Abstract
HIV-1 and its surface envelope glycoproteins (Env), gp120 and gp41, have evolved immune evasion strategies that render the elicitation of effective antibody responses to the functional Env entry unit extremely difficult. HIV-1 establishes chronic infection and stimulates vigorous immune responses in the human host; forcing selection of viral variants that escape cellular and antibody (Ab)-mediated immune pressure, yet possess contemporary fitness. Successful survival of fit variants through the gauntlet of the human immune system make this virus and these glycoproteins a formidable challenge to target by vaccination, requiring a systematic approach to Env mimetic immunogen design and evaluation of elicited responses. Here, we review key aspects of HIV-1 Env immunogenicity and immunogen re-design, based on experimental data generated by us and others over the past decade or more. We further provide rationale and details regarding the use of newly evolving tools to analyze B cell responses, including approaches to use Next Generation Sequencing for antibody lineage tracing and B cell fate mapping. Together, these developments offer opportunities to address long-standing questions about the establishment of effective B cell immunity elicited by vaccination, not just against HIV-1.
Introduction
HIV-1 Env properties, natural responses and consequences for vaccine design
Relatively few viruses exhibit continual active replication in immunocompetent mammalian hosts, as does HIV-1, the focus of this review. We addressed previously the challenge HIV-1 presents as a vaccine target (1) our appreciation of which has even increased over the ensuing years. In part, the challenge arises as a consequence of chronic replication in literally millions of human hosts, during which HIV-1 acquires features that allow it to successfully evade both innate and adaptive B cell and T cell immune responses. At a high-level perspective, HIV-1 is a member of the enveloped virus class, incorporating a host lipid bilayer and accompanying self-proteins upon budding to become “a wolf in sheep's cloaking” to the immune system. The HIV-1-encoded envelope glycoproteins function to mediate viral entry, which due to their exposed location on the surface of the virus, are under intense antibody selection pressure. To protect the functionally vital Env complex, HIV-1 evolves in a Darwinian manner by masking the vast majority of its molecular surface in N-linked glycans (see FIG 1, potential N-glycosylation sites), with significant clusters selected for on the external and apical major variable (V) regions V1 and V2, and individual sites at the base of V3. The remainder of the Env surface is comprised of the external subunit gp120 variable regions V4 and V5, resulting in a surface that is composed of either carbohydrate or variable amino acids (2), but yet adopts a meta-stable state as one requirement to maintain function. This metastability involves the gp120 “cap”, constraining the gp41 “springs” that have evolved to facilitate receptor-triggered entry. The quaternary-dependent, tightly packed trimer possesses hydrophilic V regions that tolerate amino acid or glycan heterogeneity that aid in escape of antibody-mediated neutralization without interfering with essential functions that mediate viral entry.
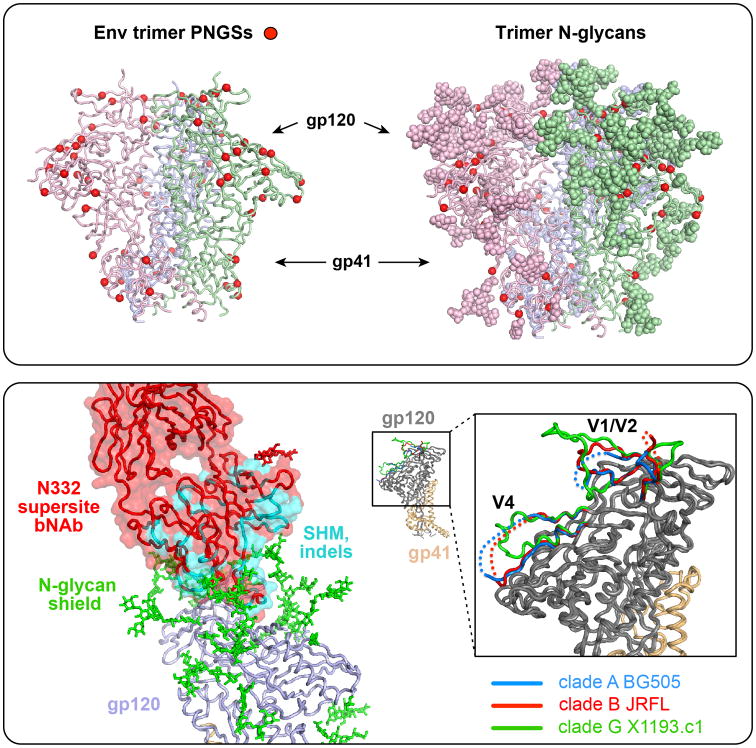
Fig 1. N-glycan shielding, trimer packing and variability.
Top box, left; Potential N- glycosylation sites (PNGS; motif is NXT/S) are shown (red balls) on the structure of the HIV trimer spike (BG505 SOSIP.664). Each of the three protomers, composed of gp120 and gp41 associated subunits are shown with matching colors for each monomer (hot pink, blue and green, left, back and right, respectively). Note the “spring-loaded” helices of gp41 can be seen in the center of the lower region of the trimer (and in the lower box, lower right inset). Right, the N-glycans are shown in a space-filling, ball rendition, “shielding” the underlying protein polypeptide surface; asparagine residues at the N-glycan base are indicated (red balls). Lower box, lower left, the bNAb PGT121 (red, surface and alpha carbon backbone) penetrates the N-glycan shield (green) with the gp120 alpha carbon trace in blue (ribbon). Somatic hypermutation and indels are indicated (cyan), achieving a relatively high level of 23% in this bNAb, many of which are needed to allow mAb penetration of the evolved dense N-glycan shield that accounts for half the molecular mass of the HIV Env trimer. Lower right, smaller image is one gp120-gp41 protomer with gp120 (gray) associated with the spring-loaded gp41 (gold); expanded box depicts the variable regions V1/V2 and V4 of gp120 from three strains derived from different subtypes as indicated. These V regions, along with N-glycan shielding, contribute to immune evasion from neutralizing antibodies by presenting variable surfaces that can accommodate additional mutation.
In large part, the ability to evolve in response to selection derives from the error-prone HIV-1 reverse transcriptase (RT) that allows the generation of many simultaneous variants from each infected CD4+CCR5+ host target cell. Due to widespread dissemination and relatively rapid rate of replication, multiple variants coexist within each infected individual, archived in resting or cycling memory T cells. This panoply of variants can mediate escape from the immune response regardless of selection pressure at any given moment during the chronic infection process, which in turn stimulates B and T cell responses to the new epitopes. This co-evolution of the virus and the immune response creates a steady-state continuum derived from overlapping waves of viral emergence, dominance and elimination by host immunity to only be replaced by the next most fit and temporally neutralization-resistant member of the evolving quasi-species within a given infected individual. The intense B cell selection pressure against every functional variant in the quasi-species consequently shapes the evolution of antibody (Ab) responses against the exposed determinants of Env, resulting in a close to ideal spacing of the N-linked glycans on the trimer spike, occluding antibody access to the underlying polypeptide surface. In addition to this evolving glycan shield, key amino acid point mutant variants and V region duplications or (3) deletions generate increasing surface variations that help evade Ab recognition. In general, protein surfaces are much more immunogenic than are the essentially self, host-generated N-linked glycans, resulting in effective shielding of conserved, functionally constrained (4, 5) determinants of Env (see FIG 1). Env N-glycans are separated at their base by 4-5 Ångströms (6) since they cannot be appended too closely by the host glycosyl-transferases. These enzymes need access to the asparagine (N) residue of the glycosylation motif to attach the proximal N-acetyl glucosamine glycan residues and for further processing in the ER and Golgi reviewed in (3). This tight glycan spacing effectively blocks access by most membrane-bound B cell receptors (BCRs), limiting B cell activation, and to antibodies, limiting recognition of underlying protein epitopes. In some chronically HIV-1-infected individuals, antibodies eventually evolve that can penetrate the armor of this effective glycan shield, demonstrating that the human immune system can generate HIV-1 broadly neutralizing antibodies (bNAbs) to several distinct regions of Env. Often the bNAbs contain very high levels of somatic hypermutation (SHM) and, relatively rare, activation-induced cytidine deaminase (AID)-mediated insertions and deletions (indels) in either or both the immunoglobulin heavy and light chain sequences, or other unusual features such as long HCDR3s, N-glycan reactivity or self-reactivity ((5, 7-11) and see FIG 1). These relatively rare qualities probably come into play more frequently during chronic infection due to the immune system-disruptive effects of the chronic HIV-1 infection, including prolonged antigenic exposure. Such remarkable, but infrequent, antibodies may be less likely to be elicited by active vaccination in healthy subjects using relatively short immunization regimens. Reports of bNAbs isolated from chronically infected subjects that display lower levels of SHM are therefore encouraging for ongoing efforts to develop a vaccine (12, 13).
For purposes of vaccine design and development, a beneficial application of the bNAbs is for “antigenic profiling. That is, to confirm that as many broadly neutralizing determinants as possible are present on a given Env immunogen that is designed to present such epitopes to the immune system. This, together with lack of recognition by most non-neutralizing antibodies to a given immunogen, is indicative of faithful mimicry of the functional spike. A general rationale for many Env immunogen design efforts is that by presenting the broadly neutralizing determinants and minimizing the exposure of unwanted non-neutralizing determinants, immunization will preferentially activate B cells specific for broadly neutralizing, cross-conserved determinants. High-resolution evaluation of Ab responses elicited by Env mimetics designed based on these criteria have recently begun in several laboratories. Another application of bNAbs is to design immunogens that bind putative germline-reverted (gL) version of such MAbs, and to evaluate the ability of such immunogens to activate B cell responses in mice engineered to express the corresponding gL-reverted Ab as a BCR in vivo. These topics are covered elsewhere in this edition (see Stamatatos and Alt papers).
