Abstract
Self-assembling coiled coils, which occur commonly in native proteins, have received significant interest for the design of new biomaterials-based medical therapies. Considerable effort over recent years has led to a detailed understanding of the self-assembly process of coiled coils, and a diverse collection of strategies have been developed for designing functional materials using this motif. The ability to engineer the interface between coiled coils allows one to achieve variously connected components, leading to precisely defined structures such as nanofibers, nanotubes, nanoparticles, networks, gels, and combinations of these. Currently these materials are being developed for a range of biotechnological and medical applications, including drug delivery systems for controlled release, targeted nanomaterials, “drug-free” therapeutics, vaccine delivery systems, and others.
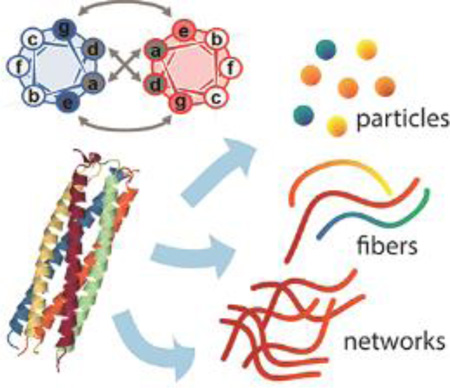
Graphical/Visual Abstract and Caption
Coiled coils are oligomerizing structures useful for designing nanomaterials such as particles, fibers, and networks within biomedical applications.
The Coiled Coil
The self-assembly of synthetic or expressed polypeptide materials has been a major approach for developing biologically active materials for tissue engineering, drug delivery, immunotherapies, controlled release, 3D cell culture, and other applications. Self-assembly is controlled by non-covalent interactions, including van der Waals forces, electrostatic effects, hydrophobicity, hydrogen bonding, metal-ligand, or π-π staking interactions.1–3 A wide range of self-assembling materials have been generated in a bottom-up fashion using different biomolecular components including nucleic acids,4–6 liposomes,7, 8 polymersomes,9–11 polypeptides,12, 13 and polypeptide-polymer conjugates14. Inspired by natural protein assembly, synthetic and expressed polypeptides have received particular attention, as they have the advantages of having well defined folded and oligomerized structures, generally straightforward synthesis processes, and the entire range of canonical, post translationally modified, and synthetic amino acids to choose from for installing function.
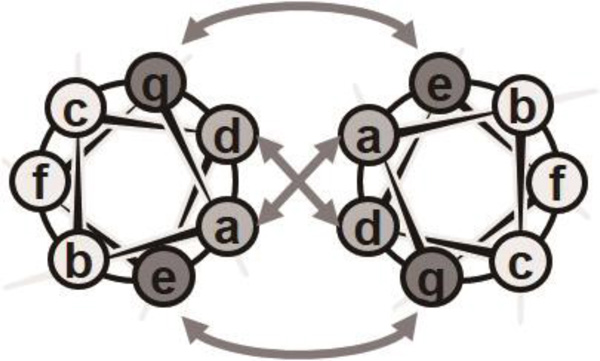
Coiled coils, consisting of two or more α-helices wrapped around each other into a superhelical bundle, are ubiquitous protein structural motifs associated with a range of functions in nature.15, 16 The amino acid sequences of peptides forming coiled coil bundles are characterized by a heptad repeating unit denoted as (abcdefg)n, where n is the number of repeats. The interaction between helices reduces the typical α-helical pitch from 3.6 to 3.5 residues per turn, creating an interfacial stripe between associating helices, where residues in the a and d positions are typically hydrophobic (Figure 1).17 This forms the core of the coiled coil via the packing of hydrophobic amino acids, and this core is stabilized by both hydrophobic and van der Waals interactions. Importantly, the type of hydrophobic amino acids present in the interface can specify the number of helices in each superhelix bundle.18, 19 The e and g positions tend to be occupied by polar/charged amino acids, together creating complementary charge pairs across the bundle, and these help stabilize the coiled coil via interstrand electrostatic interactions.20 Both the hydrophobic interactions arising from residues in the a and d positions and the electrostatic interactions between residues in the e and g positions can be utilized to influence the oligomerization state, parallel versus antiparallel topology, registry, and thermodynamic stability of bundles. Residues in the b, c, and f positions can be used to provide sufficient solubility of the peptides, as well as to control the higher-order aggregation of oligomers via the exterior surface of the coiled coil.
Figure 1.
Helical wheel diagram showing the positions of the (abcdefg) heptad repeat residues in a parallel coiled coil; the arrows indicate the hydrophobic interactions between positions a and d and the electrostatic interactions between positions e and g.
Beyond engineering the hydrophobic core and ionic attractions between e and g residues, other strategies for adjusting the specificity of coiled-coil interactions have been developed by several groups. Keating’s research group analyzed the interactions of coiled coils within 49 human basic leucine zipper domain proteins (bZIP) using a microscale protein array technique.21 They then utilized the information obtained to develop computer algorithms for predicting the specificity of coiled coil interactions.19, 22, 23 These studies revealed several guidelines for specifying the assembly of coiled coils. One was that an asparagine in an a position is strongly favored to interact with another asparagine in the same position of an opposing helix (unless the residue is close to the end of helix).19 Keating’s group also utilized a peptide microarray technique to measure the interactions of 48 synthetic coiled coils and 7 human bZIP coiled coils, discovering orthogonal pairs, orthogonal triplets, and hub-type networks.24 Crystal structures of the orthogonal pairs suggested that the interactions of the coiled coil pairs involved both polar and charged residues, including paired asparagines at a-a’ positions and partially buried water molecules between coiled coil pairs.24 Asparagine pairing has been exploited recently for the de novo design of orthogonal heterodimeric coiled coil peptides.25, 26 Interaction between buried asparagines can stabilize paired heptads, but asparagine and isoleucine/leucine interactions are strongly penalized, also enabling the construction of “in-register” assembling structures.26 Even further, Baker’s group developed an algorithm (HBNet) to find protein oligomers with all the polar atoms on side chains stabilized by hydrogen-bonding networks, and they designed a series of oligomers with two-ring topologies supported by the hydrogen-bond network.27 Most of the oligomers assembled into controlled oligomeric states with high precision, and 90% of the assemblies were stable at temperatures as high as 95°C.27 The hydrogen-bond network design resembled DNA base-pairing in some respects, and it showed great potential for generating complex protein-based materials with programmable structures. All of these design rules for controlling coiled coil oligomerization provide tremendous flexibility for creating materials with tailored morphologies and functionalities.
Coiled coils can also be engineered to assemble and disassemble conditionally. For example, by adjusting the amino acids at position a and d, and/or the intrahelical interactions between e and g residues, bundles can be made to be responsive to environmental factors including pH and temperature.28–37 As an early example, Graddis and coworkers designed a heterodimeric peptide coiled coil self-assembly system, (VSSLESE)5 (E/E35) and (VSSLKSK)5 (K/K35).30 These two peptides could form heterodimeric coiled coil assemblies at pH values ranging from 5 to 7, but E/E35 formed homodimeric coiled coils at pH 5, and K/K35 formed homodimeric coiled coils at pH 4.30 To engineer the thermal stability of coiled coils, Vinson and coworkers designed a series of heterodimerizing peptides with various pairs of electrostatic attractive amino acids in the e and g positions.37 By adjusting the charged residues and peptide lengths, they achieved a wide range of melting temperatures, between 66 °C and 90 °C.37
The ability to specify the structure and stability of coiled coils gives them unique advantages over other options for supramolecular materials such as β-sheet fibrillizing peptides, peptide amphiphiles, and proteins. The simple design rules available for coiled coils and the ability to specify oligomerization number, strand topology, lateral association, and registry allow one to create a diverse range of elegant nanostructures, examples of which will be described below. Moreover, the ability to finely tune the stability of the materials is a property that can be less accessible in other self-assembling peptide platforms. For example β-sheet fibrillizing peptides tend to form structures with high degrees of stability, hindering the degradation of these peptides in biological applications, whereas in contrast the thermal stability and pH stability of coiled coil peptides can be adjusted within a wide range.37–40 Although β-sheet polypeptides or peptide conjugates can be engineered to yield different supramolecular morphologies,41, 42 this process still lacks the inherent flexibility and predictability of coiled coil peptide design. Additionally, although many functional and biologically benign synthetic and native amyloids exist, some β-sheet nanofibers are associated with pathologies, and rules for separating pathological from non-pathological amyloids are evolving,43–46, whereas coiled coil materials avoid this concern altogether. Despite the advantages of coiled coils, they do possess some drawbacks. For example, they typically have longer amino acid sequences than other self-assembling peptide systems owing to the typical requirement of 3 or 4 heptad repeating units per peptide chain for stable self-assembly. This can contribute to relatively low yields in solid phase peptide synthesis and purification or difficulty during protein expression.
The object of this review is to highlight recent development of materials based on coiled coil peptide self-assembly, as well as steps towards therapeutic applications for these materials. Our main focus will be on synthetic peptides and their corresponding biomedical applications, but some particularly relevant protein-based systems and interesting peptide/polymer hybrid systems will also be discussed in some sections to illustrate the versatility of coiled coil-based materials design.
