Abstract
Recent genetic analysis has identified frequent mutations in ten-eleven translocation 2 (TET2), DNA methyltransferase 3A (DNMT3A), isocitrate dehydrogenase 2 (IDH2) and ras homolog family member A (RHOA) in nodal T-cell lymphomas, including angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma, not otherwise specified. We examined the distribution of mutations in these subtypes of mature T-/natural killer cell neoplasms to determine their clonal architecture. Targeted sequencing was performed for 71 genes in tumor-derived DNA of 87 cases. The mutations were then analyzed in a programmed death-1 (PD1)-positive population enriched with tumor cells and CD20-positive B cells purified by laser microdissection from 19 cases. TET2 and DNMT3A mutations were identified in both the PD1+ cells and the CD20+ cells in 15/16 and 4/7 cases, respectively. All the RHOA and IDH2 mutations were confined to the PD1+ cells, indicating that some, including RHOA and IDH2 mutations, being specific events in tumor cells. Notably, we found that all NOTCH1 mutations were detected only in the CD20+ cells. In conclusion, we identified both B- as well as T-cell-specific mutations, and mutations common to both T and B cells. These findings indicate the expansion of a clone after multistep and multilineal acquisition of gene mutations.
Introduction
Nodal T-cell lymphomas are subtypes of mature T-/natural killer-cell neoplasms, including angioimmunoblastic T-cell lymphoma (AITL); nodal peripheral T-cell lymphoma (PTCL) with T follicular helper (TFH) phenotype; peripheral T-cell lymphoma, not otherwise specified (PTCL-NOS), and follicular T-cell lymphoma. Among them, AITL is a distinct subtype, accounting for 16.0–28.7% of all mature T-/natural killer-cell neoplasms.1, 2, 3 AITL is characterized by specific clinical features, including generalized lymphadenopathy, high fever, skin rash and autoimmune-like manifestations. AITL tumor cells share characteristics with TFH cells, expressing B-cell lymphoma protein 6, a transcription factor; C-C motif chemokine receptor 5, a chemokine receptor; C-X-C motif ligand 13, a chemokine; and programmed death-1 (PD1), a member of the CD28 costimulatory membrane receptor family.4, 5 AITL tissues display prominent infiltration of inflammatory cells, follicular dendritic cell meshwork formation and branching vascular structures. Some nodal T-cell lymphomas exhibit several features reminiscent of AITL, although they do not show the typical morphology of AITL (nodal PTCL with TFH phenotype).6, 7 The massive infiltration of inflammatory cells in AITL has been explained by cytokines and chemokines being released from TFH-like tumor cells.4
Recurrent gene mutations have been identified in nodal T-cell lymphomas, including those in ten-eleven translocation 2 (TET2) in 20–83%, isocitrate dehydrogenase 2 (IDH2) in 0–45%, and ras homolog family member A (RHOA) in 17–71%, depending on the subtypes and DNA methyltransferase 3A (DNMT3A) in approximately 30%, independent of the subtypes.8, 9, 10, 11, 12, 13 Mutations in TET2 encoding a methylcytosine dioxygenase and those in DNMT3A encoding a DNA methyltransferase presumably result in epigenetic abnormalities in nodal T-cell lymphomas. IDH2 mutations also affect epigenetic modifications by inhibiting TET and histone demethylation enzymes through production of 2-hydroxyglutarate.14 Mutations in RHOA encoding a small GTPase are almost always located at the hotspot site, resulting in conversion from glycine to valine at the seventeenth position of the RHOA protein (G17V RHOA mutation). The G17V RHOA mutants could not be converted to an active GTP-bound form, although the downstream signaling of the G17V RHOA mutants in nodal T-cell lymphomas development has yet to be clarified.8, 9, 13
TET2 and DNMT3A mutations are proposed to arise in hematopoietic stem/progenitors upstream of T-lineage commitment. This hypothesis is based on the fact that identical TET2 and DNMT3A mutations were found in both tumor tissues and apparently normal blood cells in some AITL and PTCL-NOS patients.8, 10, 15, 16, 17 In contrast, the origins of the G17V RHOA mutation remain to be elucidated: it may be a tumor-specific event, considering that the allele frequencies of G17V RHOA mutations were lower than those of TET2 mutations and that G17V RHOA mutations were found in only CD4+T lymphocytes in 1 AITL and 1 PTCL-NOS case.8
Here we describe the clonal architecture of nodal T-cell lymphomas by determining the distribution of mutations in enriched tumor cells and infiltrated B cells.
Materials and methods
Patients and samples
Samples, obtained from 87 patients (Supplementary Table S1) with AITL (n=48), nodal PTCL with TFH phenotype (n=5) and either of PTCL-NOS or of nodal PTCL with TFH phenotype (PTCL-NOS/nodal PTCL with TFH phenotype, n=34),5 were used after approval was obtained from the local ethics committees of all the participating institutes.
Genomic DNA was extracted from 56 fresh frozen samples using the Puregene DNA Blood Kit (Qiagen, Hilden, Germany) and 31 periodate-lysine-paraformaldehyde (PLP)-fixed frozen samples using the QIAamp DNA FFPE Tissue Kit (Qiagen).
Targeted sequencing
Targeted sequencing was performed for 71 genes, which are listed in Supplementary Table S2. Sixty-one of the genes were previously screened by whole-exome sequencing,8 while 6 were the family genes of those whose mutations were identified by the whole-exome sequencing. The other four genes were deemed susceptible to mutations in PTCLs on the basis of the mutational profiles of other lymphoid malignancies.18, 19, 20, 21, 22, 23 All the exons of the selected genes were captured by use of a SureSelect Target Enrichment Kit (Agilent, Santa Clara, CA, USA) and then massively sequenced using HiSeq2000 (Illumina, Santa Clara, CA, USA). For each sample, all the sequencing reads were aligned to hg19 using BWA version 0.5.8 with default parameters. After all the duplicated reads and the low-quality reads and bases were removed, the allele frequencies of single-nucleotide variants and indels at each genomic position were calculated by enumerating the relevant reads using SAMtools (http://www.htslib.org). Initially, all the variants showing allele frequencies >0.02 were extracted and annotated using ANNOVAR24, 25 for further consideration, if they were found in >6 reads of >10 total reads and appeared in both the positive- and the negative-strand reads. All synonymous variants and known single-nucleotide polymorphisms in public and private databases, including dbSNP131, the 1000 genomes project as of 2012/05/21 and our in-house database, were removed. To exclude germline variants, nonsynonymous variants were excluded when the allele frequencies were from 0.45 to 0.55. Candidate mutations were validated by amplicon-based deep sequencing using Ion PGM (Life Technologies, Carlsbad, CA, USA) and/or Sanger sequencing (see below).
