Abstract
The bacterial second messenger cyclic di-3′,5′-guanosine monophosphate (c-di-GMP) is a key regulator of bacterial motility and virulence. As high levels of c-di-GMP are associated with the biofilm lifestyle, c-di-GMP hydrolysing phosphodiesterases (PDEs) have been identified as key targets to aid development of novel strategies to treat chronic infection by exploiting biofilm dispersal. We have studied the EAL signature motif-containing phosphodiesterase domains from the Pseudomonas aeruginosa proteins PA3825 (PA3825EAL) and PA1727 (MucREAL). Different dimerisation interfaces allow us to identify interface independent principles of enzyme regulation. Unlike previously characterised two-metal binding EAL-phosphodiesterases, PA3825EAL in complex with pGpG provides a model for a third metal site. The third metal is positioned to stabilise the negative charge of the 5′-phosphate, and thus three metals could be required for catalysis in analogy to other nucleases. This newly uncovered variation in metal coordination may provide a further level of bacterial PDE regulation.
Cyclic di-3′,5′-guanosine monophosphate (c-di-GMP) is a universal second messenger in eubacteria that regulates a wide variety of functions including formation and dispersal of biofilms1,2,3,4, adhesion1,5,6, motility7,8,9, synthesis of virulence factors10 and developmental transitions11,12. Cellular levels of c-di-GMP are regulated by synthesis and degradation through diguanylate cyclases (DGCs) and phosphodiesterases (PDEs), respectively. DGC domains containing the signature GGDEF motif synthesise c-di-GMP from two GTP molecules9,13,14 while EAL15,16 and HD-GYP17,18,19 domain-containing PDEs are responsible for the hydrolysis of c-di-GMP to 5′-phosphoguanylyl-(3′-5′)-guanosine (pGpG). Structural and biochemical characterisation of GGDEF, EAL and HD-GYP domains provide a good understanding of the mechanisms of c-di-GMP turnover, interactions with other proteins and the direct and indirect effects of c-di-GMP levels on cell phenotype. A number of excellent reviews comprehensively summarise the current knowledge for c-di-GMP signalling in eubacteria and more specifically Pseudomonas aeruginosa20,21,22.
Bacteria within a biofilm are 100–1000 times more tolerant to antibiotics23,24,25. Therefore, being able to revert bacteria to a planktonic phenotype may present new opportunities to remove chronic infection26. As biofilm dispersal is associated with a reduction in intracellular levels of c-di-GMP, this strategy would likely depend upon c-di-GMP hydrolysing PDEs3,5,9. EAL domain containing proteins from a number of organisms have now been investigated structurally, with all EAL domains to date presenting with a highly conserved αβ(βα)7-barrel fold27,28,29. Most EAL domains possess PDE activity; although a significant group of inactive EAL domains exist, which likely function as c-di-GMP sensors30,31,32. Hydrolysis of c-di-GMP by EAL domains has been shown to require the presence of divalent ions, either Mg2+ or Mn2+, with the most efficient reactions occurring at alkaline pH29,33.
Dimerisation-induced conformational changes have been shown to regulate enzymatic activity in EAL-PDEs33,34,35,36. Specifically, dimerisation of the EAL domain YahA from Escherichia Coli has been shown to increase substrate-turnover, while specific site directed mutagenesis preventing dimerisation results in reduced PDE activity; without altering the substrate binding affinity33. This regulation is further rationalised through observations of EAL domain dimerisation in P. aeruginosa MorA, which causes the α5 helix to unwind at the dimerisation interface; allowing the catalytic DDFGTG motif on the β5-α5 loop (previously labelled as connecting β6 and α627,34) to enter the substrate binding pocket and coordinate catalytically important metal ions in the active site35. Solution studies of Klebsiella pneumoniae BlrP1 identify this mechanism as regulatory in the context of full-length proteins, with the dimerisation interface and catalytic DDFGTG motif stabilised when activity is increased by stimulation of the regulatory BLUF domain36. Collectively these studies demonstrate how substrate-binding and metal coordination is controlled. Surprisingly, crystal structures of dimers have predominantly been reported with bound substrate, c-di-GMP28,29,37. This is not consistent with a model of activation through dimer formation and suggests that further regulatory steps must be required for the activation of EAL-PDEs.
A model organism for study of biofilm dispersal, and thus EAL-PDE behaviour, is Pseudomonas aeruginosa PAO1. The bacterial genome of this organism contains 41 proteins with PDE or DGC domains38,39, many of which regulate individual components of the biofilm phenotype40,41. As these seemingly redundant proteins have different roles related to biofilm behaviour we sought to determine conserved features of EAL-PDE activation.
Here we report as yet structurally uncharacterised EAL domains from the core genome of Pseudomonas aeruginosa, PA1727 (MucR) and PA3825. Structural analysis of MucREAL and PA3825EAL, in different metal binding states with or without substrate or product, provide evidence for the link between dimerisation and the formation of metal binding sites that are independent of the dimerisation interface. Finally we characterise a potential third metal binding site that directly interacts with the reaction product pGpG.
Results
Functional characterisation of MucR and PA3825 multi-domain proteins
We found that deletion of the PA1727 (mucR) gene leads to increased motility in P. aeruginosa; swim zones exhibited an average area of 0.762 ± 0.112 cm2 in the ∆mucR strain compared to 0.499 ± 0.064 cm2 in the wild type (n = 6, P value = 0.001, 3 biological repeat experiments). MucR is a bifunctional transmembranous enzyme containing DGC and PDE activity that regulates alginate biosynthesis and biofilm dispersal42,43,44. Both cytoplasmic DGC and PDE domains are required to regulate the levels of c-di-GMP controlling alginate synthesis43,45. The ∆mucR strain suggests that this multi-domain protein is an active DGC under test conditions, as deletion results in increased swimming motility, presumably due to a reduction in cellular c-di-GMP.
Neither swimming, swarming, nor twitching motilities were affected in the PA3825 knockout strain under conditions tested. PA3825 is a two-domain cytoplasmic protein consisting of a structurally uncharacterised N-terminal CSS domain, with a predicted fold similar to the extracellular receptors of bacterial dimeric histidine kinases, and a PDE domain. However PA3825EAL in vitro shows PDE activity in the presence of Mn2+, Fig. 1. In vitro PA3825EAL was able to convert c-di-GMP to pGpG, with a kcat of 0.35 ± 0.1 s−1, in the presence of Mn2+, consistent with observations of other active EAL-PDEs15,29,33.
Figure 1. Kinetics analysis of PA3825EAL in the presence of Mn2+.