EMERGING COILED COIL PEPTIDE SELF-ASSEMBLING MATERIALS
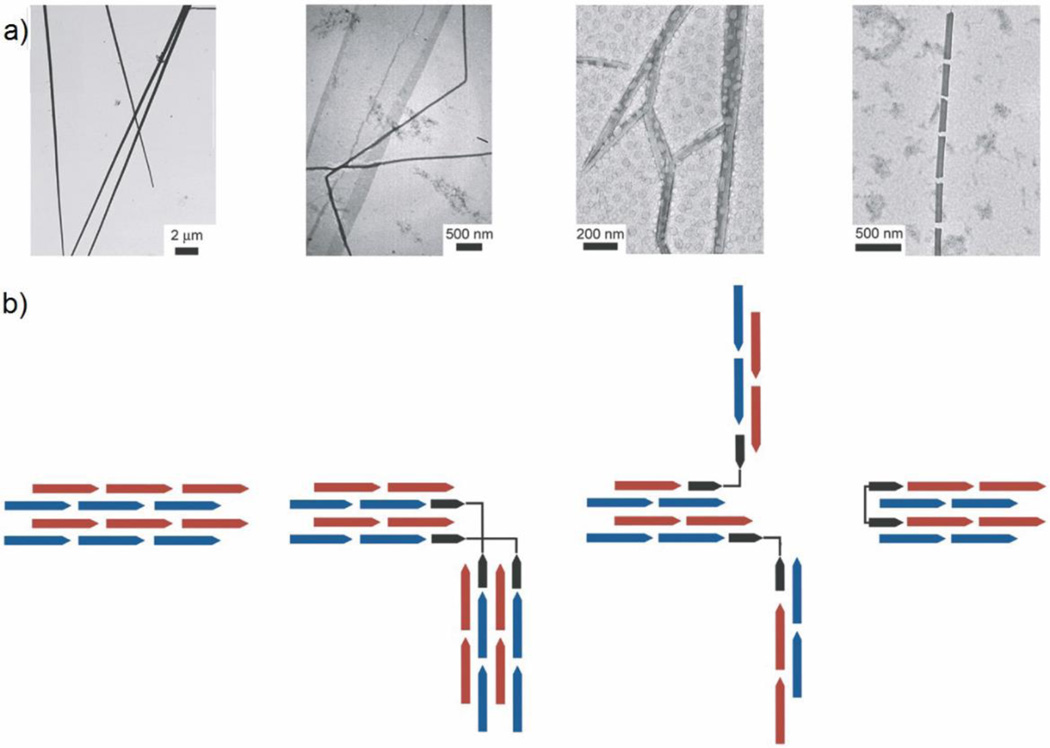
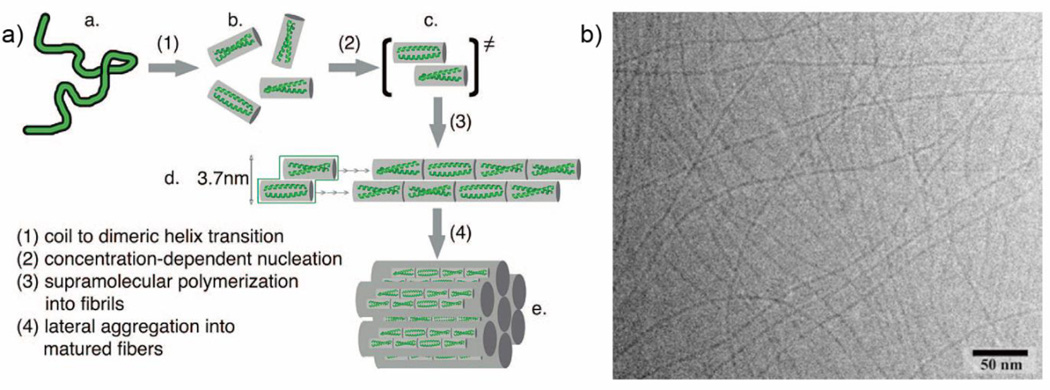
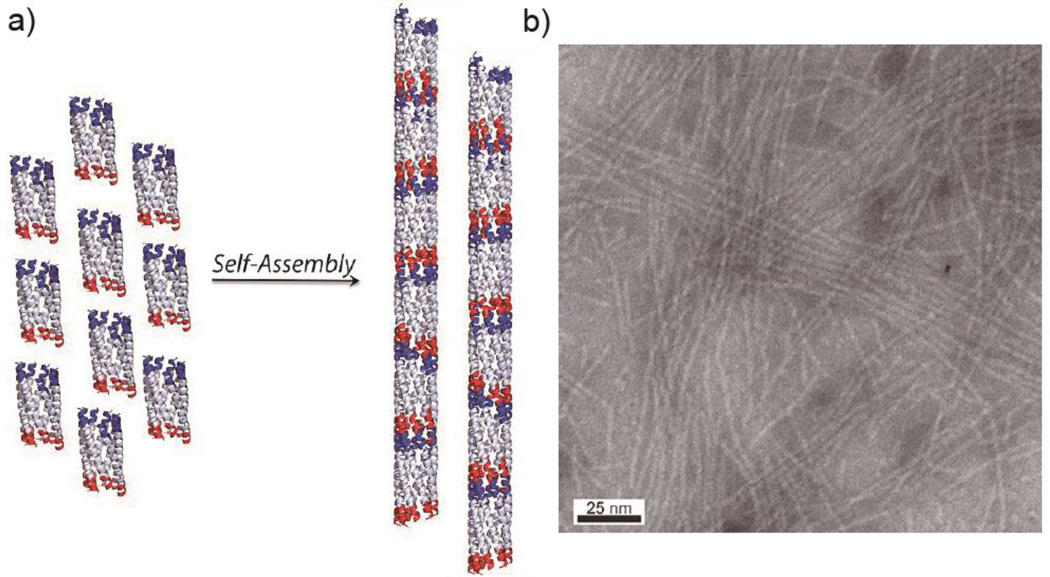
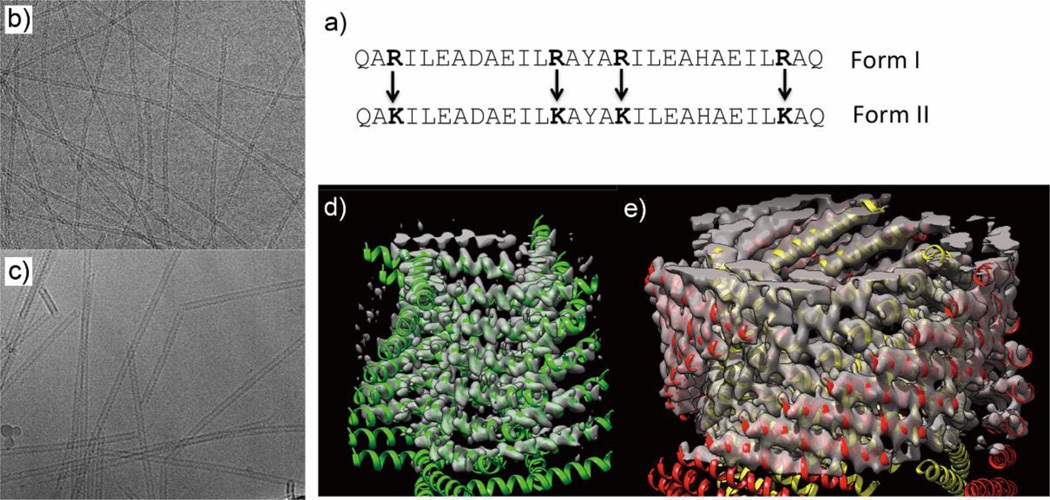
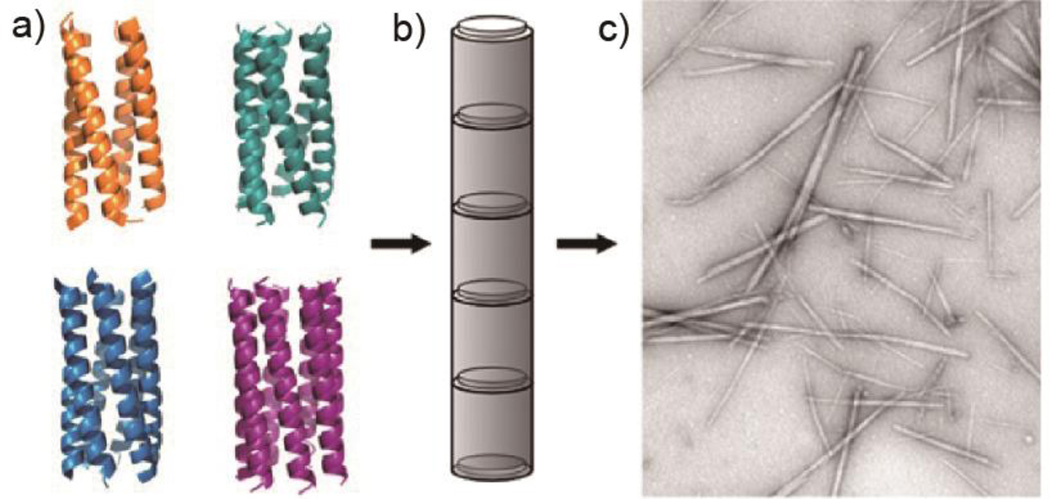
As the α-helical coiled coil motif exists in a range of native proteins, including fibrillar structures such as the cytoskeleton and extracellular matrix,47 it has naturally been an attractive synthesis target for the de novo design of oligomerizing materials. One of the first coiled coil fibrous biomaterials to be introduced was the self-assembling fiber (SAF) system designed by the Woolfson research group.48 These peptides ((KIppLKp)2(EIppLEp)2) have hydrophobic isoleucine and leucine residues at positions a and d respectively, which promote helix oligomerization (“p” stands for polar residues here). In addition, lysine and glutamic acid are incorporated at positions e and g to yield positively charged N-termini and negatively charged C-termini, which allow for staggered, “sticky ended” assembly.48, 49 This modular design offered a variety of opportunities to engineer different morphologies into the peptide fibers.50, 51 By rearranging the charged heptad units or introducing branched or dendritic linker peptides, kinks and branches could be induced in the nanofiber, along with segmented fiber morphology (Figure 2).51 Hartgerink and coworkers designed a series of 21 amino-acid peptides with isoleucine and leucine at positions a and d. Incorporating glutamic acid residues in positions e and g allowed the self-assembly process to be controlled via pH adjustment.52 Woolfson and coworkers have also designed pH-responsive coiled coils.53 More importantly, by changing the amino acids at position c, b, and f, the thickness and length of the resulting fiber could be controlled (Figure 3). For example, the inclusion of tyrosine at position f reduced the length of the nanofiber by undermining the fiber integrity and destabilizing the helix.52 Xu et al. designed a helical peptide (7HSAP1) based on GCN4-pAA54, and included a “lock washer” region to induce nanotube formation (Figure 4).55 It was hypothesized that the 7-helix bundle, GCN4-pAA, was unable to assemble into a higher-order structure because the presence of arginine at the d position broke the hydrophobic periodicity of the heptad repeating unit. To test this hypothesis, the single arginine was replaced with a hydrophobic leucine residue in the 7HSAP1 peptide. To maximize the electrostatic attraction, glutamic acid and lysine were used in positions b and c, respectively. Consequently, the obtained helical peptide was able to assemble into nanofibers with diameters of 3 nm according to cryoTEM imaging.55 The coiled coil domain of cartilage oligomeric matrix protein (COMP) has also been an attractive model for coiled coil nanofiber design.56–58 Hume et al. designed pentameric coiled coil protein fibers by engineering COMP coiled coil mutations. The fiber, with a diameter of 20−560 nm, was found to have a melting temperature of 46.4−63.5°C across different pH values.58 Conticello and coworkers recently developed a series of de novo coiled coil peptides with two hydrophobic faces, self-assembling into nanotubes in a fashion similar to TolC protein, an outer-membrane protein of E. coli (Figure 5).59, 60 The TolC protein is composed of a 12-stranded anti-parallel β-barrel and a 12-stranded anti-parallel α-helical cylindrical tube.59 The synthetic peptides recapitulated the heptad repeat of TolC, by replacing larger residues at positions a and f with alanine to lower the degree of curvature between adjacent helices, and by introducing charged residues at positions b and e to favor the formation of cylindrical structures. The nanotubes formed by the designed peptides had a unique structure where the helices ran perpendicular to the axis of the overall fiber (Figure 5).60 Woolfson and coworkers recently presented modularly designed peptide-based nanotubes (PNTs) with blunt ended α-helical barrels (Figure 6).61, 62 Without the assistance of sticky ends or lock washer building blocks, the researchers tested a wide range of coiled coil helices, including pentamers, hexamers, and a heptamer, identifying a 28-residue hexameric α-helical barrel unit (CC-Hex-T) that can self-organize end-to-end to yield nanofibers.61 Furthermore, by introducing lysine at the f position of CC-Hex-T, nanotube thickening could be reversibly adjusted by varying the pH conditions. The stability of the achieved nanotube was also improved by covalent conjugation via native chemical ligation.62
Figure 2.
Different morphologies created using the “sticky ended” self-assembling fiber (SAF) system of Woolfson. a) TEM images of (left to right) straight, kinked, branched, and segmented coiled coil fibers. b) Schematic representations of the different assembly models, where red and blue arrows represent sticky ended dimeric coiled coil peptides and black arrows represent peptides for introducing structural features into the nanofiber morphology. Adapted from Ref. 51, Copyright 2006, with permission from Elsevier.
Figure 3.
α-helical coiled coil peptide nanofibers designed by Hartgerink and coworkers. a) Proposed mechanism of self-assembly into fibers. b) Representative cryoTEM image of self-assembling nanofibers. Adapted with permission from Ref. 52, Copyright 2008, American Chemical Society.
Figure 4.