In the cohort of 87 cases, 79 were analyzed for RHOA, TET2, DNMT3A and IDH2 mutations, and the results of this analysis were described in the previous paper.7 Now, eight were new cases. We re-analyzed all the 87 samples for targeted sequencing of 71 genes.
Amplicon-based sequencing
The libraries were prepared using the Ion Plus Fragment Library Kit according to the protocol for preparing short amplicon libraries (Life Technologies). Briefly, PCR amplicons were ligated to the barcode adapters and P1 adapters and then amplified. The amplified libraries were quantitated by quantitative PCR with the Ion Library Quantitation Kit according to the manufacturer's instructions (Life Technologies). The libraries were then subjected to deep sequencing on the Ion Torrent PGM platform according to the standard protocol for 300 base-pair single-end reads (Life Technologies). The data were analyzed using Variant Caller 3.4 (Life Technologies).
Immunohistochemistry
PLP-fixed frozen samples were cut in a cryostat at −22 °C into 5-μm sections and mounted on PEN-Membrane slides (Leica, Wetzlar, Germany). The tissue sections were stained with mouse anti-human PD1 (NAT105 ab52587, Abcam, Cambridge, UK) and anti-human CD20cy (clone L26, Dako, Michigan, MI, USA) antibodies, diluted 1:2000 and 1:1000, respectively, and detected by use of the Envision+ Dual Link System-HRP (Dako). The tissue sections were then counterstained with hematoxylin (Mayer's hematoxylin, Muto Pure Chemical, Tokyo, Japan) for 20 s at room temperature. After staining, tissue sections were dehydrated with ethanol and dried at room temperature before laser microdissection (LMD).
LMD, DNA extraction and PCR
Nineteen of the 87 cases (13 AITL, 1 nodal PTCL with TFH phenotype and PTCL-NOS/nodal PTCL with TFH phenotype) were analyzed by LMD, which was performed using LMD7000 (Leica). The cells being positive for either PD1 or CD20 were dissected and collected into 0.2-ml PCR tubes (Takara, Shiga, Japan) containing 20 μl of distilled water. Stained cells at approximately 100 000 μm2 were dissected and collected for each sample. Genomic DNA was extracted using the QIAamp DNA PFFE Tissue Kit (Qiagen) following the manufacturer's protocol. Then 1 μl of DNA was used for PCR under the following conditions: 95 °C for 15 min, 60 °C for 4 min, 72 °C for 4 min, 35 to 40 cycles at 95 °C for 1 min, 60 °C for 1 min, 72 °C for 1 min, and 72 °C for 10 min using the AmpliTaq Gold 360 Kit (Applied Biosystems, Foster City, CA, USA) with each primer set (Supplementary Table S6). PCR amplicons were used for amplicon-based sequencing and Sanger sequencing.
IgH gene rearrangement analysis and subcloning of the PCR product
Multiplex PCR assays were used to detect the clonality of B cells according to the European BIOMED-2 collaborative study.26 PCR products migrating at the expected size were extracted and sequenced using the Sanger method. Subcloning was performed if the Sanger sequencing indicated a polyclonal background by use of the pGEM-T Easy Vector System I (Promega, Madison, WI, USA). At least 12 colonies were picked up and sequenced to confirm the clonal expansion. The sequence results were analyzed using the IMGT tools27 and aligned to the closest match with the germline IGHV segment. Sequencing results with a germline identity of <98% were regarded as mutated and vice versa according to previous study.28
Results
Novel recurrent mutations in nodal T-cell lymphomas
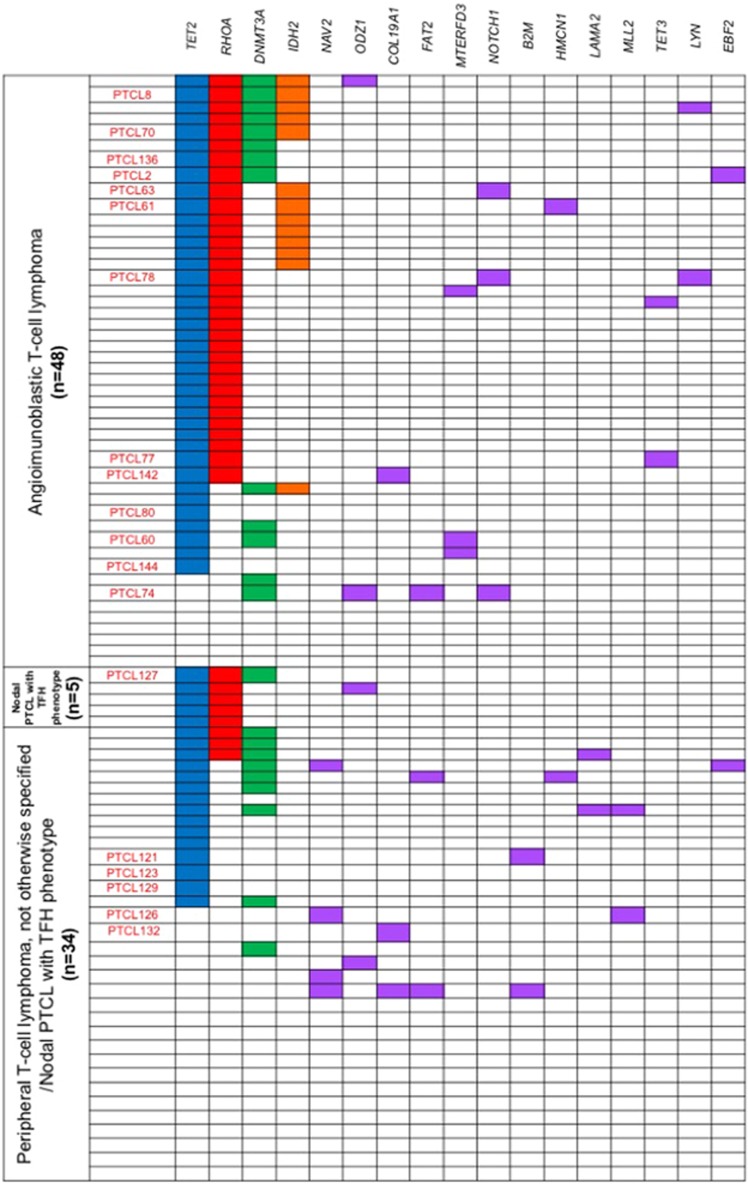
Targeted sequencing for 71 genes was performed in 87 samples (Supplementary Table S1), including AITL (n=48), nodal PTCL with TFH phenotype (n=5) and PTCL-NOS/nodal PTCL with TFH phenotype (n=34). TET2, DNMT3A, RHOA and IDH2 mutations were identified in 60 (68.7%), 23 (26.4%), 41 (47.1%) and 13 (14.9%) of 87 cases, respectively (Figure 1, Table 1, Supplementary Table S3). The mutational profiles of these 4 genes in 79 of the 87 samples are described elsewhere.7
Figure 1.
