Abstract
CapB2 is recognized as a tyrosine kinase and is likely a vital factor in extracellular polysaccharide synthesis in Staphylococcus aureus, but its pathogenicity function and regulatory mechanism remain obscure. Here, we demonstrate that CapB2 enhances bacterial virulence in a murine model. Mice infected with the wild type SA75 strain exhibited significantly larger (P < 0.05) skin lesions from days 4 to 7 of infection than those challenged with the capB2 mutant strain. The effect on virulence was reverted by restoring the capB2 mutation to the wild type. The related components of the wild type SA75 cell wall in the capB2 mutant strain (SA75ΔcapB2) were thinner than wild type SA75 strain and the capB2 mutant complemented strain (SA75ΔcapB2-C), which was determined by the transmission electron microscopy. The survival percentages of the wild type strain SA75 and SA75ΔcapB2-C were significantly higher relative to SA75ΔcapB2. The results of qRT-PCR studies also indicated that mutations in regulatory gene sarA led to a drastic increase in capB2 gene transcription, with a 326-fold increase of growth at 6 h compared with the wild type strain, suggesting that sarA is a major negative regulator of capB2 expression. Taken together, these results demonstrate that the expression of CapB2 promotes S. aureus virulence in a mouse model of skin infection, and that capB2 gene transcription is regulated negatively by SarA.
Keywords: Staphylococcus aureus, capB2, sarA, virulence factor, regulation
Introduction
Staphylococcus aureus is a common bacterial pathogen which causes a wide range of human diseases, ranging from relatively benign skin and tissue infections to complicated life threatening diseases such as osteomyelitis, septic arthritis, and bacteremia. S. aureus pathogenicity is mediated by bacterial components and secreted virulence factors such as surface-associated adhesins, capsular polysaccharide (CP), and exotoxins. Studies have shown that CP is an important virulence factor in S. aureus, and it is believed to enhance microbial virulence (Nilsson et al., 1997; Thakker et al., 1998) in host immune evasion (Nanra et al., 2013).
More than 90% of S. aureus strains express CP, and 13 capsular serotypes have been described to date (Arbeit et al., 1984; Karakawa et al., 1985; von Eiff et al., 2007). Polysaccharide capsules corresponding to serotypes 1 and 2 (CP1 and CP2) are associated with heavily encapsulated mucoid strains, but are rarely encountered among clinical isolates (Luong T.T. et al., 2002; Luong et al., 2003). By contrast, CP5 and CP8 are both predominant among clinical infection isolates (Sompolinsky et al., 1985; O’Riordan and Lee, 2004), and are associated with microencapsulated strains exhibiting non-mucoid colony morphology. The cap5 and cap8 operons both contain 16 open reading frames, cap5A1 (cap8A1) through cap5P (cap8P), with the type-specific genes situated in the central region of the operons (cap5HIJK or cap8HIJK) (Sau et al., 1997a). CapB1, which forms a complex with the C-terminal domain of transmembrane protein CapA1, is probably a regulator of chain length in CP biosynthesis (Sau et al., 1997b), and has been shown to exhibit tyrosine kinase activity (Olivares-Illana et al., 2008). Whole genome sequencing of S. aureus has demonstrated that the gene products of capA2/capB2 (located elsewhere in the genome) are highly similar to those of capA1/capB1 (Kuroda et al., 2001; Soulat et al., 2006). However, despite CapB1 sharing more than 70% sequence identity with CapB2, no kinase activity could be detected for CapB1 in vitro. Of interest, the tyrosine kinase activity of CapB2 is more efficiently activated by the transmembrane stimulatory protein CapA1 than by the corresponding CapA2 activator (Kuroda et al., 2001; Soulat et al., 2006). Once CapB2 has been activated, its tyrosine cluster is phosphorylated (Grangeasse et al., 2012), indicating that CapB2 is probably a vital factor in the synthesis and/or export of extracellular polysaccharides.
The Staphylococcal accessory regular (sar) locus encodes a DNA binding protein, SarA, which is a key pleiotropic transcriptional regulator for the synthesis of extracellular virulence proteins (Cheung and Zhang, 2002). The sarA gene can regulate virulence expression dependently of agr (Cheung and Zhang, 2002). And it can also activate certain virulence genes by binding the promoter of the target gene directly (Cheung and Zhang, 2002).
