al., 2008, Sánchez-Alcañiz et al., 2011, Tanaka et al., 2010, Tiveron et al., 2006, Wang et al., 2011), a finding that can now be interpreted as a premature response to Nrg3.
The timing of interneuron responses to the cues guiding their migration into the cortex seems tightly regulated, and it is important for the final distribution of interneurons in different regions and layers of the cortex. Premature entry in the cortical plate, as observed in Cxcr4 and Cxcr7 mutants, causes defects in the regional and laminar distribution of interneurons in the adult cortex (Li et al., 2008, López-Bendito et al., 2008, Sánchez-Alcañiz et al., 2011, Tanaka et al., 2010, Wang et al., 2011). Similarly, removal of Nrg3, a factor that facilitates CP entry, also disrupts laminar distribution in the adult cortex. Of note, the laminar defects observed in Cxcr4/Cxcr7 and Nrg3 mutants are complementary (accumulation of interneurons in deep or superficial layers for Cxcr4/Cxcr7 and Nrg3 mutants, respectively). These observations reinforce the notion that Cxcl12 and Nrg3 play sequential and largely complementary roles in the migration of cortical interneurons.
Pyramidal Cells and Microglia Regulate the Distribution of Interneurons
Studies over the last decade have revealed some important aspects on the regulation of layer acquisition by cortical interneurons. Most notably, several studies have suggested that pyramidal cells regulate the laminar distribution of cortical interneurons (Hevner et al., 2004, Lodato et al., 2011, Pla et al., 2006). In reeler mice, for example, the distribution of interneurons follow that of pyramidal cells, which is roughly inverted compared to control mice (Hevner et al., 2004, Pla et al., 2006). This phenotype does not depend on Reelin signaling, but rather on the abnormal distribution of pyramidal cells (Pla et al., 2006). Subsequent work revealed that experimental manipulations causing the formation of clusters of misplaced pyramidal cells under the white matter are sufficient to misguide interneurons into these ectopic locations (Lodato et al., 2011). These results reinforced the notion that pyramidal cells produce factors that guide the migration of cortical interneurons, and our experiments now reveal that Nrg3 is one of these molecules. In addition, recent work has shown that Cxcl12 may also play additional functions in the postnatal differentiation of cortical interneurons, affecting their laminar distribution and synapse development (Vogt et al., 2014, Wu et al., 2016).
Recent work suggests that microglia may also regulate the laminar positioning of cortical interneurons (Squarzoni et al., 2014). Microglia invade the cortex following a gradient similar to interneurons (Cunningham et al., 2013, Squarzoni et al., 2014, Swinnen et al., 2013). In the absence of microglia, or when microglia is abnormally activated, MGE-derived interneurons enter the CP prematurely and adopt abnormal laminar distribution patterns (Squarzoni et al., 2014). It remains to be tested whether microglia exert their function through the regulation of chemokines or by directly acting on signals produced by pyramidal cells.
Nrg3, Cortical Interneurons, and Psychiatric Disorders
Human genetic studies have associated variation in the NRG3 gene with attention deficit/hyperactivity disorder (ADHD) and schizophrenia (Chen et al., 2009, Kao et al., 2010, Meier et al., 2013, Morar et al., 2011, Sonuga-Barke et al., 2008). Although some of these studies await replication in larger genome-wide association studies (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014), increasing evidence suggests that pathological variants in the neuregulin signaling pathway are present in at least a subset of phenotypically distinct patients (Hatzimanolis et al., 2013). In mice, Nrg3 overexpression in the adult prefrontal cortex increases impulsivity (Loos et al., 2014), a feature that is observed in ADHD and schizophrenia patients (Aron and Poldrack, 2005, Nolan et al., 2011). Loss of Nrg3 in mice has also been linked with behavioral deficits observed in psychotic disorders (Hayes et al., 2016). Considering that the balance between excitatory and inhibitory circuits seems compromised in schizophrenia patients, and that this might be caused by subtle defects in the function of GABAergic interneurons (Lewis et al., 2012), our results suggest a possible mechanism through which changes in NRG3 expression may influence the assembly of inhibitory circuits in the cerebral cortex.
Experimental Procedures
Animals
This study was performed in strict accordance with Spanish, United Kingdom, and European Union regulations. The local ethical committees at the Instituto de Neurociencias and King’s College London approved all experimental procedures involving animals.
Nkx2-1Cre (Xu et al., 2008) and Ai9 (Rosa26 Reporter CAG-boosted tdTomato) mice (Madisen et al., 2010) were maintained in a C57BL/6 background, whereas wild-type and ubiquitously expressing GFP mice (Hadjantonakis et al., 1998) were maintained in a CD1 background. HER4heart transgenic mice, which express a human ErbB4 (HER4) cDNA under the control of the cardiac-specific myosin heavy-chain (HMC) promoter, were maintained in a mixed C57b/6 and 129/SvJ background. The generation of ErbB4 mutant mice (Gassmann et al., 1995) and HER4heart transgenic mice (Tidcombe et al., 2003) has been previously described. Lhx6-Cre;Erbb4F/F mice were generated by breeding Lhx6-Cre mice (Fogarty et al., 2007) with mice carrying loxP-flanked (F) Erbb4 alleles (Golub et al., 2004). For birth-dating experiments, pregnant females received intraperitoneal injections at E12.5 (three injections in 18 hr) or E15.5 (three injections in 12 hr) with 50 mg/kg 5-bromo-2′-deoxyuridine (BrdU) (B5002; Sigma-Aldrich).