Targeted sequencing result of 87 nodal T-cell lymphoma samples. TET2-, RHOA-, IDH2- and DNMT3A-mutated cases are indicated by blue, red, orange and green boxes, respectively. Other recurrently mutated genes are in purple. The laser-microdissected samples are indicated in red letters.
Table 1. Targeted sequencing result of 87 nodal T-cell lymphoma samples.
| Gene |
AITL (n=48) |
Nodal PTCL with TFH phenotype (n=5) |
PTCL-NOS/nodal PTCL with TFH phenotype (n=34) |
All (n=87) |
||||
|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | |
| TET2 | 36 | 75 | 5 | 100 | 19 | 55.9 | 60 | 69 |
| RHOA | 33 | 68.8 | 5 | 100 | 3 | 8.8 | 41 | 47.1 |
| DNMT3A | 11 | 22.9 | 1 | 20 | 11 | 32.4 | 23 | 26.4 |
| IDH2 | 13 | 27.1 | 0 | 0 | 0 | 0 | 13 | 14.9 |
| NAV2 | 0 | 0 | 0 | 0 | 4 | 11.8 | 4 | 4.6 |
| ODZ1 | 2 | 4.2 | 1 | 20 | 1 | 2.9 | 4 | 4.6 |
| COL19A1 | 1 | 2.1 | 0 | 0 | 2 | 5.9 | 3 | 3.4 |
| FAT2 | 1 | 2.1 | 0 | 0 | 2 | 5.9 | 3 | 3.4 |
| MTERFD3 | 2 | 4.2 | 0 | 0 | 1 | 2.9 | 3 | 3.4 |
| NOTCH1 | 3 | 6.3 | 0 | 0 | 0 | 0 | 3 | 3.4 |
| B2M | 0 | 0 | 0 | 0 | 2 | 5.9 | 2 | 2.3 |
| HMCN1 | 1 | 2.1 | 0 | 0 | 1 | 2.9 | 2 | 2.3 |
| LAMA2 | 0 | 0 | 0 | 0 | 2 | 5.9 | 2 | 2.3 |
| MLL2 | 0 | 0 | 0 | 0 | 2 | 5.9 | 2 | 2.3 |
| TET3 | 2 | 4.2 | 0 | 0 | 0 | 0 | 2 | 2.3 |
| LYN | 2 | 4.2 | 0 | 0 | 0 | 0 | 2 | 2.3 |
| EBF2 | 1 | 2.1 | 0 | 0 | 1 | 2.9 | 2 | 2.3 |
Abbreviations: AITL, angioimmunoblastic T-cell lymphoma; nodal PTCL with TFH phenotype, nodal peripheral T-cell lymhoma with T follicular helper phenotype; PTCL-NOS, peripheral T-cell lymhoma, not otherwise specified.
Thirty-four novel recurrent mutations were identified in 13 of the 71 (18.3%) genes and in 24 of the 87 (26.4%) cases (Figure 1, Table 1 and Supplementary Table S4). Mutations in genes associated with lymphoid malignancies, for example, Notch homolog 1, translocation-associated (NOTCH1), β2 microglobulin (B2M) and mixed-lineage leukemia 2 (MLL2) were identified in 3, 2 and 2 cases, respectively. Mutations in FAT atypical cadherin 2 (FAT2), a gene associating with several cancers, and those in TET3, a member of the TET gene family, were identified in 3 and 2 cases, respectively.
Nineteen of the 87 samples were analyzed by the use of LMD (Figure 1). The frequencies of TET2, DNMT3A, RHOA and IDH2 mutations in the laser-microdissected samples were similar to those found in the entire cohort (TET2 mutations, 16/19 (84.1%); DNMT3A mutations, 7/19 (36.8%); RHOA mutations, 10/19 (52.6%) and IDH2 mutations, 4/19 (21.1%)) (Table 2). A total of 26 TET2 mutations were found in 16 cases, while 2 TET2 mutations were found in 10 samples each (Supplementary Table S5). NOTCH1 and COL19A1 mutations were identified in 3 and 2 cases, respectively. Other gene mutations, including NAV2, ODZ1, FAT2, MTERFD3, B2M, HMCN1, MLL2, TET3 and LYN, were identified in a single case each.
Table 2. Mutation profiles of 19 laser microdissected samples.
| Gene |
AITL (n=13) |
Nodal PTCL with TFH phenotype (n=1) |
PTCL-NOS/nodal PTCL with TFH phenotype (n=5) |
All (n=19) |
||||
|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | |
| TET2 | 12 | 92.3 | 1 | 100 | 3 | 60 | 16 | 84.2 |
| RHOA | 9 | 69.2 | 1 | 100 | 0 | 0 | 10 | 52.6 |
| DNMT3A | 6 | 46.2 | 1 | 100 | 0 | 0 | 7 | 36.8 |
| IDH2 | 4 | 30.8 | 0 | 0 | 0 | 0 | 4 | 21.1 |
| NAV2 | 0 | 0 | 0 | 0 | 1 | 20 | 1 | 5.3 |
| ODZ1 | 1 | 7.7 | 0 | 0 | 0 | 0 | 1 | 5.3 |
| COL19A1 | 1 | 7.7 | 0 | 0 | 1 | 20 | 2 | 10.5 |
| FAT2 | 1 | 7.7 | 0 | 0 | 0 | 0 | 1 | 5.3 |
| MTERFD3 | 1 | 7.7 | 0 | 0 | 0 | 0 | 1 | 5.3 |
| NOTCH1 | 3 | 23.1 | 0 | 0 | 0 | 0 | 3 | 15.8 |
| B2M | 0 | 0 | 0 | 0 | 1 | 20 | 1 | 5.3 |
| HMCN1 | 1 | 7.7 | 0 | 0 | 0 | 0 | 1 | 5.3 |
| MLL2 | 0 | 0 | 0 | 0 | 1 | 20 | 1 | 5.3 |
| TET3 | 1 | 7.7 | 0 | 0 | 0 | 0 | 1 | 5.3 |
| LYN | 1 | 7.7 | 0 | 0 | 0 | 0 | 1 | 5.3 |
Abbreviations: AITL, angioimmunoblastic T-cell lymphoma; nodal PTCL with TFH phenotype, nodal peripheral T-cell lymhoma with T follicular helper phenotype; PTCL-NOS, peripheral T-cell lymhoma, not otherwise specified.