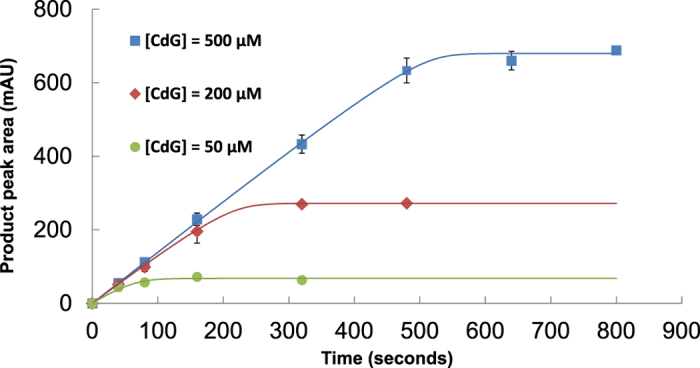
PA3825EAL (3 μM final concentration) was added to different concentrations of c-di-GMP (labelled as CdG) and the progress of each reaction measured at specified time points through separation of nucleotides using Resource-Q FPLC, followed by integration of peaks at 254 nm absorbance.
Different dimer contacts in c-di-GMP/Mg2+ bound MucREAL and PA3825EAL
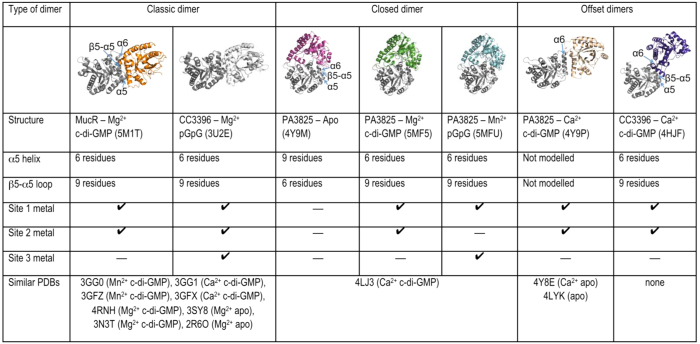
We determined the PDE domain structures of MucREAL and PA3825EAL in complex with c-di-GMP/Mg2+. Data processing and refinement statistics are reported in Table 1. Both PDE domains maintain the αβ(βα)7-barrel fold consistent with all EAL-PDE structures determined to date27. The MucREAL dimerisation interface involves helices α5 and α6 and has been observed in many other characterised PDE domains as listed in Fig. 2 (classic dimer). It is well established that significant structural rearrangements induced by dimer formation lead to a repositioning of active site residues to make the enzyme catalytically competent33,35,36.
Table 1. Crystallographic data and refinement statistics.
| Structure | PA3825EAL Apo | PA3825EAL Ca-c-di-GMP | PA3825EAL Mg-c-di-GMP | PA3825EAL Mn-pGpG | MucREAL-Mg-c-di-GMP |
|---|---|---|---|---|---|
| Data Collection | |||||
| Space Group | P43212 | C2 | P43212 | P43212 | P21 |
| Cell Axes (Å) | a = 64.3 | a = 112.2 | a = 64.6 | a = 64.8 | a = 46.4 |
| b = 64.3 | b = 59.4 | b = 64.6 | b = 64.8 | b = 116.1 | |
| c = 134.0 | c = 92.8 | c = 135.7 | c = 135.8 | c = 52.1 | |
| Angles (°) | α = β = γ = 90 | α = γ = 90 | α = β = γ = 90 | α = β = γ = 90 | α = γ = 90 |
| β = 115.0 | β = 102.5 | ||||
| Beamline | I03 | I04-1 | I02 | I02 | I04 |
| Wavelength (Å) | 0.9763 | 0.9200 | 0.9795 | 1.8785 | 0.9795 |
| Resolution (Å) | 64.34–1.60 (1.64–1.60) | 51.29–2.44 (2.50–2.44) | 29.16–1.77 (1.81–1.77) | 27.16–2.15 (2.21–2.15) | 45.27–2.27 (2.33–2.27) |
| Unique Reflections (#) | 36008 (2598) | 20405 (1494) | 28763 (2072) | 16288 (1689) | 24883 (1831) |
| Measured Reflections (#) | 373940 (26770) | 91744 (7123) | 178724 (13790) | 61259 (6341) | 170715 (12632) |
| Redundancy | 9.3 (9.2) | 4.5 (4.8) | 6.2 (6.7) | 3.8 (3.8) | 6.9 (6.9) |
| Rpim (%) | 3.1 (30.2) | 2.6 (42.2) | 2.2 (26.7) | 7.2 (40.5) | 6.9 (27.6) |
| I/σ (I) | 18.0 (3.2) | 17.3 (2.0) | 18.8 (2.6) | 6.6 (2.4) | 11.3 (2.9) |
| Completeness (%) | 100.0 (99.5) | 98.1 (98.9) | 99.5 (99.8) | 98.8 (99.2) | 99.9 (99.8) |
| Refinement | |||||
| Molecules/AU | 1 | 2 | 1 | 1 | 2 |
| Rwork/Rfree (%) | 18.3/23.2 | 23.8/28.5 | 19.9/23.2 | 20.2/26.1 | 19.5/23.8 |
| Rmsd | |||||
| Bond Length (Å) | 0.022 | 0.008 | 0.014 | 0.016 | 0.011 |
| Bond Angles (°) | 2.088 | 1.373 | 1.638 | 1.725 | 1.440 |
| Average B Factor (Å2) | |||||
| Protein | 21.4 | 61.0 | 32.9 | 36.8 | 36.6 |
| Nucleotide | - | 44.2 | 31.0 | 38.9 | 27.8 |
| Metals | - | 77.3 | 29.9 | 41.3 | 22.0 |
| Water | 28.9 | 53.1 | 35.4 | 37.7 | 36.0 |
| Ramachandran (%) | |||||
| Favoured Regions | 99.3 | 93.8 | 98.3 | 97.6 | 98.0 |
| Allowed Regions | 0.7 | 5.2 | 1.7 | 2.4 | 2.0 |
| PDB Code | 4Y9M | 5MKG | 5MF5 | 5MFU | 5M1T |
Values in parentheses refer to the highest resolution bin.
Figure 2. Analysis of dimerisation behaviour of EAL-PDE domains.
Shown are different dimer architectures of EAL-PDEs, and associated structures identified in the PDB (as of November 2016). Information on the structure of the α5-helix and the β5-α5 loop is given; a short α5-helix (6 residues) is compatible with active site formation and metal binding through the two aspartate side chains of the DDFGTG motif on the elongated β5-α5 loop. Listed are the metal binding states and the nucleotide loading states.
The structure of PA3825EAL in complex with c-di-GMP/Mg2+ shows a different dimer interface to MucR, named here as the “closed dimer” (Fig. 2). We sought to confirm the dimerisation induced activation mechanism, seen in the classic dimer33,34,36, across this different dimer interface. The β5-α5 loop of PA3825EAL, residues 160–170, undergo significant changes between Mg2+-c-di-GMP bound and metal-free-apo states. The β5-α5 loop moves towards the active site and contributes to the formation of metal binding sites via two highly conserved Asp residues (Asp160–161, Fig. 3). The restructuring of the β5-α5 loop and the functional role in metal coordination is thus conserved across different dimer interfaces.