We generated a pre-conditional allele for Nrg3 by crossing mice carrying a knockout first Nrg3 allele (Knockout Mouse Program of the University of California, Davis) (Skarnes et al., 2011) with mice ubiquitously expressing Flp (Rodríguez et al., 2000). We then produced conditional Nrg3 mutants by crossing Nex-Cre (Goebbels et al., 2006) or Nestin-Cre (Tronche et al., 1999) with mice carrying conditional Nrg3 alleles. To genotype these mice, we used the following primer sequences: CSD-loxP, 5′-GAGATGGCGCAACGCAATTAATG-3′; CSD-Nrg3-SR1: 5′-AGTGCTGGAAATAAAAGCATGGTGGG-3′; CSD-Nrg3-wtF: 5′-CATATTACATACAGAATTCAAAGATAGGC-3′; CSD-Nrg3-wtR: 5′-CCAGTGCTGGAATTTGAATACAA-3′. CSD-loxP and CSD-Nrg3-SR1 primers were used to detect the knockout first allele. CSD-Nrg3-wtF and CSD-Nrg3-wtR were used to detect the wild-type allele and the wild-type pre-conditional allele generated after Flp-mediated recombination.
In Situ Hybridization and Immunohistochemistry
For in situ hybridization, postnatal mice were perfused transcardially with 4% paraformaldehyde (PFA) in PBS, and the dissected brains were fixed overnight at 4°C in the same solution. Brains were then cut at 40 μm on a freezing microtome, and free-floating coronal sections were subsequently hybridized with digoxigenin-labeled probes as described before (Flames et al., 2007). The following cDNA probes were used in this study: Nrg3 and ErbB4 (kindly provided by Cary Lai, Indiana University, Bloomington), GAD67 (kindly provided by John Rubenstein, UCSF), Lhx6 (kindly provided by V. Pachnis, The Crick Institute, London), Cxcr4 (Invitrogen; 174412), Cxcl12 (Invitrogen; 3483088), Cdh6 and Cdh9 (kindly provided by C. Redies, University of Jena), Ephb6 (Source BioScience; EST clone IMAGp998L1511952Q), Epha6 (kindly provided by V. Borrell, Instituto de Neurociencias, Alicante), and Sema7a (Source BioScience; EST clone IMAGp998I188236Q).
For immunohistochemistry, postnatal mice were perfused transcardially with 4% PFA in PBS, and the dissected brains were fixed for 2 hr at 4°C in the same solution. Brains were sectioned at 60 μm on a vibratome or 40 μm on a freezing microtome, and free-floating coronal sections were then subsequently processed for immunohistochemistry as previously described (Pla et al., 2006). The following primary antibodies were used: rat anti-BrdU (1:200; ab6326; Abcam), rat anti-Ctip2 (1:500; ab18465; Abcam), rabbit anti-Cux1 (CDP-M222; 1:100; Santa Cruz), rabbit anti-DsRed (1:500; 632496; Clontech), chicken anti-GFP (1:1,000; GFP-1020; Aves Labs), and rabbit anti-PV (1:3,000; Swant). The following secondary antibodies were used: goat anti-chicken 488, donkey anti-rabbit 555, donkey anti-mouse 488, and goat anti-rat IgG (H+L) Alexa Fluor 555 conjugate (Molecular Probes). Cell nuclei were stained with 5 μM DAPI in PBS, and sections were mounted with Mowiol (Sigma) with NPG (Calbiochem).
In Utero Electroporation
E14.5 timed-pregnant ICR or Nkx2-1Cre;Ai9 females were deeply anesthetized, and the abdominal cavity was cut open. Embryos were exposed in the uterus, and 1 μg/μL pCAG-Gfp or Nrg3 (kindly provided by C. Lai, Indiana University, Bloomington, and subcloned into pCAGGS) plasmids were injected into the lateral ventricle of the telencephalon through the uterine wall. Square electric pulses of 45 V and 50 ms were passed through the uterus five times, spaced 950 ms, using a square pulse electroporator. The uterine horns were placed back in the abdominal cavity, which was then suture closed, and the female was allowed to recover.
Explant Cultures
For COS cell confrontation assays, COS7 cells were transfected with plasmids encoding Rfp alone, Rfp and Cxcl12, Rfp and Nrg3, Rfp and CRD-Nrg1, or Rfp and Ig-Nrg1, and cell aggregates were prepared by diluting transfected cells with Matrigel in a 1:1 proportion. After jellification, COS cell aggregates were cut with a scalpel in small rectangular prisms of approximately 400 × 400 × 800 μm and confronted to MGE explants obtained from GFP-expressing transgenic mice in 3D Matrigel pads. The cDNA used for expression of Cxcl12 was obtained from Invitrogen (clone number: 3483088; accession number: BC006640). Nrg3 was kindly provided by Cary Lai (Indiana University, Bloomington). The sequences used for expression of type I NRG1 (Ig-Nrg1) and type III NRG1 (CRD-Nrg1) correspond to the accession numbers AY648976 and AY648975, respectively. For Cxcl12 chemokine-blocking experiments, SU6656 (Sigma; 330161-87-0) was added to the medium at a final concentration of 15 μM. Previous worked has shown that Src functions downstream of Cxcr4 activation (Cabioglu et al., 2005).
