Abstract
Frequent climatic changes in conjunction with other extreme environmental factors are known to affect growth, development and productivity of diverse crop plants. Pigeonpea, a major grain legume of the semiarid tropics, endowed with an excellent deep-root system, is known as one of the important drought tolerant crop plants. Cyclin dependent kinases (CDKs) are core cell cycle regulators and play important role in different aspects of plant growth and development. The cyclin-dependent kinase regulatory subunit gene (CKS) was isolated from the cDNA library of pigeonpea plants subjected to drought stress. Pigeonpea CKS (CcCKS) gene expression was detected in both the root and leaf tissues of pigeonpea and was upregulated by polyethylene glycol (PEG), mannitol, NaCl and abscisic acid (ABA) treatments. The overexpression of CcCKS gene in Arabidopsis significantly enhanced tolerance of transgenics to drought and salt stresses as evidenced by different physiological parameters. Under stress conditions, transgenics showed higher biomass, decreased rate of water loss, decreased MDA levels, higher free proline contents, and glutathione levels. Moreover, under stress conditions transgenics exhibited lower stomatal conductance, lower transpiration, and higher photosynthetic rates. However, under normal conditions, CcCKS-transgenics displayed decreased plant growth rate, increased cell size and decreased stomatal number compared to those of wild-type plants. Real-time polymerase chain reaction revealed that CcCKS could regulate the expression of both ABA-dependent and ABA-independent genes associated with abiotic stress tolerance as well as plant growth and development. As such, the CcCKS seems promising and might serve as a potential candidate gene for enhancing the abiotic stress tolerance of crop plants.
Keywords: abiotic stress tolerance, abscisic acid, Cajanus cajan, cyclin-dependent kinase regulatory sub unit, glutathione, growth, development
Introduction
Plants often encounter extreme temperatures, drought, and high salt concentrations in soil, which are the major abiotic stresses that negatively affect plant growth and development (Krasensky and Jonak, 2012). A prerequisite for plants to acclimatize under extreme conditions is their ability to perceive stress signals and transduce them through a chain of signaling molecules, which ultimately interact with the regulatory elements of different stress-inducible genes and bring about changes in cellular gene expression (Kasuga et al., 1999; Tong et al., 2007; Tamirisa et al., 2014). An understanding of the mechanisms that regulate the expression of stress-responsive genes is essential for enhancing plant productivity under adverse environmental conditions (Priyanka et al., 2010a,b).
The development of plant organs directly depends on the frequency of cell division, parameters of the cell cycle, and number and size of the cells. These developmental processes are controlled by the molecular machinery that regulates cell cycle progression in coordination with nutritional, hormonal, developmental and environmental signals (Inze and Veylder, 2006; Tank and Thaker, 2011). A central role in the regulation of the cell cycle is played by a group of proteins known as cyclin-dependent kinases (CDKs), a family of protein kinases that were indentified for their role in regulating the cell cycle. CDKs are core cell cycle regulators that are involved in various processes of plant growth and development (Ma et al., 2015) and have been implicated in additional cellular processes such as transcription and translation. A progressive transition through different phases of the cell cycle is controlled by a conserved mechanism based on the sequential formation and activation of complexes between CDKs and their activating cyclin (CYC) subunits (De Veylder et al., 2003). CDK subunit proteins (CKS) might act as both activators and inhibitors of CDK activity (De Veylder et al., 2001a). CKS regulate CDK activity by functioning as docking factors for the CDK/CYC substrate complexes (Boruc et al., 2010a). The Schizosaccharomyces pombe SUC1 gene was identified in fusion yeast by its ability to rescue certain temperature-sensitive cdc2 mutants (Hayles et al., 1986), and its homolog, the Saccharomyces cerevisiae CKS1 gene was found to be a suppressor of CDC28 mutants (Hadwiger et al., 1989). The SUC1/CKS1 homolog was isolated from Arabidopsis using yeast two-hybrid screening and designated as AtCKS1 (De Veylder et al., 1997). The overexpression of AtCKS1 in Arabidopsis decreased the leaf size and reduced the root growth rate without any effect on endoreduplication (De Veylder et al., 2001a).
Under biotic and abiotic stress conditions, plant growth is adversely affected as a result of cell cycle inhibition due to a prolonged S phase and delayed entry into mitosis (Kitsios and Doonan, 2011). Though the molecular interactions linking the cell cycle machinery to the perception of stress signals are not fully understood, recent studies have indicated the involvement of CDKs in the plant stress responses. An Arabidopsis CYCH;1 gene was reported to be involved in drought stress response by regulating the blue light-mediated stomatal opening as well as by controlling the reactive oxygen species (ROS) homeostasis (Zhou et al., 2013). However, their precise roles in abiotic stress responses are largely unknown (Kitsios and Doonan, 2011; Komaki and Sugimoto, 2012). Therefore, further research will help in the elucidation of possible mechanisms of CDK activity under different types of abiotic stresses.
In this study, we isolated the stress-responsive Cajanus cajan cyclin-dependent kinase regulatory subunit gene (CcCKS) from pigeonpea. The overexpression of CcCKS in Arabidopsis conferred drought and salinity stress tolerance as well as marked changes in the growth and development of transgenic plants. This report clearly demonstrates that the CcCKS in Arabidopsis affords abiotic stress tolerance and regulates plant growth and development.
Materials and methods
Plant materials and stress treatments
The pigeonpea seeds (variety ICP 8744) were disinfected with 0.1% (w/v) mercuric chloride and germinated in Petri plates. Four-week-old plants were subjected to polyethylene glycol (PEG) 6000 (20% w/v), mannitol (400 mM), NaCl (1.0 M), and ABA (100 μM) for 6 and 12 h. The sterilized seeds of Arabidopsis (Col-0), after stratification for 3 days at 4°C, were grown on Murashige and Skoog (1962) at 20 ± 1°C with a 16 h photoperiod under fluorescent light (120–150 μmol/m2 sec) in a Conviron growth chamber (Tamirisa et al., 2014).
Subtraction cDNA library construction
Total RNA was isolated from 4-week-old 20% PEG-treated (−1.01 ± 0.02 Mpa) and unstressed (−0.49 ± 0.02 Mpa) plants by the guanidinium thiocyanate method (Sambrook and Russell, 2001). Poly (A+) RNA was purified from the total RNA through oligo (dT) cellulose chromatography using an mRNA isolation kit (Amersham Pharmacia Biotech, Asia Pacific Ltd, Quarry Bay, Hong Kong). The PCR select cDNA subtractive hybridization method was used to construct the cDNA library (Clontech, Mountain View, CA, USA) with 2 μg of mRNA isolated from control and PEG-treated plants (Tamirisa et al., 2014).
Isolation of full-length Cajanus cajan cyclin-dependent kinase regulatory subunit gene (CcCKS)
Total RNA was isolated from the PEG (20%) treated 4-week-old plants as described above. RT-PCR for first-strand cDNA synthesis was carried out using a reaction mixture containing Tris-HCl (10 mM), KCl (50 mM), MgCl2 (1.5 mM), dNTPs (200 μM each), MMLV reverse transcriptase (2 units), total RNA (1.0 μg), and Oligo (dT) primer. The resultant cDNA product was used as a template in a PCR reaction using gene-specific primers to amplify the CcCKS gene. The full-length coding sequence (267 bp) of CcCKS, amplified using gene-specific primers and using the drought-stressed cDNA library as a template, was ligated at the SmaI-site of the pBSSK (+) vector and transformed into E. coli (TOP10) cells. The resultant recombinant clones were selected and confirmed by restriction digestion with XhoI and XbaI enzymes and later sequenced using an automated DNA sequencer. Homology search of nucleotide and amino acid sequences was carried out using BLAST (NCBI). A maximum likelihood phylogenetic tree was constructed based on the amino acid sequences of CcCKS protein, and other CKS proteins from different plant species, using Clustal Omega and MEGA6 software. The bootstrap parameter was set at 100.
Plant expression cassettes construction and transformation of A. thaliana
Plasmid DNA containing the CcCKS gene was digested with XhoI-XbaI enzymes and was subcloned into the pRT100 vector at the 3′ end of the CaMV35S promoter. Furthermore, the gene cassette was digested with HindIII and cloned into the pBI101 vector. The pBI101 vector containing CcCKS and nptII expression units and the pBI101 vector with nptII (empty vector) were mobilized independently into the EHA105 strain of Agrobacterium through triparental mating (Wise et al., 2006). The Agrobacterium-mediated floral dip method was carried out to transform A. thaliana (Clough and Bent, 1998). Seeds from the transformed plants were germinated on MS medium supplemented with kanamycin (50 mg L−1) to select the putative transformants.