In the present study, we analyzed the role of capB2 in the virulence of S. aureus by constructed an allelic replacement mutant of S. aureus 75. Our results show that a capB2 mutant of S. aureus is less virulence in the mice model of skin infection. In addition, we investigated the capB2 expression regulated by SarA.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions
The bacterial strains and plasmids used in this study are described in Table 1. S. aureus strain SA75 was isolated from a patient with a purulent skin infection at the First Affiliated Hospital in Wenzhou. Unless otherwise stated, Escherichia coli strains were cultured in Luria-Bertani medium, and S. aureus strains were grown in tryptic soy broth (TSB) medium, at 37°C with shaking at 200 rpm. The culture media were supplemented with appropriate antibiotics when required (ampicillin at 100 mg/l, chloramphenicol at 10 mg/l and anhydrotetracycline at 50 ng/ml).
Table 1.
Bacterial strains and plasmids used in this study.
| Strains and plasmixds | Description | Source |
|---|---|---|
| Strains | ||
| S. aureus | ||
| SA75 | Wild type, CP8-positive, clinical MRSA strain isolated from an abscess | Laboratory stock |
| SA75ΔsarA | Isogenic sarA deletion mutant in SA75 | Laboratory stock |
| SA75ΔcapB2 | Isogenic capB2 deletion mutant in SA75 | This study |
| SA75ΔsarA-C | sarA mutant complemented with pRBsarA | This study |
| SA75ΔcapB2-C | capB2 mutant complemented with pRBcapB2 | This study |
| E. coli | ||
| DH5α | Clone host strain | Laboratory stock |
| DC10B | dam+Δdcm-ΔhsdRMS endA1 recA1; clone host strain | Laboratory stock |
| Plasmids | ||
| pKOR1 | Shuttle cloning vector, temp sensitive (CmrAmpr)a | Laboratory stock |
| pKcapB2 | pKOR1 containing fragments 1500-bp upstream and 1500-bp downstream of capB2 gene, for capB2 mutagenesis (CmrAmpr) | This study |
| pRB473 | Shuttle cloning vector (Cmr) | Laboratory stock |
| pRBsarA | pRB473 with sarA and its promoter (Cmr) | This study |
| pRBcapB2 | pRB473 with capB2 and its promoter (Cmr) | This study |
aCmrAmpr, chloramphenical and ampicillin resistance.
DNA Manipulations
Genomic DNA extraction was performed as described previously (Yu et al., 2014). Plasmid DNA extracted from E. coli strains was purified according to the manufacturer’s instruction (Axygen, Union City, CA, USA). To extract plasmid DNA from S. aureus strains, 20 μl lysostaphin (2 mg/ml) was used with suspension buffer S1 to re-suspend the collected cells from 10 ml overnight culture, incubated at 37°C for 1 h, and then processed following the manufacturer’s instruction (TIANGEN, Beijing, China).
Construction of S. aureus capB2 Mutant Strain by Allelic Replacement
In order to generate a capB2 mutant from SA75, genomic DNA was extracted from S. aureus SA75 and the two sets of primers listed in Table 2 were used to amplify DNA fragments 1.5-kb upstream and downstream of capB2 from SA75 chromosomal DNA. The amplified products were digested with HindIII and then ligated with T4 DNA ligase to yield a 693 bp deletion fragment (called ΔcapB2). The primers UP-F-attB2 and Dn-R-attB1 were used to amplify ΔcapB2 fragment, yielding a PCR product with attB sites at the 5′ and 3′ ends. The resulting fragments were recombined with temperature-sensitive shuttle plasmid pKOR1 to generate recombinant plasmid pKcapB2.
Table 2.
Primers used in this study.
| Primers | Oligonucleotide (5′→3′)a |
|---|---|
| capB2-up-F- BamHI | CGggatccGCACGCTCGGTGTTGTAAAG |
| capB2-up-R- HindIII | CCCaagcttCTTCTTCGTGTATTCGTCAT |
| capB2-down-F- HindIII | CCCaagcttGCATATTATGGGACTGATGA |
| capB2-down-R- XhoI | CCGctcgagTTGATCTAAGTCCTGCATGA |
| UP-F-attB2 | GGGGACCACTTTGTACAAGAAAGCTGGGTGCACGCTCGGTGTTGTAAAG |
| Dn-R-attB1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTTGATCTAAGTCCTGCATGA |
| SW-cp-F | ATAGTTTAGTAAGCGACCTG |
| XW-cp-R | TACAGGAAATCCGCATAAAT |
| Cp-BD-F | ATGACGAATACACGAAGAAG |
| Cp-BD-R | TCATGATTCATCAGTCCCAT |
| PstI-sarA-F | AAAActgcagTAAACCAAATGCTAACCCA |
| BamHI-sarA-R | CGCggatccAGTGCCATTAGTGCAAAAC |
| PstI-cap-19F | AAAActgcagATTTTTGTTACTAGTTTGT |
| BamHI-cap-19R | CGggatccTCATGATTCATCAGTCCCA |
| cap5K-F | GTCAAAGATTATGTGATGCTACTGAG |
| cap5K-R | ACTTCGAATATAAACTTGAATCAATGTTATACAG |
| cap8I-F | GCCTTATGTTAGGTGATAAACC |
| cap8I-R | GGAAAAACACTATCATAGCAGG |
| gyrB-F | ACATTACAGCAGCGTATTAG |
| gyrB-R | CTCATAGTGATAGGAGTCTTCT |
| cap-XF | ACGAATACACGAAGAAGTA |
| cap-XR | CCTCTGAAGTGATTACAATG |
| sarA-qtF | GTTATCAATGGTCACTTATGC |
| sarA-qtR | CTTGTGGTTGTTTGTAGTTT |
aRestriction sites are indicated by lowercase letters; underlined sequences denote recombination sites.