In Vitro Focal Electroporation
Coronal slice cultures were obtained as described previously (Anderson et al., 1997). A pCAGG-based dsRed plasmid was pressure injected focally into the MGE of coronal slice cultures by a Pneumatic PicoPump through a glass micropipette. Slices were then electroporated within a setup of two horizontally oriented platinum electrodes powered by a Electro-Square-Porator, as described before (Flames et al., 2004).
Time-Lapse Videomicroscopy
Slices were transferred to the stage of an upright Leica DMLFSA or inverted Leica DMIRE2 microscope coupled to a confocal spectral scanning head (Leica; TCS SL) and viewed through 10–60× water immersion or 20× oil objectives. Slices were continuously superfused with warmed (32°C) artificial cerebrospinal fluid at a rate of 1 mL/min or maintained in supplemented Neurobasal medium. To block Cxcl12 function, SU6656 (Sigma; 330161-87-0) was added to the medium at a final concentration of 15 μM.
Stripe Assay
Purified CXCL12 protein was obtained from PeproTech (250-20A) and used at 1 ng/μL. GST and EGF-Nrg3-GST were purified using standard protocols and used at 10 μg/mL. Alternating lanes, 50 μm wide, were laid down on a poly-lysine-coated plastic dish. Alexa 555-labeled anti-rabbit IgGs were added to the GST, EGF-Nrg3-GST, and CXCL12 protein solution for lane identification. The lanes were further coated with laminin. MGE explants were dissected out of GFP+ brain slices, plated on top of the protein stripes, and incubated in methylcellulose-containing Neurobasal medium for 48 hr.
FACS
We dissected the sensorimotor cortex of E17.5 embryos and P4 pups following in utero electroporation at E14.5. Cortical tissue was dissociated as described previously (Catapano et al., 2001). GFP+ cells were purified using fluorescent activated cell sorting (FACSARIA III; BD Biosciences), and the resulting pellet was kept at −80°C.
TaqMan Gene Expression Assays
We isolated GFP+ pyramidal cells by FACS at E17.5 and P4 after in utero electroporation at E14.5. mRNA was then extracted using the RNeasy Micro Kit (QIAGEN) according to the manufacturer’s instructions. RNA quality was assessed using a bioanalyzer (Agilent Technologies) and then retro-transcribed into single-stranded cDNA. The RNA was sent to Unidad Genómica (Fundación Parque Científico de Madrid) for quality control and retro-transcription. Relative gene expression levels from three independent samples were analyzed using custom designed TaqMan low-density array (TLDA) plates (Micro Fluidic Cards; Applied Biosystems). Each plate contained duplicates for all the genes shown in Table S1. Data were collected and analyzed using the threshold cycle (Ct) relative quantification method. The housekeeping gene 18 RNA was included in the array for assessing RNA quality and sample normalization.
Western Blot
Cortical lysates were prepared from P30 control and Nestin-Cre;Nrg3F/F and Nex-Cre;Nrg3F/F mutants as described before (Fazzari et al., 2010, Vullhorst et al., 2009) and blotted using mouse anti-β-Actin (1:4,000; Sigma) and rabbit anti-Nrg3 (1:500; Abcam). Signals were detected with a luminescent image analyzer (LAS-1000PLUS; Fujifilm) and quantified with Quantity One 1D Analysis Software (Bio-Rad Laboratories).
Image Analysis and Quantification
Images were acquired using fluorescence microscopes (DM5000B, CTR5000, and DMIRB from Leica, or Apotome.2 from Zeiss) coupled to digital cameras (DC500 or DFC350FX, Leica; OrcaR2, Hamamatsu), or in an inverted Leica TCS SP8 confocal microscope. All images were analyzed with ImageJ (Fiji). For the quantification of migration in MGE explants, the distance migrated by the 30 furthest cells was measured. For the quantification of short-range chemoattraction, the colocalizing area between MGE and COS cells was measured. For the analysis of the interneuron angle of migration, we draw a grid of virtual radial lines (lines perpendicular to the ventricular zone and the pial surface) and oriented each cell in relation to the most adjacent “radial line.” Cells that deviated less than 25° from radial lines were considered as radially oriented; those that deviate more than 25° were designated as tangentially oriented. We systematically exclude from this analysis those cells located in the more lateral or medial regions of the cortex, so that the curvature of the slice in those regions would not interfere with our analysis (Martini et al., 2009). For the quantification of cell migration in MGE explants, we measured the distance migrated by the 30 furthest cells and normalized the average migrated distance to the distance between MGE and COS explants. For the quantification of the colocalizing area between migrating interneurons and COS cells in the short-distance confrontation assays, we quantified the colocalizing area using ImageJ (Fiji). Stripes were quantified by counting the number of neurons contained in a virtual grid containing five black and five red lines. The same area was used for all explants. Sections from control and mutant mice were imaged during the same imaging session. Data acquisition was performed using the same laser power, photomultiplier gain, pinhole, and detection filter settings (1,024 × 1,024 resolution; 12 bits). Quantifications were done using ImageJ (Fiji). Layers were drawn following nuclear staining. For in situ hybridization, the area quantified was divided in ten equal bins, and the percentage of cells in each bin was calculated. The bins were then matched to the appropriate layers.