The extensive evolution and N-glycan shrouding of HIV-1 Env limits antibody access to conserved Env surfaces, such as at the primary receptor CD4 binding site (CD4bs). That said, the molecular surface of the CD4bs itself is devoid of N-glycans, facilitating direct protein:protein interaction between the gp120 CD4bs and CD4 (14). However, although the CD4bs is glycan-free, this surface is ringed by N-glycans on the outer domain “side” and from “above” by the V cap spike apex. These N-glycans restrict accessibility to two-domain-wide antibodies that display a larger foot print than does the one-domain-wide, Ig superfamily member, CD4. In addition, due to Env spike quaternary packing, occlusion by elements of the adjacent protomer increases difficulty of access by cell surface-expressed BCRs as well as by soluble Ab. BNAbs, such as VRC01, PGV04 and 3BCN17, are examples of successful, but relatively rare, solutions to accessing the conserved CD4bs, as illustrated in several studies (9, 15-18). Other antigenically conserved sites of Env do exist, as defined by bNAbs, for example, in the trimer apex (4, 19-21), the base of V3 (19, 22) and at the gp120-gp41 junctions (23-26). However, these regions are heavily glycan-protected and bNAbs to these regions often engage N-glycan as well as polypeptide, requiring infrequent structural Ab characteristics mediated by high levels of SHM for binding (see Fig 1, lower left panel for a structural example).
To put current efforts to design an HIV-1 vaccine into perspective, it is worth noting that no currently licensed human vaccine elicits cross-reactive breadth to protect against variable pathogens. Even the extremely effective and durable human papilloma virus (HPV) vaccine elicits limited “breadth” only by including multiple clinically relevant subtypes in the vaccine to directly match the immune response with a given human papilloma virus subtype (27). And, unlike HIV, the surface-exposed hydrophilic loops of this non-enveloped virus are immunogenic, non-glycosylated and are “in play” to achieve highly effective strain-restricted neutralizing antibodies. Similarly, the seasonal influenza vaccine provides protection against the circulating variants included in the vaccine and not through cross-reactive Ab neutralization breadth, as we have discussed previously (1). Hence, to create an effective HIV vaccine, new approaches must be explored, analyzed and honed to perfection to establish new paradigms not remotely approached by other current and successful anti-viral vaccines.
Section 1 – Properties of Env and first generation trimer mimetics
The elements of the HIV-1 Env trimer metastability involve, in part, packing of the spring-loaded gp41 helices to store free energy for the process of viral-to-cell fusion (see Fig 1, top and lower right panels). The gp41 helices, confined by the associated gp120 cap, are hair-trigged to harpoon the fusion peptide into target cells following engagement by the primary receptor, CD4, and the co-receptor, CCR5. This spring-loaded mechanism presents challenges when attempting to generate native-like soluble versions of the HIV-1 functional Env spike. For the past several decades, many trimer designs attempted to create faithful mimetics of the native spike, with limited success, until the relatively recent emergence of the so-called well-ordered trimers, addressed later in this review. Early iterations to generate soluble Env spike mimetics began with the simple approach of introducing a stop codon at the C-terminus of the gp41 ectodomain, upstream of the hydrophobic transmembrane domain, to generate soluble oligomers. However, due to the non-covalent association of the external envelope glycoprotein, gp120, with the transmembrane glycoprotein, gp41, these “gp140” molecules, comprised of gp120 and the gp41 ectodomain, were unstable, dissociating into two distinct subunits (reviewed in www.ncbi.nlm.nih.gov/pubmed/20048701). As we now know, this occurred in large part due to the spring-loaded nature of gp41 and releasing the natural transmembrane anchor point by genetic truncation. Since gp120 and gp41 derive from cellular furin cleavage of the gp160 precursor in the Golgi, the furin cleavage site, REKR, was either genetically deleted or mutated (i.e. to SEKS) to create an unnatural covalent association of gp120 to gp41. However, this approach generated gp140 oligomers that consisted of mixtures of molecules in different conformational states, primarily because the metastable Env proceeds down multiple off-target conformational paths following translation and folding in the ER and Golgi. Next, we and others attempted to generate more stable and relevant trimers by adding heterologous trimerization motifs such as modified GCN4 or foldon (28-30), or other trimerization motifs such as aspartate transcarbamoylase (31).
In the last few years, it has become increasingly apparent that many of these trimers are not faithful mimetics of the functional Env spike, although there is some disagreement regarding what are the most relevant biophysical assays to determine native-like properties of a given trimer mimetic (32, 33). In our more recent work, we rely on the analytical value of antigenic profiling where bNAbs recognize the native trimer and non-neutralizing antibodies do not. These data, coupled with differential scanning calorimetry (DSC) electron microscopy (EM) are used to analyze candidate trimer mimetics. Trimers that generate single homogeneous melting transitions by DSC and 2D classifications and 3D reconstructions of three-fold symmetry by EM, confirm well-ordered trimer integrity (34-36) as discussed below. By antigenic profiling and EM, foldon trimers derived from the YU2 and JRFL strains do not appear to faithfully mimic the native spike as they are well-recognized by non-neutralizing antibodies and do not display an ordered appearance or trimeric symmetry by negative stain EM (37). However, despite the inability of these earlier trimer designs to fully mimic properties of the native spike, we learned a great deal by studying the B cell and antibody responses they elicit in the context of immunization, establishing analytical tools that are broadly used in the field today. In non-human primates (NHPs), using the foldon trimers as immunogens, we defined a number of fundamental features of immune responses to Env vaccination, establishing a system amenable for high-resolution analysis of B cell responses following HIV trimer immunization (37-46). We discuss the specifics of our high-resolution B cell studies in more detail below (Section 3).
Section 2 – Env immunogenicity at the polyclonal Ab level
One approach to defining the polyclonal response to Env following vaccination involves mapping such responses at the polyclonal Ab level. As described more than two decades ago, the most immunodominant element on Env is the third variable region, V3, of gp120. Over the past several years, by converging mapping tools, it has become increasingly clear that much of the tier 1 neutralizing activity detected in polyclonal antisera elicited by monomeric or trimeric Env immunogens is directed to the V3 region. This is especially true for the so-called tier 1B viruses, which tend to be less sensitive to the CD4-binding-site-directed “F105-like” or the co-receptor binding site-directed “17b-like” non-broadly neutralizing monoclonal antibodies (MAbs) that often neutralize tier 1A viruses. As previously described, tier 1 isolates are not representative of most circulating clinical isolates, which are termed tier 2 or tier 3 viruses (47). The distinction is that tier 1 viruses display a more open Env trimer conformation, often as a result of in vitro replication of the virus in the absence of antibody selective pressure prior to cloning the corresponding Env sequence. Viruses with this more open Env conformation display improved in vitro fitness, likely by more efficient receptor and/or co-receptor binding. In contrast, in vivo, selection against such viruses occurs as continuous antibody pressure forces concealment of the conserved receptor-interacting Env elements as mentioned above. In a series of immunogenicity studies, we and others demonstrated that V3 peptides can deplete most neutralizing activity against tier 1 viruses such as SF162 and MW965 present in the sera of foldon trimer-immunized small animals and NHPs (48-50)). The major variable loops V1 and V2 are also relatively immunogenic (50, 51), which is not surprising as they are hydrophilic, flexible surface elements. In fact, as far back as 2006, when we immunized guinea pigs with YU2 foldon trimers in selected adjuvants, we detected the consistent elicitation of homologous YU2 tier 2 neutralizing activity. Some of the neutralizing specificity mapped to the V1 region, which we demonstrated by peptide inhibition analysis (50). Autologous tier 2 neutralization of YU2 was also observed in our previous NHP experiment where we used a long immunization interval between the second and the third boost with the YU2 foldon trimers (41).
Over the past decade, we and others have developed additional mapping tools to discern B cell and antibody responses to Env at the cellular and serological level, which are now applied to anti-sera elicited by the more recently developed well-ordered trimers (see below). These methods preceded the higher resolution approaches to study B cell responses that are now available as described in Section 3. For example, we developed a novel B cell ELISpot assay which, when combined with a series of domain-deleted Env probes (i.e., deleted of gp41, V1V2 and then V3), can be used to enumerate Ab-secreting cells specific for distinct determinants of Env (51). We demonstrated that following a single trimer inoculation in mice, most of the initial immune response focuses on determinants in gp41, but following additional Env immunizations the response shifted towards the variable regions, in particular V3. From these data, we interpret that Env immunization initially activates gp41-reactive clones that dominate the response. However, B cells recognizing V3 increase in frequency following boosting, shifting the overall reactivity towards this hydrophilic surface-exposed V3 determinant. The early gp41-directed response may be related to reports showing that gp41 reactivity is associated with cross-reactivity to microbiota, and hence may arise from recall responses by existing memory B cells (52, 53), potentially explaining their early appearance after exposure to the post-fusion gp41 conformation contained in the Env immunogen used in the human clinical trial.
To study sub-specificities contained within polyclonal samples, we asked, nearly 10 years ago, whether we could map Ab specificities in sera from HIV-infected individuals displaying broadly neutralizing activity. This work established the basis for approaches to map vaccine-elicited antibodies, a more challenging task due to lower neutralization titers elicited by protein subunit immunization. The first method we developed to map neutralizing Ab responses to the CD4bs was solid-phase differential adsorption. In this assay, we used both unmodified gp120 and gp120 possessing a mutation in the CD4bs central “CD4 loop” known to abrogate gp120 interaction with CD4, namely D368R. This mutation and the proximal E370A suffice to eliminate gp120 to CD4 binding, as can other mutations in this region including D474A. By coupling wt gp120 and the gp120 D368R variant to solid-phase Dynal paramagnetic beads, we demonstrated that two broadly neutralizing sera contained CD4bs-directed neutralizing activity to multiple tier 2 viruses (54). To strengthen our mapping process, we coupled differential gp120 absorption with V1/V2/V3 peptide “dump in” assays to increase serologic mapping capacity. For the V region peptides, these reagents can be added directly to the standardized TZM-bl neutralization assay (47, 55) to diminish or deplete virus neutralization mediated by hyperimmune serum or control MAbs (54, 56).