Schematic representation and STEM image of coiled coil “lock washer” nanotubes. a) Schematic of lock washer self-assembly using peptides derived from the 7-helix bundle of 7HSAP1, with negatively charged C-terminal residues (glutamic acid) in red and positively charged N-terminal residues (lysine) in blue. b) STEM image of 7HSAP1 nanotubes formed in MES buffer (10mM, pH 6.0) Adapted with permission from Ref. 55, Copyright 2013, American Chemical Society.
Figure 5.
Coiled coil nanotubes based upon peptides inspired by TolC protein, designed by Conticello and coworkers. a) 29-amino acid peptide sequences in two forms, one having arginines in the positions indicated (Form I) and the other containing lysines in those positions (Form II). CryoTEM image (b) and 3D structure (d) of Form I nanotubes; cryoTEM image (c) and 3D structure (e) of Form II nanotubes. Adapted from Ref. 60, Copyright 2006, with permission from Elsevier.
Figure 6.
Peptide-based nanotubes (PNTs) formed from coiled coils. a) Ribbon diagrams of coiled coil assemblies: tetramers (orange; 3R4A), pentamers (Cyan; 4PN8), hexamers (blue; 3R3K), and heptamers (purple; 4PNA). b) Schematic of helical barrel assembly into PNT. c) TEM of PNTs formed from hexamers. Adapted with permission from Ref. 61, Copyright 2015, American Chemical Society.
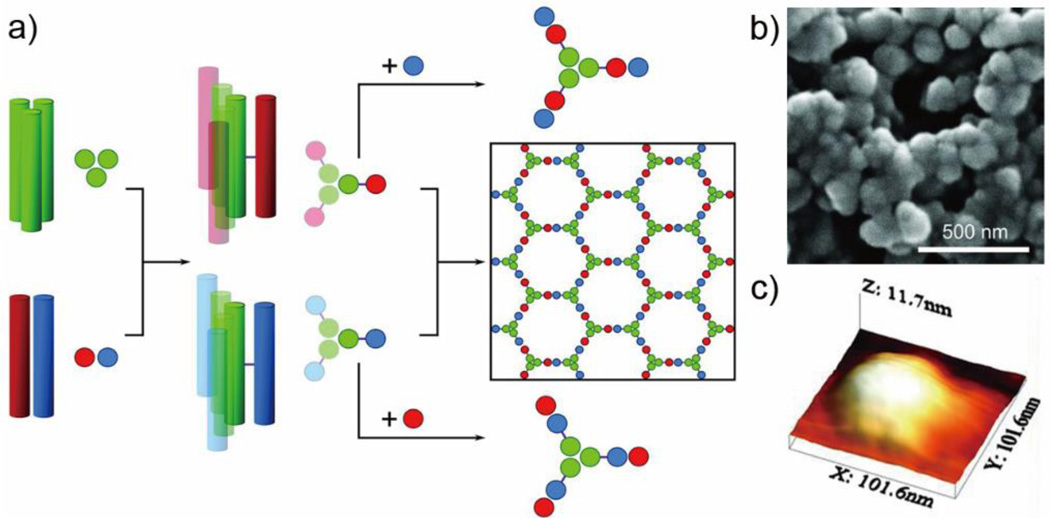
In addition to peptide fiber/nanotube construction, the coiled coil can also be used to build more complicated, three-dimensional structures. For example, the Woolfson group demonstrated a self-assembled cage using two complementary peptide-hubs.63 Homotrimeric coiled coil assemblies (CC-Tri3) were linked with disulfide bonds to either heterodimeric acidic α-helix (CC-Di-A), yielding hub A (CC-Tri3-CC-Di-A), or to heterodimeric basic α-helix (CC-Di-B), yielding hub B (CC-Tri3-CC-Di-B). Upon mixing in equal molar ratios, hub A and hub B assembled and formed closed peptide cages with a diameter of about 97 nm (Figure 7). Atomic force microscopy was employed to characterize the morphology of deposited dried particles, which appeared to be flattened disks with thicknesses of 9.2 ± 1.0 nm and diameters of 95 nm. This suggested that the spheres formed in solution were hollow and unilamellar, rather than “onion-like” in structure. The maximization of coiled coil interactions was proposed to be the driving force for formation of the closed cages, and curvature of the hubs could also facilitate cage formation.63
Figure 7.
Self-assembling cages from coiled coil peptides. a) The design scheme of self-assembling cages. Homotrimeric coiled coils (CC-Tri3, green) and dimeric coiled coils (CC-Di-A, red; CC-Di-B, blue) from Hub A and Hub B by covalently conjugating CC-Tri3 with CC-Di-A and CC-Di-B respectively. Mixing Hub A and Hub B yielded a hexagonal network, while combining Hub A with CC-Di-B or combining Hub B with CC-Di-A yields discrete structures. b) SEM images of cages assembled in PBS. c) Three-dimensional representation of a single cage as measured by AFM. Reproduced with permission from Ref 63. Copyright 2013, AAAS.
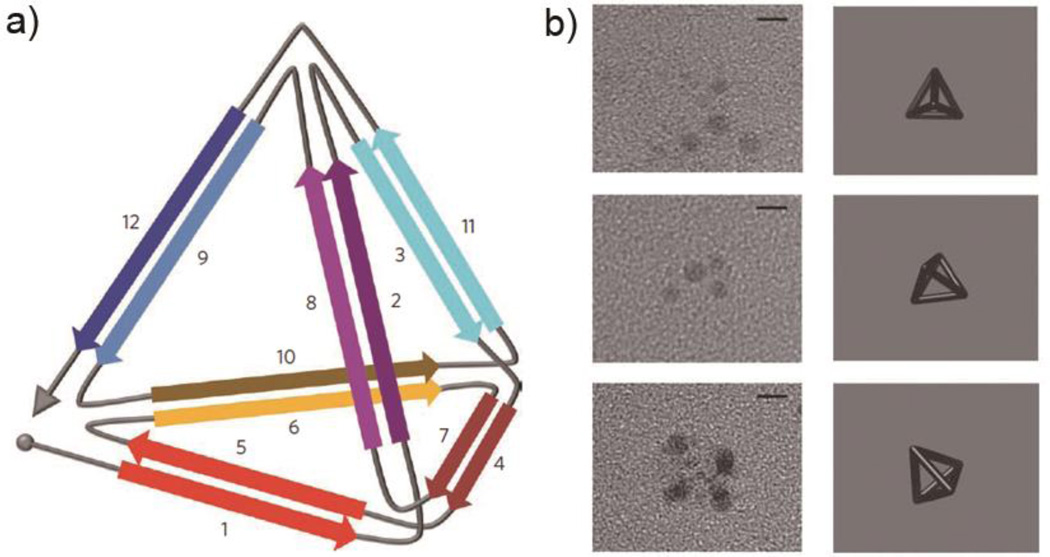
In another approach for creating defined nanoscale objects using coiled coils, Jerala and colleagues developed a strategy to form self-assembled three-dimensional tetrahedrons using single-chain coiled coil polypeptides (Figure 8).64 In this approach, an expressed protein contains 12 concatenated coiled coil dimer-forming segments connected via flexible linkers. The 12 segments form parallel homodimers (segments 1 and 5, 3 and 11), parallel heterodimers (2 and 8, 6 and 10, 9 and 12), and a parallel homodimer (4 and 7). These segments are cleverly connected in an “Eulerian trail” fashion, which is a path that connects each of the two vertices by exactly two connections. The hydrodynamic diameter of the folded tetrahedron was measured by dynamic light scattering (DLS) to be about 7 nm. This was much more compact than the unfolded peptide, which had a diameter of about 20 nm. The tetrahedron was observed using TEM with uranyl acetate, which selectively stained the vertices (Figure 8b).64
Figure 8.
Single chain polypeptide tetrahedron. a) Schematic representation of the self-assembled tetrahedron designed by Jerala and colleagues. Design elements contained antiparallel coiled coil homodimers (1–5, 3–11), parallel heterodimers (2–8, 6–10, and 9–12), and a parallel homodimer (4–7). b) TEM images and three-dimensional projections of tetrahedra with different orientations, with uranyl acetate staining of the vertices. Reproduced with permission from Ref 64, copyright 2013, Macmillan Publishers Ltd.
COILED-COIL PEPTIDE SELF-ASSEMBLY AS DRUG DELIVERY SYSTEMS
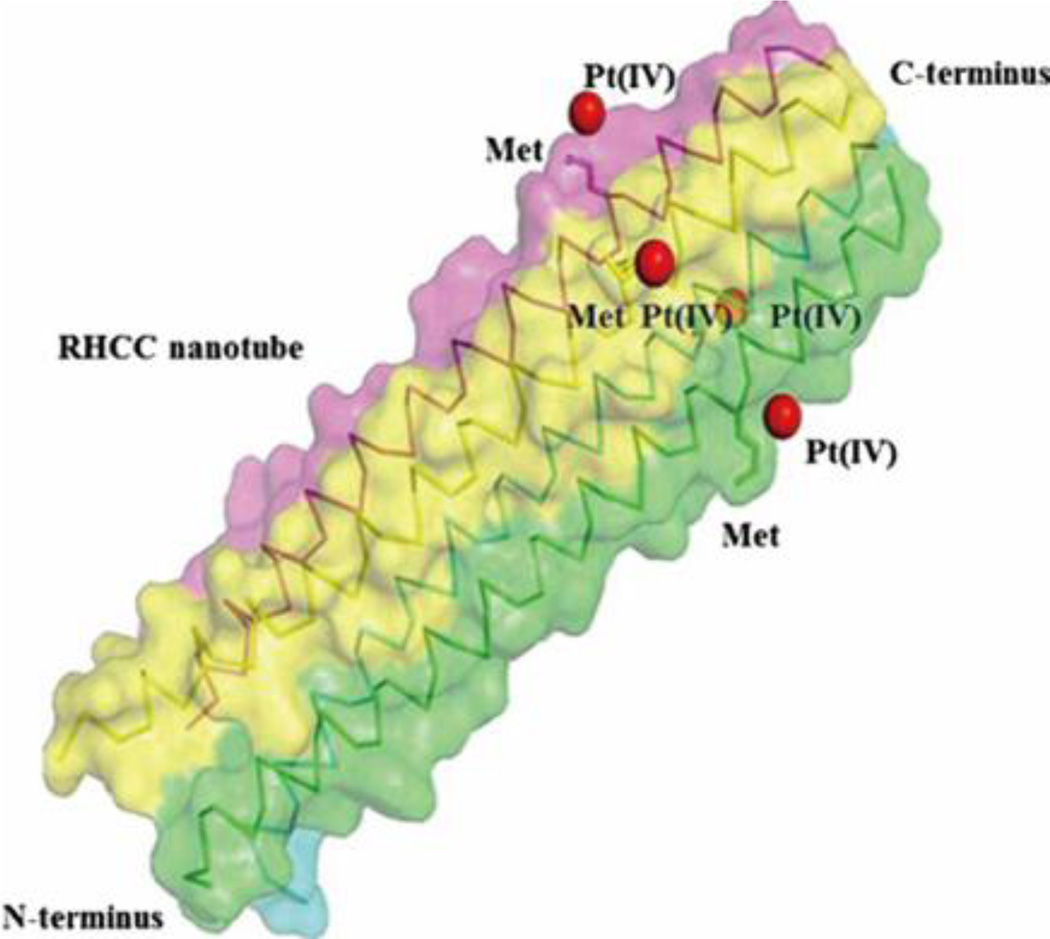
To improve the pharmacokinetic profile of a range of drugs, different carriers have been explored to prolong circulation time or to realize targeted release. Coiled coils have attracted attention for these purposes owing to their pH-sensitivity, temperature-sensitivity, and ability to carry hydrophobic drugs within their hydrophobic cores. Eriksson et al. first demonstrated that the right-handed coiled coil (RHCC) domain from Staphylothermus marinus (20 kDa) could be utilized as a carrier for the chemotherapeutic drug cisplatin (CP).65 It was determined that, on average, RHCC could incorporate about one cisplatin molecule per cavity in each coiled coil tetramer. Both RHCC and the acquired CP-RHCC complex could be internalized by various cancer cell lines, but the CP-RHCC complex showed elevated in vitro cytotoxic effects on multiple tested cell lines in comparison with cisplatin alone, while RHCC exhibited no effect on cells.65 Thanasupawat et al. recently utilized RHCC as a platinum (IV) prodrug delivery platform for glioblastoma treatment (Figure 9).66 The Pt(IV)-RHCC complex showed improved chemotherapeutic efficacy and selectivity toward human glioblastoma cells at up to 20-fold lower concentration of Pt(IV), as compared to Pt(IV) prodrug alone. Kinetically inert Pt(IV) prodrug can be biologically reduced to Pt(II) in a hypoxic extracellular tumor environment, which leads to its improved tolerability.67
Figure 9.