Statistical Analyses
Statistical analysis was carried out in SPSS (SPSS, Inc.). The p values below 0.05 were considered statistically significant. Data are presented as mean and SEM throughout the manuscript (Table S3). Normality and variance tests were first applied to all experimental data. When data followed a normal distribution, paired comparisons were analyzed with t test, whereas multiple comparisons were analyzed using either ANOVA with post hoc Bonferroni correction (equal variances) or the Welch test with post hoc Games-Howell (different variances). A χ2 test was applied to analyze the distribution of cells in either bins or layers.
Author Contributions
Conception and Design, Acquisition of Data, Analysis and Interpretation of Data, Drafting or Revising the Article, and Contribution of Unpublished Essential Data or Reagents, G.B., J.A.S.-A., and O.M.; Acquisition of Data, Analysis and Interpretation of Data, and Drafting or Revising the Article, C.O., C.G.-F., and M.V.
Acknowledgments
We thank I. Andrew, S. Bae, M.A. Casillas, M. Fernández, and T. Gil for excellent technical assistance and laboratory support; A. Caler for excellent support with FACS experiments; G. Expósito for support with imaging; L. Lim for help with quantitative methods; V. Borrell, R. Hevner, C. Lai, V. Pachnis, C. Redies, B. Rico, J.L.R. Rubenstein, and M. Tessier-Lavigne for plasmids and antibodies; and A. Barco, M.A. Nieto, and K. Nave for mouse strains. We are grateful to members of the Flames, O.M., and Rico laboratories for stimulating discussions and ideas. This work was supported by grants from European Research Council (ERC-2011-AdG 293683) and the Spanish Government (CSD2007-00023 and SAF2011-28845) to O.M. O.M. is a Wellcome Trust Investigator.
Published: January 31, 2017
Footnotes
Supplemental Information includes four figures, three tables, and two movies and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.12.089.
Supplemental Information
References
- Abe P., Mueller W., Schütz D., MacKay F., Thelen M., Zhang P., Stumm R. CXCR7 prevents excessive CXCL12-mediated downregulation of CXCR4 in migrating cortical interneurons. Development. 2014;141:1857–1863. doi: 10.1242/dev.104224. [DOI] [PubMed] [Google Scholar]
- Abe P., Molnár Z., Tzeng Y.S., Lai D.M., Arnold S.J., Stumm R. Intermediate progenitors facilitate intracortical progression of thalamocortical axons and interneurons through CXCL12 chemokine signaling. J. Neurosci. 2015;35:13053–13063. doi: 10.1523/JNEUROSCI.1488-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S.A., Eisenstat D.D., Shi L., Rubenstein J.L.R. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Aron A.R., Poldrack R.A. The cognitive neuroscience of response inhibition: relevance for genetic research in attention-deficit/hyperactivity disorder. Biol. Psychiatry. 2005;57:1285–1292. doi: 10.1016/j.biopsych.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Baudoin J.P., Viou L., Launay P.S., Luccardini C., Espeso Gil S., Kiyasova V., Irinopoulou T., Alvarez C., Rio J.P., Boudier T. Tangentially migrating neurons assemble a primary cilium that promotes their reorientation to the cortical plate. Neuron. 2012;76:1108–1122. doi: 10.1016/j.neuron.2012.10.027. [DOI] [PubMed] [Google Scholar]
- Borrell V., Marín O. Meninges control tangential migration of hem-derived Cajal-Retzius cells via CXCL12/CXCR4 signaling. Nat. Neurosci. 2006;9:1284–1293. doi: 10.1038/nn1764. [DOI] [PubMed] [Google Scholar]
- Cabioglu N., Summy J., Miller C., Parikh N.U., Sahin A.A., Tuzlali S., Pumiglia K., Gallick G.E., Price J.E. CXCL-12/stromal cell-derived factor-1alpha transactivates HER2-neu in breast cancer cells by a novel pathway involving Src kinase activation. Cancer Res. 2005;65:6493–6497. doi: 10.1158/0008-5472.CAN-04-1303. [DOI] [PubMed] [Google Scholar]
- Carteron C., Ferrer-Montiel A., Cabedo H. Characterization of a neural-specific splicing form of the human neuregulin 3 gene involved in oligodendrocyte survival. J. Cell Sci. 2006;119:898–909. doi: 10.1242/jcs.02799. [DOI] [PubMed] [Google Scholar]
- Catapano L.A., Arnold M.W., Perez F.A., Macklis J.D. Specific neurotrophic factors support the survival of cortical projection neurons at distinct stages of development. J. Neurosci. 2001;21:8863–8872. doi: 10.1523/JNEUROSCI.21-22-08863.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.L., Avramopoulos D., Lasseter V.K., McGrath J.A., Fallin M.D., Liang K.Y., Nestadt G., Feng N., Steel G., Cutting A.S. Fine mapping on chromosome 10q22-q23 implicates Neuregulin 3 in schizophrenia. Am. J. Hum. Genet. 2009;84:21–34. doi: 10.1016/j.ajhg.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C.L., Martínez-Cerdeño V., Noctor S.C. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J. Neurosci. 2013;33:4216–4233. doi: 10.1523/JNEUROSCI.3441-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Pino I., García-Frigola C., Dehorter N., Brotons-Mas J.R., Alvarez-Salvado E., Martínez de Lagrán M., Ciceri G., Gabaldón M.V., Moratal D., Dierssen M. Erbb4 deletion from fast-spiking interneurons causes schizophrenia-like phenotypes. Neuron. 2013;79:1152–1168. doi: 10.1016/j.neuron.2013.07.010. [DOI] [PubMed] [Google Scholar]