Specific existence of G17V RHOA mutations in tumor cell-enriched cells of nodal T-cell lymphomas
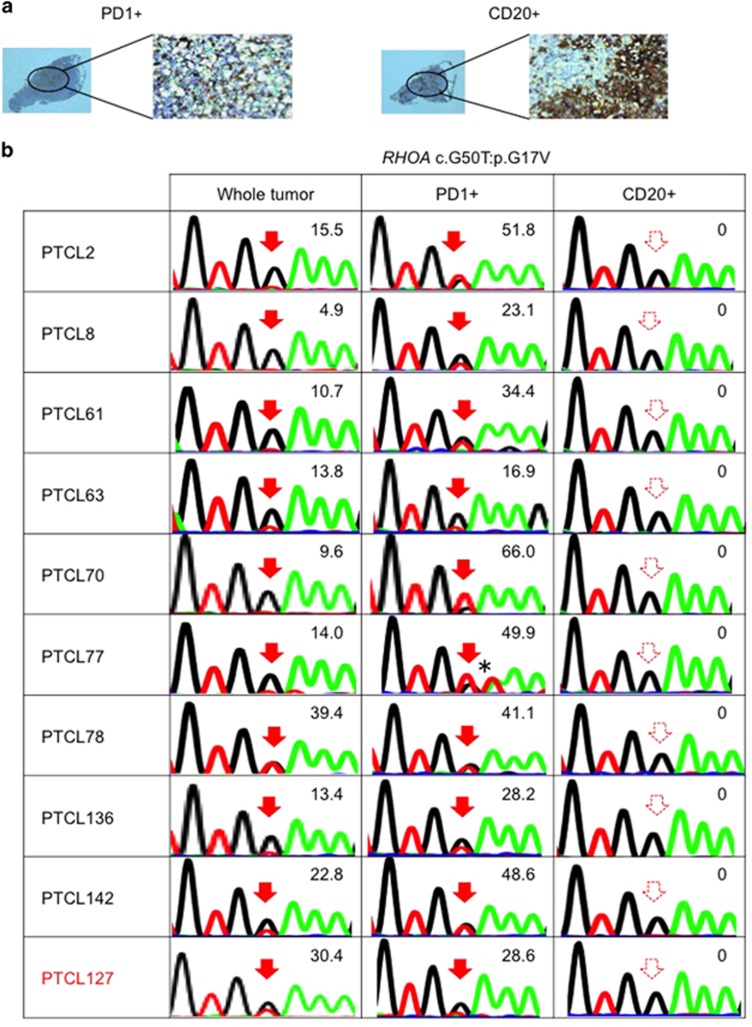
Previously, we reported that G17V RHOA mutations were detected by flow cytometry only in CD4-positive (CD4+) cells but not in other cell lineages purified from the skin tumor of a PTCL-NOS patient and the pleural effusion cells of an AITL patient.8 These preliminary results suggested that the G17V RHOA mutation may specifically exist in mature CD4+ T cells in PTCL-NOS and AITL. To gain further insight into the origin of the G17V RHOA mutation, we examined the mutation in laser-microdissected PD1+ and CD20+ B cells, which were assumed to be enriched and depleted in tumor cells, respectively, in 10 nodal T-cell lymphomas (1 nodal PTCL with TFH phenotype and 9 AITL cases). The G17V RHOA mutation was detected only in the PD1+ cells but not in the CD20+ cells in all 10 cases (Figure 2). The allele frequencies of the G17V RHOA mutations in the dissected PD1+ cells were substantially higher than those in the matched whole tumor samples in 7 of the 10 cases. The efficiency of mutation allele enrichment was not substantial in three cases (PTCL63, PTCL78 and PTCL127). In these cases, PD1+ cell selection was not sucessful enough to purify the tumor cells because of the presumed abundance of PD1+ non-tumor cells or the very high tumor cell content before the selection. Additionally, using flow cytometry, we found an AITL case showing that the G17V RHOA mutation existed in PD1+CD4+ cells sorted from bone marrow mononuclear cells (Supplementary Figure S2). This finding strongly supports our hypothesis that the acquisition of the G17V RHOA mutation is a specific event in TFH cells.
Figure 2.
RHOA mutations are specific to PD1+ cells. (a) An example of the immunostaining pattern for PD1 and CD20 in AITL. Left, PD1+ cells; right, CD20+ cells. (b) Sequences of G17V RHOA mutations in whole tumor, PD1+ cells and CD20+ cells. The numeric values indicate allele frequencies of mutations defined by amplicon-based deep sequencing. The AITL samples are indicated in black letters. The nodal PTCL with TFH phenotype sample is indicated in red letters *: RHOA c.A51T:p.G17V, silent mutation. The filled and dashed red arrows indicate mutations and no mutations, respectively.
Distribution of TET2, IDH2 and DNMT3A mutations
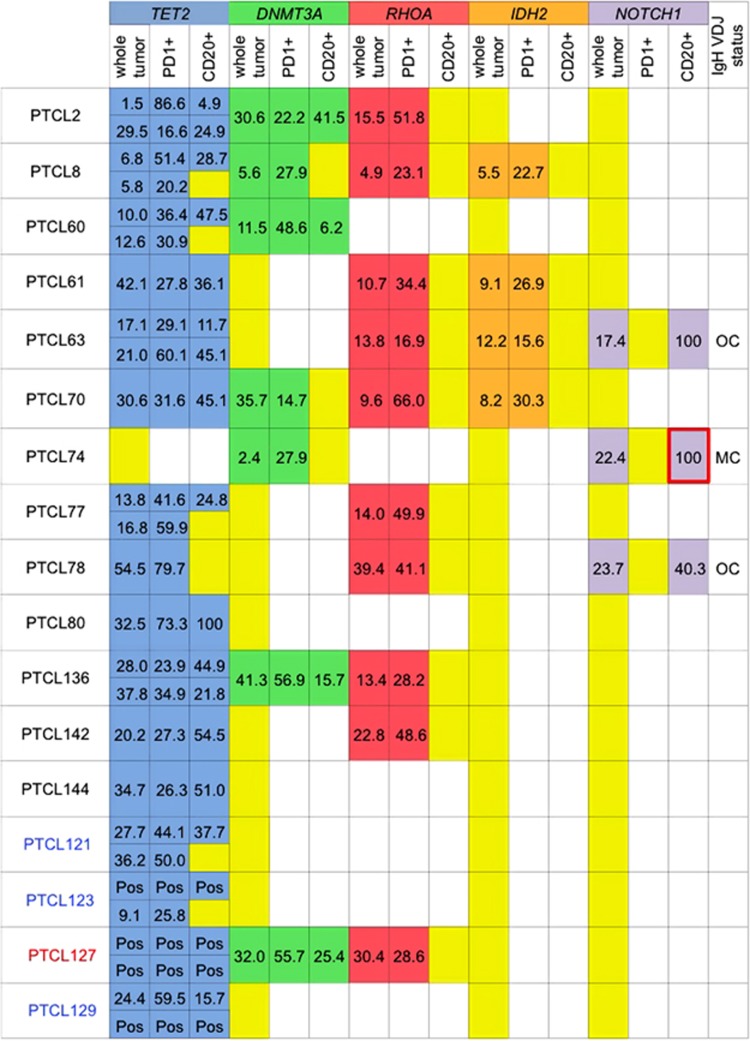
We and others have previously reported that TET2 and DNMT3A mutations were found in apparently normal blood cells, including bone marrow mononuclear cells, and in immature progenitors and blood cells of various lineages isolated from peripheral blood of a few PTCL patients.7, 9, 15, 16, 17 We examined the distribution of TET2, IDH2 and DNMT3A mutations in PD1+ and CD20+ cells. Twenty of the 26 TET2 mutations were identified in both the PD1+ and the CD20+ cells (Supplementary Table S7), and 15 of the 16 TET2-mutated samples had at least one mutation in both the PD1+ and the CD20+ cells (Figure 3). Concomitantly, DNMT3A mutations were identified in both