Molecular analysis of CcCKS transgenic Arabidopsis plants
Genomic DNA isolated from kanamycin tolerant plants was used to carry out PCR analysis. DNA isolated from untransformed plants (WT) was used as a negative control and plasmid (pBI12135SnptII-p35SCcCKS) DNA was used as a positive control. For PCR, plasmid DNA (10 ng) and genomic DNA (50 ng) were used as templates in separate reactions. The reactions were carried out using CcCKS gene-specific primers, and the amplified PCR products were analyzed on a 1.0% agarose gel.
Southern blot analysis of Arabidopsis transgenic plants
Genomic DNA (15 μg) was isolated from three CcCKS transgenic plants by the CTAB method (Saghai-Maroof et al., 1984), and was digested independently with the HindIII enzyme. The digested DNA was resolved on 0.8% agarose gel and transferred to a positively charged nylon membrane as per the manufacturer's instructions (Amersham Pharmacia Biotech Asia Pacific Ltd, Quarry Bay, Hong Kong). A Southern blot analysis was performed according to Sambrook and Russell (2001). The nptII coding region was labeled with the AlkPhos Direct Labeling and Detection System according to the manufacturer's instructions (Amersham Pharmacia Biotech Asia Pacific Ltd, Quarry Bay, Hong Kong) and used as a probe.
RT-PCR analysis CcCKS transgenic Arabidopsis plants
Total RNA was extracted from transgenic (CS1, CS2 and CS3), wild-type (WT) and vector control (VT) plants using the TRIZOL (Invitrogen, Carlsbad, CA, USA). RT-PCR was carried out as described earlier, and the amplified PCR products were analyzed on 1.0% agarose gel.
Construction of the CcCKS:GFP fusion gene construct and generation of transgenic Arabidopsis plants
The CcCKS coding sequence without a termination codon was fused with the 5′ region of green fluorescent protein (GFP) and the fusion gene was driven by the CaMV35S promoter. The expression cassettes of pCaMV35S–CcCKS:GFP/pCaMV35S:GFP were subcloned into pBI101 containing the nptII expression unit and mobilized into the Agrobacterium (EHA105 strain) through triparental mating. The transformed seedlings were selected on MS medium supplemented with kanamycin (50 mgL−1). Callus was induced from the roots of pCaMV35S:GFP and pCaMV35S–CcCKS:GFP transgenics on 2, 4-dichlorophenoxyacetic acid (2 mgL-1) containing medium. Individual callus cells were visualized under confocal microscopy (Leica SP5 Microsystems, Excitation: 488, and Emission: 520).
Functional analysis of transgenics for abiotic stress tolerance
The seeds of WT, VT and homozygous transgenic (T3) Arabidopsis plants of CS1, CS2 and CS3 were surface-sterilized, placed on MS salt medium, and allowed to grow as described above. Drought and salt tolerance of 2-week-old transgenic seedlings was tested by growing them on MS medium supplemented with mannitol (150 mM) or NaCl (125 mM) for 7 days. Later, the seedlings were transferred onto the MS salt medium and allowed to recover for 10 days under normal conditions. After 10 days of recovery, the survival rate, root length and biomass of the seedlings were recorded. Another set of stressed seedlings were transferred to the soil, grown for maturity and photographed. For drought tolerance assays, 3-week-old plants growing in pots were withheld from water for 2 weeks, and then rewatered for 1 week (Sekhar et al., 2010).
Relative water content measurements
The relative water contents (RWC) of 4-week-old transgenic plants of CS1, CS2 and CS3 along with WT and VT plants was measured. Fresh weight loss was calculated relative to the initial plant weight. To measure the fresh weight, plants were weighed immediately and left in the growth chamber (20 ± 1°C) until there was no further weight loss (desiccated weight). Finally, the plants were dried for 24 h at 70°C and the dry weights were recorded. The relative water content of the samples was measured using the formula RWC (%) = (desiccation weight—dry weight)/(fresh weight–dry weight) × 100 (Mao et al., 2010).
Osmotic potential analysis
Two-week-old plants of CS1, CS2, and CS3 along with WT and VT plants were selected and frozen with liquid nitrogen for 30 s, and preserved at −80°C. Sap was collected by squeezing the leaves using a micro pestle, and the osmotic potential of the sap was determined using a vapor pressure osmometer (Vapro, Model 5520; Wescor Inc., Logan, UT, USA; Mao et al., 2010).
Cell membrane stability analysis
Ten-day-old seedlings grown on 1X MS medium were placed on filter paper saturated with NaCl (150 mM) solution. As soon as symptoms of stress appeared (appx. after 6 h) in WT and VT plants, the seedlings were removed, rinsed thoroughly and immersed in 20 ml of double distilled water at room temperature (20 ± 1°C). The initial conductivity of samples was recorded after 2 h using a conductivity meter (Model Sension 5; HACH Company, Loveland, CO, USA). Later, samples were boiled for 30 min and, cooled to room temperature, and the final conductivity was measured. CMS was calculated using the formula CMS (%) = 1—initial electrical conductivity/electrical conductivity after boiling × 100 (Mao et al., 2010).
Measurement of the leaf chlorophyll content
Leaf disks of CS1, CS2, and CS3 transgenic lines along with WT and VT plants were floated independently on water (control), 150 mM mannitol and 125 mM NaCl solutions at room temperature (20 ± 1°C) for 72 h. Leaf disks were then used to measure the chlorophyll content spectrophotometrically as described by Tamirisa et al. (2014).
Measurement of the proline content
Fifteen-day-old CcCKS transgenic lines along with WT and VT seedlings were exposed to 150 mM mannitol and 125 mM NaCl for 3 days. Plant tissues were homogenized in 3% aqueous sulfosalicylic acid and the homogenates were centrifuged at 1000 rpm for 5 min. equal volumes of glacial acetic acid and ninhydrin were added to the supernatant collected. The proline content was quantified as described by Bates et al. (1973).
Measurement of the malonaldehyde (MDA) content
Tissue (50 mg) was collected from stressed and unstressed transgenics, WT and VT seedlings and homogenized with 2 ml of 0.1% (w/v) cold trichlorocactic acid (TCA) on ice. The homogenates were centrifuged at 14,000 × g for 10 min at 4°C, after which 250 μl of supernatant was mixed with 1.5 ml of TCA/thiobarbituric acid (0.25% thiobarbituric acid containing 10% TCA) reagent. The MDA content was estimated according to Shin et al. (2012).
Detection of superoxide radicals in WT and transgenic plants
The nitroblue tetrazolium (NBT) staining method was used for super oxide detection. Three-week-old transgenic, WT and VT plants were used for NBT staining. Plants were germinated on MS basal medium and later transferred to medium containing 150 mM mannitol (drought stress) and 125 mM NaCl (salt stress) for 3 days, along with unstressed seedlings. The treated seedlings were vacuum-infiltrated with 0.1 mg/ml NBT in 25 mM HEPES buffer, pH 7.6. The samples were incubated in the dark for 2 h in a growth chamber and later stained samples were transferred to 80% ethanol and incubated at 70°C for 10 min to remove chlorophyll. For each treatment, 20 seedlings of each line were used.
Effect of ABA on seed germination and seedling growth
Seeds of transgenic and WT plants were placed on MS medium containing different concentrations of ABA (0–2.0 μM) to record the germination frequency. Further, germinated seedlings were allowed to grow for 20 days to assess their sensitivity to ABA.
Measurement of stomatal aperture sizes
Rosette leaves of 3-week-old plants were detached and floated (abaxial side down) on solution containing 10 mM MES-KOH (pH 6.15), 30 mM KCl and 1 mM CaCl2 and were incubated under light for 2 h. Later, they were treated with ABA-containing solution for 2 h. In each treatment, 50 stomatal aperture sizes were measured under confocal microscopy.
Root growth rate
For root growth rate analysis, the seeds were germinated on MS medium and placed vertically in the growth chamber. The position of the root tip was marked everyday on the backside of the Petri plates to determine the daily growth. After 20 days, the plates were scanned using a WinRHIZO standard systems desktop optical scanner. The daily length increase over the entire growing period was determined by measuring the distance between successive marks along the root axis using the WinRHIZO software. By dividing the daily growth by the precise time interval between the corresponding marks, the average growth rate was calculated (De Veylder et al., 2001a).
Flow cytometric analysis
The DNA content was estimated by flow cytometric analysis. Plant materials (50 mg) of trangenic and WT plants were taken and chopped using a razor blade into 1 ml of ice-cold Otto I buffer in a Petri plates. The solution with the chopped plant tissue (suspension) was filtered through 40 μm nylon mesh; later, nuclei were pelleted by centrifugation (150 g/5 min). The nuclei were resuspended by gentle shaking and 100 μl of fresh Otto I buffer was added. The samples were incubated for 60 min. After incubation, Otto II buffer (1 ml) which includes propidium iodide (50 μg/ml) and RNase (50 μg/ml) was added. The samples were left at room temperature and analyzed after 20 min using a Guava EasyCyte flow cytometer (Millipore, France; Dolezel and Bartos, 2005).
Pollen staining
Pollen staining was performed using the pollen collected from newly dehiscent flowers as described by Alexander (1969).