Figure 3. The structure of PA3825EAL in complex with c-di-GMP/Mg2+.
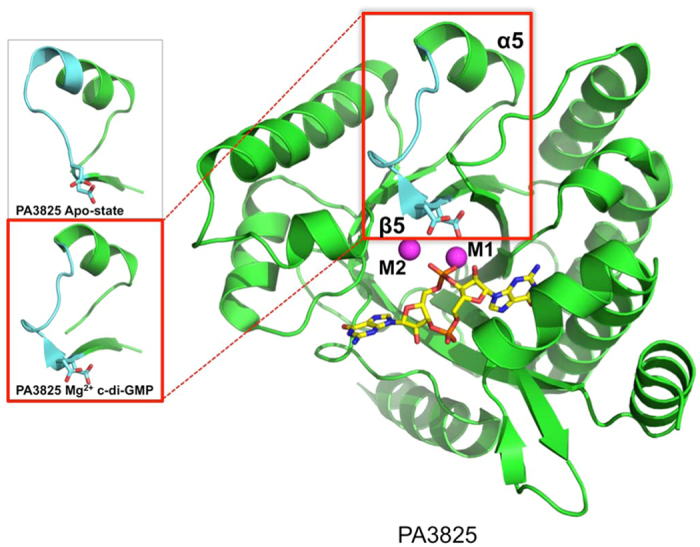
C-di-GMP is shown as yellow sticks and Mg2+ ions are represented by magenta spheres. Zoomed views of the β5-α5 loop, highlighted in light-blue, demonstrate the variation in the length of the α5 helix between the apo form and substrate forms of PA3825EAL. The structural shift in the β5-α5 loop allows Asp160 and Asp 161 to coordinate catalytic metals indicated as M1 and M2.
The basis of EAL-PDE inhibition by Ca2+
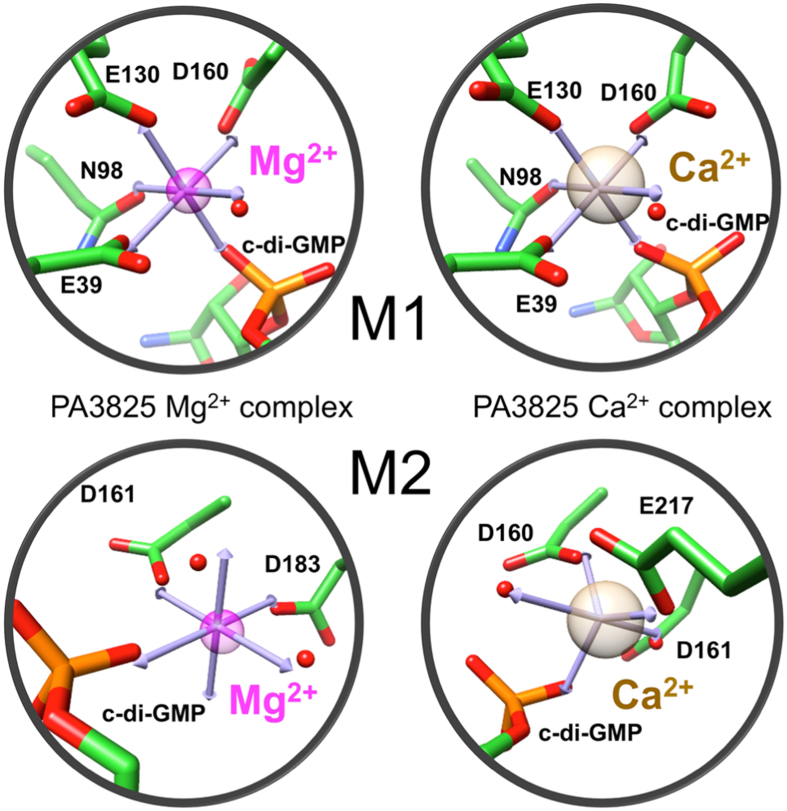
Ca2+ is a well known inhibitor of EAL-PDE activity15,16,29. To better understand the reasons for the inhibitory role of Ca2+, we determined PA3825EAL structures in the presence of Mg2+ or Ca2+ ions with bound c-di-GMP (Fig. 4). The Mg2+ and Ca2+ complexes show metals in the catalytic centre, annotated as M1 and M2, coordinating the phosphate group of the scissile phosphodiester in c-di-GMP.
Figure 4. Perturbed metal geometry with altered activity in PA3825EAL.
Idealised coordination geometries of the Mg2+- and Ca2-c-di-GMP PA3825EAL complexes show the M1 metal ions coordinated in octahedral coordination. The M2 site metal in the Mg2+ -c-di-GMP PA3825EAL complex is coordinated in an octahedral geometry, in comparison to the M2 metal site within the Ca2+ -c-di-GMP PA3825EAL complex, which is coordinated in a trigonal bipyramidal geometry. Metal ions are shown as coloured transparent spheres (Mg2+ in magenta, Ca2+ in tan) with coordinating residues and nucleotide displayed in stick form with coordinating waters shown as red spheres.
The Mg2+ and Ca2+ metals in the M1 site superimpose well between the two complexes, (Fig. 4). In both cases, the metal is coordinated in an octahedral geometry by Glu39, Asn98, Asp130, Asp160, a non-bridging oxygen from the scissile phosphate (P1) of c-di-GMP and a putative in-line attacking water molecule (Fig. 4). Significant differences are seen in the positioning of the Asp160 side chain between metal bound complexes, with the second oxygen of Asp160 contacting a peptide nitrogen atom with Mg2+ bound; while the Ca2+ bound complex shows Asp160 bridging between the M1 and M2 metals.
The M2 metal shows octahedral coordination for Mg2+ but trigonal bipyramidal coordination geometry for Ca2+ (Fig. 4). Mg2+ is coordinated by Asp161, Asp183, a non-bridging phosphate (P1) oxygen and two waters (Fig. 4). This contrasts with the calcium complex, where Asp183 is not involved in coordination but instead Asp160 and Glu217 make contact to the Ca2+ in the M2 position. Both Asp160 and Asp161 are part of the conserved catalytic sequence motif DDFGTG. The different positioning of Asp160 and different coordination geometry of Ca2+, together with physicochemical properties, thus contribute to the inhibitory effect by calcium46,47.