Side view of the RHCC protein tetramer containing Pt(IV) prodrug, where the RHCC protein is shown by van der Waals spheres and ribbon representation, with different colors for each helical chain, and the Pt(IV) prodrugs are represented by red spheres. Reproduced with permission from Ref 66, copyright 2015, Elsevier.
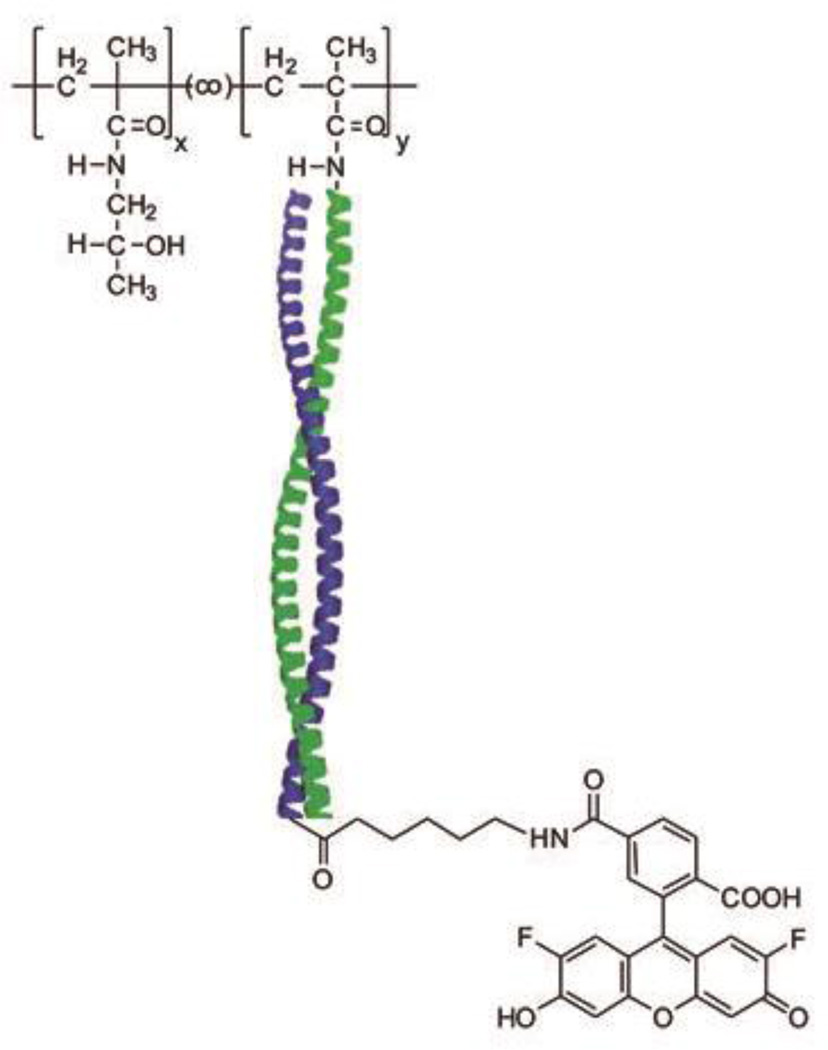
Apostolovic et al. demonstrated that coiled coils could be employed to construct a polymer-drug conjugate system (Figure 10).68, 69 The E3 ((IAALEKE)2IAALEKG) / K3 ((IAALKEK)2IAALKEG) coiled coil dimer was found to be stable at pH 7, but unfolded at pH 5,70 making it a promising linker to release drug in the late endosome or lysosome once internalized via endocytosis. Polymer-drug conjugates were prepared by mixing MTX-functionalized E3 peptide and K3 conjugated to N-(2-hydroxypropyl)methacrylamide (HPMA) copolymers.69 Fluorescence-activated cell sorting (FACS) experiments showed the uptake of polymer-drug conjugates by B16F10 cells. In vitro Förster Resonance Energy Transfer (FRET) experiments also suggested that cargo molecules were released intracellularly after endocytosis.68
Figure 10.
Schematic representation of the structure of noncovalent poly(N-(2-hydroxypropyl)methacrylamide) (PHPMA)-based polymer/fluorescent dye complex. Oregon Green is the model cargo for delivery. The subscripts x and y represent the mole fractions of HPMA and peptide methacrylate repeat units in the final copolymers. Reproduced with permission from Ref. 68, Copyright 2010, American Chemical Society.
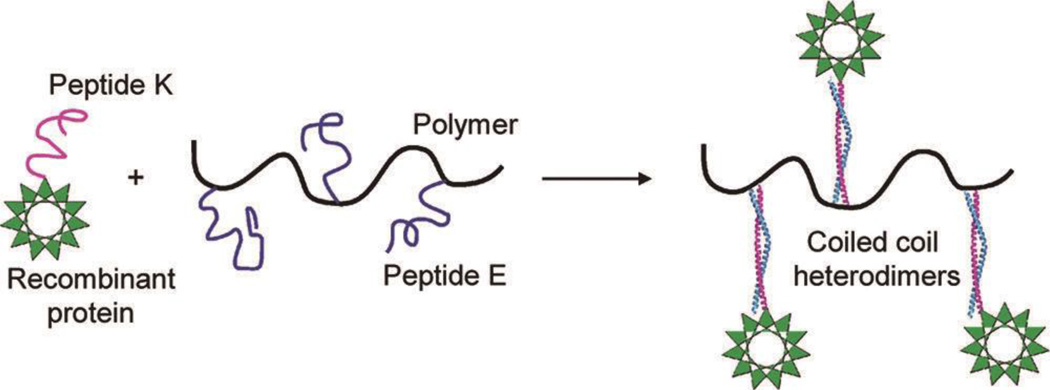
Pechar and coworkers designed peptide E ((VAALEKE)4) and peptide K ((VAALKEK)4) and utilized the resultant heterodimeric coiled coils to non-covalently bind HPMA polymers with recombinant proteins (Figure 11).71–73 Peptide K was incorporated into the recombinant single-chain variable fragment (scFv) of antibody M75 at the C-terminus.71 The ratio between scFv-K and HPMA polymers was found to be close to 1:1 for the complex.71 The scFv fragment could bind efficiently to its cognate antigen CA IX according to enzyme-linked immunosorbent assay (ELISA), allowing it to be utilized as a targeting moiety to deliver doxorubicin (DOX) to mouse BCL1 leukemia cells.72 The IC50 value of the obtained DOX-polymers for BCL1 was significantly reduced from over 140 µg/L to about 2 µg/L because of the conjugation of scFv-K.72
Figure 11.
Schematic illustration of a polymer/protein delivery complex with peptide E and peptide K as the non-covalently binding linker. Reproduced with permission from Ref. 71, Copyright 2011, American Chemical Society.
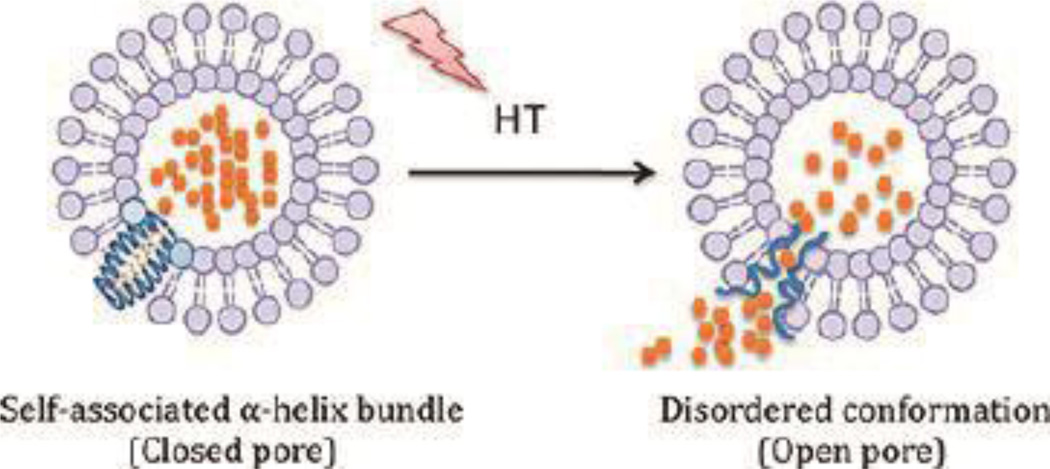
The temperature-sensitive nature of coiled coils has also been utilized in peptide-lipid hybrid liposome systems for DOX delivery.74 The leucine zipper peptide (VSSLESKVSSLESKVSKLESKKSKLESKVSKLESKVSSLESK-NH2) was incorporated into liposome bilayers to form a temperature-responsive release trigger (Figure 12). At the coiled coil melting temperature of about 42 °C, pores formed to release DOX. This was illustrated with localized hyperthermia after intravenous administration, where the amount of DOX accumulated at a heated tumor site was 3-fold higher than controls.
Figure 12.
Schematic of controlled drug release for a peptide-lipid hybrid system triggered by hyperthermia (HT), with DOX represented by orange dots. Reproduced with permission from Ref. 74, Copyright 2012, American Chemical Society.
These examples illustrate the range of different drug delivery and release technologies that can be envisioned and constructed using coiled coils.