The resultant plasmid containing the ΔcapB2 gene deletion was transferred first to E. coli DH5α and then to E. coli DC10B, and subsequently electroporated into strain SA75. The allelic exchange procedure was performed as described previously (Bae and Schneewind, 2006). Putative transformants were screened by PCR with LATaq DNA polymerase, using primer pairs SW-cp-F/XW-cp-R and Cp-BD-F/Cp-BD-R. Deletion of the desired gene was verified by PCR, quantitative reverse transcription-PCR (qRT-PCR), and sequencing.
Construction of Complemented Strains
For complementation of the sarA gene and capB2 gene, the deleted genes and their own putative promoters region were amplified by PCR with Iproof High-Fidelity DNA polymerase using primers PstI-sarA-F and BamHI-sarA-R for sarA, and primers PstI-cap-19F and BamHI-cap-19R for capB2. The PCR products were digested with BamHI and PstI, and the fragments were ligated into shuttle plasmid pRB473 to generate plasmid pRBsarA and pRBcapB2. The resultant plasmids were transferred into E. coli DH5α and DC10B successively, and finally electroporated into mutant strains SA75ΔsarA and SA75ΔcapB2. Successful uptake of the complementation plasmid was confirmed by restriction mapping, PCR and sequencing of PCR fragments. The presence of sarA transcription or capB2 transcription within the transformants was verified by qRT-PCR.
Capsular Genotype Detection
Capsular genotype was detected by PCR as described previously (Verdier et al., 2007). Genomic DNA from strain SA75 was used as template, with primers cap5k-F and cap5k-R for detection of capsular type 5, and primers cap8I-F and cap8I-R for detection of capsular type 8. PCR product sizes were 361 and 173 bp for capsular types 5 and 8, respectively, and were confirmed by sequencing.
Growth Curve
Staphylococcus aureus wild type strain SA75 and its derivative strains were incubated overnight in 5 ml TSB at 37°C with shaking at 200 rpm. The overnight cultures were diluted in 30 ml TSB to obtain the same starting optical density (OD) at 562 nm. The growth of each strain was monitored by Microplate Manager 6 (Bio-Rad, USA) software at 1 h intervals for a total of 12 h.
S. aureus Survival Analyze in Human Blood
Blood samples from healthy donors were collected in 10 mg/ml heparin anticoagulation tubes. The ability of S. aureus SA75 and SA75ΔcapB2 to survive in human blood was determined under post-logarithmic growth phase. The strains were analyzed after washed twice and suspended to an OD of 0.6 at 562 nm. Then, 1 ml of heparinized human blood was inoculated with 100 μl aliquot of these suspensions and was incubated at 37°C. After 7 h, bacteria cells were appropriate diluted to detect the endpoint numbers of CFU. The survival rate of the bacteria cells was determined by comparing to the initial inoculum.
Mouse Model of Skin Infection Assay
Animal studies were approved by the Institutional Animal Care and Use Committee at Wenzhou Medical University. We confirm that all animal care and methods were performed in accordance with the guidelines and regulations approved by the Administration of Affairs Concerning Experimental Animals in China. The skin abscess model was performed as described previously (Li et al., 2012). Female, 4- to 6-week old, BALB/C-nu mice were used in the study. The mice were housed for 7 days before inoculation with food and water ad libitum. S. aureus cultures were grown to post-exponential phase, washed with sterile phosphate-buffered saline (PBS), and then suspended in PBS to achieve a concentration of 1.0 × 105 CFU/100 μl. The bacterial numbers were confirmed by plate counts. After the mice were anesthetized with diethyl ether, each mouse was injected subcutaneously with 100 μl of either S. aureus suspension or PBS. Skin lesions developing on test animals were observed daily, and the abscess area calculated using the formula length (L) × width (W). The values of L and W for each infected mouse were determined by caliper. Dermonecrosis was observed and recorded for 10 days, following which all animals were euthanized.