Structure of PA3825EAL in complex with pGpG reveals a third metal binding site
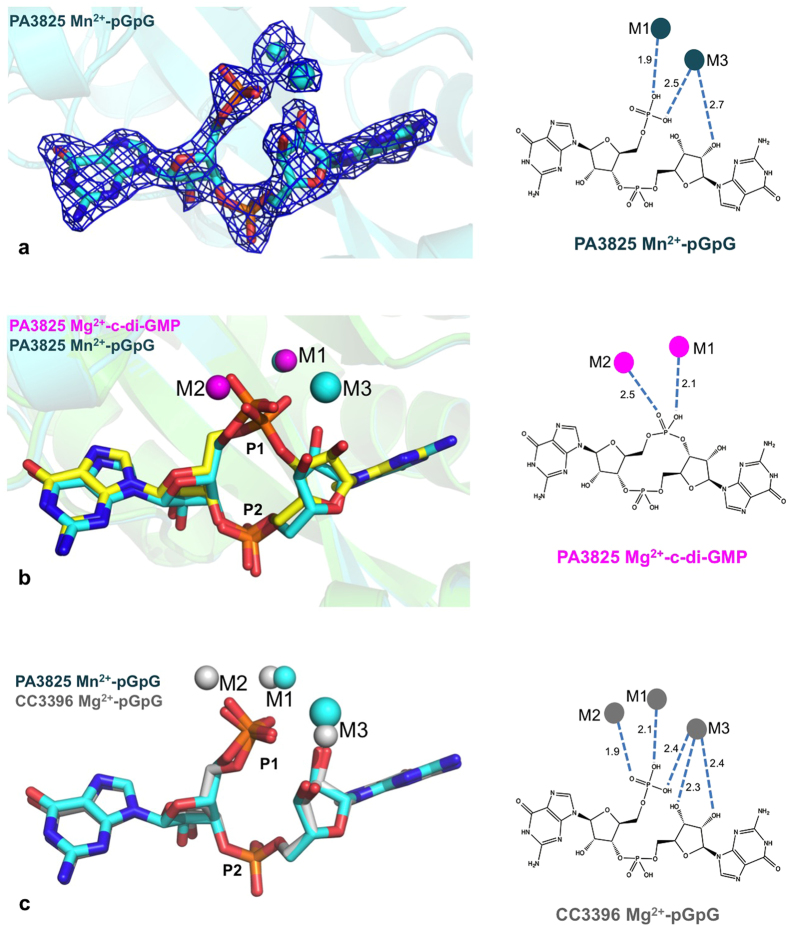
Most PDB structures to date are reported either as apo-enzyme or as c-di-GMP substrate complexes, Table 2. We were fortuitous to observe PA3825EAL with bound product pGpG, from co-crystallisation with c-di-GMP and Mn2+ (Fig. 5a). In this complex, the M1 site in the pGpG-bound structure is superimposable to the M1 metal in Mg2+ and Ca2+ complexes, the M2 site is vacant, but we observed a solvent binding site that interacts directly with the bound pGpG that presents metal like coordination geometry. We propose this as a potential third metal binding site, designated as M3.
Table 2. Classification of all EAL domain structures in the PDB to date.
| No metal C-di-GMP | No metal No substrate | 1 metal (M1) No substrate | 1 metal (M1) C-di-GMP | 2 metals (M1 & M2) C-di-GMP | pGpG |
|---|---|---|---|---|---|
| Monomeric | Monomeric | Dimeric | Dimeric | Dimeric | Dimeric |
| FimX-EAL (3 HV8) | DosP-EAL (4HU3) | Mg2+ | Mg2+ | Mn2+ | Mn2+ |
| FimX-EAL (4FOJ) | LapD-EAL (3PFM) | TBD1265-EAL (2R6O) | MorA-EAL (4RNH) | Blrp1 (3GFZ) | PA3825-EAL (5MFU) |
| FimX-EAL (4FOU) | FimX-EAL (3HV9) | Ca2+ | Ca2+ | Blrp1 (3GG0) | Mg2+ |
| FimX-EAL (4FOK) | CC3396-EAL (3S83) | PA3825-EAL (4Y8E) | Blrp1_B (3GFX) | Mg2+ | CC3396-EAL |
| FimX-EAL (4F3H) | YkuI_AB (2W27) | PA3825-EAL (5MF5) | no metal | ||
| FimX-EAL (4F48) | Dimeric | Tetrameric | MucR-EAL (5M1T) | FimX-EAL (4AFY) | |
| LapD-EAL (3PJW) | PA3825-EAL (4Y9M) | Mg2+ | TBD1265_EAL (3N3T) | ||
| LapD-EAL (3PJX) | YkuI (2BAS) | RocR (3SY8) | Ca2+ | ||
| LapD-EAL (3PJU) | YdiV-EAL (3TLQ) | PA3825-EAL (4Y9P) | |||
| Imo0131-EAL (4Q6J) | CC3396-EAL (4HJF) | ||||
| Dimeric | MorA-EAL (4RNJ) | Blrp1_A (3GFX) | |||
| LapD-EAL (3PJT) | MorA-EAL (4RNI) | Blrp1 (3GG1) | |||
| FimX-EAL (4AG0) | YahA (4LJ3) |
Inactive EAL domains are indicated in italics.
Figure 5. Identification of the M3 position in the pGpG complex.
(a) Electron density of pGpG and metal ions bound to PA3825EAL, contoured at 1.3 σ. (b) Structural overlay of the substrate and product complexes of PA3825EAL. The substrate c-di-GMP is shown as yellow sticks; Mg2+ ions (magenta spheres) occupy the M1 and M2 binding sites. The product pGpG is shown as blue sticks; Mn2+ ions (blue spheres) occupy the M1 and M3 binding sites. (c) Structural comparison of the product complexes of PA3825EAL and CC3396EAL (PDB code 3U2E). In PA3825EAL manganese and sodium ions are present in the M1 and M3 positions, respectively, while in CC3396EAL magnesium ions are present in the M1, M2 and M3 positions. Schematic representations on the right show the interactions between the coordinated metal ions and the bound substrate or product, with distances labelled in Å.
Diffraction data collected above the manganese absorption edge confirmed the presence of Mn2+ in the M1 position in the pGpG/Mn2+ PA3825EAL complex (Supplementary Fig S1) but showed no signal for manganese at the M3 metal like centre. Nonetheless, this metal-like M3 site is coordinated by the P1 phosphate non-bridging oxygen and the O2′ ribose oxygen (Fig. 5b). Consistent with the absence of any anomalous signal in the M3 site, we have placed and refined sodium in this location which is abundant in the crystallisation condition and consistent with the geometry observed48.