- Fazzari P., Paternain A.V., Valiente M., Pla R., Luján R., Lloyd K., Lerma J., Marín O., Rico B. Control of cortical GABA circuitry development by Nrg1 and ErbB4 signalling. Nature. 2010;464:1376–1380. doi: 10.1038/nature08928. [DOI] [PubMed] [Google Scholar]
- Flames N., Marín O. Developmental mechanisms underlying the generation of cortical interneuron diversity. Neuron. 2005;46:377–381. doi: 10.1016/j.neuron.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Flames N., Long J.E., Garratt A.N., Fischer T.M., Gassmann M., Birchmeier C., Lai C., Rubenstein J.L., Marín O. Short- and long-range attraction of cortical GABAergic interneurons by neuregulin-1. Neuron. 2004;44:251–261. doi: 10.1016/j.neuron.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Flames N., Pla R., Gelman D.M., Rubenstein J.L., Puelles L., Marín O. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J. Neurosci. 2007;27:9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty M., Grist M., Gelman D., Marín O., Pachnis V., Kessaris N. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J. Neurosci. 2007;27:10935–10946. doi: 10.1523/JNEUROSCI.1629-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann M., Casagranda F., Orioli D., Simon H., Lai C., Klein R., Lemke G. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature. 1995;378:390–394. doi: 10.1038/378390a0. [DOI] [PubMed] [Google Scholar]
- Goebbels S., Bormuth I., Bode U., Hermanson O., Schwab M.H., Nave K.A. Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis. 2006;44:611–621. doi: 10.1002/dvg.20256. [DOI] [PubMed] [Google Scholar]
- Golub M.S., Germann S.L., Lloyd K.C. Behavioral characteristics of a nervous system-specific erbB4 knock-out mouse. Behav. Brain Res. 2004;153:159–170. doi: 10.1016/j.bbr.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Hadjantonakis A.K., Gertsenstein M., Ikawa M., Okabe M., Nagy A. Generating green fluorescent mice by germline transmission of green fluorescent ES cells. Mech. Dev. 1998;76:79–90. doi: 10.1016/s0925-4773(98)00093-8. [DOI] [PubMed] [Google Scholar]
- Hatzimanolis A., McGrath J.A., Wang R., Li T., Wong P.C., Nestadt G., Wolyniec P.S., Valle D., Pulver A.E., Avramopoulos D. Multiple variants aggregate in the neuregulin signaling pathway in a subset of schizophrenia patients. Transl. Psychiatry. 2013;3:e264. doi: 10.1038/tp.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes L.N., Shevelkin A., Zeledon M., Steel G., Chen P.L., Obie C., Pulver A., Avramopoulos D., Valle D., Sawa A., Pletnikov M.V. Neuregulin 3 knockout mice exhibit behaviors consistent with psychotic disorders. Mol. Neuropsychiatry. 2016;2:79–87. doi: 10.1159/000445836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Miranda L.R., Cariboni A., Faux C., Ruhrberg C., Cho J.H., Cloutier J.F., Eickholt B.J., Parnavelas J.G., Andrews W.D. Robo1 regulates semaphorin signaling to guide the migration of cortical interneurons through the ventral forebrain. J. Neurosci. 2011;31:6174–6187. doi: 10.1523/JNEUROSCI.5464-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner R.F., Daza R.A., Englund C., Kohtz J., Fink A. Postnatal shifts of interneuron position in the neocortex of normal and reeler mice: evidence for inward radial migration. Neuroscience. 2004;124:605–618. doi: 10.1016/j.neuroscience.2003.11.033. [DOI] [PubMed] [Google Scholar]
- Kao W.T., Wang Y., Kleinman J.E., Lipska B.K., Hyde T.M., Weinberger D.R., Law A.J. Common genetic variation in Neuregulin 3 (NRG3) influences risk for schizophrenia and impacts NRG3 expression in human brain. Proc. Natl. Acad. Sci. USA. 2010;107:15619–15624. doi: 10.1073/pnas.1005410107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausberger T., Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321:53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavdas A.A., Grigoriou M., Pachnis V., Parnavelas J.G. The medial ganglionic eminence gives rise to a population of early neurons in the developing cerebral cortex. J. Neurosci. 1999;19:7881–7888. doi: 10.1523/JNEUROSCI.19-18-07881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis D.A., Curley A.A., Glausier J.R., Volk D.W. Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci. 2012;35:57–67. doi: 10.1016/j.tins.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Adesnik H., Li J., Long J., Nicoll R.A., Rubenstein J.L.R., Pleasure S.J. Regional distribution of cortical interneurons and development of inhibitory tone are regulated by Cxcl12/Cxcr4 signaling. J. Neurosci. 