Electron Microscopy Study on Abscess Tissue
Overnight cultures of S. aureus SA75, capB2 mutant and the complemented strains were harvested and washed twice with sterile PBS, and then suspended to achieve a concentration of 1.0 × 108 CFU/100 μl. Mouse model of subcutaneous abscess was performed as above. The abscess tissue was obtained at the fourth day of infection. Samples were fixed and processed further for transmission electron microscopy.
RNA Extraction, cDNA Synthesis, and Quantitative Reverse Transcription-PCR (qRT-PCR)
The quantitative Real-Time PCR was performed according to the MIQE guidelines. For RNA isolation, S. aureus strains were diluted 1:100 in TSB from overnight cultures and grown to post-exponential phase at 37°C with shaking at 200 rpm. Total RNA was isolated and purified using a PureLink RNA Mini Kit (Invitrogen, Carlsbad, CA, USA); 2 μg purified RNA was then used to obtain cDNA using a PrimeScript RT reagent kit (TaKaRa, Japan) following the manufacturer’s instructions. The resultant cDNA was amplified using a SsoFas EvaGreen Supermix kit (Bio-Rad, USA) with the Bio-Rad CFX96 Manager software. Oligonucleotide primers used are listed in Table 2. Wild type strain SA75 was used as a control (relative expression = 1), and gyrB was used as a reference gene to investigate genes of interest. RNA transcript levels were calculated by the method of delta delta Ct (ΔΔCt) (Montgomery et al., 2012). Data analysis was carried out using Bio-Rad CFX software. Each reaction was performed in triplicate.
Statistical Analysis
Skin abscesses in the mouse infection model were compared with one-way analysis of variance (ANOVA) using SPSS Statistics 17.0. Results were considered statistically significant if P-values were < 0.05. Relative expression levels resulting from qRT-PCR were analyzed using GraphPad Prism 5.0 software.
Results
Capsular Genotype and Growth Curve Analysis
In order to rule out the potential influence of the capB2 deletion on bacterial growth rate, the growth curves of strains SA75, SA75ΔcapB2, and SA75ΔcapB2-C were monitored hourly. The growth curves of the three strains were measured in TSB medium at the same starting OD of 562 nm, with TSB medium used as a blank control. There were no substantial differences in growth curves among all three strains. The capB2 gene was expressed at the indicated time point (3, 6, 9, 12 h) and assessed in strain SA75, with expression shown to be highest at the 6 h time point.
Deletion of capB2 in S. aureus
The capB2 deletion was introduced by allelic replacement into S. aureus strain SA75, yielding the mutant strain SA75ΔcapB2. High-efficiency cloning strain E. coli DC10B was used as an intermediate host instead of S. aureus RN4220 in the study in order to improve the transformation efficiency. To confirm the deletion of capB2,SA75ΔcapB2 was amplified with primers SW-cp-F and XW-cp-R using LA Taq DNA polymerase. The band size in strain SA75ΔcapB2 reflected a 693 bp deletion compared to the wild type strain. The relative expression of capB2 in strain SA75ΔcapB2 was dramatically decreased relative to that of strain SA75 (Figure 1).
FIGURE 1.
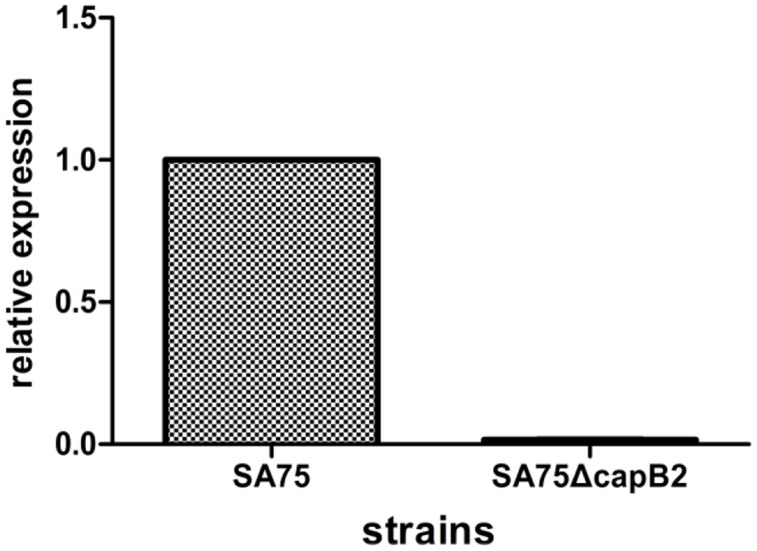
Deletion of capB2 in S. aureus. Gene gyrB was used as the reference gene; the expression of capB2 in strain SA75 was regarded as 1.