The M3 metal site displays a tetrahedral coordination sphere consisting of Asp 160, a water molecule, the ribose O2′ oxygen and a P1 hydroxyl oxygen from pGpG. To identify whether the M3 site is unique we searched the PDB and identified two EAL-pGpG complexes, Table 2. The first, FimX49 (PDB code 4AFY) is an inactive PDE and lacks the DDFGTG motif required for metal coordination49; the guanine bases in the FimX-pGpG complex are positioned in different positions from either the substrate complex or the product complexes of PA3825EAL, which in themselves superpose well. The second, CC3396EAL from Caulobacter crescentus (PDB code 3U2E), shows pGpG adopting a near identical conformation to the pGpG moiety in PA3825EAL (Fig. 5c). This structure also identifies a third metal binding site which aligns with the proposed M3 binding site in PA3825EAL. CC3396EAL was refined and deposited with Mg2+ modelled at all three metal binding sites. These magnesium ions directly interact with the bound pGpG through coordination to oxygen atoms of the phosphate and the ribose around the scissile bond (Fig. 5c). In CC3396EAL the M3 metal binding site coordination differs to that of PA3825EAL in that the O3′ oxygen from the pGpG sugar ring and waters take the place of the conserved aspartate, allowing octahedral coordination of the d-metal Mg2+. In PA3825EAL a tetrahedral coordination is observed for the s-metal Na+ complex. Superposition of the two complexes shows that the Na+ of PA3825EAL is further removed from the scissile bond, compared with the Mg2+ observed in CC3396EAL, consistent with ionic radii and metal co-ordination geometries (Fig. 5c). The M1 and M2 metal sites of CC3396EAL correspond to the characteristic M1 and M2 metal sites discussed above. In PA3825EAL, the M2 site is empty, but Glu217 (contributed by the β7-α7 loop in close vicinity to the β5-α5 loop) replaces interactions with the phosphate moiety normally made by M2. While PA3825EAL lacks a metal in the M2 site and possesses one in the M3 site, the comparison with CC3396EAL, in which all three metal sites are occupied, suggests M3 site is a bona fide metal binding site in PA3825EAL.
Discussion
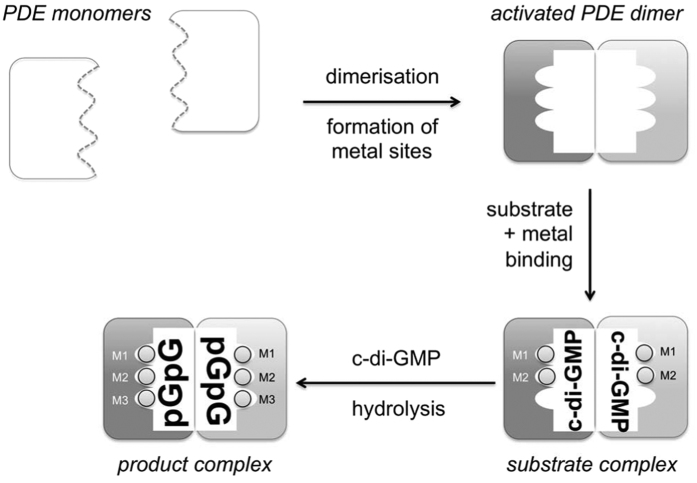
To identify features of EAL-PDE activation conserved across different phenotypic responses we have studied the EAL domains of the multi-domain proteins PA3825 and MucR from P. aeruginosa. While EAL-PDE activity is modulated by dimerisation through length variation of helix α5 and correct positioning of the metal coordinating motif DDFGTG35, we show conservation of this mechanism across different dimer interfaces (Fig. 2). The data presented also allows the identification of a third metal site M3 which is a hallmark of the product state. This may explain the observation of c-di-GMP substrate complexes despite apparent dimerisation and well-formed active sites with coordinated M1 and M2 metals. We summarise the structural changes that discriminate substrate and product bound states of EAL-PDEs in a scheme (Fig. 6).
Figure 6. Proposed regulatory steps in EAL-PDE catalysed c-di-GMP hydrolysis.
Dimerisation induces a conformational change in the β5-α5 loop, allowing conserved aspartate residues to coordinate metal ions in the M1 and M2 position and c-di-GMP to bind. A third metal ion binds in the M3 position in the pGpG product complex.
The substrate- and product-bound forms of PA3825EAL are characterised by changes around the α5 helix, consistent with an activation step from the apo/ground state induced by dimerisation. The model put forward requires dimerisation for activity that by itself would not be sufficient for gaining full activity. Summarising knowledge gained from structure determinations of different EAL PDEs suggests that binding of metal ions or ligands is always concurrent with a shortening of the α5-helix and linked to the requirement of protein-protein interactions (Fig. 2 and Table 2)33,35,36. Observation of the third metal binding site now requires rationalising.
Historically, mutagenesis and structural studies on RocR and YkuL led to catalytic proposals involving only a single-metal27,28. A number of c-di-GMP bound EAL structures with catalytic centres containing pairs of either Mg2+, Mn2+ or Ca2+ ions then lead to the proposals of two metal-ion catalysis16,29,37,50. Observed differences in inter-metal distances within a range of metal-substrate BlrP1 complexes at various pHs, with different in vitro activity, finally lead to the suggestion of metal ion migration within the active site regulating activity. It was proposed that metals move closer to one another to activate a water molecule for phosphodiester bond hydrolysis29. Further, deuterium exchange data showed that activation through a sensor domain (BLUF) and active site changes, including ordering of the α5 region, are consistent with preformed dimers36. These data together now point at further changes required in the active site for the catalytic cycle of phosphodiester bond hydrolysis.
Observation of the M3 metal site close to the hydrolysed phosphodiester bond in pGpG may now provide a first hint to rationalise the unexplained observations around enzyme activation. While metals in M1 and M2 are required for activation of the nucleophile, a metal in the M3 site is too far removed from the nucleophile to perform a similar function. Instead, the M3 metal is observed in the product state where it is ideally poised to stabilise the negatively charged transition state during hydrolysis of c-di-GMP to pGpG; where pGpG directly contributes to formation of the M3 metal site via its non-bridging phosphate oxygen (terminal phosphate group of pGpG). The M3 site is consistent with the pGpG-bound state of Caulobacter crescentus EAL PDE CC3396EAL (PDB code 3U2E). The presence of a metal in the M3 site is concurrent with bound pGpG, providing a further example for requirement of a metal in this position.
Parallels exist between the proposed three-metal EAL-PDE and nuclease P1, along with other endonucleases. Binding of three Zn2+ ions has been shown in high-resolution structures for nuclease P151 and endonuclease IV52,53. Quantum mechanics calculations since have supported functional roles for all three metals54,55, with the third metal ion positioned close to the hydrolysed bond to stabilise the negatively charged leaving group. The analogy holds for magnesium ions as well, as recent kinetic analysis of T5 flap endonuclease across a range of ion concentrations demonstrated the requirement for three magnesium ions in catalysis56. The EAL type PDEs thus might also require three metals in its catalytic cycle, which has parallels to a subset of HD-GYP type PDEs19,57.