2008;28:1085–1098. doi: 10.1523/JNEUROSCI.4602-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Chou S.J., Hamasaki T., Perez-Garcia C.G., O’Leary D.D. Neuregulin repellent signaling via ErbB4 restricts GABAergic interneurons to migratory paths from ganglionic eminence to cortical destinations. Neural Dev. 2012;7:10. doi: 10.1186/1749-8104-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato S., Rouaux C., Quast K.B., Jantrachotechatchawan C., Studer M., Hensch T.K., Arlotta P. Excitatory projection neuron subtypes control the distribution of local inhibitory interneurons in the cerebral cortex. Neuron. 2011;69:763–779. doi: 10.1016/j.neuron.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos M., Mueller T., Gouwenberg Y., Wijnands R., van der Loo R.J., Birchmeier C., Smit A.B., Spijker S., Neuro-BSIK Mouse Phenomics Consortium Neuregulin-3 in the mouse medial prefrontal cortex regulates impulsive action. Biol. Psychiatry. 2014;76:648–655. doi: 10.1016/j.biopsych.2014.02.011. [DOI] [PubMed] [Google Scholar]
- López-Bendito G., Sánchez-Alcañiz J.A., Pla R., Borrell V., Picó E., Valdeolmillos M., Marín O. Chemokine signaling controls intracortical migration and final distribution of GABAergic interneurons. J. Neurosci. 2008;28:1613–1624. doi: 10.1523/JNEUROSCI.4651-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysko D.E., Putt M., Golden J.A. SDF1 reduces interneuron leading process branching through dual regulation of actin and microtubules. J. Neurosci. 2014;34:4941–4962. doi: 10.1523/JNEUROSCI.4351-12.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín O. Cellular and molecular mechanisms controlling the migration of neocortical interneurons. Eur. J. Neurosci. 2013;38:2019–2029. doi: 10.1111/ejn.12225. [DOI] [PubMed] [Google Scholar]
- Marín O., Rubenstein J.L.R. A long, remarkable journey: tangential migration in the telencephalon. Nat. Rev. Neurosci. 2001;2:780–790. doi: 10.1038/35097509. [DOI] [PubMed] [Google Scholar]
- Marín O., Rubenstein J.L. Cell migration in the forebrain. Annu. Rev. Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- Marín O., Yaron A., Bagri A., Tessier-Lavigne M., Rubenstein J.L. Sorting of striatal and cortical interneurons regulated by semaphorin-neuropilin interactions. Science. 2001;293:872–875. doi: 10.1126/science.1061891. [DOI] [PubMed] [Google Scholar]
- Martini F.J., Valiente M., López Bendito G., Szabó G., Moya F., Valdeolmillos M., Marín O. Biased selection of leading process branches mediates chemotaxis during tangential neuronal migration. Development. 2009;136:41–50. doi: 10.1242/dev.025502. [DOI] [PubMed] [Google Scholar]
- Mei L., Nave K.A. Neuregulin-ERBB signaling in the nervous system and neuropsychiatric diseases. Neuron. 2014;83:27–49. doi: 10.1016/j.neuron.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei L., Xiong W.C. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat. Rev. Neurosci. 2008;9:437–452. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier S., Strohmaier J., Breuer R., Mattheisen M., Degenhardt F., Mühleisen T.W., Schulze T.G., Nöthen M.M., Cichon S., Rietschel M., Wüst S. Neuregulin 3 is associated with attention deficits in schizophrenia and bipolar disorder. Int. J. Neuropsychopharmacol. 2013;16:549–556. doi: 10.1017/S1461145712000697. [DOI] [PubMed] [Google Scholar]
- Miyoshi G., Fishell G. GABAergic interneuron lineages selectively sort into specific cortical layers during early postnatal development. Cereb. Cortex. 2011;21:845–852. doi: 10.1093/cercor/bhq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morar B., Dragović M., Waters F.A., Chandler D., Kalaydjieva L., Jablensky A. Neuregulin 3 (NRG3) as a susceptibility gene in a schizophrenia subtype with florid delusions and relatively spared cognition. Mol. Psychiatry. 2011;16:860–866. doi: 10.1038/mp.2010.70. [DOI] [PubMed] [Google Scholar]
- Neddens J., Buonanno A. Selective populations of hippocampal interneurons express ErbB4 and their number and distribution is altered in ErbB4 knockout mice. Hippocampus. 2010;20:724–744. doi: 10.1002/hipo.20675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nóbrega-Pereira S., Kessaris N., Du T., Kimura S., Anderson S.A., Marín O. Postmitotic Nkx2-1 controls the migration of telencephalic interneurons by direct repression of guidance receptors. Neuron. 2008;59:733–745. doi: 10.1016/j.neuron.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan K.A., D’Angelo D., Hoptman M.J. Self-report and laboratory measures of impulsivity in patients with schizophrenia or schizoaffective disorder and healthy controls. Psychiatry Res. 2011;187:301–303. doi: 10.1016/j.psychres.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson C., Law A.J. Transient overexposure of neuregulin 3 during early postnatal development impacts selective behaviors in adulthood. PLoS One. 2014;9:e104172. doi: 10.1371/journal.pone.0104172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pla R., Borrell V., Flames N., Marín O. Layer acquisition by cortical GABAergic interneurons is independent of Reelin signaling. J. Neurosci. 2006;26:6924–6934. doi: 10.1523/JNEUROSCI.0245-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polleux F., Whitford K.L., Dijkhuizen P.A., Vitalis T., Ghosh A. Control of cortical interneuron migration by neurotrophins and PI3-kinase signaling. Development. 2002;129:3147–3160. doi: 10.1242/dev.129.13.3147. [DOI] [PubMed] [Google Scholar]