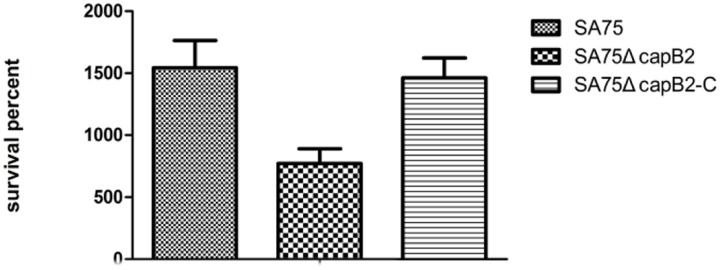
Absence of capB2 Decreased S. aureus Viability in Human Blood
To determine the contribution of capB2 in the pathogenicity of S. aureus, we compared the relative viability of S. aureus SA75 and SA75ΔcapB2 in human blood. After 7 h incubation, the survival percentages of the wild type strain SA75 and the complemented strain SA75ΔcapB2-C were significantly higher relative to SA75ΔcapB2 (Figure 2).
FIGURE 2.
The suvival rate of S. aureus derivatives in human blood. The number of CFU was detected at 0 and 7 h to calculate the rates of survival for S. aureus strains SA75, SA75ΔcapB2, and SA75ΔcapB2-C exposed to human blood.
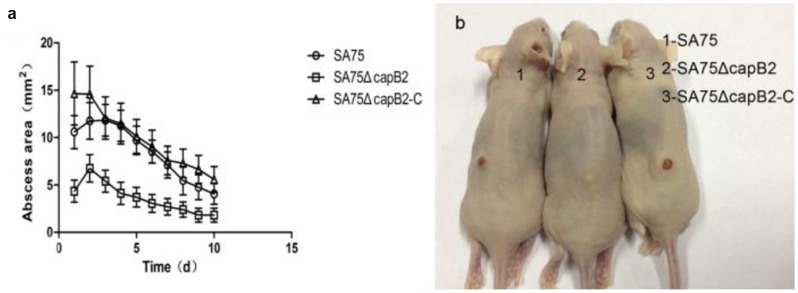
Deletion of capB2 Decreases Virulence of S. aureus SA75 in a Skin Infection Model
To determine the role of capB2 in the pathogenesis of S. aureus, 4-to-6 week old BALB/C-nu mice were subcutaneously injected with 1.0 × 105 CFU of strains SA75, SA75ΔcapB2, or SA75ΔcapB2-C. As shown in Figure 3b, the resulting abscess sizes in mice inoculated with the capB2 mutant strain were smaller than the lesion sizes among animals infected with SA75 or the complemented strain SA75ΔcapB2-C. The lesions remained until the end point of the experiment (Figure 3a). The overall rates of skin lesions were similar between the wild type and the complemented strain.
FIGURE 3.
Results of mouse skin infection model experiments. (a) Comparison of abscess size (area) of BALB/C-nu mice infected with wild type (SA75), isogenic capB2 mutant (SA75ΔcapB2), and capB2-complemented (SA75ΔcapB2-C) strains strains; (b) Skin lesions resulting from S. aureus infection (picture taken at day 10 of infection). Differences in the size/area of skin abscesses were compared by one-way analysis of variance (ANOVA) using SPSS Statistics 17.0.
As shown in Figure 3a, animals challenged with 1.0 × 105 CFU of SA75 had significantly larger (P < 0.05) skin lesions from the fourth day to the seventh day of infection than animals challenged with an equivalent amount of SA75ΔcapB Likewise, significant differences between the SA75ΔcapB and the SA75ΔcapB-C strains were observed from the second day to the ninth day of infection. These data demonstrate that the presence of capB2 in S. aureus promotes bacterial virulence.
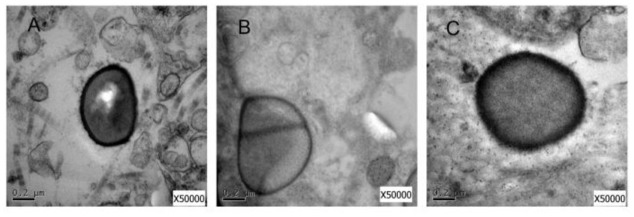
Electron Microscopy Study
To analyze the influence of the deletion of capB2 gene in S. aureus SA75, we used the transmission electron microscopy to detect the difference between S. aureus SA75, SA75ΔcapB and SA75ΔcapB-C strains. As shown in Figure 4, the related components of S. aureus SA75 cell wall in SA75ΔcapB was thinner than SA75 and SA75ΔcapB-C.