COILED-COIL PEPTIDE NANOFIBER DECORATION FOR BIOLOGICAL APPLICATIONS
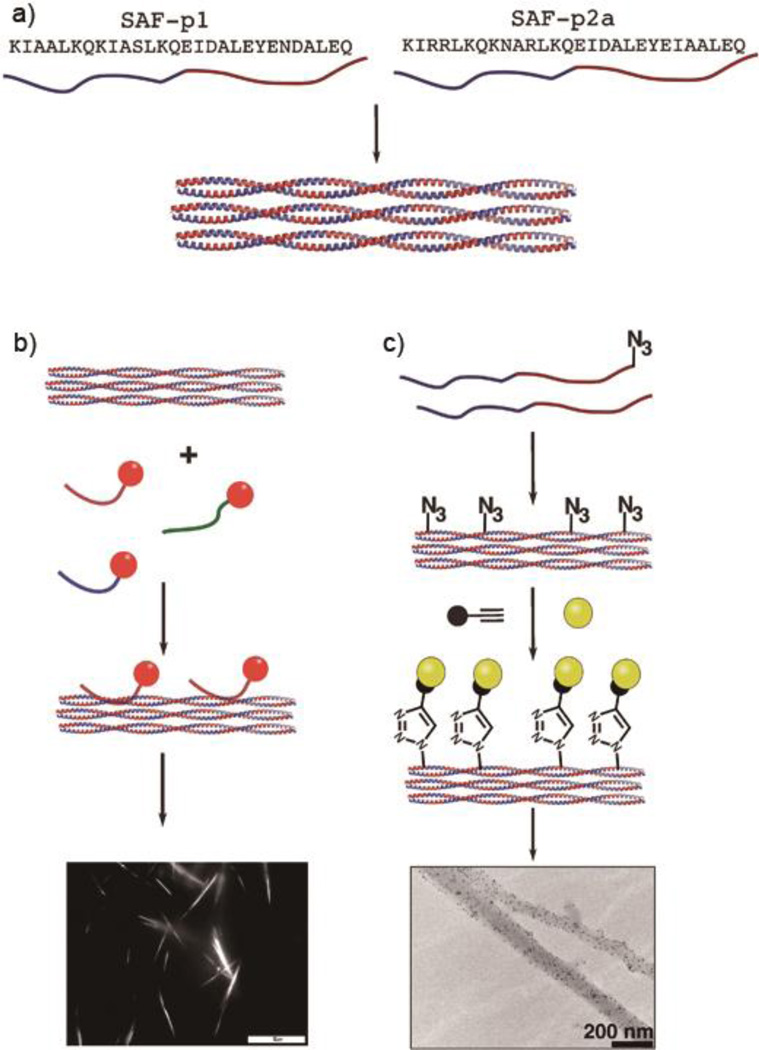
A key strategy-probably the most common strategy- for targeting nanomaterials to specific tissues or engaging them with specific biology is to functionalize them with peptide or protein ligands. However, functionalizing fibrillar coiled coil assemblies for biological applications presents some challenges, as the majority of the backbone hydrogen bonding in α-helical coiled coil fibers is restrained within local structure. Additionally, compared to other supramolecular systems such as peptide amphiphiles, β-sheet fibrillar peptides, or micellar systems, the peptide termini in most fibrous coiled coil assemblies are buried and poorly available for functionalization with bulky or diverse functional peptides. To address this problem, Woolfson and coworkers developed several strategies to decorate their aforementioned SAF systems (Figure 13).48, 50, 51, 53 Mahmoud et al. demonstrated a non-covalent labelling strategy using a negatively charged peptide (DEDEDE) as an anchor to bind to SAF, while neither neutral peptide (AQAQAQ) nor positively charged peptide (KRKRKR) exhibited strong binding ability. Noncovalent modification with fluorophores or gold nanoparticles was visualized using light microscopy and TEM, respectively.75 To avoid the potential drawback of nonspecific binding with the anchor peptide, a biocompatible “click” reaction was also employed by introducing azide functional groups directly within the SAF.76 The resulting peptides could be functionalized with a variety of functional peptides using copper-catalysed azide-alkyne click chemistry, yielding up to 100% conjugation. This approach was used to conjugate an RGDS cell-adhesion motif onto a coiled coil fiber hydrogel for the culture of PC 12 cells and enables the introduction of considerably diverse functionality.53, 77
Figure 13.
Schematic of different decoration strategies for self-assembling peptide fibers (SAFs). a) Peptide fibers are assembled by mixing equal amounts of SAF-p1 and SAF-p2a at pH 7.4. b) Non-covalent decoration using short peptides (SAF-tags). Rhodamine was attached to the N-terminus of the tag and visualized via light microscopy. c) “Click” chemistry was employed for post-assembly functionalization of fibers incorporating azide groups. The decoration was carried out by reacting the surface azide moieties with biotin-alkyne, followed by the addition of 5 nm streptavidin nanogold. The decorated nanofiber was confirmed with TEM imaging. Reproduced from Ref 76 with permission, Copyright 2010, The Royal Society of Chemistry.
COILED-COIL PEPTIDE SELF-ASSEMBLY IN A “DRUG-FREE” THERAPEUTIC SYSTEM
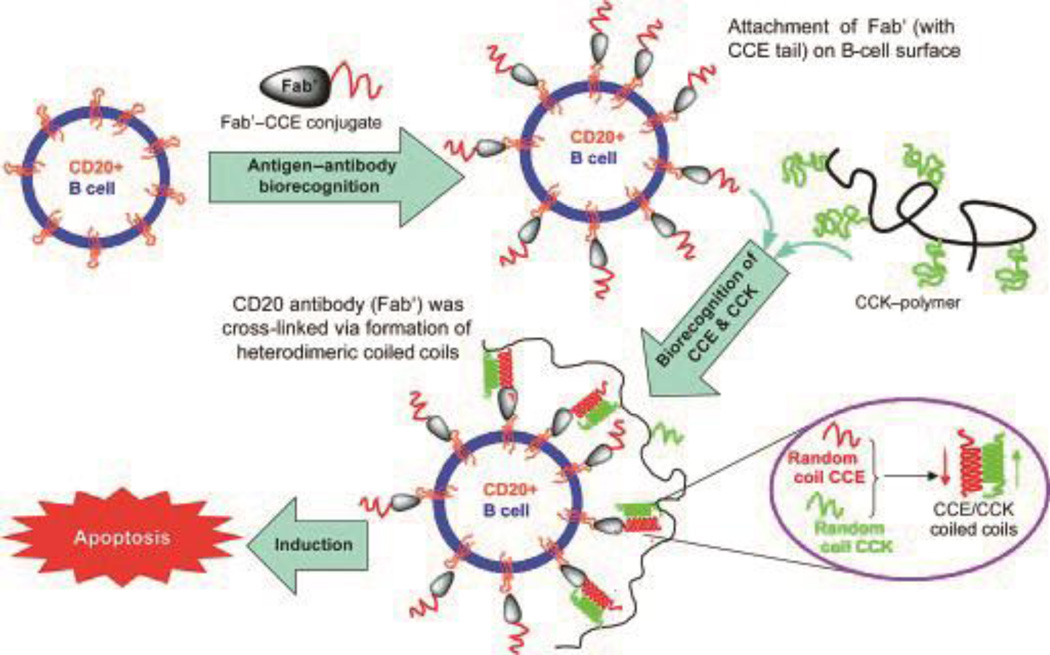
The Kopeček group pioneered a unique research area in “drug-free” macromolecular therapeutics using a pair of oppositely charged pentaheptad peptides (CCE and CCK) (Figure 14).78–81 CD20 is one of the most reliable human B-cell markers.82, 83 Although CD20 is expressed on both normal B-cells and malignant B-cells, it is still considered to be a safe target for immunotherapeutic B cell depletion because it is not expressed on stem cells or progenitor cells, and B cells can be restored after depletion.83–86 The anti-CD20 antibody Rituximab was approved in 1997 to treat low-grade non-Hodgkin’s lymphoma (NHL).87 The mechanisms of anti-CD20 mAb are not clearly understood, but are likely related to three immunological processes: direct induction of apoptosis, antibody-dependent cell-mediated cytotoxicity (ADCC), and complement-dependent lysis (CDC).88 One natural process to trigger apoptosis is through cell receptor clustering (crosslinking). Crosslinking of CD20 receptors on B cells with crosslinked antibodies showed improved therapeutic outcome in treating human B cell lymphoma.89–91 Based upon this biological rationale, Wu et al. designed a pair of oppositely charged 35 amino acid coiled coil peptides, CCE and CCK. Subsequently, CCK was conjugated with HPMA polymer ((CCK)9-P), and CCE peptide was attached to a Fab' fragment of anti-CD20 antibody (Fab'-CCK).78 Exposure of a CD20+ human B-NHL cell line (Raji B cells) to Fab'-CCE led to the decoration of the cell surface with CCE peptide. Upon treating decorated Raji cells with (CCK)9-P, CD20 receptors were crosslinked due to the attraction between CCK and CCE.78 A significant percentage of apoptotic induction activity was found after treating Raji cells with either premixed Fab'-CCE/(CCK)9-P (0.5 µM/25 µM) or with Fab'-CCE(0.5 µM) followed by (CCK)9-P (25 µM), as assessed using three different assays: a caspase 3 activity assay, an annexin V/propidium iodide assay, and a TUNEL (terminal deoxynucleotidyl transferase dUTP (deoxyuridine triphosphate) nick end labeling) assay.78 In vivo anticancer efficacy of the “drug-free” therapeutics was also evaluated in a Raji B-lymphoma-bearing mouse model.79 Both premixed treatments and consecutive administration of Fab'-CCE and (CCK)9-P were able to eradicate lymphoma cells in both bone marrow and blood. Zhang et al. further demonstrated that the Fab'-CCE and P-(CCK)x coiled-coil conjugates could assemble at the cell surface, resulting in cancer cell apoptosis, which was imaged with several techniques, including confocal microscopy 2-channel FMT, 3D confocal microscopy, and 4-color FACS.81
Figure 14.
Schematic of “drug-free” therapeutics using polymer-coiled coil conjugates. The cell surfaces of human Burkitt′s NHL B cells were decorated with one half of a coiled coil binding partner (CCE peptide) by binding a CCE peptide/antiCD20 Fab' conjugate to cell-surface CD20. Subsequent binding with HPMA copolymers presenting complementary CCK coils led to cross-linking of the CD20 receptors and subsequent apoptosis. Reproduced from Ref 78 with permission, Copyright 2010, Wiley.