Since then, we developed modified gp120 cores, i.e. gp120 deleted of V1, V2 and V3 and the N- and C-termini, with and without mutations in the CD4bs to map neutralizing antibody responses directed toward this region by attachment to a solid phase. In addition, we generated the TriMut cores, which contain 3 substitutions in the bridging sheet that eliminate CD4 binding but maintain recognition by most CD4bs-directed mAbs, whether they are broadly neutralizing or not (39). An isogenic TriMut contains mutations (D368R/E370A) (57), and more recently, residue D474A (58) in the CD4bs that eliminate binding by most CD4bs-directed antibodies. The advantage of the TriMut proteins for mapping neutralizing activity is that they can be pre-incubated with mAbs or antisera for differential adsorption. Following this step, the mixtures can be added directly into the TZM-bl neutralization assay without interfering with viral entry because they no longer bind CD4 due to the substitutions in the bridging sheet. If the TriMut depletes neutralization, but the isogenic CD4bs-modified mutant TriMut protein does not, this outcome indicates that neutralization is directed to the CD4bs. Using these and other mapping tools, it is possible to determine the major neutralizing specificities present in Env-elicited hyperimmune antisera.
In addition, all but the CD4bs-directed antibodies present in polyclonal sera can be adsorbed out using the D368R Env mutant in excess, allowing CD4bs-directed antibodies to flow through a solid-phase column conjugated with this protein variant. The antibodies present in the flow through can then be bound to gp120 for purification of the CD4bs-specific antibody fraction. Of late, we and others use the CD4bs-modified Envs to analyze neutralizing serum antibody activity specific for a given autologous tier 2 virus (49, 59). The data indicate that most of the vaccine-elicited autologous tier 2 neutralization in these experiments is not trimer-specific, and may not be CD4bs-directed, as both the D368R monomer and trimer adsorb out autologous neutralization for JRFL, 16055 and BG505(49). However, the polyclonal mapping data are only indicative until more definitive results are obtained by isolating monoclonal Abs (MAbs) that recapitulate the serum neutralizing Ab specificity, as described below. Iterations on the approach of using modified Env subunits for polyclonal serum mapping were recently described using engineered gp120 outer domains (eOD), which represent approximately half the mass of core gp120. These subunits were modified with the D368R CD4bs modification, along with other point mutations for mapping purposes. In addition, the point mutants were combined with chimeric eODs generated by genetic swapping of elements derived from different strains to map tier 2 autologous neutralizing activity to specific OD subdomains such as V4 or V5 (60). Going forward, these tools will be valuable to map neutralizing Ab activity to specific elements of Env in ongoing and future pre-clinical and clinical HIV-1 vaccine studies.
Additionally, point-mutated viruses provide additional tools to map Env-directed neutralizing activity. For example, we originally designed an N-glycan deletion at residue 301, located at the base of the V3 loop. Elimination of this N-glycan renders some viruses more sensitive to non-broadly neutralizing Abs (61, 62) and can be used for mapping antisera or mAbs. In another study from the Binley group, a glycan knock-in in the JRFL Env, which normally lacks an N-linked glycan at residue N197, renders the virus resistant to serum neutralization from high-titer rabbit possessing JRFL tier 2 autologous neutralizing activity. Conversely, other viruses that are resistant to neutralization by this anti-serum, and contain an N-glycan at residue 197, become sensitive to this serum if they are genetically deleted of their respective N-glycans at residue N197 (63). These data suggest that gaps in the N-glycan shield might contribute to tier 2 autologous neutralizing activity. In a recent study, McCoy and colleagues isolated tier 2 BG505 autologous neutralizing mAbs, elicited by BG505 SOSIP trimers, which are directed to a “hole” in the N-glycan shield at residue 241. The MAb specificity was confirmed by assessing neutralizing capacity of viruses with or without N-glycan or non-BG505 residues at this site. These results are consistent with the model that some autologous neutralizing Ab activity is directed at such N-glycan gaps in the shield (64). However, since most viruses possess an N-glycan at this position, it is not yet clear if this type of tier 2 autologous neutralizing specificity can evolve into greater breadth of neutralization.
Section 3 - Env immunogenicity assessed by isolation of monoclonal Abs
More detailed and definitive information about the quality of vaccine-induced B cell responses is obtainable by analyzing antibody responses at the single B cell and MAb level. During the past years, we have established methods to isolate MAbs from non-human primates (NHPs), providing a higher resolution view of B cell responses induced by Env immunization (38, 42, 62). An overview of the B cell and Ab analysis platforms we employ to characterize vaccine-induced responses is shown in FIG 2. So far, we have focused this work on rhesus macaques, which are used to model human immunology in the evaluation of infection- and vaccine-induced immune responses. The high level of genetic homology between humans and macaques makes iterative testing and analyses of vaccine candidates in macaques highly relevant for the development of human vaccines. In addition to macaque genetic homology to humans, which is about 93% at the nucleotide level (65), work by us and others demonstrates that there is considerable similarity between macaque and human immune cell subsets (66-70), (71, 72). This greatly facilitates the translation of protocols developed for human studies to the NHP model as many antibody-based reagents are cross-reactive between the two species. Conveniently, the NIH Nonhuman Primate Reagent Resource summarizes the properties of an increasing number of cross-reactive MAbs to facilitate work in the NHP model. In the cases where Ab cross-reactivity to macaque markers is insufficient or lacking, we have evaluated alternative markers, such as in our recent study of rhesus macaque bone marrow plasma cells (46).
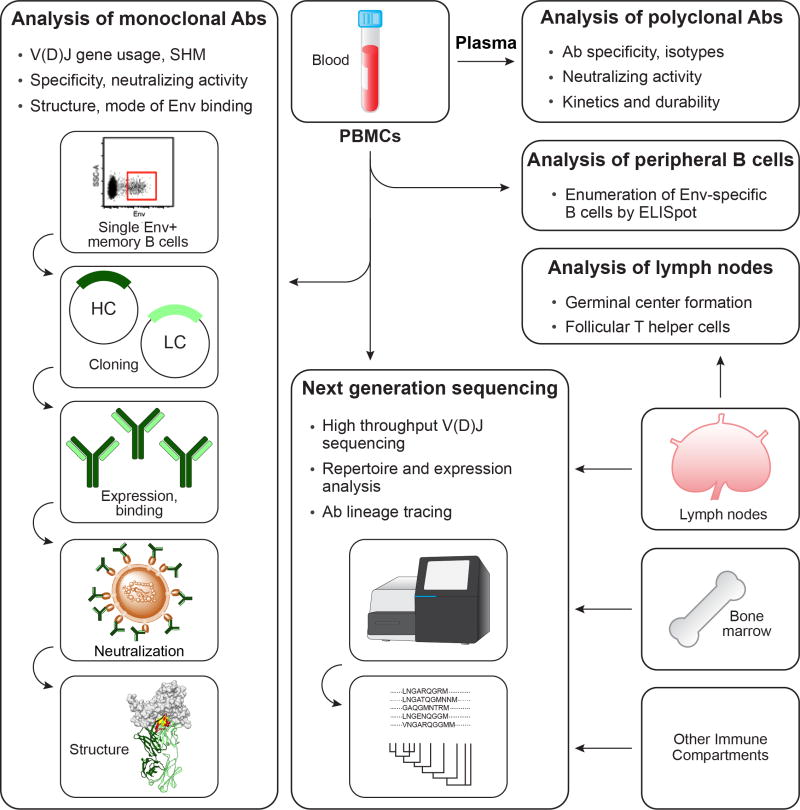
Fig 2. Overview of B cell and Ab analysis.
Peripheral blood is the primary immune compartment analyzed for Env-specific B cell and Ab responses following immunization. In a typical sampling, the blood is separated into plasma and PBMCs and both components are frozen into several aliquots. From the plasma, the titer of circulating Env-binding Abs is determined by ELISA while neutralizing Ab activity is determined by a standardized HIV-1 neutralization assays, usually the TZM-bl assay. In addition to Env-binding Ab titers, the duration of the response and the magnitude of recall responses following boosting are assessed. In addition, Ab isotype and avidity may be measured. From the PBMCs, Env-specific memory B cell and plasma cell responses are measured by B cell ELISpot analysis (not shown) and antigen-specific B cells are isolated by FACS-based sorting of single cells for subsequent cloning of MAbs. Plasmids encoding matching Ab heavy and light chains are transfected into 293F cells and expressed Abs are purified and evaluated for Env binding by ELISA and for their capacity to neutralize diverse strains of HIV-1. In the cases, interesting Ab specificities are identified such MAbs are analyzed for their fine epitope specificities and structural properties. Draining lymph nodes may be analyzed for germinal center and Tfh responses to provide additional qualitative information about the vaccine-induced response. Finally, B cells from any immune compartment from which cells can be isolated, exemplified here by blood, bone marrow and draining lymph nodes, may be analyzed in a comprehensive manner by NGS of Ab V(D)J transcripts. This may be done from V(D)J libraries generated from sorted Env-specific B cells for repertoire and expression analysis, or from bulk cells for tracing of specific Ab lineages identified through prior MAb cloning.