FIGURE 4.
Electron microscopy study. (A) Transmission electron micrograph of cell walls of S. aureus SA75 (B) the capB2 mutant strain (C) and the complement strain.
Effect of sarA on capB2 Expression
In our previous work, we constructed a sarA mutant strain. To eliminate the possibility of a second mutant, we constructed a sarA-complemented strain containing a plasmid bearing the sarA gene. Since the transcriptional expression of capB2 was highest at the 6 h time point, we selected that time point to assess expression differences among the three strains. The effect of sarA on expression of capB2 was assessed by qRT-PCR. As shown in Figure 5, deletion of sarA had a dramatic effect on capB2 expression, with a 326-fold increase at 6 h of growth compared with that of the wild type strain. The difference in capB2 expression was thus reversed by complementation of sarA. These results suggest that sarA represses the expression of capB2.
FIGURE 5.
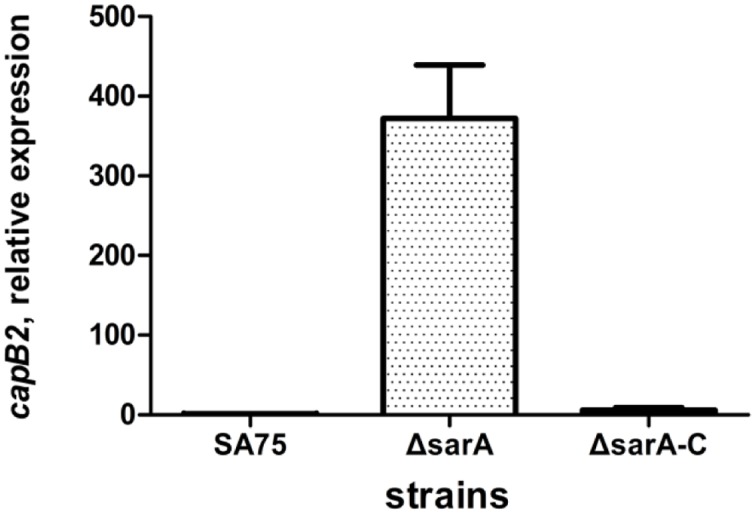
Effect of sarA on expression of capB2. Relative transcription of the capB2 gene was detected by qRT-PCR. Reference gene gyrB was used as the relative expression gene; the expression of capB2 in strain SA75 was regarded as 1.
Discussion
Capsular polysaccharide produced by S. aureus has been acknowledged as an important virulence factor for bacterial infection. Methods for S. aureus capsule serotyping include enzyme-linked immune sorbent assay (ELISA) inhibition, colony immune blotting, and double immune diffusion assay (the gold standard). However, all of them are based on the availability of specialized antibodies, and are not suitable for rapid capsule serotyping. Grunert et al. (2013) have developed a high-throughput method to identify capsular serotypes 5, 8 and NT (non-typable). DNA sequence analysis of S. aureus has determined that the cap5HIJK and cap8HIJK genes are the primary capsular type-specific genes. This information allowed us to identify the capsular genotype of SA75 using type-specific oligonucleotide primers (Verdier et al., 2007) designed against the cap5K and cap8I genes, with the results demonstrating that SA75 belongs to capsular type CP8.
Since the skin infection model was shown to be more sensitive, the role of CapB2 in S. aureus pathogenicity was evaluated using the skin infection model. Notably, the capB2 deletion decreased virulence in the mouse model of S. aureus skin infection, but full virulence was restored by transformation with the complementation plasmid containing the capB2 gene. Neither the capB2 mutant strain nor the complementation strain showed any difference in bacterial growth compared with the wild type strain when grown in TSB medium. Of important, the mutant of capB2 in S. aureus strain resulted in difference in the related components of cell wall. The role of CP in the pathogenesis of S. aureus infection has been reported by a number of studies (Baddour et al., 1992; Luong and Lee, 2002; O’Riordan and Lee, 2004; Nanra et al., 2013), which found that expression of CP5 or CP8 can enhance bacterial virulence both in vitro and in vivo. Taken together, we conclude that the decrease in virulence of the capB2 mutant strain is likely due to decreased synthesis of CP rather than the effect on overall growth.