Measurement of the glutathione (GSH) content
The GSH levels were measured in the intact roots and guard cells of Arabidopsis after conjugation with monochlorobimane (MCB), to produce a fluorescent glutathione S-bimane (GSB) adduct. Seedlings that were treated with 150 mM mannitol for 2 days were carefully removed from the agarose surface, and the roots were placed in a drop of 100 μM monochlorobimane. After labeling for 15 min, the roots were washed briefly in distilled water and mounted on a microscope slide with a cover slip. To measure the GSH levels in the guard cells, excised leaves from treated plants were incubated in a staining solution containing 100 μM of MCB for 2 h at room temperature. Photographs were taken using a fluorescent microscope (Olympus BX41fluorescence microscope). The captured images were used to measure the number of pixels/ fluorescence intensity using ImageJ software.
Histological analysis
The cell density, stomatal density and stomatal index were determined as described by De Veylder et al. (2001a,b). Leaves of the same age and same relative position were sampled for analysis, and the leaf surface imprint method was used as described by Yu et al. (2008). Three leaves were sampled from each plant, and 10 of each WT, VT and transgenic plants were sampled.
Scanning electron microscopy
Cotyledons from 3-week-old seedlings were processed and analyzed using scanning electron microscopy, as described De Veylder et al. (2002). Seedlings were fixed overnight in 4% paraformaldehyde and 1% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2), followed by post-fixation in 2% osmiumtetroxide, and a graded ethanol series. These seedlings were mounted on stubs, sputter-coated with gold and examined using a scanning electron microscope (Zeiss, Germany).
Measurement of stomatal conductance, transpiration, photosynthetic rates, and water use efficiency
The stomatal conductance, transpiration and photosynthetic rates were measured using a portable photosynthesis system (Li-Cor LI-6400XT). Four-week-old plants growing on soil were treated separately with 150 mM mannitol and 125 mM NaCl for 3 days. All of the photosynthetic measurements were taken at a constant air flow rate of 500 mmol s−1. Furthermore, the concentration of CO2 (400 mmol mol−1 using the system's CO2 injector), temperature (22°C), and photosynthetic photon flux density (1,200 mmol (photon) m−2 s−1) were maintained. Water use efficiency was calculated using the photosynthetic and transpiration measurements. Three measurements were taken on each plant, and 10 plants from WT, VT and each transgenic line were used for measurements.
Quantitative real time PCR (qRT-PCR)
qRT-PCR was performed using RNA isolated from stressed and unstressed pigeonpea plants as described earlier. RNA was isolated from WT and transgenic plants subjected to mannitol (150 mM) stress for 2 days as well as from unstressed plants as described earlier. First strand cDNA was synthesized from RNA samples and used as a template for qRT-PCR. qRT-PCR analysis was performed using SYBR green master mix with the Applied Biosystems 7500 Real Time PCR system (USA). The genes were selected based on the literature available and the specific primers were designed. Each reaction was performed thrice, and the relative expression ratio was calculated using 2−ΔΔct method with actin as the reference gene. The primers that were used for qRT-PCR are listed in Supplementary Table 1 (Tamirisa et al., 2014).
Statistical analysis
All the experiments were done in triplicate using 20 seedlings in each treatment. Fresh tissue was collected for all the experiments. Mean, standard error and t-test were computed with the help of pre-loaded software in Excel, for statistical calculations.
Results
Characterization of the CcCKS gene
A cDNA clone of 267 bp sequence (KU522130), encoding a polypeptide of 88 amino acids, was designated as the cyclin-dependent kinase regulatory subunit gene (CKS). Phylogenetic tree analysis revealed that the CcCKS protein was clustered in the same monophyletic group as putative CKS proteins from Glycine max, Medicago truncatula and Arabidopsis lyrata (Supplementary Figure S1). Further, the CcCKS protein revealed high similarity with CDKs regulatory subunits of G. max (99%), M. truncatula (97%), A. lyrata (95%), Nicotiana tomentosiformis (95%), Zea mays (90%), A. thaliana (90%), and Brassica rapa (87%). To examine the subcellular localization of the CcCKS protein, pCaMV35S-CcCKS:GFP fusion and pCaMV35S:GFP gene (control) constructs were introduced into Arabidopsis and the individual callus cells were visualized under laser-scanning confocal microscopy. Cells expressing the CcCKS:GFP fusion protein showed the fluorescence both in the nucleus and the cytoplasm (Supplementary Figure S2).
Expression profile of CcCKS under abiotic stress conditions in pigeonpea
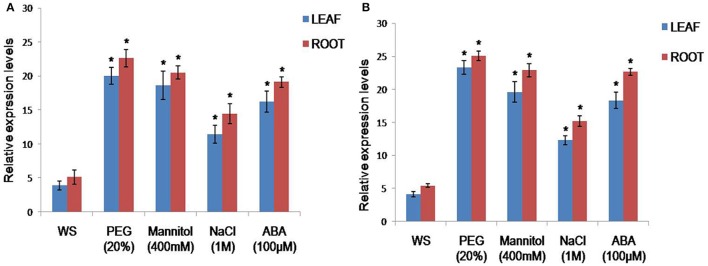
To analyse the stress-inducible nature of the CcCKS gene, real-time PCR was done using the total RNA isolated from pigeonpea plants after treatment under different stress conditions. These results revealed that increased levels of CcCKS transcripts were detected in both the roots and leaves of PEG-, mannitol-, NaCl- , and ABA- treated plants compared to untreated plants (Figure 1).
Figure 1.
Analysis of Cajanus cajan cyclin dependent kinase regulatory subunit gene (CcCKS) expression in pigeonpea under different treatment conditions by qRT- PCR. Comparison of the relative transcript levels of CcCKS gene under (PEG) 6000 (20% w/v), Mannitol (400 mM), NaCl (1.0 M), and ABA (100 μM) stress for 6 h (A) and 12 h (B). Actin has been used as reference gene. The vertical column indicates the relative transcript level. Bar represents mean and error bars represents SE from three independent experiments. Asterisks indicate significant differences in comparison with the WT at P < 0.05. WS represents without stress.
Development of CcCKS-transgenic plants of A. thaliana
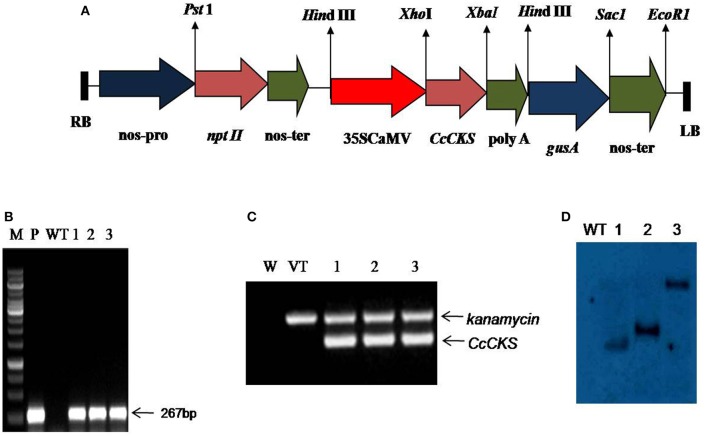
Putative transgenic plants, obtained with plasmid constructs containing CcCKS (Figure 2A), were selected on MS medium containing kanamycin (50 mg L−1). PCR using CcCKS gene-specific primers amplified an ~250 bp fragment from the genomic DNA of transgenic plants, while no such band was observed in WT (Figure 2B). RT-PCR revealed the presence of CcCKS transcripts in pCaMV35S-CcCKS lines (Figure 2C). A Southern blot analysis of the genomic DNA isolated from transgenic lines demonstrated the existence of the transgene in the genome of Arabidopsis, when probed with nptII (Figure 2D). Homozygous transgenic lines of CS1, CS2 and CS3, containing a single T-DNA insertion, were chosen for subsequent experiments.
Figure 2.
T-DNA region of pBI101 containing CcCKS expression cassette and molecular confirmation of CcCKS transgenic Arabidopsis plants. (A) Restriction map of T-DNA region of pBI101 containing CcCKS expression unit driven by CaMV35S promoter and nptII selection marker. (B) PCR analysis for the presence of CcCKS gene in the genomic DNA of transgenic Arabidopsis plants. Lane M: 1.0 kb DNA size standard marker, Lane P, Positive control; Lane WT, wild type; Lanes 1–3, PCR amplification of CcCKS from transgenic plants. (C) RT PCR analysis showing the expression of kanamycin and CcCKS genes in transgenic Arabidopsis. Lane WT, wild-type; Lane VT, vector control; Lanes 1, 2, and 3: different transgenic lines. (D) Southern blot analysis of transgenic Arabidopsis plants carrying CcCKS gene. Lane WT, wild type plant; Lanes 1, 2, and 3: transgenic plants of three different lines (CS1, CS2, and CS3).