COILED-COILED PEPTIDE SELF-ASSEMBLY FOR IMMUNOLOGICAL APPLICATIONS
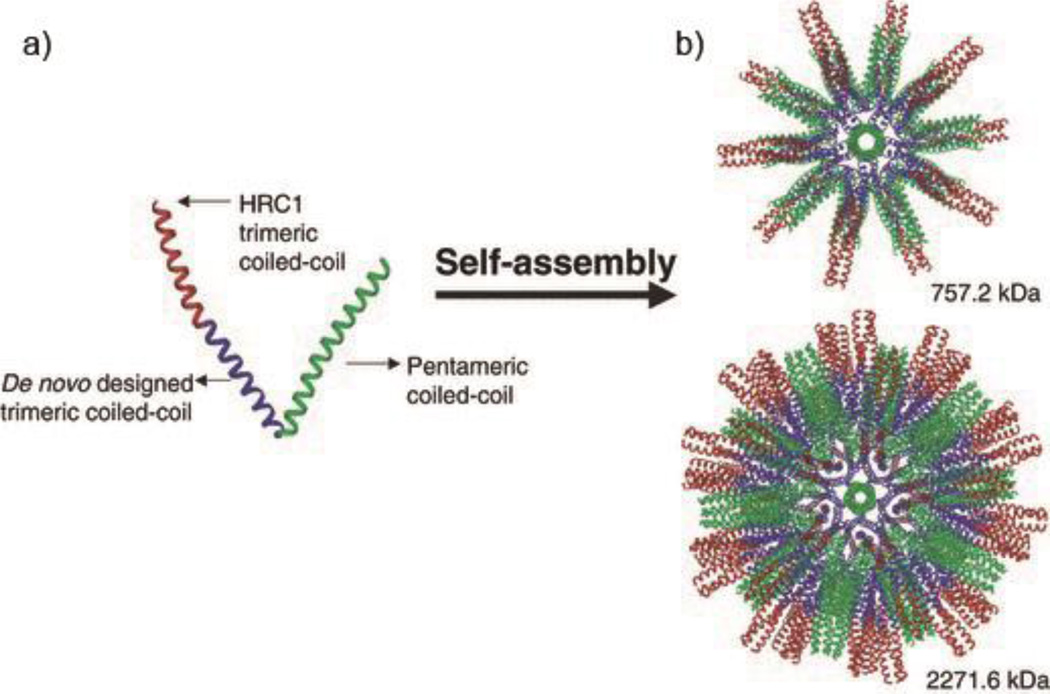
Vaccine development remains challenging for many diseases including HIV,92 influenza,93 and recent notable diseases such as Zika and Ebola. Subunit vaccines have received interest for new vaccine strategies owing to their synthetic, engineerable nature. A key strategy for the design of subunit vaccines is to create multivalent platforms, which increase immunogenicity of antigens, and coiled coils have received recent interest in this regard. Inspired by virus capsids, Burkhard and colleagues developed self-assembling protein nanoparticles (SAPNs) by designing a building block consisting of a pentameric coiled coil sequence genetically linked with a trimeric coiled coil peptide.94 The design of this building block is based on the symmetry elements found in an icosahedron, which has fivefold, threefold, and twofold rotational symmetry axes. By incorporating both the threefold symmetry (from the trimeric coiled coil) and fivefold symmetry (from the pentameric coiled coil) into the fusion protein, the obtained peptide chain could assemble into a spherical nanoparticle displaying icosahedral symmetry, with a diameter ranging from 25nm to 30 nm. A trimeric coiled coil B cell epitope (HRC1) for severe acute respiratory syndrome (SARS) was fused to the building block peptide at the C-terminus, such that it was repetitively displayed on the surface of the SAPNs. The antibodies elicited by this platform were proven to be conformation-specific according to ELISA, and they were capable of neutralizing virus.94 It was also confirmed that multivalent presentation by the self-assembling platform is essential to elicit protective immune responses, as no significant level of antibody production was detected from non-assembled epitopes.95 This coiled coil assembling system has been developed towards various other diseases including HIV and malaria, and is capable of raising epitope-specific humoral responses even in adjuvant-free formulations.95–98
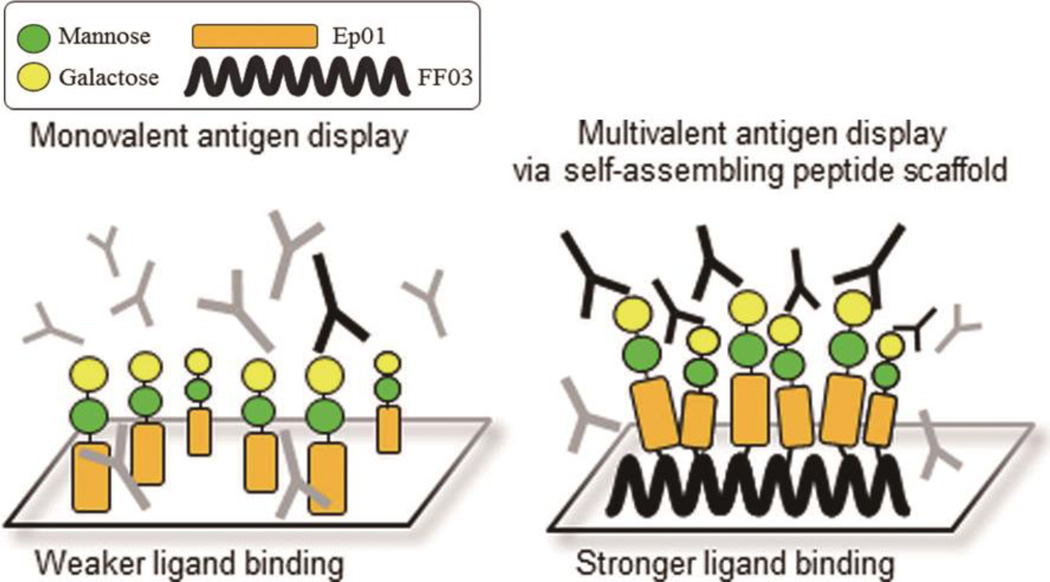
Towards improved diagnostics for infectious diseases and other conditions, Koksch and coworkers recently designed a coiled coil fibrilization system (FF03) for presenting multiple antigens for the sensitive detection of antibodies and other analytes.99 By selectively deprotecting a lysine residue at the f position of the coiled coil peptide FF03 on resin, the researchers were able to extend a side chain peptide epitope (Ep01) through solid phase peptide synthesis (SPPS). The obtained peptide conjugates were further functionalized with two carbohydrate moieties, mannose and galactose. Due to its multivalent presentation, the FF03-Ep01 conjugate peptide showed significantly higher antibody binding compared to Ep01 alone. Additionally, when antigenic Leishmania disaccharide was co-presented with FF03-Ep01 on the scaffold, the scaffold could detect polyclonal antibodies against the parasite from canine or human sources.99 This recent work illustrates the potential of coiled coil peptides for use within sensitive multivalent bioassays and diagnostics.
In other work investigating immune responses raised by coiled coil supramolecular materials, our group designed a hydrogel-forming system inspired by the coiled coil domain of fibrin.100, 101 The triblock peptide-PEG-peptide (γKEI-PEG-γKEI) molecule elicited a moderate antibody response while neither the native peptide nor γKEI elicited detectable responses, indicating that higher-order self-assembly can impact the immunogenicity of oligomerized peptide-based materials.101 This raises translational considerations for many of the non-native coiled coil systems discussed above. It remains to be seen whether mild immune responses raised by highly oligomerized supramolecular materials will or will not be compatible with their intended applications. Going further, any guiding principles for predicting, managing, or adjusting immune responses against supramolecular materials, coiled coil or otherwise, are only beginning to be elucidated.
Conclusion
Although the design of coiled coil materials has been increasingly refined over the past several years, and a range of creative materials and structures are now ripe for translation, progress towards medical technologies is still in its early stages. Above we discussed a range of recent strategies developed to rationally design self-assembling nanomaterials from coiled coil peptides. Through this work the coiled coil has been shown to be quite useful for producing nanofibers, nanotubes, networks, and even nanoparticles in some cases. We examined the emerging applications of coiled coil peptide in the fields of bioconjugation, drug delivery, unique “drug-free” therapeutics, and immune therapies. Coiled coil peptide self-assembly remains an exciting area of research, and it will be interesting to see how these elegant and useful structures are applied in the coming years.
Figure 15.
Schematic of self-assembling polypeptide nanoparticles (SAPN). a) Ribbon representation of a building block incorporating a trimeric coiled coil, a pentameric coiled coil, and a trimeric coiled coil B cell epitope (HRC1). b) 3D models of the nanoparticles formed from 60 peptides (top), and 180 peptides (bottom). Reproduced from Ref 94 with permission, Copyright 2008, Wiley.
Figure 16.
Schematic illustration of coiled coils used for multivalent antigen display in diagnostics and bioassays. Conjugation of a peptide epitope (Ep01) to coiled coil peptide fibers (FF03) produces multivalent platforms with enhanced antibody detection capabilities compared with the monovalent system. Reproduced from Ref 99 with permission, Copyright 2015, American Chemical Society.
Table 1.
Summary of coiled coil peptides for drug delivery systems
Acknowledgments
Work on self-assembling peptide biomaterials in our group is supported by the National Institutes of Health (NIBIB, 7R01EB009701; NIAID, 5R01AI118182). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Sean Kelly for assistance in preparing the manuscript.
Footnotes
Neither of the authors have financial conflicts of interest to disclose.
Contributor Information
Yaoying Wu, Department of Biomedical Engineering, Duke University.
Joel H. Collier, Department of Biomedical Engineering, Duke University
References
- 1.Sun WY, Yoshizawa M, Kusukawa T, Fujita M. Multicomponent metal-ligand self-assembly. Curr Opin Chem Biol. 2002;6:757–764. doi: 10.1016/s1367-5931(02)00358-7. [DOI] [PubMed] [Google Scholar]
- 2.Fujita M, Tominaga M, Hori A, Therrien B. Coordination assemblies from a Pd(II)-cornered square complex. Acc Chem Res. 2005;38:369–378. doi: 10.1021/ar040153h. [DOI] [PubMed] [Google Scholar]
- 3.Stupp SI. Self-assembly and biomaterials. Nano Lett. 2010;10:4783–4786. doi: 10.1021/nl103567y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ke YG, Ong LL, Shih WM, Yin P. Three-dimensional structures self-assembled from DNA bricks. Science. 2012;338:1177–1183. doi: 10.1126/science.1227268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwardson TGW, Carneiro KMM, McLaughlin CK, Serpell CJ, Sleiman HF. Site-specific positioning of dendritic alkyl chains on DNA cages enables their geometry-dependent self-assembly. Nat Chem. 2013;5:868–875. doi: 10.1038/nchem.1745. [DOI] [PubMed] [Google Scholar]
- 6.Ke YG. Designer three-dimensional DNA architectures. Curr Opin Struct Biol. 2014;27:122–128. doi: 10.1016/j.sbi.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Allen TM, Cullis PR. Liposomal drug delivery systems: From concept to clinical applications. Adv Drug Deliv Rev. 2013;65:36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 8.Bozzuto G, Molinari A. Liposomes as nanomedical devices. Int J Nanomedicine. 2015;10:975–999. doi: 10.2147/IJN.S68861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLaughlin CK, Hamblin GD, Hänni KD, Conway JW, Nayak MK, Carneiro KMM, Bazzi HS, Sleiman HF. Three-dimensional organization of block copolymers on “DNA-Minimal” scaffolds. J Am Chem Soc. 2012;134:4280–4286. doi: 10.1021/ja210313p. [DOI] [PubMed] [Google Scholar]
- 10.Wang L, Chierico L, Little D, Patikarnmonthon N, Yang Z, Azzouz M, Madsen J, Armes SP, Battaglia G. Encapsulation of biomacromolecules within polymersomes by electroporation. Angew Chem Int Ed Engl. 2012;51:11122–11125. doi: 10.1002/anie.201204169. [DOI] [PubMed] [Google Scholar]
- 11.Colley HE, Hearnden V, Avila-Olias M, Cecchin D, Canton I, Madsen J, MacNeil S, Warren N, Hu K, McKeating JA, et al. Polymersome-mediated delivery of combination anticancer therapy to head and neck cancer cells: 2D and 3D in vitro evaluation. Mol Pharm. 2014;11:1176–1188. doi: 10.1021/mp400610b. [DOI] [PubMed] [Google Scholar]
- 12.de la Rica R, Matsui H. Applications of peptide and protein-based materials in bionanotechnology. Chem Soc Rev. 2010;39:3499–3509. doi: 10.1039/b917574c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Zheng F, Grigoryan G. Design and designability of protein-based assemblies. Curr Opin Struct Biol. 2014;27:79–86. doi: 10.1016/j.sbi.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Pechar M, Pola R. The coiled coil motif in polymer drug delivery systems. Biotechnol Adv. 2013;31:90–96. doi: 10.1016/j.biotechadv.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Rose A, Meier I. Scaffolds, levers, rods and springs: diverse cellular functions of long coiled-coil proteins. Cell Mol Life Sci. 2004;61:1996–2009. doi: 10.1007/s00018-004-4039-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rose A, Schraegle SJ, Stahlberg EA, Meier I. Coiled-coil protein composition of 22 proteomes - differences and common themes in subcellular infrastructure and traffic control. BMC Evol Biol. 2005;5:66–86. doi: 10.1186/1471-2148-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen C, Parry DAD. α-Helical coiled coils and bundles: How to design an α-helical protein. Proteins. 1990;7:1–15. doi: 10.1002/prot.340070102. [DOI] [PubMed] [Google Scholar]