the PD1+ and CD20+ cells in four of the seven DNMT3A-mutated samples (Figure 3, Supplementary Table S8). In myeloid malignancies, TET2 and IDH2 mutations are known to be mutually exclusive.14, 29 However, we and others reported that IDH2 mutations often coexist with TET2 mutations in PTCL.8, 10, 30 IDH2 mutations were identified in PD1+ cells but not in CD20+ cells in all 4 TET2- and IDH2-comutated samples (PTCL8, PTCL61, PTCL63 and PTCL70) (Figure 3). Each of these samples had at least one TET2 mutation in both the PD1+ and CD20+ cells and the G17V RHOA mutation only in the PD1+ cells. That is, TET2, IDH2 and G17V RHOA mutations coexisted in the PD1+ cells in these cases. In addition, we also found the coexistence of IDH2, TET2 and G17V RHOA mutations in PD1+CD4+ cells sorted from the bone marrow mononuclear cells of an AITL patient (Supplementary Figure S2).
Figure 3.
Distributions of TET2/DNMT3A/RHOA/IDH2/NOTCH1 mutations and IgH VDJ status. Allele frequencies of TET2/DNMT3A/RHOA/IDH2/NOTCH1 mutations in whole tumor, PD1+ cells and CD20+ cells are shown. The blue boxes represent positive TET2 mutations; the green boxes, positive DNMT3A mutations; the red boxes, positive RHOA mutations; the orange boxes, positive IDH2 mutations; the purple boxes, positive NOTCH1 mutations; the yellow boxes, no mutations; and the white boxes, not examined. The numeric values indicate allele frequencies of mutations defined by deep sequencing, except for that in the box surrounded by bold red lines which was estimated by Sanger sequencing. IgH VDJ status indicates the IgH VDJ rearrangement status in whole-tumor-derived DNA. The AITL samples are indicated in black letters. The nodal PTCL with TFH phenotype sample is indicated in red letters. The PTCL-NOS/nodal PTCL with TFH phenotype sample is indicated in blue letters. MC, monoclonality; OC, oligoclonality; Pos: positivity was evaluated only by Sanger sequencing. Multiple TET2 mutations were identified in PTLC2, 8, 60, 63, 77, 136, 121, 123, 127 and 129.
B-cell-specific mutations in nodal T-cell lymphomas
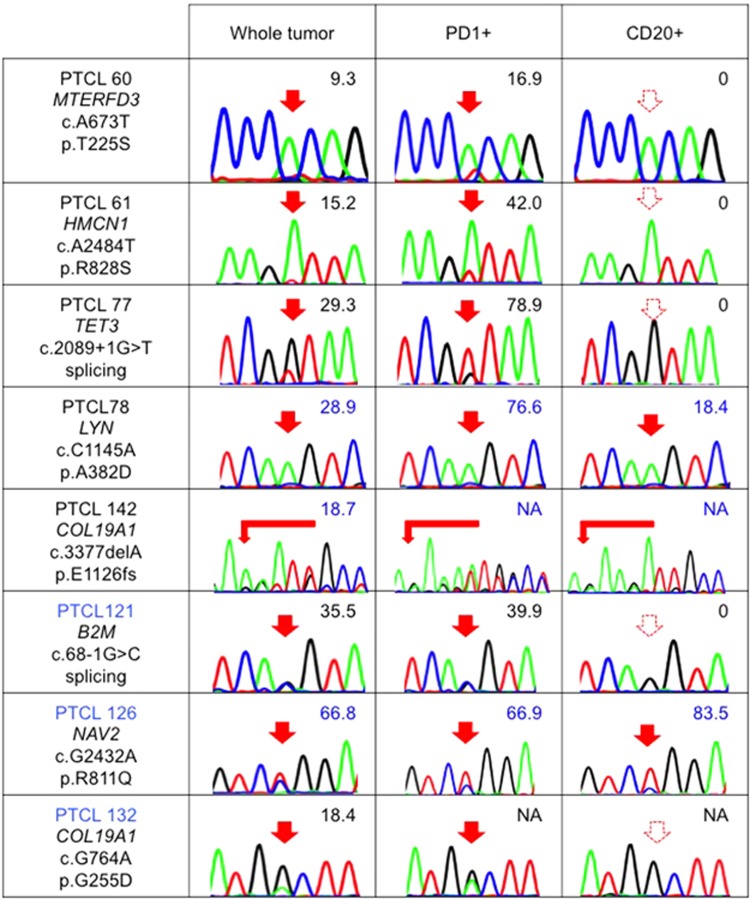
To clarify the cellular origin of newly identified gene mutations, we also checked the distribution of these mutations in PD1+ and CD20+ cells (Table 2). We identified B2M, COL19A1, HMCN1, MTERFD3 and TET3 mutations only in PD1+ cells but not in CD20+ cells. COL19A1, LYN, NAV2 and NOTCH2NL mutations were identified in both the PD1+ and CD20+ cells (Figure 4).
Figure 4.
Distribution of newly identified gene mutations in nodal T-cell lymphomas. The results of Sanger sequencing and/or amplicon-based deep sequencing for some newly identified gene mutations in whole tumor, PD1+ cells and CD20+ cells are shown. The numeric values indicate allele frequencies of mutations defined by deep sequencing. The AITL samples are indicated in black letters. The PTCL-NOS/nodal PTCL with TFH phenotype sample is indicated in blue letters. NA, not analyzed by deep sequencing. The filled and dashed red arrows indicate mutations and no mutations, respectively.