Functional analysis of CcCKS-transgenic lines for abiotic stress tolerance
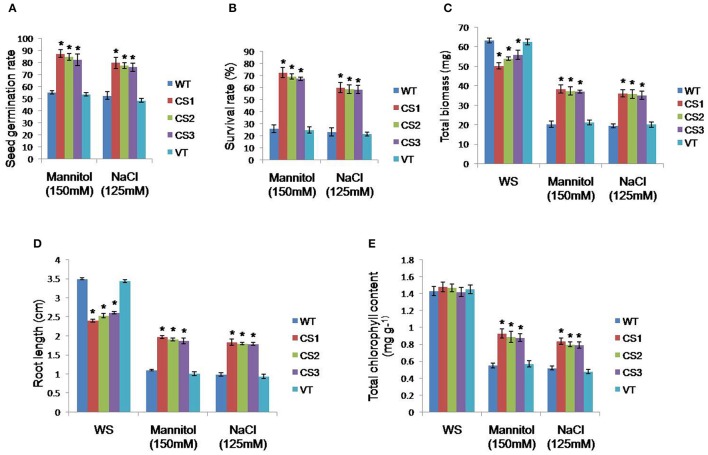
No variation was observed in the seed germination of transgenic, VT or WT seeds under normal conditions. However, under mannitol (150 mM) and NaCl (125 mM) stress, higher germination rates were found in transgenic plants than in WT or VT plants (Figure 3A). To characterize the stress tolerance ability of transgenic plants, 2-week-old seedlings were subjected to drought (150 mM mannitol) and salt (125 mM NaCl) stress treatments for 7 days along with VT and WT seedlings. Transgenic lines of CS1, CS2, and CS3 exhibited increased survival rate, biomass and root lengths compared to VT and WT plants (Figures 3A–D).
Figure 3.
Evaluation of CcCKS-transgenics against different abiotic stresses. Seed germination rate (A) of WT, VT, and transgenic plants under normal, 150 mM mannitol and 125 mM NaCl stress. Two-week-old seedlings of WT, VT, and transgenics were grown on 150 mM mannitol and 125 mM NaCl for 7 days and were later allowed to recover on MS plates. Data on survival rate (B), total biomass (C) and root length (D) were recorded after 10 days of recovery. In each treatment, 20 seedlings of WT, VT, and two transgenic lines were used. Chlorophyll content (E) was determined from the leaf disks of control and transgenics after 72 h of incubation in water, 150 mM mannitol and 125 mM NaCl solutions independently at room temperature (20 ± 1°C). Bar represents mean and error bars represents SE from three independent experiments. Asterisks indicate significant differences in comparison with the WT at P < 0.05. WS, without stress; CS1, CS2, and CS3: three different transgenic lines; WT, wild type; VT, vector transformed plants.
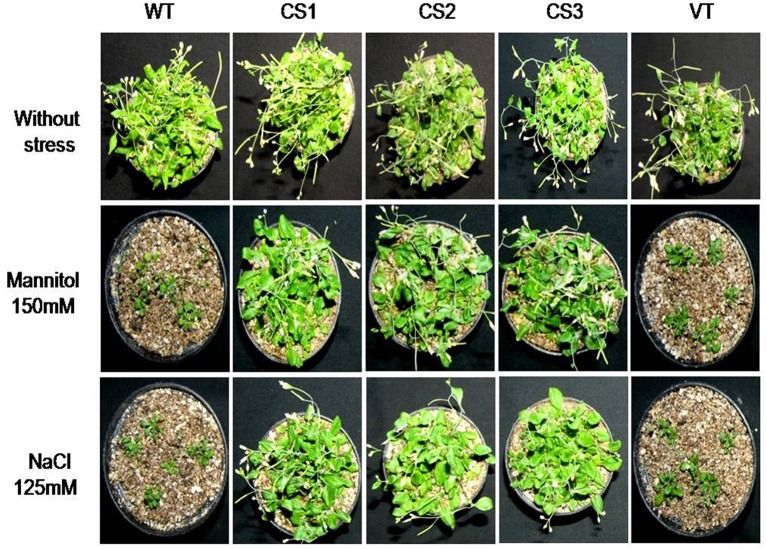
Under normal conditions, no significant differences in chlorophyll contents were observed between the control and transgenic plants. The transgenic plants showed higher mean chlorophyll contents under both mannitol and NaCl stresses compared to those of WT and VT plants (Figure 3E). Further, the transgenic plants expressing CcCKS gene could successfully complete their reproductive cycle, while the control plants failed to reach the reproductive phase under drought and salt stress conditions (Figure 4). The transgenic plants subjected to low water conditions also showed greater tolerance compared to the WT and VT plants (Supplementary Figure S3A). Although water losses were observed in transgenic and control plants, the final relative water content of transgenics was significantly higher than that of WT and VT plants (Supplementary Figure S3B). When the osmotic potential of transgenic, VT, and WT plants, grown under well-watered conditions was measured, the CS1, CS2, and CS3 lines showed higher osmotic potential than in VT and WT plants (Supplementary Figure S3C).
Figure 4.
Evaluation of transgenic plants expressing CcCKS gene against different abiotic stress conditions. Two-week-old seedlings of WT, VT, and CcCKS-transgenics were subjected to 150 mM mannitol, 125 mM NaCl for 7 days. In each treatment 20 seedlings were used. Treated seedlings were allowed to recover for 7 days at 20 ± 1°C temperature. Later, seedlings were transferred to soil and allowed to grow for 3 weeks under normal conditions, and were photographed. WT, wild type; VT, vector transformed; CS1, CS2, and CS3: three different transgenic lines.
Estimation of the proline, malonaldehyde (MDA), and glutathione levels and the cell membrane stability in CcCKS-transgenic plants
To understand the physiological basis of stress tolerance of CcCKS-transgenic plants, the proline content was estimated under normal and stress conditions. Under normal conditions, transgenic lines showed enhanced proline levels, while there was no significant difference between WT and VT plants. When 15-day-old transgenics were subjected to mannitol (150 mM) and NaCl (125 mM) stresses, they could accumulate higher proline contents (Figure 5A), whereas, WT and VT plants showed lower levels of proline under similar stress conditions (Figure 5A).
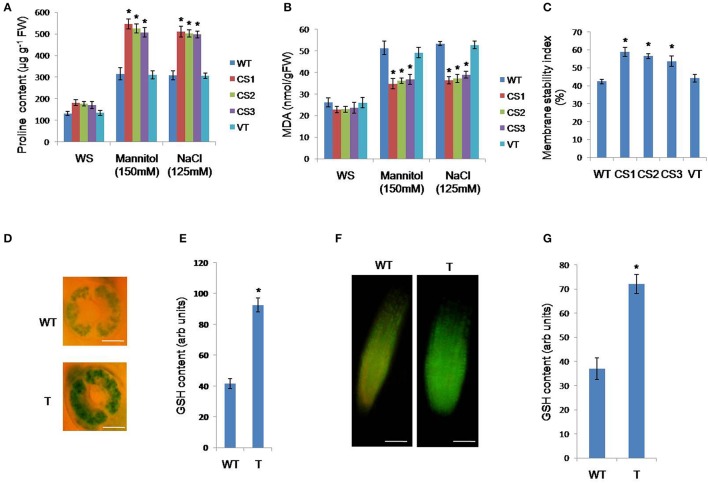
Figure 5.
Measurement of proline, MDA, osmotic potential, cell membrane stability and relative water contents of transgenic plants. Two-week-old seedlings of WT, VT, and transgenics were grown on 150 mM mannitol for 3 days, for estimation of proline (A), MDA (B). Cell membrane stability (C) of transgenics, WT and VT seedlings were treated with 150 mM NaCl. Two-week-old A. thaliana seedlings, grown on MS medium, were subjected to mannitol (150 mM) for 2 days. GSH content in guard cells (D,E) and root meristem (F,G) of WT and transgenic plant. T represents CcCKS-transgenic and WT represents wild-type. Bar represents mean, and error bars represents SE from three independent experiments. For each treatment, 20 seedlings were used. Asterisks indicate significant differences in comparison with the WT at P < 0.05. WT, wild type; VT, vector transformed; CS1, CS2, and CS3: three different transgenic lines. (D) Scale bar 50 μm; (F). Scale bar 100 μm.
The MDA contents in transgenic plants was significantly lower than those of WT and VT plants when subjected to mannitol and NaCl stress conditions (Figure 5B). However, there was no significant difference between WT and VT plants under similar stress conditions (Figure 5B).
To analyze the cell membrane stability, transgenic, WT, and VT plants were treated with NaCl (150 mM). Transgenic plants showed significantly higher membrane stability than that of WT and VT plants. However, there was no difference between WT and VT plants under similar stress conditions (Figure 5C).
Monochlorobimane (MCB) staining was performed to measure the GSH contents in the roots and guard cells of Arabidopsis. Under stress conditions, the fluorescence intensity of GSH in the guard cells of transgenic plants was much higher than that of WT plants (Figures 5D,E). Similarly, GSH fluorescence intensity in the roots of transgenic plants was higher than that of WT plants (Figures 5F,G).