- 18.Harbury P, Zhang T, Kim P, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 19.Grigoryan G, Keating AE. Structural specificity in coiled-coil interactions. Curr Opin Struct Biol. 2008;18:477–483. doi: 10.1016/j.sbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou NE, Kay CM, Hodges RS. The role of interhelical ionic interactions in controlling protein-folding and stability - de-novo designed synthetic 2-stranded alpha-helical coiled-coils. J Mol Biol. 1994;237:500–512. doi: 10.1006/jmbi.1994.1250. [DOI] [PubMed] [Google Scholar]
- 21.Newman JRS, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- 22.Fong JH, Keating AE, Singh M. Predicting specificity in bZIP coiled-coil protein interactions. Genome Biol. 2004;5:1–10. doi: 10.1186/gb-2004-5-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grigoryan G, Keating AE. Structure-based prediction of bZIP partnering specificity. J Mol Biol. 2006;355:1125–1142. doi: 10.1016/j.jmb.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 24.Reinke AW, Grant RA, Keating AE. A Synthetic Coiled-Coil Interactome Provides Heterospecific Modules for Molecular Engineering. Journal of the American Chemical Society. 2010;132:6025–6031. doi: 10.1021/ja907617a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gradišar H, Jerala R. De novo design of orthogonal peptide pairs forming parallel coiled-coil heterodimers. J Pept Sci. 2011;17:100–106. doi: 10.1002/psc.1331. [DOI] [PubMed] [Google Scholar]
- 26.Aronsson C, Dånmark S, Zhou F, Öberg P, Enander K, Su H, Aili D. Self-sorting heterodimeric coiled coil peptides with defined and tuneable self-assembly properties. Sci Rep. 2015;5:14063. doi: 10.1038/srep14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyken SE, Chen Z, Groves B, Langan RA, Oberdorfer G, Ford A, Gilmore JM, Xu C, DiMaio F, Pereira JH, et al. De novo design of protein homo-oligomers with modular hydrogen-bond network–mediated specificity. Science. 2016;352:680–687. doi: 10.1126/science.aad8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki K, Doi T, Imanishi T, Kodama T, Tanaka T. The Conformation of the α-Helical Coiled Coil Domain of Macrophage Scavenger Receptor Is pH Dependent. Biochemistry. 1997;36:15140–15146. doi: 10.1021/bi971655o. [DOI] [PubMed] [Google Scholar]
- 29.Vandermeulen GWM, Tziatzios C, Duncan R, Klok H-A. PEG-based hybrid block copolymers containing α-helical coiled coil peptide sequences: control of self-assembly and preliminary biological evaluation. Macromolecules. 2005;38:761–769. [Google Scholar]
- 30.Graddis TJ, Myszka DG, Chaiken IM. Controlled formation of model homo- and heterodimer coiled coil polypeptides. Biochemistry. 1993;32:12664–12671. doi: 10.1021/bi00210a015. [DOI] [PubMed] [Google Scholar]
- 31.Marti DN, Jelesarov I, Bosshard HR. Interhelical ion pairing in coiled coils: solution structure of a heterodimeric leucine zipper and determination of pKa values of glu side chains. Biochemistry. 2000;39:12804–12818. doi: 10.1021/bi001242e. [DOI] [PubMed] [Google Scholar]
- 32.Petka WA, Harden JL, McGrath KP, Wirtz D, Tirrell DA. Reversible hydrogels from self-assembling artificial proteins. Science. 1998;281:389–392. doi: 10.1126/science.281.5375.389. [DOI] [PubMed] [Google Scholar]
- 33.Kohn WD, Kay CM, Hodges RS. Salt effects on protein stability: two-stranded α-helical coiled-coils containing inter- or intrahelical ion pairs1. J Mol Biol. 1997;267:1039–1052. doi: 10.1006/jmbi.1997.0930. [DOI] [PubMed] [Google Scholar]
- 34.Su JY, Hodges RS, Kay CM. Effect of chain length on the formation and stability of synthetic .alpha.-Helical coiled coils. Biochemistry. 1994;33:15501–15510. doi: 10.1021/bi00255a032. [DOI] [PubMed] [Google Scholar]
- 35.Betz SF, DeGrado WF. Controlling topology and native-like behavior of de novo-designed peptides: design and characterization of antiparallel four-stranded coiled coils. Biochemistry. 1996;35:6955–6962. doi: 10.1021/bi960095a. [DOI] [PubMed] [Google Scholar]
- 36.Slovic AM, Lear JD, DeGrado WF. De novo design of a pentameric coiled-coil: decoding the motif for tetramer versus pentamer formation in water-soluble phospholamban. J Pept Res. 2005;65:312–321. doi: 10.1111/j.1399-3011.2005.00244.x. [DOI] [PubMed] [Google Scholar]
- 37.Moll JR, Ruvinov SB, Pastan I, Vinson C. Designed heterodimerizing leucine zippers with a range of pIs and stabilities up to 10(−15) M. Protein Sci. 2001;10:649–655. doi: 10.1110/ps.39401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang S, Holmes T, Lockshin C, Rich A. Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane. Proc Natl Acad Sci U S A. 1993;90:3334–3338. doi: 10.1073/pnas.90.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Apostolovic B, Danial M, Klok H-A. Coiled coils: attractive protein folding motifs for the fabrication of self-assembled, responsive and bioactive materials. Chem Soc Rev. 2010;39:3541–3575. doi: 10.1039/b914339b. [DOI] [PubMed] [Google Scholar]
- 40.Tian YF, Hudalla GA, Han H, Collier JH. Controllably degradable β-sheet nanofibers and gels from self-assembling depsipeptides. Biomater Sci. 2013;1 doi: 10.1039/C3BM60161G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hauser CAE, Maurer-Stroh S, Martins IC. Amyloid-based nanosensors and nanodevices. Chem Soc Rev. 2014;43:5326–5345. doi: 10.1039/c4cs00082j. [DOI] [PubMed] [Google Scholar]
- 42.Matsuura K, Murasato K, Kimizuka N. Artificial peptide-nanospheres self-assembled from three-way junctions of β-sheet-forming peptides. J Am Chem Soc. 2005;127:10148–10149. doi: 10.1021/ja052644i. [DOI] [PubMed] [Google Scholar]
- 43.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 44.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 45.Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid – from bacteria to humans. Trends Biochem Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 46.Otzen D. Functional amyloid. Prion. 2010;4:256–264. doi: 10.4161/pri.4.4.13676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beck K, Brodsky B. Supercoiled protein motifs: the collagen triple-helix and the α-helical coiled coil. J Struct Biol. 1998;122:17–29. doi: 10.1006/jsbi.1998.3965. [DOI] [PubMed] [Google Scholar]
- 48.Pandya MJ, Spooner GM, Sunde M, Thorpe JR, Rodger A, Woolfson DN. Sticky-end assembly of a designed peptide fiber provides insight into protein fibrillogenesis. Biochemistry. 2000;39:8728–8734. doi: 10.1021/bi000246g. [DOI] [PubMed] [Google Scholar]
- 49.Woolfson DN. The design of coiled-coil structures and assemblies. In: David ADP, John MS, editors. Adv Protein Chem. Vol. 70. Academic Press; 2005. pp. 79–112. [DOI] [PubMed] [Google Scholar]
- 50.Ryadnov MG, Woolfson DN. Engineering the morphology of a self-assembling protein fibre. Nat Mater. 2003;2:329–332. doi: 10.1038/nmat885. [DOI] [PubMed] [Google Scholar]
- 51.Ryadnov MG, Woolfson DN. MaP peptides: programming the self-assembly of peptide-based mesoscopic matrices. J Am Chem Soc. 2005;127:12407–12415. doi: 10.1021/ja052972i. [DOI] [PubMed] [Google Scholar]
- 52.Dong H, Paramonov SE, Hartgerink JD. Self-assembly of alpha-helical coiled coil nanofibers. J Am Chem Soc. 2008;130:13691–13695. doi: 10.1021/ja8037323. [DOI] [PubMed] [Google Scholar]
- 53.Banwell EF, Abelardo ES, Adams DJ, Birchall MA, Corrigan A, Donald AM, Kirkland M, Serpell LC, Butler MF, Woolfson DN. Rational design and application of responsive α-helical peptide hydrogels. Nat Mater. 2009;8:596–600. doi: 10.1038/nmat2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J, Zheng Q, Deng Y, Cheng C-S, Kallenbach NR, Lu M. A seven-helix coiled coil. Proc Natl Acad Sci U S A. 2006;103:15457–15462. doi: 10.1073/pnas.0604871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu C, Liu R, Mehta AK, Guerrero-Ferreira RC, Wright ER, Dunin-Horkawicz S, Morris K, Serpell LC, Zuo X, Wall JS, et al. Rational design of helical nanotubes from self-assembly of coiled-coil lock washers. J Am Chem Soc. 2013;135:15565–15578. doi: 10.1021/ja4074529. [DOI] [PubMed] [Google Scholar]
- 56.Gunasekar SK, Haghpanah JS, Montclare JK. Assembly of bioinspired helical protein fibers. Polym Adv Technol. 2008;19:454–468. [Google Scholar]
- 57.Gunasekar SK, Asnani M, Limbad C, Haghpanah JS, Hom W, Barra H, Nanda S, Lu M, Montclare JK. N-terminal aliphatic residues dictate the structure, stability, assembly, and small molecule binding of the coiled-coil region of cartilage oligomeric matrix protein. Biochemistry. 2009;48:8559–8567. doi: 10.1021/bi900534r. [DOI] [PubMed] [Google Scholar]
- 58.Hume J, Sun J, Jacquet R, Renfrew PD, Martin JA, Bonneau R, Gilchrist ML, Montclare JK. Engineered coiled-coil protein microfibers. Biomacromolecules. 2014;15:3503–3510. doi: 10.1021/bm5004948. [DOI] [PubMed] [Google Scholar]
- 59.Calladine CR, Sharff A, Luisi B. How to untwist an α-helix: structural principles of an α-helical barrel. J Mol Biol. 2001;305:603–618. doi: 10.1006/jmbi.2000.4320. [DOI] [PubMed] [Google Scholar]
- 60.Egelman EH, Xu C, DiMaio F, Magnotti E, Modlin C, Yu X, Wright E, Baker D, Conticello VP. Structural plasticity of helical nanotubes based on coiled-coil assemblies. Structure. 2015;23:280–289. doi: 10.1016/j.str.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burgess NC, Sharp TH, Thomas F, Wood CW, Thomson AR, Zaccai NR, Brady RL, Serpell LC, Woolfson DN. Modular design of self-assembling peptide-based nanotubes. J Am Chem Soc. 2015;137:10554–10562. doi: 10.1021/jacs.5b03973. [DOI] [PubMed] [Google Scholar]
- 62.Thomas F, Burgess NC, Thomson AR, Woolfson DN. Controlling the assembly of coiled-coil peptide nanotubes. Angew Chem Int Ed Engl. 2016;55:987–991. doi: 10.1002/anie.201509304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fletcher JM, Harniman RL, Barnes FR, Boyle AL, Collins A, Mantell J, Sharp TH, Antognozzi M, Booth PJ, Linden N, et al. Self-assembling cages from coiled-coil peptide modules. Science. 2013;340:595–599. doi: 10.1126/science.1233936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gradišar H, Božič S, Doles T, Vengust D, Hafner-Bratkovič I, Mertelj A, Webb B, Šali A, Klavžar S, Jerala R. Design of a single-chain polypeptide tetrahedron assembled from coiled-coil segments. Nat Chem Biol. 2013;9:362–366. doi: 10.1038/nchembio.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eriksson M, Hassan S, Larsson R, Linder S, Ramqvist T, Lövborg H, Vikinge T, Figgemeier E, Müller J, Stetefeld J, et al. Utilization of a right-handed coiled-coil protein from archaebacterium staphylothermus marinus as a carrier for cisplatin. Anticancer Res. 2009;29:11–18. [PubMed] [Google Scholar]