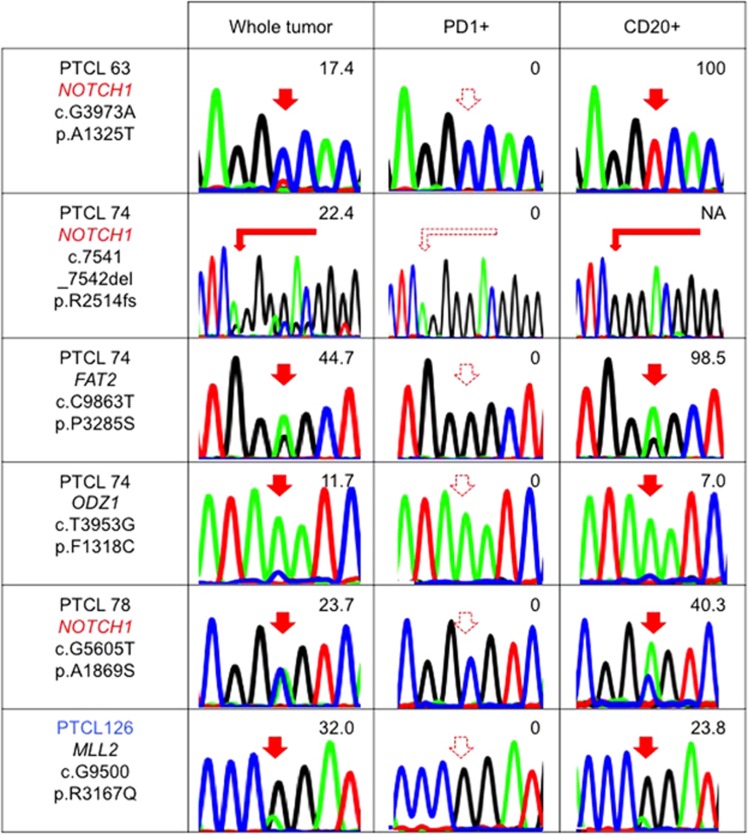
Interestingly, three NOTCH1 and one FAT2, MLL2 and ODZ1 mutations each were found only in the CD20+ but not in the PD1+ cells in four samples (PTCL 63, PTCL70, PTCL78 and PTCL128) (Figure 5). Especially, all three NOTCH1 mutations identified by targeted sequencing were identified only in the CD20+ cells with high allele frequencies. The NOTCH1 gene encodes a transmembrane protein. One of the NOTCH1 mutations was a frameshift mutation residing in the PEST domain of the Notch1 protein. This would be an activating mutation, because deletion of the PEST domain enhances Notch signaling after ligand binding.19 The other two mutations were located in one of the epidermal growth factor-like and in the ankyrin repeat domains (Supplementary Figure S1). One of the NOTCH1-mutated samples simultaneously had two TET2 mutations and G17V RHOA mutation (PTCL 63, Supplementary Table S9). In this case, both TET2 mutations were detected in both the PD1+ and CD20+ cells, while the G17V RHOA mutation was confined to the PD1+ cells. We used the multiplex PCR method26 to also check the clonality of immunoglobulin genes in the samples with B-cell-specific mutations. Interestingly, only one sample showed monoclonal rearrangement while the others showed oligoclonal rearrangement (Table 3).
Figure 5.
B-cell-specific mutations in nodal T-cell lymphomas. The results of Sanger sequencing and/or amplicon-based deep sequencing for some newly identified gene mutations in whole tumor, PD1+ cells and CD20+ cells are shown. The numeric values indicate allele frequencies of mutations defined by deep sequencing. The AITL samples are indicated in black letters. The PTCL-NOS/nodal PTCL with TFH phenotype sample is indicated in blue letters. NA, not analyzed by deep sequencing. The filled and dashed red arrows indicate mutations and no mutations, respectively. NOTCH1 is marked by red letters because this is repetitive.
Table 3. VDJ rearrangement status of B-cell clones in B-cell-specific mutated samples.
| Sample | Diagnosis | Number of colonies having the identical VDJ gene usage/total number of colonies analyzed | Common VDJ gene usage | Identity of V gene (%) | Amino-acid sequences of junctions |
|---|---|---|---|---|---|
| PTCL63 | AITL | 2/12 | V3-21/D2-2/J5 | 72.2 | CARSTQTYYQLLWNG#NWFDPWa |
| PTCL74 | AITL | NAb | V1-2/J1 or J2 or J3 | 84.4 | Not identified at http://www.imgt.org |
| PTCL78 | AITL | 2/12 | V3-23/J6/D4-17 | 72.2 | CAKGNDYGDSYYYGMDVW |
| 2/12 | V3/J6/D3-10 | 77.2 | CARDRGYYYYGMDVW | ||
| PTCL126 | PTCL-NOS/nodal PTCL with TFH phenotype | 2/12 | V6-1/J6/D3-3 | 71.0 | CARTTPSTIFGVVTAGYYYYGMDVW |
Out of frame junction.
NA, not applicable because direct sequencing demonstrated monoclonality.
Discussion
By determining the distribution of the mutations, we elucidated the clonal architecture of nodal T-cell lymphomas. RHOA mutations were identified only in PD1+ cells in 100% cases, while TET2 and DNMT3A mutations were identified in both the PD1+ cells and CD20+, tumor-cell-depleted cells in the majority of cases. In addition, IDH2 mutations were actually found only in the PD1+ cells and coexisted with TET2 mutations. These data suggest that, in nodal T-cell lymphoma development, multistep tumorigenesis may progress in association with the differentiation of blood cells/lymphocytes. Surprisingly, some of the mutations resided in a B-cell-specific manner.
Recent genetic studies have revealed that, in several hematological cancers, several gene mutations existed in preleukemic hematopoietic cells as well as in tumor cells;31 examples are TET2 and/or DNMT3A mutations in acute myeloid leukemia32, 33, 34 and NOTCH1 and SF3B1 mutations in chronic lymphocytic leukemia.35, 36 Moreover, somatic mutations have been demonstrated in elderly individuals without hematological malignancies: DNMT3A, ASXL1, and TET2 mutations frequently observed in hematological malignancies were the most frequent in these cohorts.37, 38, 39, 40 Similarly, our data indicated that in nodal T-cell lymphomas, premalignant cells having TET2 and/or DNMT3A mutations may differentiate not only into T-lineage tumor cells but also into B cells. In contrast, the G17V RHOA mutations specifically existed in the T cells of nodal T-cell lymphomas in all 13 cases (11 cases have been described in this paper, while 2 were previously described elsewhere8), indicating that the G17V RHOA mutation is the event after the B- and T-cell specification. This could happen right after the T/B specification, after differentiation into TFH cells or even after malignant transformation establishing a subclone. One possibility is that the G17V RHOA mutation occurs in TET2-mutated premalignant cells and facilitates the selective differentiation of TET2-mutated premalignant cells into tumor cells with the TFH phenotype. This needs to be proven in the future.