Detection of reactive oxygen species by NBT staining
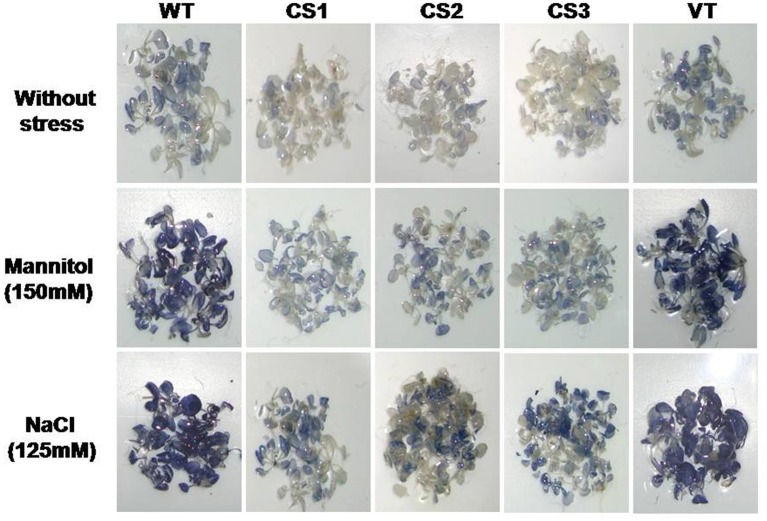
The superoxide concentration in the 2-week-old seedlings, treated with mannitol (150 mM) and NaCl (125 mM), was examined using nitroblue tetrazolium (NBT) staining. Under unstressed conditions, transgenic and control seedlings revealed a low intensity of staining. However, under stress conditions, the transgenics exhibited a lower staining intensity than that of WT and VT seedlings, as visualized by the purple formazan deposit (Figure 6).
Figure 6.
Superoxide detection in transgenic plants subjected to different abiotic stresses. Two-week-old A. thaliana seedlings grown on MS medium were subjected to mannitol (150 mM) and NaCl (125 mM) for 72 h. For each treatment, 20 seedlings were used. NBT staining was done for superoxide detection. WT: wild type; VT: vector control; CS1, CS2, and CS3: three different transgenic lines.
ABA sensitivity of CcCKS-transgenic plants and their stomatal aperture sizes
The seed germination rate of transgenic plants was severely inhibited by ABA compared with the WT plants (Supplementary Figure S4A). Transgenics when treated with ABA, showed a greater reduction in the stomatal aperture size compared to that of WT plants (Supplementary Figure S4B). Furthermore, transgenic seedlings showed greater hypersensitivity to ABA than that of WT plants (Supplementary Figure S4C).
Stomatal conductance, transpiration, photosynthetic rates, and water use efficiency under stress treatments
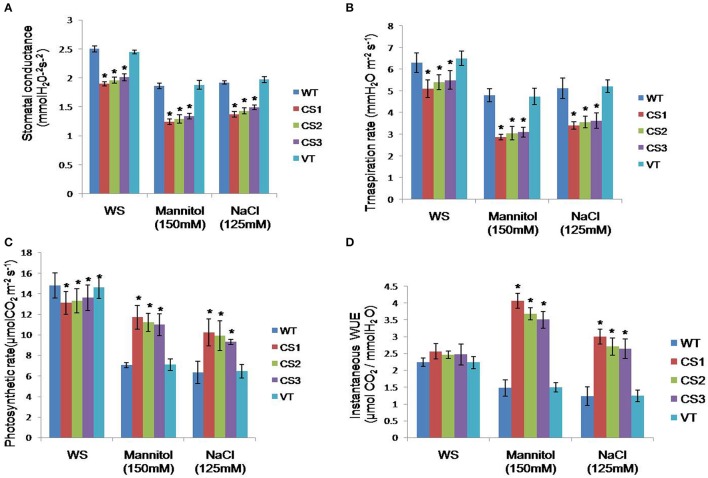
Under normal conditions, the transgenic lines showed lower stomatal conductance, transpiration and photosynthetic rates compared to WT plants. However, transgenics subjected to mannitol (150 mM) and NaCl (125 mM) stresses, exhibited lower stomatal conductance, lower transpiration, higher photosynthetic rates and water use efficiency. By contrast, WT and VT plants showed higher stomatal conductance, greater transpiration and lower photosynthetic rates under similar stress conditions (Figures 7A–D).
Figure 7.
Measurement of stomatal conductance, photosynthesis, transpiration and water use efficiency of transgenic plants. Four weeks old plants grown on soil were treated with 150 mM mannitol (drought stress) and 125 mM NaCl (salt stress) separately for 3 days. Stomatal conductance (A), transpiration (B), photosynthetic rates (C) and water use efficiency (D) were measured using a portable photosynthesis system (Li-Cor LI-6400XT). Three measurements were made on each plant, and 10 plants from WT as well as each transgenic line were used for measurements. WT represents wild type; VT represents vector transformed; CS1, CS2, and CS3 represent transgenic lines. Bar represents mean error bars represents SE from three independent experiments. Asterisks indicate significant differences in comparison with the WT at P < 0.05.
Pollen viability
Fertile pollen grains were observed in both transgenic and WT plants without any differences in the pollen fertility as evidenced by Alexander staining (Supplementary Figure S5A).
Flow cytometric analysis
To determine whether the cell cycle was affected in the CcCKS overexpressing plants, flow cytometric analysis was performed on the nuclei isolated from the roots of WT and transgenic plants. Wild-type roots of Arabidopsis showed the typical patterned peaks ranging from 2C to 8C, and the C values of transgenic lines were similar to those of WT plants (Supplementary Figure S5B).
Developmental changes in CcCKS transgenics in the root growth rate, leaf size, pavement cell number, stomatal number, stomatal index, and stomatal morphology
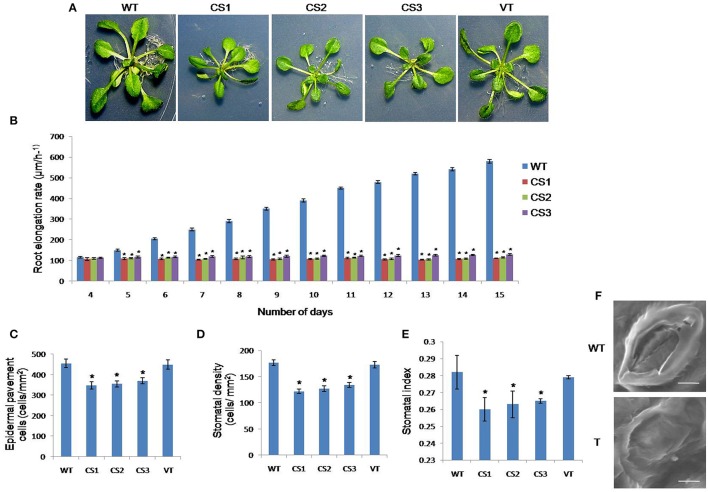
Three weeks after seed germination, WT, VT, and CcCKS transgenic plants exhibited distinct observable differences in their morphological features. Transgenic plants showed smaller rosettes and leaves compared to WT plants (Figure 8A). There were no observable differences in the root length of transgenic and WT seedlings during the initial 3 days of root growth. Later, CcCKS transgenic lines showed an approximately steady growth rate, while WT plants showed accelerated root growth during the 15 day growth period analyzed (Figure 8B). The pavement cell density on the adaxial surface in similar leaf areas from CS1, CS2, and CS3 transgenics was lower than that of WT and VT plants (Figure 8C). The size of cells leaving the meristem was greater in CcCKS transgenics (85 ± 7.87 μm) than in WT (47 ± 3.54 μm). The results clearly showed significant differences between transgenics and WT plants (P < 0.05). Moreover, the mature cell length in the transgenic roots was slightly longer (191 ± 8.96 μm) than in WT cells (175 ± 10.1 μm).
Figure 8.
Effect of CcCKS on growth and development of transgenic Arabiodpsis plants. (A) Rosette sizes of three weeks old WT and CcCKS independent transgenic lines grown on MS medium. (B) Primary root elongation rates of transgenic lines and WT plants. (C) Density of epidermal pavement cells, (D) Stomatal density, (E) Stomatal index, (F) Scanning electron microscope images of normal (WT) and aberrant stomatal cells in transgenic (T). WT: wild type; CS1, CS2, and CS3: 35S transgenic lines. Bar represents mean error bars represents SE from three independent experiments. Asterisks indicate significant differences in comparison with the WT at P < 0.05. Scale (F) bar 2μm.
Compared with WT and VT plants, transgenics revealed a substantially lower stomatal density and stomatal index (Figures 8D,E). The morphology of stomata in WT and CcCKS-transgenic Arabidopsis plants appeared normal. However, scanning electron micrographs clearly demonstrated a few (3%) abnormal stomata in the transgenic plants (Figure 8F).