A number of years ago, we developed strategies to clone antigen-specific MAbs from immunized rhesus macaques, adapted from B cell analysis of HIV-infected humans (73). To achieve this objective, we developed antibody staining panels for flow cytometry-based cell sorting of macaque B cells and optimized primers for efficient isolation of macaque Ab transcripts from single cells (68). This work resulted in the publication of a series of papers describing the properties of vaccine-induced MAbs directed against different epitope regions of Env (37, 40, 42, 62). In our experimental approach, we first assess the Env specificity of the cloned MAbs by ELISA and, depending on the availability of probes, we perform initial mapping of their sub-specificities. For example, the capacity of a given MAb to bind both trimeric (gp140) and monomeric Env (gp120) indicates that the specificity is not trimer-dependent, nor directed against gp41. Binding to additional probes, such as those lacking various structural determinants or modified in specific regions by alanine (Ala) point mutations or by N-glycan knock in mutations, provides additional information about the MAb sub-specificities. If the MAbs possess neutralizing capacity, the specificity can be further mapped by Ala scans on pseudo-virus Env (40, 62). In this instance, Ala scanning mutations in the context of virus-encoded Env and measurement of neutralizing Ab activity is a much more sensitive assay to evaluate the impact of specific point mutations than are the same mutations introduced in soluble gp120 and evaluated by ELISA binding. The isolation of MAbs is now a standard analytical tool for our immunization experiments performed in NHPs and this approach is generally gaining momentum in the field, accelerating our understanding of Env vaccine-induced B cell responses in primates and other species.
The availability of MAbs provides opportunities to examine the genetic composition of individual antibodies and to investigate functional and structural properties of Env-specific Abs. In brief, cloning of MAbs involves the isolation of recombined, variable (V), diversity (D), and joining (J) gene segments from matching Ab heavy (V(D)J) and light (VJ) chains and cloning the sequences into expression vectors using procedures initially developed for the isolation of human MAbs (74). The D gene and the V-D-J junction of the heavy chain encode the complementary determining region 3 (HCDR3), while the V-J junction of the light chain corresponds to the LCDR3. The CDR3s, together with the V-gene encoded CDR1 and 2 regions of both the heavy and light chains, usually comprise most Ab contacts with antigen. V(D)J sequences are subsequently joined in frame to a sequence encoding the heavy chain constant (C) region in the cloning vector (usually IgG1) to encode full-length IgG upon co-transfection with the matching light chain. Following co-transfection of the heavy and light chain expression plasmids, recombinant MAbs are produced in 293F cells resulting in large quantities of MAb that can be purified by protein G affinity chromatography as described (44, 62).
In our standard approach, we sort antigen-specific, single memory B cells from peripheral blood 14 days after immunization using fluorescein-conjugated Env probes, which allow us to derive Env-specific Abs from more than 90% of the sorted cells (42, 45, 75). An alternative approach is to isolate single B cells in the absence of antigen-specific sorting from peak plasmablast responses which, in primates, occur approximately 7 days after boosting (76). However, the latter approach usually results in lower frequency of specific MAbs obtained after cloning, perhaps due to bystander B cell activation during boosting. From our own studies using antigen-specific sorting of memory B cells, we demonstrate that Env immunization elicits a highly polyclonal Ab response with a highly diverse repertoire of rearrangements in the pool of antibodies targeting Env. In one study, we sequenced heavy chain V(D)J transcripts from over 600 individual Env-specific B cells and found that 502 unique clonotypes from 606 functional sequences. This indicates that Env immunization elicits a high diversity of genetically unrelated Ab specificities (75). Each of these clonotypes may differ in the fine specificity of Env binding, either by recognizing different regions of Env or by recognizing the same epitope region in different ways. These results demonstrate that the primate immune system responds with a broad B cell repertoire to protein immunization, employing its full potential to generate antibodies that bind the vaccine antigen using as many V(D)J configurations as possible.
In the same study, we show that heavy chain V gene segments of all families, except VH6 (a small gene family), were found in the Env-specific populations, although at variable frequencies. Quite likely, Abs using VH6 gene segments would be identified if a greater number of cells and/or more individuals were analyzed, in combination with higher-throughput approaches to clone MAbs. Interestingly, the relative VH gene usage of the Env-specific IgG+ memory B cells is very similar to the distribution observed in the total IgG+ memory B cell repertoire from the same NHP, which represents all B cell responses to foreign antigens that this animal has generated and archived during its lifetime up to the time of analysis (75). The only difference we observed when comparing V gene usage in the total IgG+ memory B cell repertoire with that in the Env-specific IgG+ memory B cell repertoire was an overrepresentation of the VH5.46 segment in the Env-specific group. By cloning the heavy chains from a number of these VH5.46-using Env-specific B cells, along with their corresponding light chains, we produced MAbs and analyzed their epitope specificity. Many of these MAbs recognized the V3 region of gp120, suggesting a preferential use of this V segment for V3-specific responses. However, other VH genes were also used to generate V3-directed MAbs, suggesting a great diversity of clonotypes even within the pool of V3-specific MAbs (42). When analyzing other subsets of Env-specific MAbs, including those against gp41, the CD4bs or other regions of the trimer, similar results emerge. Each of these subsets consists of many different clonotypes and only rarely do we isolate MAbs that are somatic variants of each other. This suggests that each clonal Ab family is quite small (42), even though some expansions are observed at late compared to earlier time points (45). Highly polyclonal B cell responses are also detected in humans vaccinated with tetanus toxoid, another protein subunit-based vaccine (77, 78). One might conjecture that this may be typical of vaccination regimens using multiple boosts with invariant subunit antigens. Such vaccination results in efficient activation of many B cells that, after some affinity maturation, reach an affinity ceiling that is no longer driven by selective competition for antigen (79).
The degree to which different immunization protocols promote Ab SHM is of central interest to the HIV-1 vaccine field. As mentioned earlier, many bNAbs isolated from chronically infected individuals are highly modified by SHM and that this level of affinity maturation is required for their neutralizing activity (5, 80). One report describing the cloning of approximately 500 MAbs from chronic HIV-1 infection demonstrates that high levels of SHM are not only observed in bNAbs, but also in non-neutralizing Abs, suggesting that it is a consequence of the chronic infection (73). This is especially true in HIV-1 infection where the selecting antigen is highly variable, and new viral variants present modified epitopes, lowering affinity at these sites, until new Ab variants with increased affinity are again selected. This arms race between the virus and the host B cell response is well documented and is a hallmark of HIV-1 infection (25, 81, 82). High levels of SHM are also seen in other chronic infections and some settings of autoimmunity (83), consistent with ongoing antigen exposure and selection of the highest affinity B cells. This is consistent with the oligoclonal nature of the Ab response in chronic HIV-infection where a repertoire size of approximately 50 clonotypes, with a bias towards the use of the heavy chain variable (VH) 1 family gene segment is observed (73).
Investigators do not generally observe a similar level of SHM for vaccine-induced B cell responses, at least not when presenting invariant and non-replicating antigens to the immune system (78, 84). In our studies we find that most of the cloned MAbs display relatively low SHM levels, around 5%, based on assignment to germline V genes available in current public databases (42, 62), a topic that will be addressed in more detail below. This level of SHM is similar to other studies of vaccinations (78). In our studies, we demonstrate that several vaccine-induced CD4bs-directed MAbs isolated from memory B cells after only two trimer immunizations, bind Env with relatively high affinities, in the nanomolar range, even in their germline-reverted state. Gene assignment indicates that quite low levels of SHM are sufficient to convert a non-neutralizing germline-reverted Ab to a neutralizing Ab (44). The level of SHM required for positive selection of a given B cell in the GC reaction will differ depending on the starting affinity of the BCR to its cognate antigen. Overall, our studies of individual B cells from vaccinated NHPs demonstrate that the Env-specific B cell response induced by immunization is distinctly different from that observed in chronically HIV-infected individuals, both in terms of SHM and extent of clonal expansions. Additional and higher through-put studies are required to obtain an even more detailed understanding of the diversity of Env-directed B cell responses induced by immunization to contribute to the circulating Ab or B cell memory pool.
In addition to the genetic analysis, the availability of cloned recombinant MAbs allow studies of Ab function and structure. Our studies of vaccine-induced B cell responses provide the first definitive information that vaccine-induced Abs that target the highly conserved primary receptor binding site of HIV-1 gp120, the CD4bs, bind with an angle of approach that differs from that of bNAbs isolated from chronically infected individuals, such as VRC01 or PGV04 (37, 40). Specifically, we found that in this study, the vaccine-elicited MAbs attempts to access the CD4bs from a top angle approach, unlike the side approach used by VRC01. We deduced the angle of approach by high resolution crystal structure of the MAb Fab, modeling and docking of negative stain EM of Fab:gp120 complexes into the known BG505 SOSIP high-resolution structures. The angle of approach by the vaccine-induced CD4bs-directed MAbs likely explains their limited neutralization efficacy on primary viruses as the variable regions at the apex of the functional spike present a physical barrier that obstructs Ab access to the CD4bs. Our results also demonstrate that unlike the VRC01-class of Abs, the vaccine-induced CD4bs-specific MAbs depend on their HCDR3s for binding their epitopes. Thus, in this regard, they are more similar to the infection-induced broadly neutralizing MAbs CH103 or VRC13 that depend on their HCDR3 region for cognate antigen binding (12, 15). This information has motivated us to develop and design more well-ordered mimics of the functional spike, to restrict access from the top of the spike and to use other strategies to target the CD4bs as described in Section 6 below.