IDH2 mutations were also specifically identified in the tumor-cell-enriched cells, suggesting that IDH2 mutations are also tumor-cell-specific events in AITL, although the number of samples was not large enough to allow a definite conclusion. We have previously showed that the IHD2-mutated cases were almost a subcohort of G17V RHOA-mutated cases.8 This result could be interpreted that the acquisition of IDH2 mutations may be the event occurring after the acquisition of RHOA mutation and thus the IDH2-mutated cells may, at least in some cases such as PTCL70, constitute a subclone in the RHOA-mutated clone. TET2- and IDH2-comutated AITL samples were reported to have more extensive histone modification profiles than those with TET2 mutations without an IDH2 mutation, while the difference in genome-wide cytosine methylation profiles between these samples was only moderate.30
Our data showed that B cells that have infiltrated AITL tissues also have gene mutations: the multilineal mutations represented by those in TET2 and DNMT3A, and B-cell-specific mutations represented by those in NOTCH1 and other genes. Monoclonal or oligoclonal expansion of B cells has been found in up to 30% of AITL cases.41, 42, 43 Furthermore, approximately 10% of AITL cases develop B-cell malignancies during their clinical course.42, 44, 45, 46, 47 Some lymphoma cells are infected by Epstein–Barr virus. In such cases, Epstein–Barr virus is proposed to contribute to the transformation of B cells.45, 46 This hypothesis, however, needs to be re-evaluated because Epstein–Barr virus was not detected in a substantial proportion of B-cell malignancies accompanying AITL.48 TET2 mutations are found in diffuse large B-cell lymphomas.49 Tet2-deficient mice show the expansion of both B- and T-cell lineages in addition to prominent myeloproliferation.15 Combinational loss of Tet1 and Tet2 provokes B-cell malignancies in mice.50 Activating NOTCH1 mutations were reported in diffuse large B-cell lymphomas,51 chronic lymphocytic leukemia,52 mantle cell lymphoma22 and follicular lymphoma.53 In our cohort, all three NOTCH1 mutations were defined only in B cells with very high allele frequencies (Figure 5, Supplementary Table S9) and two of the three samples showed oligoclonality of B cells (Table 3). This implies that the origin of NOTCH1 mutation is earlier than the acquisition of hypermutation of the CDR3 region in the immunoglobulin gene. Anyway, acquisition of these mutations in B-cell lineage may account for the frequent occurrence of B-cell lymphomas in AITL. Moreover, our data alert us to the need for careful interpretation of the mutational profiles of PTCLs because some of the mutations may not exist in tumor cells.
In conclusion, our findings illustrate the concept of multistep and multilineal tumorigenesis in nodal T-cell lymphomas (Supplementary Figure S3). Understanding the pathogenesis will lead us to better management of nodal T-cell lymphomas in future.
Acknowledgments
We thank Tamaki Takahashi and Yukari Sakashita for their technical assistances. We also thank Ms Flaminia Miyamasu, associate professor of English for Medical Purposes, Medical English Communications Center, University of Tsukuba, for her editorial assistance. This work was supported by Grants-in-Aid for Scientific Research (KAKENHI: 25461407 to MS-Y; and 25112703, 15H01504 and 16H02660 to SC) from the Ministry of Education, Culture, Sports, and Science of Japan.This work was also supported by The Project for Cancer Research and Therapeutic Evolution (P-CREATE) from The Japan Agency for Medical Research and Development, AMED and by research grants from the Daiichi Sankyo Foundation of Life Science, the Naito Foundation, the Cell Science Research Foundation, KANAE Foundation for the Promotion of Medical Science and Shiseido Female Researcher Science Grant to MS-Y; and The Uehara Memorial Foundation, Leukemia Research Fund, Takeda Science Foundation, and Kobayashi Foundation for Cancer Research to SC.
Author contributions
TBN, MS-Y and YA performed experiments and data analysis; KY, YS, KC, HT, SM and SO performed the targeted sequencing; NN, KT, HM, MN and KO were responsible for pathological diagnosis; DM, JK, TM, NN, KT, HM and MN established the LMD procedure; KI and KO collected specimens and were involved in planning the project; TBN, MS-Y and SC generated figures and tables and wrote the manuscript; SC lad the entire project; and all authors participated in discussions and interpretation of the data and results.
Footnotes
Supplementary Information accompanies this paper on Blood Cancer Journal website (http://www.nature.com/bcj)
The authors declare no conflict of interest.
Supplementary Material
References
- Wang SS, Vose MJ. Epidemiology and Prognosis of T-cell Lymphoma. Humana Press, 2013, pp 25–38. [Google Scholar]
- Park S, Ko YH. Peripheral T cell lymphoma in Asia. Int J Hematol 2014; 99: 227–239. [DOI] [PubMed] [Google Scholar]
- Wang SS, Flowers CR, Kadin ME, Chang ET, Hughes AM, Ansell SM et al. Medical history, lifestyle, family history, and occupational risk factors for peripheral T-cell lymphomas: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr 2014; 2014: 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leval L, Gaulard P. Pathobiology and molecular profiling of peripheral T-cell lymphomas. Hematology Am Soc Hematol Educ Program 2008, 272–279. [DOI] [PubMed]
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016; 127: 2391–2405. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Pinilla SM, Atienza L, Murillo C, Perez-Rodriguez A, Montes-Moreno S, Roncador G et al. Peripheral T-cell lymphoma with follicular T-cell markers. Am J Surg Pathol 2008; 32: 1787–1799. [DOI] [PubMed] [Google Scholar]
- Lemonnier F, Couronne L, Parrens M, Jais JP, Travert M, Lamant L et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood 2012; 120: 1466–1469. [DOI] [PubMed] [Google Scholar]
- Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet 2014; 46: 171–175. [DOI] [PubMed] [Google Scholar]
- Palomero T, Couronne L, Khiabanian H, Kim MY, Ambesi-Impiombato A, Perez-Garcia A et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet 2014; 46: 166–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odejide O, Weigert O, Lane AA, Toscano D, Lunning MA, Kopp N et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood 2014; 123: 1293–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Rau R, Goodell MA. DNMT3A in haematological malignancies. Nat Rev Cancer 2015; 15: 152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Iqbal J, Lemonnier F, Kucuk C, de Leval L, Jais JP et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood 2012; 119: 1901–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Sung MK, Lee SH, Kim S, Lee H, Park S et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet 2014; 46: 371–375. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010; 18: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 2011; 20: 25–38. [DOI] [PubMed] [Google Scholar]
- Nguyen TB, Sakata-Yanagimoto M, Nakamoto-Matsubara R, Enami T, Ito Y, Kobayashi T et al. Double somatic mosaic mutations in TET2 and DNMT3A--origin of peripheral T cell lymphoma in a case. Ann Hematol 2015; 94: 1221–1223. [DOI] [PubMed] [Google Scholar]
- Couronne L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med 2012; 366: 95–96. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 2009; 137: 216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004; 306: 269–271. [DOI] [PubMed] [Google Scholar]
- Pancewicz J, Taylor JM, Datta A, Baydoun HH, Waldmann TA, Hermine O et al. Notch signaling contributes to proliferation and tumor formation of human T-cell leukemia virus type 1-associated adult T-cell leukemia. Proc Natl Acad Sci USA 2010; 107: 16619–16624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011; 475: 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kridel R, Meissner B, Rogic S, Boyle M, Telenius A, Woolcock B et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood 2012; 119: 1963–1971. [DOI] [PubMed] [Google Scholar]
- Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 2011; 476: 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet 2012; 49: 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 2003; 17: 2257–2317. [DOI] [PubMed] [Google Scholar]
- Lefranc M-P IMGT, the International ImMuno GeneTics Information System (cited). Available from http://www.imgt.org/.