Similar to other virulence proteins, CP and CapB2 are also regulated by a complex regulatory network. Luong T. et al. (2002) investigated the regulation of S. aureus CP8 expression by the global regulators agr and sarA, and found that the CP8 expression was positively regulated by agr and sarA, with agr being the major activator. However, our results demonstrate that capB2 expression was dramatically repressed by sarA, suggesting that SarA is a negative regulator of capB2 expression. The difference in regulation may be due to the complex regulatory network present in S. aureus, with the sarA interface within the regulatory network controlling the expression of virulence-associated proteins (Arvidson and Tegmark, 2001). In addition, the expression of CP is also influenced by several regulatory proteins. The regulation of CP by the two-component system KdpDE has been reported by Zhao et al. (2010), who found that CP expression was reduced in the kdpDE mutant strain, and that positive regulation occurred via the phosphorylation of KdpE. Furthermore, Lei and Lee (2015) found that RbsR (a Lacl family of repressor) positively regulates cap gene expression by directly binding to the cap operon promoter region, as evidenced by electrophoretic mobility shift assays (EMSA). In brief, the complexity of virulence regulatory networks in S. aureus is such that a single virulence protein may be controlled by a remarkable number of regulators.
In summary, CapB2 is a probable virulence gene in S. aureus infection, with deletion of capB2 resulting in decreased virulence in a mouse model of skin infection. SarA, on the other hand, is a negative regulator of capB2 expression, but further in-depth studies are required to determine if it directly regulates capB2.
Ethics Statement
This study was reviewed by the Ethics Committee of the first Affiliated Hospital of Wenzhou Medical University.
Author Contributions
DL performed the experiments, wrote the main manuscript text and analyzed the data. YG, SW, ZW, JL, and XQ helped to conduct and perform the experiments. ZC, LH, and XZ contributed to preparing the reagents/materials and supervised the project. LW and FY conceived the experiments and helped with the discussion of results. All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank José R. Mediavilla from International Center for Public Health Rutgers, the State University of New Jersey, USA, for revising the manuscript.
References
- Arbeit R. D., Karakawa W. W., Vann W. F., Robbins J. B. (1984). Predominance of two newly described capsular polysaccharide types among clinical isolates of Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 2 85–91. 10.1016/0732-8893(84)90002-6 [DOI] [PubMed] [Google Scholar]
- Arvidson S., Tegmark K. (2001). Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291 159–170. 10.1078/1438-4221-00112 [DOI] [PubMed] [Google Scholar]
- Baddour L. M., Lowrance C., Albus A., Lowrance J. H., Anderson S. K., Lee J. C. (1992). Staphylococcus aureus microcapsule expression attenuates bacterial virulence in a rat model of experimental endocarditis. J. Infect. Dis. 165 749–753. 10.1093/infdis/165.4.749 [DOI] [PubMed] [Google Scholar]
- Bae T., Schneewind O. (2006). Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55 58–63. 10.1016/j.plasmid.2005.05.005 [DOI] [PubMed] [Google Scholar]
- Cheung A. L., Zhang G. (2002). Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front. Biosci. 7:d1825–d1842. 10.2741/cheung [DOI] [PubMed] [Google Scholar]
- Grangeasse C., Nessler S., Mijakovic I. (2012). Bacterial tyrosine kinases: evolution, biological function and structural insights. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367 2640–2655. 10.1098/rstb.2011.0424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunert T., Wenning M., Barbagelata M. S., Fricker M., Sordelli D. O., Buzzola F. R., et al. (2013). Rapid and reliable identification of Staphylococcus aureus capsular serotypes by means of artificial neural network-assisted Fourier transform infrared spectroscopy. J. Clin. Microbiol. 51 2261–2266. 10.1128/JCM.00581-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakawa W. W., Fournier J. M., Vann W. F., Arbeit R., Schneerson R. S., Robbins J. B. (1985). Method for the serological typing of the capsular polysaccharides of Staphylococcus aureus. J. Clin. Microbiol. 22 445–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M., Ohta T., Uchiyama I., Baba T., Yuzawa H., Kobayashi I., et al. (2001). Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357 1225–1240. 10.1016/S0140-6736(00)04403-2 [DOI] [PubMed] [Google Scholar]