Conclusion
Understanding the regulation of PDE containing proteins is essential to study biofilm development and dispersal mechanisms. New structural data shows three metal sites in EAL-type phosphodiesterases. In the product complex, a metal directly interacts with the hydrolysed phosphodiester, and while the precise role of the newly identified M3 metal in catalysis requires further investigation, a role in stabilisation of the negative charge during catalysis is likely. We further describe a model of inhibition of EAL type PDEs using Ca2+, and demonstrate that variation of metal binding is fully consistent with the previously described enzyme activation mechanism by dimerisation, even involving different dimer interfaces. Understanding the regulatory layers of EAL-PDE activation may provide an angle to using PDEs as drug targets in biofilm dispersal.
Experimental Procedures
Bacterial strains, media, and growth conditions
Pseudomonas aeruginosa and Escherichia coli strains used in this study are listed in Table S158,59. All overnight cultures were routinely grown at 37 °C in LB medium. Antibiotics were used at the following concentrations: 30 μg/ml of gentamicin (Gm), 200 μg/ml of carbenicillin for P. aeruginosa and 15 μg/ml of Gm, 100 μg/ml of ampicillin for E. coli S17-1.
Construction of isogenic P. aeruginosa mutants
Isogenic mutants were constructed by replacing part of the mucR and PA3825 coding regions (as identified from the Pseudomonas genome database60) with a gentamicin (Gm) resistance cassette as previously described61. Briefly, two flanking up- and downstream regions (approximately 350–400 bp) from the mucR and PA3825 genes were amplified by standard PCR using primers listed in Table S2. Up- and downstream mucR and PA3825 DNA fragments were digested with EcoRI and HindIII and ligated to the Gm cassette (amplified from pPS85659) to give DNA fragments with the Gm cassette positioned between the up- and downstream regions of the gene of interest. The constructed fragments were then inserted into the SmaI site on pEX100 T, generating plasmids pEX100 T::∆mucR::Gm and pEX100 T::∆PA3825::Gm containing sacB genes for counter selection. Plasmids were introduced into E. coli S17-1 by chemical transformation and then transferred into PAO1 by conjugation. Transconjugants were first selected on Pseudomonas isolation agar containing 30 μg/ml Gm and then patched onto LB + 5% sucrose + 30 μg/ml Gm agar and LB + 200 μg/ml carbenicillin agar. Colonies that only grew on LB + 5% sucrose + 30 μg/ml Gm agar but not on LB + 200 μg/ml carbenicillin were selected as double-recombinant mutants. Mutants were confirmed by PCR, followed by sequencing.
Motility assays
To assess swarming motility, 3 μl overnight LB culture (absorbance at 600 nm, A600, 1.3–1.5) was inoculated onto 0.5% (w/v) agar with 8 g/L nutrient broth (Oxoid) and 5 g/L glucose (poured at 55 °C and dried under laminar flow for 40 mins) and incubated at 37 °C for 24 hrs.
To assess swimming motility, bacteria grown overnight on LB agar were inoculated onto the surface of a swimming plate (10 g/L tryptone (Oxoid)/5 g/L NaCl) containing 0.3% (w/v) agarose, poured at 55 °C and dried under laminar flow for 15 mins) with a sterile toothpick and incubated at 30 °C for 16–18 hrs.
To assess twitching motility, bacteria grown overnight on LB agar were stab inoculated into the bottom of 1% (w/v) LB agar (poured at 55 °C and dried under laminar flow for 1 hrs) with a sterile toothpick and incubated at 37 °C for 24 hrs.
Cloning, Expression and Purification
A DNA fragment for the EAL domain of PA3825 (residues 255–517) was PCR-amplified from the genome of P. aeruginosa strain PAO1 and subcloned into popinF vectors using in-fusionTM cloning, and subsequently transformed into E. coli Lemo21 (DE3) cells in the presence of 50 μg/ml ampicillin. Cells were grown at 37 °C to an A600~0.6, induced with 0.15 mM IPTG and left at 18 °C for 16 h after induction. Cultures were subsequently spun at 5500 g for 10 minutes at 4 °C and the pellets resuspended in buffer A (0.1 M Tris-HCL pH 8.0, 0.5 M NaCl, 1 mM TCEP, 20 mM imidazole, 2% glycerol) plus 10 mM MgCl2, 10 μg/ml DNAse and 300 μg of lysozyme per gram of pellet. Cells were lysed by sonication in two cycles of 45 and 30 seconds and centrifuged for 15 minutes at 30,600 g at 4 °C. Cleared lysate was loaded onto a Ni-NTA affinity column pre-equilibrated in buffer A. Affinity chromatography was carried out in an AKTA purifier (GE Healthcare) according to the manufacturers’ instructions. The N-terminal His6-tag was cleaved overnight at 4 °C with a His6-tagged 3 C protease, which was subsequently removed by passing the sample back onto the Ni-NTA column. Fractions containing the untagged EAL domain were pooled together and concentrated to a final volume of 6 ml and loaded onto a Superdex 200 16/60 gel filtration column equilibrated with size exclusion chromatography (SEC) buffer (20 mM Tris-HCL, pH 8, 300 mM NaCl, 1 mM TCEP).
DNA coding for the EAL domain of MucR (residues 425–685) was also PCR–amplified from the genome of P. aeruginosa PAO1 and subcloned into pET-28a vector using HndIII and NdeI restriction sites (Novagen). This plasmid was then transformed into E. coli BL21 DE3 competent cells, grown in LB medium at 37 °C to an A600 of 0.6, before induction with 1 mM IPTG and incubated at 37 °C for a further 2 hours. Cultures were subsequently spun at 4,000 g for 20 mins at 4 °C and the pellets resuspended in buffer B (50 mM HEPES pH 7.5, 300 mM NaCl, 2 mM β-Mercaptoethanol and 5% glycerol). Cells were lysed by sonication for 5 minutes and centrifuged for 40 minutes at 30,000 g. The supernatant was loaded onto 4 ml of Ni-NTA resin (Qiagen), equilibrated with buffer B containing 20 mM imidazole, followed by washing with buffer B containing 80 mM imidazole and elution using buffer B containing 300 mM imidazole. The eluate was concentrated using spin-concentrators (Generon) before being loaded onto a S75 16/600 size exclusion column (GE) equilibrated in 50 mM HEPES pH 7.5, 300 mM NaCl, 2 mM MgCl2 and 2 mM β-Mercaptoethanol.
Enzymatic Assay
Enzymatic assays quantified the conversion of c-di-GMP to pGpG by ion exchange chromatography with a 1 ml Resource-Q column pre-equilibrated in 5 mM ammonium bicarbonate. Enzymatic reactions were carried out at room temperature as previously described33. PA3825EAL was stripped of all metals with 50 mM EDTA, later removed by dialysis. Enzymatic assays were carried out in the presence of excess substrate and Mn2+ at 5 times excess. To monitor reaction progress, at different time points 100 μl of reaction solution was loaded onto the column and eluted over an 18 CV linear ammonium bicarbonate gradient (5 mM to 1 M). Elution was monitored by absorption at 254 nm. Nucleotide percentage was determined by integrating the nucleotide specific peaks. Global Kinetic Explorer software (KinTek, Austin, TX, USA) was used to fit the entire set of progress curves measured over a range of substrate concentrations.