Analysis of expression levels of selected genes in CcCKS-transgenic plants by qRT-PCR
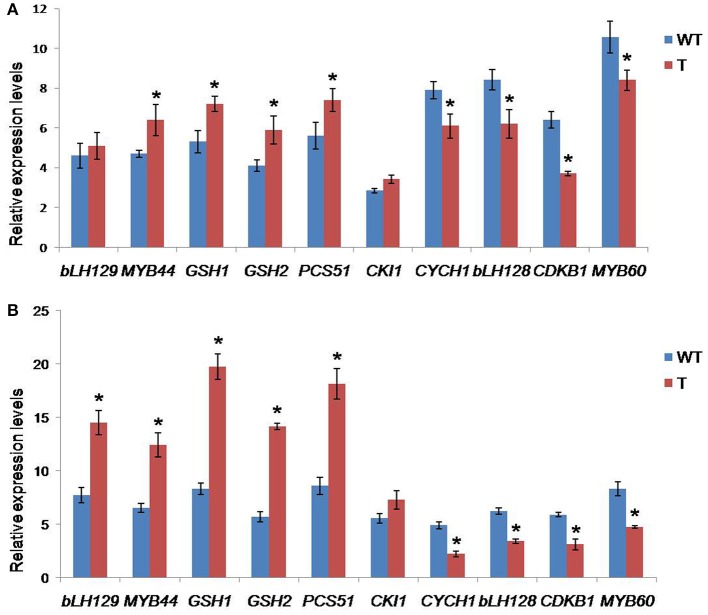
Under stress conditions, real time PCR analysis revealed substantial increases in the expression levels of certain genes, viz., delta-1-pyrroline 5-carboxylase synthetase (P5C1), bHLH129 transcription factor, MYB44 transcription factor, glutathione synthetase (gsh2), and glutamate cysteine ligase (gsh1), in transgenic plants compared to those of WT plants. However, the genes encoding cyclin H1, cyclin-dependent kinase B1, MYB60 transcription factor and bHLH128 transcription factor were significantly downregulated in transgenics under similar stress conditions compared to WT plants (Figure 9).
Figure 9.
qRT-PCR analysis for the expression profiles of selected genes in wild type and transgenic Arabidopsis under drought stress. Comparisons of the relative transcript levels of different genes in CcCKS-transgenic (T) and wild-type (WT) plants under normal (A) and 150 mM mannitol (B) stress conditions. Actin was used as a reference gene. The vertical column indicates the relative transcript level. Bar represents mean and error bars represents SE from three independent experiments. Asterisks indicate significant differences in comparison with the WT at P < 0.05.
Discussion
A stress-responsive cyclin-dependent kinase regulatory subunit gene (CKS) was isolated from the cDNA library constructed using the RNA isolated from drought stressed pigeonpea plants. The CcCKS protein structure analysis indicated its similarity to other CKS proteins of G. max, M. truncatula, A. lyrata, Z. mays, N. tomentosiformis, and B. rapa.
Higher levels of CcCKS expression were detected both in the leaves and roots under drought, ABA and salt stress treatments, compared to unstressed plants, indicating the stress-responsive nature of the gene. It was previously reported that the maize cyclin-dependent kinase regulatory subunit gene (ZmCKS2) showed variable expression levels in different parts of the seedlings, and was upregulated by drought and MeJA treatments and downregulated by cold or ABA treatments (Bao et al., 2010).
The localization of the CcCKS:GFP fusion protein in both the nucleus and the cytoplasm (Supplementary Figure S2) suggests that the CcCKS protein can enter the nucleus and it might interact with different proteins to regulate the expression of various genes involved in the plant development and stress response. Arabidopsis CKS1 and CKS2 subunits were highly and homogenously expressed in the nucleus and cytoplasm (Boruc et al., 2010b).
Under normal conditions, CcCKS transgenic plants were smaller in size with a significant reduction in fresh weight, in addition to smaller rosettes and leaves. Transgenics also exhibited fewer epidermal cells per unit leaf area (Figure 8). In transgenics, the cells leaving the root meristem were larger than that of WT plants. Moreover, the mature cells in the transgenic roots were longer than in WT plants. The difference in plant growth rate observed in transgenics compared to WT might be due to the fewer cells being produced and not because of the inhibition of cell expansion. In yeast, the expression of Arabidopsis the cyclin-dependent kinase regulatory subunit gene (AtCKS1) inhibited cell division due to its binding with CDKs (De Veylder et al., 1997). Transgenic Arabidopsis plants overexpressing AtCKS1 showed reduced leaf size and root growth rates. Moreover, changes in both cell cycle duration and meristem size strongly affected the root elongation rate in transgenic plants (De Veylder et al., 2001a). The overexpression of the CDK inhibitor gene (ICK1) resulted in fewer numbers of cells in transgenic Arabidopsis plants, and the cells were larger than those of WT plants (Wang et al., 2000).
A flow cytometric analysis of CcCKS-transgenic plants demonstrated the absence of any observable effects in ploidy levels. A well coordinated progression of mitotic and endoreduplication cycles is essential for normal plant growth, development and their response to environmental changes (Komaki and Sugimoto, 2012). Furthermore, no differences were observed in the pollen fertility between transgenic and WT plants. These results strongly suggest that CcCKS overexpression did not affect male fertility or endoreduplication in the transgenic lines.
The CcCKS-transgenic plants showed higher seed germination, survival rates, biomass, root growth, and chlorophyll content under drought and salt stress conditions. Furthermore, they could grow to maturity and set seed while the WT and VT plants failed to reach the flowering stage (Figures 3, 4, Supplementary Figure S3). These results clearly demonstrate that the transgenics were able to withstand drought and salinity treatments. Arabidopsis cdkg2 mutants showed higher percentages of seed germination, seedling growth and root elongation than those of WT under NaCl stress conditions (Ma et al., 2015).
Increased levels of proline content and the upregulation of P5CS1 gene involved in proline biosynthesis might have contributed to the osmotic potential adjustment, greater stress tolerance ability, and better growth of transgenics under stress conditions (Figure 5). Proline plays vital roles as a free radical scavenger, protecting macromolecules against denaturation, in cell-cycle control, and in differentiation and programmed cell death, in addition to modulating the expression of numerous genes that influence plant growth and development under normal conditions (Mattioli et al., 2009; Szabados and Savoure, 2010; Biancucci et al., 2015). Furthermore, proline deficiency downregulates major cyclin genes at the transcriptional level, causing cell cycle arrest and the suppression of cell proliferation (Wang et al., 2014). Lower levels of MDA and higher cell membrane stability and relative water content were observed in CcCKS transgenic lines under stress conditions (Figure 5). MDA is an indicator of lipid peroxidation due to the accumulation of ROS under stress condition (Davey et al., 2005). These results are further corroborated by lower levels of superoxide in the transgenic plants than in WT and VT plants as evidenced by the NBT staining (Figure 6).
Under stress conditions, higher levels of GSH as well as the upregulation of γ-glutamyl cysteine synthetase and glutathione synthetase genes involved in the biosynthesis of GSH were observed in transgenic plants compared to WT plants (Figure 5). Hence, the higher GSH levels might have resulted in the detoxification of ROS, leading to lower MDA levels in transgenic plants. Glutathione is the most important antioxidant in plants and is essential for defense against abiotic and biotic stresses. It also functions in redox-signaling, the modulation of defense gene expression and the regulation of enzymatic activities (Zechmann, 2014). Furthermore, the involvement GSH was reported in the regulation of cell division in the apical root meristem and in protecting proteins during seed dehydration (Sánchez-Fernández et al., 1997; Cairns et al., 2006). Transgenic Arabidopsis plants expressing tomato glutathione S-transferase (LeGSTU2) revealed enhanced resistance to NaCl and mannitol stresses (Xu et al., 2015).
Under stress conditions, the transgenics showed a significant reduction in the stomatal conductance, lower transpiration and higher photosynthetic rates compared to WT plants. In contrast, under normal conditions, the transgenics showed lower stomatal conductance, transpiration and photosynthetic rates compared to WT plants. In general, stomatal density affects water, and CO2 exchange. Decreased stomatal aperture results in a reduced transpiration rate in order to maintain the balance between water absorption and loss for better photosynthesis (Han et al., 2013).
Increased expression levels of bHLH129 and decreased transcript levels of bHLH128 were found in the transgenic plants. Basic helix-loop-helix (bHLH) transcription factors are one of the largest transcription factor families that regulate various aspects of plant growth and development, in addition to responses to abiotic stresses (Liu et al., 2014). Increased levels of MYB44 transcripts were observed in transgenic plants than in WT plants. MYB44 transgenic Arabidopsis plants efficiently prevented the accumulation of destructive ROS (Persak and Pitzschke, 2014).