The availability of MAbs also allows assessment of their neutralizing capacities using panels of pseudotyped viruses in the TZM-bl assay. In one recent study describing the properties of over 50 MAbs induced by immunization with the YU2 foldon trimers, we demonstrate that only the V3 and CD4bs-directed MAbs exhibited neutralizing activity (42). Collectively, MAbs of these two specificities fully recapitulated the neutralizing activity observed in the unfractionated plasma samples, suggesting that they are the two major specificities. Interestingly, for some viruses, the unfractionated plasma does not display measurable neutralizing activity, while some of the MAbs isolated from the same vaccine recipient do. This demonstrates the value of MAb isolation where higher concentrations of individual Abs can be used to reveal novel activities. An additional value of studying HIV-1 neutralization by isolation of MAbs is that this analysis directly reveals which viruses are sensitive to which Ab specificities. For example, tier 1 viruses, which are often created as they evolve rapidly in vitro when antibody-mediated selection pressure is removed from the replicative environment. This permits the viral Env to expose itheir V3 and CD4bs regions to become sensitive to Abs that recognize this “unshielded” or more open conformation of the trimeric HIV spike. In contrast, the globally circulating tier 2 viruses are much more resistant, displaying little exposure of the V3 region. The CD4bs is somewhat exposed on tier 2 viruses; however, access by most Abs is restricted by conformational constraints and flanking glycosylation, allowing limited angles of approach as discussed above. Thus, by isolating vaccine-induced MAbs, we have learned a considerable amount about the limits and limitations of inducing neutralizing antibodies by active vaccination. We anticipate that in the near future, this type of analysis will likely generate even more informative results that point the direction toward an effective HIV vaccine.
Section 4 – Use of next generation sequencing in studies of vaccine-induced Ab responses
Recent years have witnessed a revolution in the use of Next Generation Sequencing (NGS) technologies, opening new doors for genomics and expression profiling. NGS is also increasingly used to study immune receptor repertoires, providing novel and highly detailed information about expressed Ab repertoires and the development of antigen-specific responses (85). After an initial few years focused on 454 pyrosequencing, subsequent long read platforms such as Illumina's MiSeq system have emerged as the main technologies currently applied to the deep sequencing of V(D)J transcripts generated from IgM or IgG libraries. Use of the Illumina MiSeq 2 × 300 bp kits provides sufficient read lengths to cover the entire V(D)J segment of rearranged antibody sequences, allowing V gene usage encoded by bulk B cells or by selected B cell populations to be determined from several hundred thousand to millions of sequences. One application of considerable interest in the vaccine field is to trace specific Ab lineages in NGS datasets to study the evolution of antigen-specific Ab responses to a given immunogen. This requires prior isolation and characterization of MAbs to ensure that functional, antigen-specific lineages are studied. Because each Ab possesses a unique HCDR3, this region acts as a molecular tag for the heavy chain of an Ab, permitting bioinformatic identification of the lineage from large NGS data sets. Studies of clonal families in defined B cell populations, such as peripheral memory B cells and bone marrow-resident plasma cells, provide information about clonal expansions, levels of SHM, persistence over time and the biological fate of each studied lineage (86), (87). Optimized protocols for Ab library preparation and well-validated computational pipelines are required for such analysis and have been developed in several laboratories including ours.
NGS analysis of Ab sequences following immunization of animal has highlighted a number of specific technological hurdles that needed to be overcome to enable full use of this approach. Sequence analysis of Abs requires the accurate determination of both the allelic usage and the degree of SHM involved. This entails three main requirements: the ability to encompass the entire V(D)J sequence and part of the constant region; the ability to sequence to an accuracy that enables SHM to be distinguished from technical sequence errors; and the ability to accurately assign an individual sequence to the appropriate germline reference allele. As mentioned previously, the development of long read technologies such as Illumina's MiSeq 2 × 300 bp system and the PacBio system have largely overcome the issue of sequence distance, the two remaining issues are still a subject of intense research. Technical sequence error is an implicit factor in current NGS systems. Even the most error free systems, such as the Illumina MiSeq, have error frequencies of approximately 1%, and other systems have even higher errors (88). The current most popular method to deal with this issue is the use of unique molecular identifiers (UMI) – which are random or semi-random strings of sequences that are unique to each cDNA molecule. These UMIs can be attached at the 5′ end of cDNA generated during RACE cDNA synthesis (89), or at the 3′ end of the cDNA during gene-specific cDNA synthesis (90). Subsequent analysis of the NGS data using these UMI's enables the identification of multiple copies of antibody sequences that derive from the same initial cDNA molecule. This enables the generation of consensus sequences in which the random technical errors are removed (FIG 3, left panel).
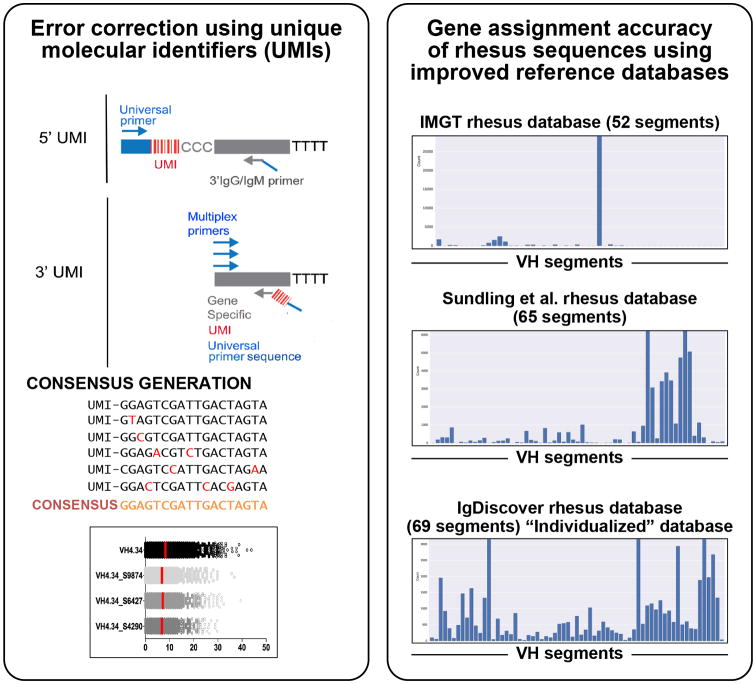
Fig 3. Next generation sequencing of Ab repertoires.
Left box: Unique molecular identifiers (UMIs), either 5′ or 3′, are attached during the cDNA synthesis step using RACE or multiplex PCR amplification, respectively. Ab sequences derived from the same starting cDNA molecule are identified using the UMI sequence, enabling the generation of a consensus sequence that has random technical sequence errors removed. The lower panel demonstrates the combination of UMI consensus generation and identification of novel alleles expressed in an individual. Ab sequences originally assigned to VH4.34 are now assigned to three related novel germline sequences. Consensus generation and correct assignment results in a lower average SHM frequency. Right box, top panel: The advantage of comprehensive germline databases is shown in these three assignments from a single NGS sequence library. The upper panel shows the result when using a restricted and incomplete rhesus database, such as the current (August 2016, 52 alleles) IMGT rhesus VH database. Middle panel: The Sundling et al. 2012 database (65 VH sequences) shows improved levels of assignment. Lower panel: An individualized database containing all the expressed VH germline gene segments identified from that specific animal using IgDiscover, shows a much-improved degree of coverage.
The final major issue; that of the accurate assignment to the germline reference allele; is of great concern to the NGS Ab sequencing community. Correct assignment of NGS generated Ab sequences requires the availability of comprehensive and accurate reference databases. With the arguable exceptions of those for humans and mouse, reference databases of sufficient accuracy and completeness are currently unavailable for most species used in immunological research. The reason underlying the lack of such databases is partly contingent on how species-specific genomic reference sequences have been commonly created. Assembling short read NGS sequences into long interlinked genomic “contigs”, the major methodology used in current reference genomes, is often inadequate for genomic regions containing high degrees of segmental duplication or highly similar gene segments. Complete sequencing of the Ig loci requires both longer reads and higher levels of coverage than is necessary for most other genomic segments. In addition, allelic genetic diversity at the Ig loci means that a single reference genome, even if complete for that individual, may only provide the resources to map a fraction of Ig allelic diversity in that species (91). For example, our own work has revealed an unexpected heterogeneity of germline V genes between rhesus macaques and the presence of more than two different alleles in a given animal (40, 44). In addition, genomic mapping of Ig gene segments suffers from issues relating to functionality of candidate sequences. Pseudogenic variable sequences are common throughout the Ig loci and are frequently difficult to distinguish from genuine functional alleles at the genomic level (92). Several groups have begun to address this issue by making use of expressed sequences rather than genomic sequences (93). Functionality of alleles can be tested by the identification of identical sequences expressed within independently rearranged antibody sequences (94). Importantly new techniques have been applied towards the use of expressed Ab sequences in identification of novel germline alleles that are missing from current reference databases (95) and in the use of NGS Ab libraries in the generation of entire species-specific databases and personalized/individualized repertoires using our recently developed IgDiscover.
We developed the IgDiscover tool out of necessity for improving the available rhesus macaque germline database, resulting in the development of a broadly applicable germline allele identification tool that can be used for any species. The IgDiscover program enables the de novo construction of species-specific V gene databases, and is currently being utilized in our laboratory to produce NGS suitable reference databases for several important immunological model organisms. The approach is based on IgM antibody libraries, which contain mixtures of germline V sequences and those subjected to SHM, with both groups exhibiting additional low rate sequence variation introduced by PCR or Illumina-based sequencing errors. IgDiscover works through the identification of clusters of expressed sequences within the NGS data, and applies a series of algorithms to identify those sequences that are germline-specific while simultaneously excluding false positives. The resulting output for each library is an individualized germline repertoire of that subject, enabling the correct assignment of Ab sequences and, in conjunction with UMI-based error correction procedures, a highly accurate estimation of SHM. The production of individualized repertoires has revealed high degrees of genetic diversity at the Ig loci in macaques, suggesting that a comprehensive macaque reference database, and probably other species, will require V gene sequences from many individuals rather than the current prevailing technique of using sequences from just a single or small number of individuals. An example of how the use of three different VH gene databases impacts gene assignment of NGS data obtained from one rhesus macaque illustrates the limitations of the current IMGT database (52 VH segments from the reference Indian origin rhesus macaque (65), or even the VH database published in Sundling et al. 2012 (62) (65 VH segments from the reference Indian origin rhesus macaque), compared to when assignment was performed using an individualized database created with IgDiscover from the specific rhesus macaque from which the NGS data was obtained (69 VH segments) (FIG 3, right panel).