- Kikushige Y, Ishikawa F, Miyamoto T, Shima T, Urata S, Yoshimoto G et al. Self-renewing hematopoietic stem cell is the primary target in pathogenesis of human chronic lymphocytic leukemia. Cancer Cell 2011; 20: 246–259. [DOI] [PubMed] [Google Scholar]
- McKenney AS, Levine RL. Isocitrate dehydrogenase mutations in leukemia. J Clin Invest 2013; 123: 3672–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, McKeithan TW, Gong Q, Zhang W, Bouska A, Rosenwald A et al. IDH2R172 mutations define a unique subgroup of patients with angioimmunoblastic T-cell lymphoma. Blood 2015; 126: 1741–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S. Guest editorial: preleukemia/prelymphoma-what is old, what is new? Int J Hematol 2015; 102: 511–512. [DOI] [PubMed] [Google Scholar]
- Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 2014; 506: 328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan M, Snyder TM, Corces-Zimmerman MR, Vyas P, Weissman IL, Quake SR et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med 2012; 4: 149ra118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlush LI, Zandi S, Itzkovitz S, Schuh AC. Aging, clonal hematopoiesis and preleukemia: not just bad luck? Int J Hematol 2015; 102: 513–522. [DOI] [PubMed] [Google Scholar]
- Damm F, Mylonas E, Cosson A, Yoshida K, Della Valle V, Mouly E et al. Acquired initiating mutations in early hematopoietic cells of CLL patients. Cancer Discov 2014; 4: 1088–1101. [DOI] [PubMed] [Google Scholar]
- Kikushige Y, Miyamoto T. Hematopoietic stem cell aging and chronic lymphocytic leukemia pathogenesis. Int J Hematol 2014; 100: 335–340. [DOI] [PubMed] [Google Scholar]
- Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet 2012; 44: 1179–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014; 371: 2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014; 371: 2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Lu C, Wang J, McLellan MD. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 2014; 20: 1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggemann M, White H, Gaulard P, Garcia-Sanz R, Gameiro P, Oeschger S et al. Powerful strategy for polymerase chain reaction-based clonality assessment in T-cell malignancies. Report of the BIOMED-2 Concerted Action BHM4 CT98-3936. Leukemia 2007; 21: 215–221. [DOI] [PubMed] [Google Scholar]
- Higgins JP, van de Rijn M, Jones CD, Zehnder JL, Warnke RA. Peripheral T-cell lymphoma complicated by a proliferation of large B cells. Am J Clin Pathol 2000; 114: 236–247. [DOI] [PubMed] [Google Scholar]
- Tan BT, Warnke RA, Arber DA. The frequency of B- and T-cell gene rearrangements and epstein-barr virus in T-cell lymphomas: a comparison between angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma, unspecified with and without associated B-cell proliferations. J Mol Diagn 2006; 8: 466–475, quiz 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zettl A, Lee SS, Rudiger T, Starostik P, Marino M, Kirchner T et al. Epstein-Barr virus-associated B-cell lymphoproliferative disorders in angloimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma, unspecified. Am J Clin Pathol 2002; 117: 368–379. [DOI] [PubMed] [Google Scholar]
- Hawley RC, Cankovic M, Zarbo RJ. Angioimmunoblastic T-cell lymphoma with supervening Epstein-Barr virus-associated large B-cell lymphoma. Arch Pathol Lab Med 2006; 130: 1707–1711. [DOI] [PubMed] [Google Scholar]
- Willenbrock K, Brauninger A, Hansmann ML. Frequent occurrence of B-cell lymphomas in angioimmunoblastic T-cell lymphoma and proliferation of Epstein-Barr virus-infected cells in early cases. Br J Haematol 2007; 138: 733–739. [DOI] [PubMed] [Google Scholar]
- Attygalle AD, Kyriakou C, Dupuis J, Grogg KL, Diss TC, Wotherspoon AC et al. Histologic evolution of angioimmunoblastic T-cell lymphoma in consecutive biopsies: clinical correlation and insights into natural history and disease progression. Am J Surg Pathol 2007; 31: 1077–1088. [DOI] [PubMed] [Google Scholar]
- Suefuji N, Niino D, Arakawa F, Karube K, Kimura Y, Kiyasu J et al. Clinicopathological analysis of a composite lymphoma containing both T- and B-cell lymphomas. Pathol Int 2012; 62: 690–698. [DOI] [PubMed] [Google Scholar]
- Asmar F, Punj V, Christensen J, Pedersen MT, Pedersen A, Nielsen AB et al. Genome-wide profiling identifies a DNA methylation signature that associates with TET2 mutations in diffuse large B-cell lymphoma. Haematologica 2013; 98: 1912–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Chen L, Dawlaty MM, Pan F, Weeks O, Zhou Y et al. Combined loss of Tet1 and Tet2 promotes B cell, but not myeloid malignancies, in mice. Cell Rep 2015; 13: 1692–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcaini L, Rossi D, Lucioni M, Nicola M, Bruscaggin A, Fiaccadori V et al. The NOTCH pathway is recurrently mutated in diffuse large B-cell lymphoma associated with hepatitis C virus infection. Haematologica 2015; 100: 246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villamor N, Conde L, Martinez-Trillos A, Cazorla M, Navarro A, Bea S et al. NOTCH1 mutations identify a genetic subgroup of chronic lymphocytic leukemia patients with high risk of transformation and poor outcome. Leukemia 2013; 27: 1100–1106. [DOI] [PubMed] [Google Scholar]
- Karube K, Martinez D, Royo C, Navarro A, Pinyol M, Cazorla M et al. Recurrent mutations of NOTCH genes in follicular lymphoma identify a distinctive subset of tumours. J Pathol 2014; 234: 423–430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.