- 66.Thanasupawat T, Bergen H, Hombach-Klonisch S, Krcek J, Ghavami S, Del Bigio MR, Krawitz S, Stelmack G, Halayko A, McDougall M, et al. Platinum (IV) coiled coil nanotubes selectively kill human glioblastoma cells. Nanomedicine. 2015;11:913–925. doi: 10.1016/j.nano.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 67.Shi Y, Liu S-A, Kerwood DJ, Goodisman J, Dabrowiak JC. Pt(IV) complexes as prodrugs for cisplatin. J Inorg Biochem. 2012;107:6–14. doi: 10.1016/j.jinorgbio.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 68.Apostolovic B, Deacon SPE, Duncan R, Klok H-A. Hybrid polymer therapeutics incorporating bioresponsive, coiled coil peptide linkers. Biomacromolecules. 2010;11:1187–1195. doi: 10.1021/bm901313c. [DOI] [PubMed] [Google Scholar]
- 69.Apostolovic B, Deacon SP, Duncan R, Klok H-AA. Cell uptake and trafficking behavior of non-covalent, coiled-coil based polymer-drug conjugates. Macromol Rapid Commun. 2011;32:11–18. doi: 10.1002/marc.201000434. [DOI] [PubMed] [Google Scholar]
- 70.Apostolovic B, Klok H-A. pH-Sensitivity of the E3/K3 heterodimeric coiled coil. Biomacromolecules. 2008;9:3173–3180. doi: 10.1021/bm800746e. [DOI] [PubMed] [Google Scholar]
- 71.Pechar M, Pola R, Laga R, Ulbrich K, Bednarova L, Malon P, Sieglova I, Kral V, Fabry M, Vanek O. Coiled coil peptides as universal linkers for the attachment of recombinant proteins to polymer therapeutics. Biomacromolecules. 2011;12:3645–3655. doi: 10.1021/bm200897b. [DOI] [PubMed] [Google Scholar]
- 72.Pola R, Laga R, Ulbrich K, Sieglova I, Kral V, Fabry M, Kabesova M, Kovar M, Pechar M. Polymer therapeutics with a coiled coil motif targeted against murine BCL1 leukemia. Biomacromolecules. 2013;14:881–889. doi: 10.1021/bm3019592. [DOI] [PubMed] [Google Scholar]
- 73.Pechar M, Pola R, Laga R, Braunová A, Filippov SK, Bogomolova A, Bednárová L, Vaněk O, Ulbrich K. Coiled coil peptides and polymer-peptide conjugates: synthesis, self-assembly, characterization and potential in drug delivery systems. Biomacromolecules. 2014;15:2590–2599. doi: 10.1021/bm500436p. [DOI] [PubMed] [Google Scholar]
- 74.Al-Ahmady ZS, Al-Jamal WT, Bossche JV, Bui TT, Drake AF, Mason AJ, Kostarelos K. Lipid-peptide vesicle nanoscale hybrids for triggered drug release by mild hyperthermia in vitro and in vivo. ACS nano. 2012;6:9335–9346. doi: 10.1021/nn302148p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mahmoud ZN, Grundy DJ, Channon KJ, Woolfson DN. The non-covalent decoration of self-assembling protein fibers. Biomaterials. 2010;31:7468–7474. doi: 10.1016/j.biomaterials.2010.06.041. [DOI] [PubMed] [Google Scholar]
- 76.Woolfson DN, Mahmoud ZN. More than just bare scaffolds: towards multi-component and decorated fibrous biomaterials. Chem Soc Rev. 2010;39:3464–3479. doi: 10.1039/c0cs00032a. [DOI] [PubMed] [Google Scholar]
- 77.Mehrban N, Abelardo E, Wasmuth A, Hudson KL, Mullen LM, Thomson AR, Birchall MA, Woolfson DN. Assessing cellular response to functionalized α-helical peptide hydrogels. Adv Healthc Mater. 2014;3:1387–1391. doi: 10.1002/adhm.201400065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu K, Liu J, Johnson RN, Yang J, Kopeček J. Drug-free macromolecular therapeutics: induction of apoptosis by coiled-coil-mediated cross-linking of antigens on the cell surface. Angew Chem Int Ed Engl. 2010;49:1451–1455. doi: 10.1002/anie.200906232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu K, Yang J, Liu J, Kopeček J. Coiled-coil based drug-free macromolecular therapeutics: In vivo efficacy. J Control Release. 2012;157:126–131. doi: 10.1016/j.jconrel.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chu T-W, Kopeček J. Drug-free macromolecular therapeutics – a new paradigm in polymeric nanomedicines. Biomater Sci. 2015;3:908–922. doi: 10.1039/C4BM00442F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang R, Yang J, Chu T-W, Hartley JM, Kopeček J. Multimodality imaging of coiled-coil mediated self-assembly in a “drug-free” therapeutic system. Adv Healthc Mater. 2015;4:1054–1065. doi: 10.1002/adhm.201400679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stashenko P, Nadler LM, Hardy R, Schlossman SF. Characterization of a human B lymphocyte-specific antigen. J Immunol. 1980;125:1678–1685. [PubMed] [Google Scholar]
- 83.Anderson K, Bates M, Slaughenhoupt B, Pinkus G, Schlossman S, Nadler L. Expression of human B cell-associated antigens on leukemias and lymphomas: a model of human B cell differentiation. Blood. 1984;63:1424–1433. [PubMed] [Google Scholar]
- 84.Maloney DG, Grillo-López AJ, White CA, Bodkin D, Schilder RJ, Neidhart JA, Janakiraman N, Foon KA, Liles T-M, Dallaire BK, et al. IDEC-C2B8 (Rituximab) Anti-CD20 Monoclonal Antibody Therapy in Patients With Relapsed Low-Grade Non-Hodgkin's Lymphoma. Blood. 1997;90:2188–2195. [PubMed] [Google Scholar]
- 85.Maloney DG. Anti-CD20 antibody therapy for B-cell lymphomas. N Engl J Med. 2012;366:2008–2016. doi: 10.1056/NEJMct1114348. [DOI] [PubMed] [Google Scholar]
- 86.Cheson BD, Leonard JP. Monoclonal antibody therapy for B-cell non-hodgkin's lymphoma. N Engl J Med. 2008;359:613–626. doi: 10.1056/NEJMra0708875. [DOI] [PubMed] [Google Scholar]
- 87.Zaja F, Iacona I, Masolini P, Russo D, Sperotto A, Prosdocimo S, Patriarca F, de Vita S, Regazzi M, Baccarani M. B-cell depletion with rituximab as treatment for immune hemolytic anemia and chronic thrombocytopenia. Haematologica. 2002;87:189–195. [PubMed] [Google Scholar]
- 88.Boross P, Leusen JHW. Mechanisms of action of CD20 antibodies. Am J Cancer Res. 2012;2:676–690. [PMC free article] [PubMed] [Google Scholar]
- 89.Vallat LD, Park Y, Li C, Gribben JG. Temporal genetic program following B-cell receptor cross-linking: altered balance between proliferation and death in healthy and malignant B cells. Blood. 2007;109:3989–3997. doi: 10.1182/blood-2006-09-045377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ghetie M-A, Podar EM, Ilgen A, Gordon BE, Uhr JW, Vitetta ES. Homodimerization of tumor-reactive monoclonal antibodies markedly increases their ability to induce growth arrest or apoptosis of tumor cells. Proc Natl Acad Sci U S A. 1997;94:7509–7514. doi: 10.1073/pnas.94.14.7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ghetie M-A, Bright H, Vitetta ES. Homodimers but not monomers of Rituxan (chimeric anti-CD20) induce apoptosis in human B-lymphoma cells and synergize with a chemotherapeutic agent and an immunotoxin. Blood. 2001;97:1392–1398. doi: 10.1182/blood.v97.5.1392. [DOI] [PubMed] [Google Scholar]
- 92.Picker LJ, Deeks SG. HIV: Antibodies advance the search for a cure. Nature. 2013;503:207–208. doi: 10.1038/nature12703. [DOI] [PubMed] [Google Scholar]
- 93.Krammer F, Palese P. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov. 2015;14:167–182. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- 94.Pimentel TAPF, Yan Z, Jeffers SA, Holmes KV, Hodges RS, Burkhard P. Peptide nanoparticles as novel immunogens: design and analysis of a prototypic severe acute respiratory syndrome vaccine. Chem Biol Drug Des. 2009;73:53–61. doi: 10.1111/j.1747-0285.2008.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kaba SA, Brando C, Guo Q, Mittelholzer C, Raman S, Tropel D, Aebi U, Burkhard P, Lanar DE. A nonadjuvanted polypeptide nanoparticle vaccine confers long-lasting protection against rodent malaria. J Immunol. 2009;183:7268–7277. doi: 10.4049/jimmunol.0901957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schroeder U, Graff A, Buchmeier S, Rigler P, Silvan U, Tropel D, Jockusch BM, Aebi U, Burkhard P, Schoenenberger C-A. Peptide nanoparticles serve as a powerful platform for the immunogenic display of poorly antigenic actin determinants. J Mol Biol. 2009;386:1368–1381. doi: 10.1016/j.jmb.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 97.Babapoor S, Neef T, Mittelholzer C, Girshick T, Garmendia A, Shang H, Khan MI, Burkhard P. A novel vaccine using nanoparticle platform to present immunogenic M2e against avian influenza infection. Influenza Res Treat. 2011;2011:12. doi: 10.1155/2011/126794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wahome N, Pfeiffer T, Ambiel I, Yang Y, Keppler OT, Bosch V, Burkhard P. Conformation-specific display of 4E10 and 2F5 epitopes on self-assembling protein nanoparticles as a potential HIV vaccine. Chem Biol Drug Des. 2012;80:349–357. doi: 10.1111/j.1747-0285.2012.01423.x. [DOI] [PubMed] [Google Scholar]
- 99.Zacco E, Anish C, Martin CE, V Berlepsch H, Brandenburg E, Seeberger PH, Koksch B. A self-assembling peptide scaffold for the multivalent presentation of antigens. Biomacromolecules. 2015;16:2188–2197. doi: 10.1021/acs.biomac.5b00572. [DOI] [PubMed] [Google Scholar]
- 100.Jing P, Rudra JS, Herr AB, Collier JH. Self-assembling peptide-polymer hydrogels designed from the coiled coil region of fibrin. Biomacromolecules. 2008;9:2438–2446. doi: 10.1021/bm800459v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rudra JS, Tripathi PK, Hildeman DA, Jung JP, Collier JH. Immune responses to coiled coil supramolecular biomaterials. Biomaterials. 2010;31:8475–8483. doi: 10.1016/j.biomaterials.2010.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further Reading
- De Santis E, Ryadnov MG. Peptide self-assembly for nanomaterials: the old new kid on the block. Chem Soc Rev. 2015;44:8288–8300. doi: 10.1039/c5cs00470e. [DOI] [PubMed] [Google Scholar]
- Matsuura K. Rational design of self-assembled proteins and peptides for nano- and micro-sized architectures. RSC Advances. 2014;4:2942–2953. [Google Scholar]