Crystallisation and Data Collection
For all crystallisation experiments, PA3825EAL was concentrated to 15–20 mg/ml (quantified by measuring absorbance at 280 nm, A280). Crystallisation was carried out by sitting drop vapour diffusion at a 1:1 protein/reservoir ratio. Primitive tetragonal crystals grew at 4 °C in 0.8 M sodium phosphate monobasic, 1.2 M potassium phosphate (dibasic) and 0.1 M sodium acetate pH 4.5 (overall pH of the final solution was found to be 7.1). Centred monoclinic crystals were grown at 4 °C in 6–11% isopropanol, 0.1 M MES pH 6.5, 0.1 M sodium acetate pH 4.5 and 0.2 M calcium acetate (overall pH 5.2). CaCl2, MgCl2 or MnCl2, respectively, were added at a concentration of 0.2 M to the crystallisation reservoir before drops were dispensed and 10 mM c-di-GMP was added directly to the protein sample to co-crystallise PA3825EAL tetragonal crystals. In the case of metal-di-GMP ternary complexes, metal-loaded PA3825EAL crystals, prepared as described above, were soaked with 10 mM c-di-GMP for 1 hour at 20 °C. The tetragonal crystal of PA3825EAL containing the bound product pGpG were instead prepared by co-crystallisation of the protein with the substrate, by adding 10x excess of CdG to PA3825EAL prior setting up crystallisation drops in the presence of MnCl2 (see above); under these conditions, only one crystal was obtained, which appeared after several weeks. All crystals were very briefly soaked in the appropriate crystallisation solution containing 25% ethylene glycol before flash freezing in liquid nitrogen. Single crystal diffraction datasets for metal-free-apo and metal-c-di-GMP complex crystals were collected at 100 K on beamlines I02 and I04-1 at Diamond Light Source, UK (Table 1).
MucREAL was concentrated to 13 mg/ml (quantified by measuring A280) and crystallised in 0.1 μL sitting drops. Crystals grew at 21 °C in 100 mM Morpheus Buffer System 1 pH 6.5, 120 mM Morpheus ethylene glycols and 37.5% of a MPD, PEG 1 K and PEG 3350 precipitant mix (Molecular Dimensions). Data were collected on I04 (Diamond Light Source) at 100 K (Table 1).
Structure Solution and Refinement
Diffraction datasets were processed with the xia2 pipeline at Diamond with 5% of reflections removed for cross-validation62,63,64,65,66,67. All structures were solved by molecular replacement using either Molrep68 or MrBump69. Both MucREAL and the apo structure of PA3825EAL were solved using residues 493–743 of the EAL domain from Thiobacillus denitrificans (PDB code 2R6O)37. PA3825EAL metal-c-di-GMP and metal-pGpG complexes were solved using the refined apo structure as the search model. Further refinement of structures was performed with REFMAC570, with model building carried out in COOT71. Model validation was performed using Molprobity72. Figures of all structures were prepared with the UCSF chimera package or PyMol version 1.3r171,72,73,74.
Additional Information
How to cite this article: Bellini, D. et al. Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases. Sci. Rep. 7, 42166; doi: 10.1038/srep42166 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We acknowledge the BBSRC, the Wellcome Trust (WT093314/Z10/Z) and Diamond Light Source for support. We thank Diamond MX village and MPL staff for help and support. We thank Chris Holes and Stuart Findlow for support at the Macromolecular Crystallisation Facility of the Centre for Biological Sciences, Southampton.
Footnotes
The authors declare no competing financial interests.
Author Contributions D.B., S.H., A.H. and Y.C. collected experimental data. D.B., S.H., A.H., C.W.P., R.W.S., Y.C., I.T. and M.A.W. analysed data. D.B., S.H., A.H., A.W., J.S.W., I.T. and M.A.W. wrote the paper. M.A.W. and I.T. managed and coordinated the project.
References
- D’Argenio D. A., Calfee M. W., Rainey P. B. & Pesci E. C. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184, 6481–6489 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirillina O., Fetherston J. D., Bobrov A. G., Abney J. & Perry R. D. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol. Microbiol. 54, 75–88 (2004). [DOI] [PubMed] [Google Scholar]
- Tischler A. D. & Camilli A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53, 857–69 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud N. et al. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J. Bacteriol. 191, 7333–42 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormann K. M. et al. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 188, 2681–2691 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A. & Wood T. K. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog. 5, 1–15 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge P., Paul R., Goymer P., Rainey P. & Jenal U. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 47, 1695–1708 (2003). [DOI] [PubMed] [Google Scholar]
- Huang B., Whitchurch C. B. & Mattick J. S. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 185, 7068–76 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm R., Morr M., Kader A., Nimtz M. & Römling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53, 1123–1134 (2004). [DOI] [PubMed] [Google Scholar]
- Tischler A. D. & Camilli A. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 73, 5873–5882 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R. et al. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18, 715–27 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd C. D. & O’Toole G. A. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu. Rev. Cell Dev. Biol. 28, 439–62 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. et al. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. USA 101, 17084–9 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deepthi A., Liew C. W., Liang Z. X., Swaminathan K. & Lescar J. Structure of a diguanylate cyclase from Thermotoga maritima: Insights into activation, feedback inhibition and thermostability. PLoS One 9, 1–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M., Christen B., Folcher M., Schauerte A. & Jenal U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280, 30829–37 (2005). [DOI] [PubMed] [Google Scholar]
- Tamayo R., Tischler A. D. & Camilli A. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 280, 33324–30 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan R. P. et al. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 103, 6712–7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Stelitano V. et al. C-di-GMP hydrolysis by Pseudomonas aeruginosa HD-GYP phosphodiesterases: analysis of the reaction mechanism and novel roles for pGpG. PLoS One 8, e74920 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini D. et al. Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre. Mol. Microbiol. 91, 26–38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U., Galperin M. Y. & Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha D.-G. & O’Toole G. A. C-di-GMP and its effects on biofilm formation and dispersion: a Pseudomonas aeruginosa review. Microb. Spectr. 3, MB -003–2014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini M. & Filloux A. Biofilms and c-di-GMP signaling: Lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 291, 12547–12555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan R. & Costerton J. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceri H. et al. The Calgary biofilm device: New technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37, 1771–1776 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorachit M., Lam K., Jayanetra P. & Costerton J. W. Resistance of Pseudomonas pseudomallei growing as a biofilm on silastic disks to ceftazidime and cotrimoxazole. Antimicrob. Agents Chemother. 37, 2000–2002 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly D. L., Bellini D., Walsh M. A., Dow J. M. & Ryan R. P. Targeting cyclic di-GMP signalling: A strategy to control biofilm formation? Curr. Pharm. Des. 21, 12–24 (2015). [DOI] [PubMed] [Google Scholar]