Since the human population is also genetically diverse, it is important to address this diversity at the immuno-genetic level. Currently, use of IgDiscover is creating individualized immunological databases from cohorts of individuals from around the world, including those of European, African and Asian ethnicities, with an aim to extend this into a large-scale screen involving thousands of individuals. The resulting database will be a unique resource to identify previously unidentified associations between immunological profiles, disease susceptibility and/or immunization efficiency. The field of immune receptor sequencing is evolving extremely rapidly and, if used with the appropriate controls and correction filters, will offer opportunities to address basic immunological questions and obtain statistically robust results in a manner not previously possible.
A further application of NGS is Ab lineage tracing. In order to identify the series of specific mutations that enables affinity-matured Abs to bind to their target epitopes at high affinities we need to be able to determine which sequence alterations are specific to a given Ab lineage. In NGS Ab studies this has been performed by creating phylogenetic trees that show the evolution of related sequences. Due to the previously mentioned issues of technical sequence error and incorrect germline assignment the ability to trace Ab lineage development was previously limited. However, with the application of UMI-based error correction and the use of individualized V gene databases our ability to trace specific lineages can be expected to be significantly improved.
Section 5 – Considerations of epitope-specific vaccine responses
One concern in the field is that the elicitation of antibodies to immunodominant regions of Env, such as V3, may distract responses away from bNAb epitopes, and that V3-directed antibodies might decrease effective boosting by sequestration of antigen. Emanating from this, is the thought that by reducing V3 exposure and thereby B cell recognition of this structure, Ab responses might preferentially shift to conserved neutralizing determinants. To explore the influences of V3 immunodominance, we performed an experiment where we genetically engineered three unnatural potential N-linked glycan sites (PNGS) to the V3 region of the foldon trimers and immunized the recombinant trimers into mice. Compared to wt trimer-immunized mice, we see a complete elimination of anti-V3 antibody responses but no changes in the magnitude of the antibody responses to other regions of Env and no detectable increases in tier 1 or tier 2 neutralizing antibodies. Similarly, we performed full genetic deletion of the major V1V2 and V3 regions of gp120 to generate gp120 cores. The rationale behind the core design was to both eliminate the immunodominant elements and to direct potentially B cell responses to the conserved, and now better exposed, CD4 binding region displayed on the engineered gp120 core. However, simply deleting the variable regions and the potentially immune-distracting N- and C-termini, did not result in the elicitation of neutralizing antibodies. As we now know, in part this is because the monomeric core, and its high-resolution structure, does not fully recapitulate the complete epitope for most CD4bs-directed bNAbs. The more recent high-resolution trimer structures reveal that some CD4bs-directed bNAbs possess cross-protomer contacts as part of their complete epitope (96, 97). Another potential issue with this core immunogen is potential competition for the CD4bs by the more common non-neutralizing BCRs that can access this site in the less-restricted context of the core. Such competition is associated with non-desired germline engagement of the more common non-neutralizing BCRs, perhaps to the detriment of the less frequent bNAb precursors. Besides suboptimal activation by the core, and its non-oligomeric state, the extensive SHM and indels often needed for the cross-neutralization displayed by bNAbs may be unlikely to be driven by repetitive core immunization alone.
Several recent studies suggest that elicitation of non-broadly neutralizing V3 responses, although not desirable in terms of neutralizing clinical isolates, may not interfere with the elicitation of tier 2 autologous neutralizing antibodies. For example, in one study performed in guinea pigs, the investigators tested the immunogenicity of a complex consisting of the trimer apex-specific bNAb, PGT145, bound to Env in complex, compared to Env alone (60). These investigators demonstrate that the elicitation of V3-directed antibodies is reduced for the PGT145-Env complex resulting in a corresponding loss of tier 1 neutralizing activity, but there is no increase in autologous tier 2 neutralization. In rabbits, a separate study performed by different investigators where they engineered Env by site-directed substitutions to prevent exposure of the V3 determinant yields similar results (98). These data are consistent with the notion that there is no substantial competition between V3-specific and other, more desirable anti-Env specificities capable of neutralizing tier 2 viruses during polyclonal B cell activation and affinity maturation elicited by vaccination.
In addition to V1V2V3 deletion, we stabilized the cores by structure-based design using a combination of internal cysteine pairs and pocket filling mutations in collaboration with the Kwong group. The stabilized cores, when increasingly locked into the CD4-bound conformation, become better recognized in vitro by the co-receptor binding-site-directed (CD4-induced (CD4i)) MAb, 17b. In vivo, in outbred rabbits, cores progressively stabilized in the CD4-bound conformation dramatically enhance the elicitation of co-receptor binding site-directed antibodies compared to strain-matched, non-stabilized core protein (99). These Abs are detected by their ability to cross-neutralize CD4-sensitized HIV-2, because the co-receptor binding site is the only cross-conserved neutralization determinant between HIV-1 and HIV-2 (100). This is a clear proof-of-principle that we can impact the Env-specific immune response by altering the biophysical properties of this conformationally flexible immunogen, an important principle also used in RSV F pre-fusion conformation redesign that enhanced stabilization and immunogenicity (101). Co-receptor antibodies require either the described core structural stabilization or the presence of primate CD4 in the immunologic host. In fact, in NHPs, where the affinity between cell-surface CD4 and Env is high, co-receptor binding site-specific antibodies are elicited (102), suggesting that, in vivo, some fraction of Env binds CD4-expressing cells. To investigate this possibility, we evaluated side-by-side otherwise isogenic trimers that could or could not engage primate CD4. We used trimers possessing mutations in the gp120 bridging sheet that no longer bind CD4 in a high-affinity manner versus unmodified Env trimers that do efficiently bind CD4 (39). As predicted, the mutant trimers are unable to induce responses against the CD4i site, while neutralizing Ab responses to other regions of Env, including the CD4bs, are unaffected. In another experimental setting, performed in rabbits transgenic for human CD4 compared to wt controls, we demonstrate that CD4i antibodies are induced only in the human CD4-expressing transgenic rabbits (103). In addition, at the serological level of analysis, we did not detect a loss of HIV tier 1 neutralizing capacity in the animals that generated the CD4i antibodies, consistent with a lack of cross-competition for divergent specificities on this polyvalent antigen. From these latter studies, we conclude that the efficient elicitation of the co-receptor binding site antibodies requires the presence of primate CD4 in vivo, but that the elicitation of antibodies to this non-neutralizing HIV region does not diminish neutralizing responses to other elements of the HIV Env.
In our more recent experiments, layered upon structure-based gp120 core stabilization by internal cysteines and pocket-filling substitutions, we performed glycan masking of structurally stabilized cores to dampen responses to non-neutralizing determinants. In principle, this should preferentially present the molecular surface of the conserved CD4bs to the immune system. However, even by this elegant, structure-based redesign of the CD4-confirmationally stabilized core, we do not detect any gain in the elicitation of neutralizing antibodies to the CD4bs (6). In fact, in this study and in previous work (48), we have explored the concept of priming with the conformationally stabilized core, derived from the HXBc2 strain, and then boosting with YU2 strain-derived trimers. The experimental rationale was to potentially focus the immune response on cross-conserved determinants between the two forms of Env and reduce priming of responses to the variable regions present only on the intact trimer. While this did not translate into measurably improved neutralizing Ab responses in those studies, similar experiments in the context of the new generation well-ordered trimers are warranted, especially when applied to higher resolution studies of individual B cells activated in the induced immune response.
Besides epitope-specific stabilization or glycan-masking, another vaccine approach is to target portions of gp41 better exposed following receptor engagement, such as the gp41 membrane proximal external region (MPER) or other occluded regions of gp41. Following our crystal structure of the mAb, 2F5, in complex with its MPER peptide epitope, we attempted to scaffold the 2F5-bound epitope on non-HIV acceptor protein scaffolds (104). Once we had generated a 2F5 well-recognized series, we tested three such 2F5 epitope scaffolds in prime:boost immunogenicity experiments in small animals. This approach does elicit anti-MPER peptides antibodies in two separate published studies. In the first, we isolated a monoclonal antibody from one of the immunized mice that recognized the MPER peptide in a manner similar to that of 2F5, indicating that scaffolding works in principle but does not induce HIV neutralizing responses (105). In the second study, we demonstrated that the elicitation of MPER-directed antibodies in the serum is more efficient if cross-conserved T cell help is included in each immunogen in this multi-step prime:boost process. However, again, the elicited MPER-specific anti-sera does not neutralize HIV, even tier 1 viruses (104). Other investigators, targeting the same or other regions of gp41 by HR1 and HR2 peptide assemblies or expressed gp41 trimers containing the MPER region have demonstrated some success, reporting the elicitation of tier 1 neutralizing antibodies to not-yet-defined determinants within gp41 (106, 107).
Section 6 - Next generation well-ordered Env trimer immunogens
The advent of the so-called well-ordered, native-like trimers began with the introduction of unnatural cysteine residues in each of the gp120 and gp41 subunits to covalently link gp120 to gp41 that, following cleavage, allow a more native and stable configuration (SOS = 501C to 604C disulfide bond). That this linkage was accomplished, using some insight and relatively limited screening was, in retrospect, a brilliant although somewhat fortuitous discovery of this important SOS linkage by James Binley (108). However, although this cysteine pair, described in 2002, did covalently link gp120 to gp41, these substitutions alone generated few soluble, stable trimers. As we now understand, this is because of the spring-loaded nature of gp41; once the transmembrane region anchor points are released by genetic truncation to form soluble Env, the “spring is sprung”. To solve this issue, reduction in the tendency of Env to “flip” to the end-stage 6-helix bundle conformation of gp41 was needed. This was initially accomplished by the I559P substitution in gp41, in the SOS context, to form SOSIPs conceived by Rogier Sanders (109). This key substitution, along with additional modifications, resulted in soluble, cleaved, well-ordered clade A trimers (33). More recently, using alternative cleavage-independent designs consisting of gp120-to-gp41peptide-linked trimers, result in well-ordered clade B and C trimers ((35) and see FIG 4). The uncleaved trimers, in particular our own native flexibly linked (NFL) design (35), are the best currently available. We continue to improve the NFLs by structure-guided modifications as described below. Other investigators have reported reasonable uncleaved covalently linked facsimiles (32, 110) just prior to our more comprehensive analysis, performed in parallel (35). Because the SOSIP trimers, led by the BG505-derived designs, will be described elsewhere in this volume by Sanders and Moore as immunogens and structurally by Ward and Wilson, here, we will focus here on a description of the NFL trimer platform that we generally use in most our own immunogenicity and structural studies.