- Pozas E., Ibáñez C.F. GDNF and GFRalpha1 promote differentiation and tangential migration of cortical GABAergic neurons. Neuron. 2005;45:701–713. doi: 10.1016/j.neuron.2005.01.043. [DOI] [PubMed] [Google Scholar]
- Rakić S., Kanatani S., Hunt D., Faux C., Cariboni A., Chiara F., Khan S., Wansbury O., Howard B., Nakajima K. Cdk5 phosphorylation of ErbB4 is required for tangential migration of cortical interneurons. Cereb. Cortex. 2015;25:991–1003. doi: 10.1093/cercor/bht290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico B., Marín O. Neuregulin signaling, cortical circuitry development and schizophrenia. Curr. Opin. Genet. Dev. 2011;21:262–270. doi: 10.1016/j.gde.2010.12.010. [DOI] [PubMed] [Google Scholar]
- Rodríguez C.I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., Dymecki S.M. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Sánchez-Alcañiz J.A., Haege S., Mueller W., Pla R., Mackay F., Schulz S., López-Bendito G., Stumm R., Marín O. Cxcr7 controls neuronal migration by regulating chemokine responsiveness. Neuron. 2011;69:77–90. doi: 10.1016/j.neuron.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A., Mao C.A., Colasante G., Nini A., Klein W.H., Broccoli V. Tbr2-positive intermediate (basal) neuronal progenitors safeguard cerebral cortex expansion by controlling amplification of pallial glutamatergic neurons and attraction of subpallial GABAergic interneurons. Genes Dev. 2010;24:1816–1826. doi: 10.1101/gad.575410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes W.C., Rosen B., West A.P., Koutsourakis M., Bushell W., Iyer V., Mujica A.O., Thomas M., Harrow J., Cox T. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonuga-Barke E.J., Lasky-Su J., Neale B.M., Oades R., Chen W., Franke B., Buitelaar J., Banaschewski T., Ebstein R., Gill M. Does parental expressed emotion moderate genetic effects in ADHD? An exploration using a genome wide association scan. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2008;147B:1359–1368. doi: 10.1002/ajmg.b.30860. [DOI] [PubMed] [Google Scholar]
- Squarzoni P., Oller G., Hoeffel G., Pont-Lezica L., Rostaing P., Low D., Bessis A., Ginhoux F., Garel S. Microglia modulate wiring of the embryonic forebrain. Cell Rep. 2014;8:1271–1279. doi: 10.1016/j.celrep.2014.07.042. [DOI] [PubMed] [Google Scholar]
- Stumm R.K., Zhou C., Ara T., Lazarini F., Dubois-Dalcq M., Nagasawa T., Höllt V., Schulz S. CXCR4 regulates interneuron migration in the developing neocortex. J. Neurosci. 2003;23:5123–5130. doi: 10.1523/JNEUROSCI.23-12-05123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumm R., Kolodziej A., Schulz S., Kohtz J.D., Höllt V. Patterns of SDF-1alpha and SDF-1gamma mRNAs, migration pathways, and phenotypes of CXCR4-expressing neurons in the developing rat telencephalon. J. Comp. Neurol. 2007;502:382–399. doi: 10.1002/cne.21336. [DOI] [PubMed] [Google Scholar]
- Swinnen N., Smolders S., Avila A., Notelaers K., Paesen R., Ameloot M., Brône B., Legendre P., Rigo J.M. Complex invasion pattern of the cerebral cortex bymicroglial cells during development of the mouse embryo. Glia. 2013;61:150–163. doi: 10.1002/glia.22421. [DOI] [PubMed] [Google Scholar]
- Tanaka D.H., Yanagida M., Zhu Y., Mikami S., Nagasawa T., Miyazaki J., Yanagawa Y., Obata K., Murakami F. Random walk behavior of migrating cortical interneurons in the marginal zone: time-lapse analysis in flat-mount cortex. J. Neurosci. 2009;29:1300–1311. doi: 10.1523/JNEUROSCI.5446-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka D.H., Mikami S., Nagasawa T., Miyazaki J., Nakajima K., Murakami F. CXCR4 is required for proper regional and laminar distribution of cortical somatostatin-, calretinin-, and neuropeptide Y-expressing GABAergic interneurons. Cereb. Cortex. 2010;20:2810–2817. doi: 10.1093/cercor/bhq027. [DOI] [PubMed] [Google Scholar]
- Tasic B., Menon V., Nguyen T.N., Kim T.K., Jarsky T., Yao Z., Levi B., Gray L.T., Sorensen S.A., Dolbeare T. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham T.N., Lazarini F., Franceschini I.A., Lachapelle F., Amara A., Dubois-Dalcq M. Developmental pattern of expression of the alpha chemokine stromal cell-derived factor 1 in the rat central nervous system. Eur. J. Neurosci. 2001;13:845–856. doi: 10.1046/j.0953-816x.2000.01451.x. [DOI] [PubMed] [Google Scholar]