- Lei M. G., Lee C. Y. (2015). RbsR Activates Capsule but Represses the rbsUDK Operon in Staphylococcus aureus. J. Bacteriol. 197 3666–3675. 10.1128/JB.00640-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Du X., Villaruz A. E., Diep B. A., Wang D., Song Y., et al. (2012). MRSA epidemic linked to a quickly spreading colonization and virulence determinant. Nat. Med. 18 816–819. 10.1038/nm.2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong T., Sau S., Gomez M., Lee J. C., Lee C. Y. (2002). Regulation of Staphylococcus aureus capsular polysaccharide expression by agr and sarA. Infect. Immun. 70 444–450. 10.1128/IAI.70.2.444-450.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong T. T., Lee C. Y. (2002). Overproduction of type 8 capsular polysaccharide augments Staphylococcus aureus virulence. Infect. Immun. 70 3389–3395. 10.1128/IAI.70.7.3389-3395.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong T. T., Newell S. W., Lee C. Y. (2003). Mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185 3703–3710. 10.1128/JB.185.13.3703-3710.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong T. T., Ouyang S., Bush K., Lee C. Y. (2002). Type 1 capsule genes of Staphylococcus aureus are carried in a staphylococcal cassette chromosome genetic element. J. Bacteriol. 184 3623–3629. 10.1128/JB.184.13.3623-3629.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery C. P., Boyle-Vavra S., Roux A., Ebine K., Sonenshein A. L., Daum R. S. (2012). CodY deletion enhances in vivo virulence of community-associated methicillin-resistant Staphylococcus aureus clone USA300. Infect. Immun. 80 2382–2389. 10.1128/IAI.06172-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanra J. S., Buitrago S. M., Crawford S., Ng J., Fink P. S., Hawkins J., et al. (2013). Capsular polysaccharides are an important immune evasion mechanism for Staphylococcus aureus. Hum. Vaccin. Immunother. 9 480–487. 10.4161/hv.23223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson I. M., Lee J. C., Bremell T., Ryden C., Tarkowski A. (1997). The role of staphylococcal polysaccharide microcapsule expression in septicemia and septic arthritis. Infect. Immun. 65 4216–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares-Illana V., Meyer P., Bechet E., Gueguen-Chaignon V., Soulat D., Lazereg-Riquier S., et al. (2008). Structural basis for the regulation mechanism of the tyrosine kinase CapB from Staphylococcus aureus. PLoS Biol. 6:e143 10.1371/journal.pbio.0060143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Riordan K., Lee J. C. (2004). Staphylococcus aureus capsular polysaccharides. Clin. Microbiol. Rev. 17 218–234. 10.1128/CMR.17.1.218-234.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sau S., Bhasin N., Wann E. R., Lee J. C., Foster T. J., Lee C. Y. (1997a). The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology 143(Pt 7), 2395–2405. 10.1099/00221287-143-7-2395 [DOI] [PubMed] [Google Scholar]
- Sau S., Sun J., Lee C. Y. (1997b). Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J. Bacteriol. 179 1614–1621. 10.1128/jb.179.5.1614-1621.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sompolinsky D., Samra Z., Karakawa W. W., Vann W. F., Schneerson R., Malik Z. (1985). Encapsulation and capsular types in isolates of Staphylococcus aureus from different sources and relationship to phage types. J. Clin. Microbiol. 22 828–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulat D., Jault J. M., Duclos B., Geourjon C., Cozzone A. J., Grangeasse C. (2006). Staphylococcus aureus operates protein-tyrosine phosphorylation through a specific mechanism. J. Biol. Chem. 281 14048–14056. 10.1074/jbc.M513600200 [DOI] [PubMed] [Google Scholar]
- Thakker M., Park J. S., Carey V., Lee J. C. (1998). Staphylococcus aureus serotype 5 capsular polysaccharide is antiphagocytic and enhances bacterial virulence in a murine bacteremia model. Infect. Immun. 66 5183–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier I., Durand G., Bes M., Taylor K. L., Lina G., Vandenesch F., et al. (2007). Identification of the capsular polysaccharides in Staphylococcus aureus clinical isolates by PCR and agglutination tests. J. Clin. Microbiol. 45 725–729. 10.1128/JCM.01572-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eiff C., Taylor K. L., Mellmann A., Fattom A. I., Friedrich A. W., Peters G., et al. (2007). Distribution of capsular and surface polysaccharide serotypes of Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 58 297–302. 10.1016/j.diagmicrobio.2007.01.016 [DOI] [PubMed] [Google Scholar]
- Yu F., Lu C., Liu Y., Sun H., Shang Y., Ding Y., et al. (2014). Emergence of quinupristin/dalfopristin resistance among livestock-associated Staphylococcus aureus ST9 clinical isolates. Int. J. Antimicrob. Agents 44 416–419. 10.1016/j.ijantimicag.2014.06.020 [DOI] [PubMed] [Google Scholar]
- Zhao L., Xue T., Shang F., Sun H., Sun B. (2010). Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence. Infect. Immun. 78 3506–3515. 10.1128/IAI.00131-10 [DOI] [PMC free article] [PubMed] [Google Scholar]