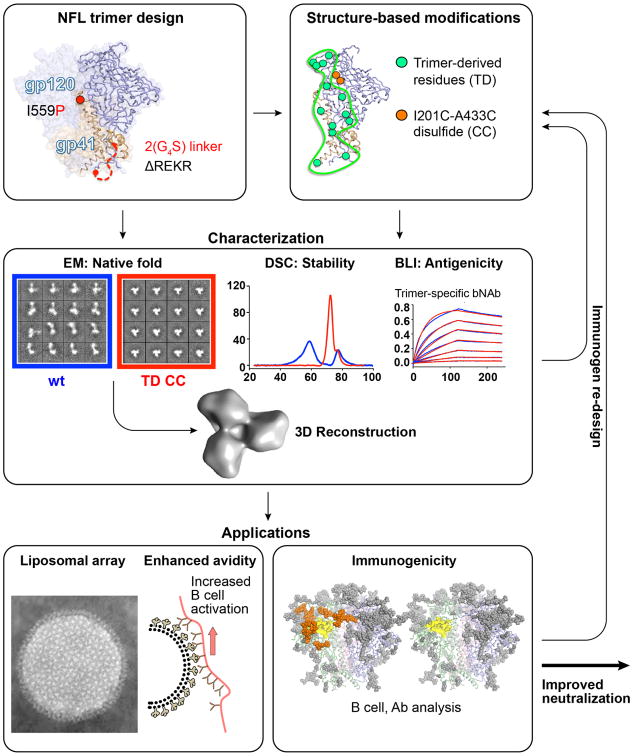
Fig 4. Immunogen design, re-design and applications process flow diagram.
Top box; the basic elements of the native flexibly linked (NFL) trimer design are shown: the G4S2 linker insertion between gp120 and gp41 (red dashed line), deletion of the natural REKR cleavage-site at the C-terminus of gp120 and the I559P substitution in gp41 (red ball). Right, the trimer-derived (TD) mutations (green balls), in this case 16055 NFL, engineered to increase trimer stability along with the internal engineered disulfide bond (CC, orange). Middle box; characterization process of new trimers is shown, left; EM negative stain 2D classifications before and after the TD substitutions and a 3D reconstruction (Andrew Ward laboratory), center; DSC thermal transitions before (blue) and after (red) the TD substitutions, right; Octet binding kinetics of PGT145 to 16055 NFL TD trimers. Lower box, left; NFL trimers arrayed on liposomes at high-density, potential enhanced avidity and increased B cell activation are depicted (117); right box, genetic deletion of CD4bs(yellow)-proximal N-glycans (orange) are shown on the BG505 SOSIP.664 structure, left versus right (deleted). Arrows between the boxes indicate flow and if a trimer fails at characterization or does not demonstrate improved immunogenicity, the re-cycling back to re-design occurs. Lower right arrow indicates if improved immunogenicity is detected (ie, increased tier 2 neutralization breadth) in a relevant animal model, then this trimer will be considered as a candidate for human testing.
The NFLs, in their first iteration, consist of gp120, deleted of its REKR furin cleavage site, a genetic insertion of a 10 residue flexible linker (G4S)2 from the C-terminus of gp120 appended to the N-terminus of gp41 (the fusion peptide; see FIG 4, top left). This effectively results in a 6 net residue insert between these two subunits to allow native-like association normally found in the natural non-covalent context. Truncation before the transmembrane region of gp41, as for SOSIP, allows soluble trimer expression. Following expression and purification, we (and others) analyze new native-like candidates, including the initial NFL design, by negative stain EM, antigenic profiling by trimer-specific and other bNAbs, as well as by differential scanning calorimetry (DSC) to confirm both stability and homogeneity (see FIG 4, middle).
One important issue is that neither the NFL nor the SOSIP designs efficiently generate high levels of native-like trimers from most Env sequences by transfection of 293T cells with DNA expression plasmids followed by transient expression, purification and analysis. This is perhaps not surprising, as the engineered mutations described above were not evolved to be tolerated in the context of natural viral Env. The subtype A Env, BG505, is the one outstanding exception to this issue as this Env forms a high percentage of native-like trimers in either the SOSIP or NFL context. For many other Envs, however, the oligomeric population is a mixture of well-ordered trimers, but also non-well-ordered trimers, aggregates, dimers and monomers. One solution to isolate well-ordered trimers from this complex mixture is to select the relatively low percentage of native-like trimers with a trimer-specific bNAb such as PGT145 or VRC26. This allows the generation of a more diverse array of ordered trimers (111), but does limit native-like spike yields. Another solution to this problem is to use a non-neutralizing antibody such the CD4bs-directed mAb, F105, to perform solid-phase negative-selection to remove disordered trimers and allow the native-like trimers to remain in the solution in supernatant derived from transiently expressing cells (34). In our system, initially focusing on subtype B JRFL and subtype C, 16055, we isolated the NFL trimers from the supernatant by lectin-affinity chromatography, followed by size-exclusion chromatography (SEC) and, when necessary, by negative selection. To overcome the low efficiency to form well-ordered trimers derived from most Envs in the NFL or SOSIP context, we modified the initial NFL design by structure-guided enhancements. Our goal was to increase greatly the propensity of the NFL (or SOSIP) initial designs to form well-ordered trimers with at a higher frequency, thereby maintaining higher yields. Accordingly, using the available BG505 SOSIP structure, we back-reverted trimer-derived (TD) residues that differed between 16055 and JRFL compared to the BG505 sequence (see FIG 4, top right). For 16055, substitution of 8 residues near the trimer axis of gp120 and gp41, permitted the generation of almost exclusively well-ordered NFL trimers in the major size-exclusion peak. These modifications generated native-like trimers approaching the levels of homogeneity displayed by BG505 NFL as determined by EM (see FIG 4, middle). For 16055, the Tm increases by 9°C as determined by DSC. We then added a cysteine-pair linkage in the bridging sheet region (CC = 201-433) that permits the trimer to resist CD4-induced conformational changes following interaction with gp120. This disulfide further increased the Tm, improving trimer homogeneneity, and trimer thermostability, while maintaining reasonable yields ((36) and FIG 4, middle).
Since we and others invest a considerable effort to stabilize HIV-1 Env trimers, one might ask, what is the immunological impact of improved Env spike mimic stability? It is currently too soon to answer to this question comprehensively since detailed evaluations of Ab responses elicited by different forms of new generation well-folded, stable trimers have been only recently initiated. However, in one recent study (49), we demonstrate that trimers with inherent higher thermostability, as determined by DSC, display more stable integrity in adjuvant at physiological temperature. We also show that glutaraldehyde cross-linking increases trimer thermostability in vitro, in buffer or adjuvant, without causing trimer aggregation. Finally, we determine that trimer thermostability correlates with the elicitation of tier 2 autologous neutralizing antibodies in vivo, an encouraging result for the immunologic outcome of structure-guided, stabilized trimers.
The availability of more stable NFL trimers allows single-variable changes, without altering other aspects of these well-ordered trimers, to better perform new immunogenicity experiments not possible previously. For example, one rationale to overcome the immune-dampening properties of the N-glycan shield, is to genetically delete targeted N-linked glycans (PNGS) shrouding known broadly neutralizing determinants to allow an array of B cells to gain a foothold against the now exposed polypeptide surface as a form of germline or “naïve repertoire” activation. Perhaps the best site to address the question whether gaps in the N-glycan shield can be exploited to elicit some degree of neutralizing breadth by immunogen modification followed by vaccination is the CD4bs. The conserved CD4bs molecular surface, a known bNAb target, although glycan-shrouded, is devoid of N-glycans to allow protein:protein interaction with the primary receptor, CD4. Therefore, this region might be the best target to test whether deletion of selected N-glycans may better expose an already glycan-free protein surface (see FIG 4, lower right) to enhance immunogenicity. Using the stabilized NFL trimer foundation, one can test whether targeted N-glycan proximal to the CD4bs might potentially give naïve B cells sufficient affinity to initiate responses that would not be activated by the fully glycosylated trimer, while maintaining full trimer integrity to not expose non-neutralizing determinants. In principle, subsequent boosting with ordered trimers possessing the restored N-glycans may then allow B cell to mature and account for the full glycan pattern, permitting shield penetration by a subset of novel CD4bs-directed B cells. In a general sense, selective N-glycan deletion of Env immunogens has been attempted previously (112), but this approach may fare better in the modern era of well-ordered trimers, as was recently reported at residue N197 (113). Targeted N-glycan deletion, in combination with better tools to assess initial B cell activation and subsequent affinity maturation processes, will aid in analyzing the initial B cell response. This information should help in guiding different trimer prime or boost strategies toward the elicitation of more effective neutralizing antibodies penetrating the N-glycan shield.