In transgenic plants, lower levels of CYCLIN H-1 and MYB60 transcripts were found under stress conditions. In Arabidopsis, CYCH;1 showed predominant expression in guard cells while its expression was downregulated by dehydration (Zhou et al., 2013). Furthermore, it regulated drought stress responses through an ABA-independent pathway (Zhou et al., 2013). Similarly, MYB60 inhibited stomatal closure and root growth during drought stress and its ectopic expression in Arabidopsis resulted in greater susceptibility to the stress (Oh et al., 2011). Lower levels of cyclin-dependent kinase B1-1 (CDKB1) transcripts were observed in transgenic plants than in WT plants. A decrease in the stomatal number, stomatal index, and formation of abnormal stomata in transgenic plants might be attributed to the down-regulation of the CDKB1 gene. Arabidopsis CDKB1-1 gene was found highly expressed in guard cells and stomatal precursor cells, and thus played an important role in the stomatal development (Boudolf et al., 2004).
ABA mediates a variety of physiological processes including responses to drought, salt and cold stress (Zhu et al., 2007). In transgenic plants, the upregulation of gene transcripts involved in both ABA-dependent and ABA-independent pathways reveal the ability of CcCKS to regulate their expression. Some of the CDKs in plants play important roles in regulating the expression of genes by participating in the transcription process and by interacting with the other activators (Cui et al., 2007).
Transgenics were hypersensitive to ABA and had smaller stomatal apertures compared to WT plants treated with ABA. These results clearly indicate that CcCKS functions in the ABA signal transduction pathway. Under stress conditions, the ABA level increases in plants and is known to the mediate transcriptional regulation of stress-responsive gene expression (Kang et al., 2002). ABA also associates the abiotic stress response with plant development since it appears to play a direct role in repressing and upregulating some cell cycle genes (Kitsios and Doonan, 2011).
In this study, we cloned a stress responsive CcCKS gene from the cDNA library of pigeonpea. The overexpression of CcCKS in Arabidopsis caused enhanced tolerance to both drought and salinity stresses. The transgenic plants also showed marked changes in plant growth and development. An overview of the results suggests that CcCKS possibly acts upstream of genes involved in stress tolerance as well as plant growth and development, and thus plays an important role in the crosstalk between the signaling pathways that integrate abiotic stress tolerance with growth and development.
Author contributions
VK, DV, and ST: Conceived and designed the experiments; ST: Performed the experiments; ST, DV, and VK: Analyzed the data and wrote the paper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project is supported by grants from the Osmania University, Hyderabad, India. We thank Prof. T. Papi Reddy of Department of Genetics, Osmania University, for his helpful suggestions. ST is grateful to the University Grants Commission, New Delhi, for the award of a Research Fellowship.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00165/full#supplementary-material
Primers sequences used in the study.
Phylogenetic tree of CcCKS protein.
Expression cassettes of pCaMV35S-CcCKS-GFP fusion gene and subcellular localization of CcCKS-GFP fusion protein in Arabidopsis callus cells.
Evaluation of CcCKS transgenics against water withhold conditions. (A) Three weeks old plants growing in pots were withheld from water for two weeks, and then rewatered for one week. Photographs of plants were taken before drought stress, at the end of drought stress and after rewatering. WT represents wild type; VI represents vector transformed; CS1, CS2 and CS3 represent transgenic lines. (B) Comparison of relative water contents of detached rosettes of WT, VI and transgenic plants. (C) Measurement of osmotic potential of transgenic lines, WT and VT plants under normal and 150 mM mannitol.
Hypersensitivity response of wild type and CcCKS transgenic Arabidopsis to ABA. (A) Germination rates of wild type (WT) and transgenic (T) Arabidopsis seedlings on ABA containing medium, (B) Stomatal aperture sizes of wild type (WT) and transgenic (T) Arabidopsis plants treated with ABA. (C) Morphology of wild type (WT) and transgenic (T) seedlings grown on ABA containing medium.
Pollen fertility and ploidy level in transgenic and wild type plants. (A) Pollen fertility of wild type and CcCKS transgenics plants. T represents CcCKS-transgenic and WT represents wild-type. Pink color shows fertile pollen grains. (B) Ploidy levels in wild-type and CcCKS transgenics. WT represents wild-type, CS1, CS2 and CS3 represent transgenic lines. (Scale bar 25μm).
References
- Alexander M. P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44, 117–122. 10.3109/10520296909063335 [DOI] [PubMed] [Google Scholar]
- Bao Z. Z., Min L., Hui Y. L., Deng F. Z., Ying H. L., Su Y. S., et al. (2010). Isolation and expression analysis of drought-induced gene ZmCKS2 in maize. Acta Agron. Sin. 36, 945–952. 10.1016/S1875-2780(09)60055-9 [DOI] [Google Scholar]
- Bates L. S., Waldran R., Teare I. D. (1973). Rapid determination of free proline for water studies. Plant Soil 39, 205–208. 10.1007/BF00018060 [DOI] [Google Scholar]
- Biancucci M., Mattioli R., Moubayidin L., Sabatini S., Costantino P., Trovato M. (2015). Proline affects the size of the root meristematic zone in Arabidopsis. BMC Plant Biol. 15:263. 10.1186/s12870-015-0637-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J., Mylle E., Duda M., De Clercq R., Rombauts S., Geelen D., et al. (2010b). Systematic localization of the Arabidopsis core cell cycle proteins reveals novel cell division complexes. Plant Physiol. 152, 553–565. 10.1104/pp.109.148643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J., Van den Daele H., Hollunder J., Rombauts S., Mylle E., Hilson P., et al. (2010a). Functional modules in the Arabidopsis core cell cycle binary protein–protein interaction network. Plant Cell 22, 1264–1280. 10.1105/tpc.109.073635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V., Barrôco R., de Engler J. A., Verkest A., Beeckman T., Naudts M., et al. (2004). B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16, 945–955. 10.1105/tpc.021774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns N. G., Pasternak M., Wachter A., Cobbett C. S., Meyer A. J. (2006). Maturation of Arabidopsis seeds is dependent on glutathione biosynthesis within the embryo. Plant Physiol. 141, 446–455. 10.1104/pp.106.077982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cui X., Fan B., Scholz J., Chen Z. (2007). Roles of Arabidopsis cyclin-dependent kinase C complexes in cauliflower mosaic virus infection, plant growth, and development. Plant Cell 19, 1388–1402. 10.1105/tpc.107.051375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M., Stals E., Panis B., Keulemans J., Swennen R. (2005). High-throughput determination of malondialdehyde in plant tissues. Anal. Biochem. 347, 201–207. 10.1016/j.ab.2005.09.041 [DOI] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Beemster G. T., de Almeida Engler J., Ormenese S., Maes S., et al. (2002). Control of Proliferation, Endoreduplication and Differentiation by the Arabidopsis E2Fa/DPa transcription factor. EMBO J. 21, 1360–1368. 10.1093/emboj/21.6.1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Beemster G. T., Krols L., Terras F., Landrieu I., et al. (2001b). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13, 1653–1668. 10.1105/tpc.13.7.1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Beemster G. T., Beeckman T., Inzé D. (2001a). CKS1At overexpression in Arabidopsis thaliana inhibits growth by reducing meristem size and inhibiting cell-cycle progression. Plant J. 25, 617–626. 10.1046/j.1365-313x.2001.00996.x [DOI] [PubMed] [Google Scholar]
- De Veylder L., Joubès J., Inzé D. (2003). Plant cell cycle transitions. Curr. Opin. Plant Biol. 6, 536–543. 10.1016/j.pbi.2003.09.001 [DOI] [PubMed] [Google Scholar]
- De Veylder L., Segers G., Glab N., Casteels P., Van Montagu M., Inzé D. (1997). The Arabidopsis Cks1At protein binds to the cyclin-dependent kinases Cdc2aAt and Cdc2bAt. FEBS Lett. 412, 446–452. 10.1016/S0014-5793(97)00822-3 [DOI] [PubMed] [Google Scholar]
- Dolezel J., Bartos J. (2005). Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. 95, 99–110. 10.1093/aob/mci005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadwiger J. A., Wittenberg C., Mendenhall M. D., Reed S. I. (1989). The Saccharomyces cerevisiae CKS1 gene, a homolog of the Schizosaccharomyces pombe suc1+ gene, encodes a subunit of the Cdc28 protein kinase complex. Mol. Cell Biol. 9, 2034–2041. 10.1128/MCB.9.5.2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Tang S., An Y., Zheng D. C., Xia X. L., Yin W. L. (2013). Overexpression of the poplar NF-YB7 transcription factor confers drought tolerance and improves water use efficiency in Arabidopsis. J. Exp. Bot. 64, 4589–4601. 10.1093/jxb/ert262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles J., Beach D., Durkacz B., Nurse P. (1986). The fission yeast cell cycle control gene cdc2: isolation of a sequence suc1 that suppresses cdc2 mutant function. Mol. Gen. Genet. 202, 291–293. 10.1007/BF00331653 [DOI] [PubMed] [Google Scholar]