- Rao F., Yang Y., Qi Y. & Liang Z.-X. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J. Bacteriol. 190, 3622–31 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minasov G. et al. Crystal structures of YkuI and its complex with second messenger cyclic Di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains. J. Biol. Chem. 284, 13174–84 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barends T. R. M. et al. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459, 1015–8 (2009). [DOI] [PubMed] [Google Scholar]
- Navarro M. V. A. S., De N., Bae N., Wang Q. & Sondermann H. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure 17, 1104–1116 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y. et al. Binding of cyclic diguanylate in the non-catalytic EAL domain of FimX induces a long-range conformational change. J. Biol. Chem. 286, 2910–2917 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell P. D., Monds R. D. & O’Toole G. A. LapD is a bis-(3′,5′)-cyclic dimer GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0–1. Proc. Natl. Acad. Sci. USA 106, 3461–3466 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundriyal A. et al. Inherent regulation of EAL domain-catalyzed hydrolysis of second messenger cyclic di-GMP. J. Biol. Chem. 289, 6978–90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F. et al. The functional role of a conserved loop in EAL domain-based cyclic di-GMP-specific phosphodiesterase. J. Bacteriol. 191, 4722–31 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phippen C. W. et al. Formation and dimerization of the phosphodiesterase active site of the Pseudomonas aeruginosa MorA, a bi-functional c-di-GMP regulator. FEBS Lett. 588, 4631–4636 (2014). [DOI] [PubMed] [Google Scholar]
- Winkler A. et al. Characterization of elements involved in allosteric light regulation of phosphodiesterase activity by comparison of different functional BlrP1 states. J. Mol. Biol. 426, 853–868 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchigvintsev A. et al. Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases. J. Mol. Biol. 402, 524–38 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasakara H. et al. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. USA 103, 2839–44 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan R. P. et al. HD-GYP domain proteins regulate biofilm formation and virulence in Pseudomonas aeruginosa. Environ. Microbiol. 11, 1126–36 (2009). [DOI] [PubMed] [Google Scholar]
- Balasubramanian D., Schneper L., Kumari H. & Mathee K. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 41, 1–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J. H. et al. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. MBio 1, e00183–10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y., Gaidenko T. a., Mulkidjanian a. Y., Nakano M. & Price C. W. MHYT, a new integral membrane sensor domain. FEMS Microbiol. Lett. 205, 17–23 (2001). [DOI] [PubMed] [Google Scholar]
- Hay I. D., Remminghorst U. & Rehm B. H. a. MucR, a novel membrane-associated regulator of alginate biosynthesis in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 75, 1110–20 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Heine S., Entian M., Sauer K. & Frankenberg-Dinkel N. NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by an MHYT domain-coupled phosphodiesterase. J. Bacteriol. 195, 3531–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Hay I. D., Rehman Z. U. & Rehm B. H. a. Membrane-anchored MucR mediates nitrate-dependent regulation of alginate production in Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 99, 7253–7265 (2015). [DOI] [PubMed] [Google Scholar]
- Kuppuraj G., Dudev M. & Lim C. Factors Governing Metal-Ligand Distances and Coordination Geometries of Metal Complexes Factors. J. Physocal Chem. 113, 2952–2960 (2009). [DOI] [PubMed] [Google Scholar]
- Grzybkowski W. Nature and Properties of Metal Cations in Aqueous Solutions. Polish J. Environ. Stud. 15, 655–663 (2006). [Google Scholar]
- Schmidt A. J., Ryjenkov D. A. & Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: Enzymatically active and inactive EAL domains. J. Bacteriol. 187, 4774–4781 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romier C., Dominguez R., Lahm A., Dahl O. & Suck D. Recognition of single-stranded DNA by nuclease P1: High resolution crystal structures of complexes with substrate analogs. Proteins Struct. Funct. Genet. 32, 414–424 (1998). [PubMed] [Google Scholar]
- Hosfield D. J., Guan Y., Haas B. J., Cunningham R. P. & Tainer J. A. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: Double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell 98, 397–408 (1999). [DOI] [PubMed] [Google Scholar]
- Mol C. D., Hosfield D. J. & Tainer J. a. Abasic site recognition by two apurinicrapyrimidinic endonuclease families in DNA base excision repair: the 3. Mutat. Res. (2000). [DOI] [PubMed] [Google Scholar]
- Ivanov I., Tainer J. a. & McCammon J. A. Unraveling the three-metal-ion catalytic mechanism of the DNA repair enzyme endonuclease IV. Proc. Natl. Acad. Sci. USA 104, 1465–1470 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao R. Z., Yu J. G. & Himo F. Phosphate mono- and diesterase activities of the trinuclear zinc enzyme nuclease P1 - Insights from quantum chemical calculations. Inorg. Chem. 49, 6883–6888 (2010). [DOI] [PubMed] [Google Scholar]
- Syson K. et al. Three metal ions participate in the reaction catalyzed by T5 flap endonuclease. J. Biol. Chem. 283, 28741–28746 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway B. W. Genetics of Pseudomonas. Bacteriol. Rev. 33, 419–443 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J. & Schweizer H. P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally -located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86 (1998). [DOI] [PubMed] [Google Scholar]
- Miner K. D. & Kurtz D. M. Active site metal occupancy and cyclic di-GMP phosphodiesterase activity of Thermotoga maritima HD-GYP. Biochemistry. 55, 970–979 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor G. L. et al. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39, D596–600 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.-H. & Schweizer H. P. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5, 30 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G. Xia2: An expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010). [Google Scholar]
- Winter G., Lobley C. M. C. & Prince S. M. Decision making in xia2. Acta Crystallogr. Sect. D Biol. Crystallogr. 69, 1260–1273 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D Biol. Crystallogr. 62, 72–82 (2006). [DOI] [PubMed] [Google Scholar]
- Evans P. R. & Murshudov G. N. How good are my data and what is the resolution? Acta Crystallogr. Sect. D Biol. Crystallogr. 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A. & Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 22–25 (2010). [DOI] [PubMed] [Google Scholar]
- Keegan R. M. & Winn M. D. MrBUMP: An automated pipeline for molecular replacement. Acta Crystallogr. Sect. D Biol. Crystallogr. 64, 119–124 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W. G. & Cowtan K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H. et al. Validation of metal-binding sites in macromolecular structures with the CheckMyMetal web server. Nat. Protoc. 9, 156–170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Paganin J., Nonin-Lecomte S. & Réty S. Crystal structure of an EAL domain in complex with reaction product 5′-pGpG. PLoS One 7, e52424 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V. B. et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F. et al. UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- Schrödinger L. The PyMOL molecular graphics system, version 1.3r1. (2010).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.