- Tidcombe H., Jackson-Fisher A., Mathers K., Stern D.F., Gassmann M., Golding J.P. Neural and mammary gland defects in ErbB4 knockout mice genetically rescued from embryonic lethality. Proc. Natl. Acad. Sci. USA. 2003;100:8281–8286. doi: 10.1073/pnas.1436402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting A.K., Chen Y., Wen L., Yin D.M., Shen C., Tao Y., Liu X., Xiong W.C., Mei L. Neuregulin 1 promotes excitatory synapse development and function in GABAergic interneurons. J. Neurosci. 2011;31:15–25. doi: 10.1523/JNEUROSCI.2538-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiveron M.C., Rossel M., Moepps B., Zhang Y.L., Seidenfaden R., Favor J., König N., Cremer H. Molecular interaction between projection neuron precursors and invading interneurons via stromal-derived factor 1 (CXCL12)/CXCR4 signaling in the cortical subventricular zone/intermediate zone. J. Neurosci. 2006;26:13273–13278. doi: 10.1523/JNEUROSCI.4162-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay R., Lee S., Rudy B. GABAergic interneurons in the neocortex: From cellular properties to circuits. Neuron. 2016;91:260–292. doi: 10.1016/j.neuron.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F., Kellendonk C., Kretz O., Gass P., Anlag K., Orban P.C., Bock R., Klein R., Schütz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- van den Berghe V., Stappers E., Vandesande B., Dimidschstein J., Kroes R., Francis A., Conidi A., Lesage F., Dries R., Cazzola S. Directed migration of cortical interneurons depends on the cell-autonomous action of Sip1. Neuron. 2013;77:70–82. doi: 10.1016/j.neuron.2012.11.009. [DOI] [PubMed] [Google Scholar]
- Villar-Cerviño V., Kappeler C., Nóbrega-Pereira S., Henkemeyer M., Rago L., Nieto M.A., Marín O. Molecular mechanisms controlling the migration of striatal interneurons. J. Neurosci. 2015;35:8718–8729. doi: 10.1523/JNEUROSCI.4317-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels T.P., Abbott L.F. Gating multiple signals through detailed balance of excitation and inhibition in spiking networks. Nat. Neurosci. 2009;12:483–491. doi: 10.1038/nn.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt D., Hunt R.F., Mandal S., Sandberg M., Silberberg S.N., Nagasawa T., Yang Z., Baraban S.C., Rubenstein J.L. Lhx6 directly regulates Arx and CXCR7 to determine cortical interneuron fate and laminar position. Neuron. 2014;82:350–364. doi: 10.1016/j.neuron.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vullhorst D., Neddens J., Karavanova I., Tricoire L., Petralia R.S., McBain C.J., Buonanno A. Selective expression of ErbB4 in interneurons, but not pyramidal cells, of the rodent hippocampus. J. Neurosci. 2009;29:12255–12264. doi: 10.1523/JNEUROSCI.2454-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J., Kern-Veits B., Huf J., Stolze B., Bonhoeffer F. Recognition of position-specific properties of tectal cell membranes by retinal axons in vitro. Development. 1987;101:685–696. doi: 10.1242/dev.101.4.685. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li G., Stanco A., Long J.E., Crawford D., Potter G.B., Pleasure S.J., Behrens T., Rubenstein J.L. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron. 2011;69:61–76. doi: 10.1016/j.neuron.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonders C.P., Anderson S.A. The origin and specification of cortical interneurons. Nat. Rev. Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Wu P.R., Cho K.K., Vogt D., Sohal V.S., Rubenstein J.L. The cytokine CXCL12 promotes basket interneuron inhibitory synapses in the medial prefrontal cortex. Cereb. Cortex. 2016 doi: 10.1093/cercor/bhw230. Published online August 6, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Tam M., Anderson S.A. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J. Comp. Neurol. 2008;506:16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- Xue M., Atallah B.V., Scanziani M. Equalizing excitation-inhibition ratios across visual cortical neurons. Nature. 2014;511:596–600. doi: 10.1038/nature13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau H.J., Wang H.F., Lai C., Liu F.C. Neural development of the neuregulin receptor ErbB4 in the cerebral cortex and the hippocampus: preferential expression by interneurons tangentially migrating from the ganglionic eminences. Cereb. Cortex. 2003;13:252–264. doi: 10.1093/cercor/13.3.252. [DOI] [PubMed] [Google Scholar]
- Yizhar O., Fenno L.E., Prigge M., Schneider F., Davidson T.J., O’Shea D.J., Sohal V.S., Goshen I., Finkelstein J., Paz J.T. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarbalis K., Choe Y., Siegenthaler J.A., Orosco L.A., Pleasure S.J. Meningeal defects alter the tangential migration of cortical interneurons in Foxc1hith/hith mice. Neural Dev. 2012;7:2. doi: 10.1186/1749-8104-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A., Muñoz-Manchado A.B., Codeluppi S., Lönnerberg P., La Manno G., Juréus A., Marques S., Munguba H., He L., Betsholtz C. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- Zhang D., Sliwkowski M.X., Mark M., Frantz G., Akita R., Sun Y., Hillan K., Crowley C., Brush J., Godowski P.J. Neuregulin-3 (NRG3): a novel neural tissue-enriched protein that binds and activates ErbB4. Proc. Natl. Acad. Sci. USA. 1997;94:9562–9567. doi: 10.1073/pnas.94.18.9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.