- Inze D., Veylder L. (2006). Cell cycle regulation in plant development Annu. Rev. Genet. 40, 77–105. 10.1146/annurev.genet.40.110405.090431 [DOI] [PubMed] [Google Scholar]
- Kang J. Y., Choi H. I., Im M. Y., Kim S. Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14, 343–357. 10.1105/tpc.010362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. 10.1038/7036 [DOI] [PubMed] [Google Scholar]
- Kitsios G., Doonan J. H. (2011). Cyclin dependent protein kinases and stress responses in plants. Plant Signal. Behav. 6, 204–209. 10.4161/psb.6.2.14835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki S., Sugimoto K. (2012). Control of the plant cell cycle by developmental and environmental cues Plant Cell Physiol. 53, 953–964. 10.1093/pcp/pcs070 [DOI] [PubMed] [Google Scholar]
- Krasensky J., Jonak C. (2012). Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 63, 1593–1608. 10.1093/jxb/err460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Tai H., Li S., Gao W., Zhang M., Xie C., et al. (2014). bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 201, 1192–1204. 10.1111/nph.12607 [DOI] [PubMed] [Google Scholar]
- Ma X., Qiao Z., Chen D., Yang W., Zhou R., Zhang W., et al. (2015). CYCLIN DEPENDENT KINASE G2 regulates salinity stress response and salt mediated flowering in Arabidopsis thaliana. Plant Mol. Biol. 88, 287–299. 10.1007/s11103-015-0324-z [DOI] [PubMed] [Google Scholar]
- Mao X., Zhang H., Tian S., Chang X., Jing R. (2010). TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis. J. Exp. Bot. 61, 683–696. 10.1093/jxb/erp331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli R., Costantino P., Trovato M. (2009). Proline accumulation in plants: not only stress. Plant Signal Behav. 4, 1016–1018. 10.4161/psb.4.11.9797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. A. (1962). Revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Oh J. E., Kwon Y., Kim J. H., Noh H., Hong S. W., Lee H. (2011). A dual role for MYB60 in stomatal regulation and root growth of Arabidopsis thaliana under drought stress. Plant Mol. Biol. 77, 91–103. 10.1007/s11103-011-9796-7 [DOI] [PubMed] [Google Scholar]
- Persak H., Pitzschke A. (2014). Dominant repression by Arabidopsis transcription factor MYB44 causes oxidative damage and hypersensitivity to abiotic stress. Int. J. Mol. Sci. 15, 2517–2537. 10.3390/ijms15022517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyanka B., Sekhar K., Reddy V. D., Rao K. V. (2010a). Expression of pigeonpea hybrid-proline-rich protein encoding gene (CcHyPRP) in yeast and Arabidopsis affords multiple abiotic stress tolerance. Plant Biotechnol. J. 8, 76–87. 10.1111/j.1467-7652.2009.00467.x [DOI] [PubMed] [Google Scholar]
- Priyanka B., Sekhar K., Sunitha T., Reddy V. D., Rao K. V. (2010b). Characterization of expressed sequence tags (ESTs) of pigeonpea (Cajanus cajan L.) and functional validation of selected genes for abiotic stress tolerance in Arabidopsis thaliana. Mol. Genet. Genomics 283, 273–287. 10.1007/s00438-010-0516-9 [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof M. A., Soliman K. M., Jorgensen R. A., Allard R. W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location and population dynamics. Proc. Natl. Acad. Sci. U.S.A. 81, 8014–8019. 10.1073/pnas.81.24.8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2001). Molecular cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sánchez-Fernández R., Fricker M., Corben L. B., White N. S., Sheard N., Leaver C. J., et al. (1997). Cell proliferation and hair tip growth in the Arabidopsis root are under mechanistically different forms of redox control. Proc. Natl. Acad. Sci. U.S.A. 94, 2745–2750. 10.1073/pnas.94.6.2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhar K., Priyanka B., Reddy V. D., Rao K. V. (2010). Isolation and characterization of a pigeonpea cyclophilin (CcCYP) gene, and its over-expression in Arabidopsis confers multiple abiotic stress tolerance. Plant Cell Environ. 33, 1324–1338. 10.1111/j.1365-3040.2010.02151.x [DOI] [PubMed] [Google Scholar]
- Shin L. J., Lo J. C., Yeh K. C. (2012). Copper chaperone antioxidant protein1 is essential for copper homeostasis. Plant Physiol. 159, 1099–1110. 10.1104/pp.112.195974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabados L., Savoure A. (2010). Proline: a multifunctional amino acid. Trends Plant Sci. 15, 89–97. 10.1016/j.tplants.2009.11.009 [DOI] [PubMed] [Google Scholar]
- Tamirisa S., Vudem D. R., Khareedu V. R. (2014). Overexpression of pigeonpea stress-induced cold and drought regulatory gene (CcCDR) confers drought, salt, and cold tolerance in Arabidopsis. J. Exp. Bot. 65, 4769–4781. 10.1093/jxb/eru224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank J. G., Thaker V. S. (2011). Cyclin dependent kinases and their role in regulation of plant cell cycle. Biol. Plantarum. 55, 201–212. 10.1007/s10535-011-0031-9 [DOI] [Google Scholar]
- Tong S., Ni Z., Peng H., Dong G., Sun Q. (2007). Ectopic overexpression of wheat TaSrg6 gene confers water stress tolerance in Arabidopsis. Plant Sci. 172, 1079–1086. 10.1016/j.plantsci.2007.03.011 [DOI] [Google Scholar]
- Wang G., Zhang J., Fan X., Sun X., Qin H., Xu N., et al. (2014). Proline responding1 plays a critical role in regulating general protein synthesis and the cell cycle in maize. Plant Cell 26, 2582–2600. 10.1105/tpc.114.125559 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang H., Zhou Y., Gilmer S., Whitwill S., Fowke L. C. (2000). Expression of the plant cyclin dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J. 24, 613–623. 10.1046/j.1365-313x.2000.00899.x [DOI] [PubMed] [Google Scholar]
- Wise A. A., Lu Z., Binns A. N. (2006). Three methods for the introduction of foreign DNA into Agrobacterium. Methods Mol. Biol. 343, 43–53. 10.1385/1-59745-130-4:43 [DOI] [PubMed] [Google Scholar]
- Xu J., Xing X. J., Tian Y. S., Peng R. H., Xue Y., Zhao W., et al. (2015). Transgenic Arabidopsis plants expressing tomato glutathione S-transferase showed enhanced resistance to salt and drought stress. PLoS ONE 10:e0136960. 10.1371/journal.pone.0136960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Chen X., Hong Y. Y., Wang Y., Xu P., Ke S. D., et al. (2008). Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20, 1134–1151. 10.1105/tpc.108.058263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechmann B. (2014). Compartment specific importance of glutathione during abiotic and biotic stress. Front. Plant Sci. 5:566. 10.3389/fpls.2014.00566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. F., Jin Y. H., Yoo C. Y., Lin X. L., Kim W. Y., Yun D. J., et al. (2013). CCYCLIN;1 regulated drought stress responses and blue light-induced stomatal opening by inhibiting reactive oxygen species accumulation in Arabidopsis. Plant Physiol. 162, 1030–1041. 10.1104/pp.113.215798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S. Y., Yu X. C., Wang X. J., Zhao R., Li Y., Fan C. R., et al. (2007). Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19, 3019–3036. 10.1105/tpc.107.050666 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers sequences used in the study.
Phylogenetic tree of CcCKS protein.
Expression cassettes of pCaMV35S-CcCKS-GFP fusion gene and subcellular localization of CcCKS-GFP fusion protein in Arabidopsis callus cells.
Evaluation of CcCKS transgenics against water withhold conditions. (A) Three weeks old plants growing in pots were withheld from water for two weeks, and then rewatered for one week. Photographs of plants were taken before drought stress, at the end of drought stress and after rewatering. WT represents wild type; VI represents vector transformed; CS1, CS2 and CS3 represent transgenic lines. (B) Comparison of relative water contents of detached rosettes of WT, VI and transgenic plants. (C) Measurement of osmotic potential of transgenic lines, WT and VT plants under normal and 150 mM mannitol.
Hypersensitivity response of wild type and CcCKS transgenic Arabidopsis to ABA. (A) Germination rates of wild type (WT) and transgenic (T) Arabidopsis seedlings on ABA containing medium, (B) Stomatal aperture sizes of wild type (WT) and transgenic (T) Arabidopsis plants treated with ABA. (C) Morphology of wild type (WT) and transgenic (T) seedlings grown on ABA containing medium.
Pollen fertility and ploidy level in transgenic and wild type plants. (A) Pollen fertility of wild type and CcCKS transgenics plants. T represents CcCKS-transgenic and WT represents wild-type. Pink color shows fertile pollen grains. (B) Ploidy levels in wild-type and CcCKS transgenics. WT represents wild-type, CS1, CS2 and CS3 represent transgenic lines. (Scale bar 25μm).