Section 7 - Particulate array of well-ordered trimers and other alternative vaccine approaches
Two highly effective licensed human anti-viral vaccines, presented to the immune system in the form of particles, are the hepatitis B vaccine (HBV) and the HPV vaccine, mentioned earlier in this review. The irony regarding both these subunit, particulate vaccines is the biology of these two viruses, which allow the production of proteinaceous particles through their natural assembly processes. Both the outer coats of the non-enveloped HBV (preS1) and HPV (L1) form particles in the absence of capsid, presumably for immune evasion during natural infection, and it is these non-replicating particle subunits that provide the basis for these human vaccines. The field has attempted to generate HIV-1 VLPs with limited success, with incorporation of functional Env spikes a common bottleneck. In fact, HIV-1 tends to dampen immune responses by limiting the levels of surface Env spikes incorporated into individual virions. Further, the incorporation of non-native trimers into VLPs, or spike degradation into non-functional and non-native states once incorporated add additional complexity in regards to HIV-based VLP generation. The Binley group has solved the latter aspect by exploiting protease-mediated degradation of non-native trimers on VLPs, while leaving the native, well-ordered trimers intact (114). As immunogens, these VLPs elicit infrequent tier 2 neutralizing antibodies (63), but are relatively laborious to generate.
Previously, we generated proteoliposomes that arrayed detergent-extracted Env oligomers isolated from transiently expressing 293T cells (115). However, these trimers, solubilized from multiple membrane compartments (ER, Golgi, trans-Golgi and the plasma membrane), are heterogeneous in conformation and a significant fraction exist in an immature, uncleaved state. More recently, the Zwick group extracted chemically stabilized Env trimers in detergent as immunogens, resulting in the elicitation of detectable but low titer neutralizing responses in rabbits (116). In light of recent developments with our well-ordered trimers, which display a desirable antigenic profile and improved homogeneity, we sought to exploit particulate array to present these relatively high-yield, native-like trimers to the immune system. Accordingly, using either well-ordered NFL or SOSIP trimers, we developed protocols for high-density conjugation of the soluble stable trimers on fully synthetic liposomes ((117) and see FIG 4, lower left). The prototype trimer-conjugated liposomes are arrayed with trimers that contain C-terminal His-tags on each protomer of the soluble Env trimer mimetic. The trimers are efficiently conjugated to liposomes containing 4% NTA-nickel lipids, allowing chelation of the nickel adducts by the His-tag tails, resulting in high-affinity, non-covalent repetitive arrays. We analyzed these high-density arrays by cryo- and negative-staining EM (FIG 4) and demonstrate that the trimers maintain a favorable antigenic profile and enhanced B cell activation in vitro and in vivo compared to the same concentration of the soluble trimers (117). In the presence of exogenous adjuvant, the trimer-conjugated liposomes elicit increased levels of tier 2 autologous neutralizing antibodies compared to the same quantity of soluble trimers in adjuvant. Optimization of trimer-conjugated liposomes with the now expanded array of well-ordered trimers from multiple Envs and clades will be a focus of future immunogenicity efforts in our laboratories. Future developments include covalent linkage of trimers containing C-terminal free cysteines to maleimide-containing liposomes, and potentially the derivatization of VLPs or other protein particles by similar chemistries. Env trimer array on other nanoparticles has been achieved by genetic fusion using ferritin or other particulate substrates (118, 119).
The classical “text book” explanation of why high-density particulate immunogens might better activate B cells is by enhancing BCR cross-linking through avidity gain, increasing signaling through BCR capping and co-clustering of the associated signaling partner, CD19. However, in the alternative model proposed by Michael Reth (120) there are distinct clusters of IgD and IgM on naive B cells. Both IgD and IgM from the same B cell possess variable regions encoded for by the same distinct V(D)J heavy and light chain rearrangements that occur in the bone marrow. The signaling molecule CD19 is associated with IgD and helps maintain a non-responsive B cell state until encounter with antigen. Antigen binding, in this case by high-density array, disrupts IgD and IgD-CD19 association, allowing translocation of CD19 with IgM and signal transduction of the signaling to activate antigen-specific B cells. Whatever the precise mechanism, as we recently demonstrate, high-density particulate array of HIV-1 Env trimers certainly enhances B cell activation (117) and should be exploited in the future for HIV-1 or other viral surface subunit proteins.
Section 8 – Initial B cell activation and establishment of durable responses
As discussed above, the difficulty to elicit Abs capable of broadly neutralizing tier 2 viruses by vaccination is a testimony to the extreme Ab neutralization resistance evolved by circulating HIV-1 strains. Studies of bNAbs isolated from chronically HIV-infected individuals demonstrate that such Abs display self-reactivity more frequently than do Abs elicited in healthy subjects (121, 122). Fortunately, auto-reactive Abs are not easily induced by vaccination as this would be detrimental to the organism. In fact, our immune systems have evolved multiple mechanisms to limit selection of self-reactive B cells, including deletion of auto-reactive clones during B cell development in the bone marrow and, during the transitional B cell stages in the periphery. Evolved as well is the deletion of auto-reactive CD4+ T cells during thymic selection and negative selection of auto-reactive B cell clones that may be generated during the germinal center reaction. During chronic infections, some of these checkpoints are disrupted (reviewed in (123)), which may explain why self- or poly-reactive Abs are more frequent in HIV-infected individuals compared to healthy subjects (121). Loss of checkpoints and/or disrupted feedback regulation may also explain the altered or abnormal B cell populations in HIV-infected persons including higher frequencies of transitional B cells, plasmablasts and exhausted memory B cells (124-127).
A few years ago, motivated by the difficulty to elicit bNAbs by Env vaccination, the tight control of auto-reactive B cells, coupled with the aforementioned bNAb auto-reactivity, we investigated whether transient alteration of the naive B cell repertoire prior to vaccination would impact the B cell response to Env immunization. The rationale was to model in part what might occur during chronic HIV-1 infection regarding expansion of the B cell repertoire. To perform this experiment, we treated mice with exogenous recombinant B Lymphocyte Stimulator (BLyS, also called BAFF) for 10 days prior to vaccination with the aim to expand the naïve repertoire and rescue self-reactive B cells that otherwise would be deleted due to limiting BAFF levels at this peripheral selection check-point (128). Upon analysis of the B cell compartments of BAFF-treated and PBS control-treated mice, we detected increased frequencies of splenic transitional B cells as well as an altered ratio of marginal zone to follicular B cells in the BAFF-pretreated mice, consistent with the expected biological activity of the treatment. Immunizing the mice with the YU2 foldon trimers, we detected increased neutralizing capacity against several tier 1 viruses in these BAFF pre-treated mice compared to the controls. This initial study suggests that transient manipulation of the naïve repertoire in the context of immunization may alter the quality of the elicited neutralizing Ab response. To fully define the effect of exogenous BAFF treatment on the naïve B cell repertoire, more detailed studies including deep sequencing of B cells under the different conditions is required. Accordingly, we are currently pursuing this concept in through a series of experiments in mice and NHPs in collaboration with Michael Cancro's laboratory at the University of Pennsylvania.
Activation of B cells encoding Ab specificities with the potential to evolve into effective neutralizing Ab responses is an area of intense investigation. In addition to studies of different immunogens, this involves evaluation of different vaccine delivery platforms and adjuvants to improve upon previous results in the field. These efforts also include reductionist approaches, such as using mice engineered to express germline-reverted versions of bNAbs to attempt to demonstrate principles of B cell activation and then to determine the best boost to mature these B cells along a “successful” path toward neutralizing breadth (111, 129-131). These approaches and the elicited B cell responses are reviewed elsewhere in this volume (Alt) and will therefore not be covered here. Some other twists on Env immunogens currently under investigation include epitope scaffolding, Env mini-subunits or trimers are to increase B cell germline engagement guided by bNAbs using Env mini-proteins, prime:boosting, recapitulation of Env lineages associated with infection-mediated neutralization breadth and other approaches. The most promising case for the so-called germline engagement approach involve the VRC01-like bNAbs that are directed against the highly conserved CD4 binding site on the surface of gp120. This class of bNAbs is genetically restricted, using limited and related VH gene segments that encode much of the HCDR2-mediated reactive surface that broadly recognizes the conserved CD4bs. Immunization with engineered outer domains that bind germline-reverted VRC01 with high affinity activates B cells encoding the VRC01 germline-reverted heavy chain mice when administered as oligomeric arrays on the surface of proteinaceous particles. As mentioned, there are as well non-genetically restricted CD4bs-directed bNAbs that successfully target the CD4bs in a HCDR3-dependent manner, such as CH103 or VRC13 (15, 17). These are the types of Abs we generally aim to elicit in our efforts to target the CD4bs through Env trimer vaccination as it has the potential to exploit the full diversity of the B cell repertoire to target this known broadly neutralizing determinant.
While the capacity to elicit cross-neutralizing tier 2 responses is a primary objective for an effective HIV-1 vaccine, the durability of the response is also important. Long-lasting responses are the goal of all prophylactic vaccines, but how well different vaccines achieve this objective is not yet well understood. Current knowledge supports the view that plasma cells residing in the bone marrow are responsible for the maintenance long-lasting serum Ab responses induced by infection or vaccination (reviewed in (132). For an effective HIV vaccine, due to its relatively rapid dissemination into available target cells, constitutive production to generate circulating Ab by long-lived plasma cells is likely needed to provide first-line defense against incoming virus for protection. Passive transfer studies with recombinant bNAbs in NHPs indicate what titers of circulating Ab are required to achieve protection against SHIV challenge (133-137). For some vaccines, such as that to HBV, it may be sufficient to maintain durable circulating memory B cell responses to provide effective recall responses upon pathogen re-exposure, but this is likely not a rapid enough response for HIV.
In practice, very long study cycles (decades) are required to formally demonstrate if a given vaccine induces durable B cell responses. Thus, alternative approaches are required to more rapidly evaluate durability of responses, such as characterization of Env-specific plasma cell in long-lived immune compartments. To establish baseline information about the kinetics of vaccine-induced Ab responses in blood as determined by ELISA, as well as to determine antigen-specific B cells in blood and bone marrow by B cell ELISpot analysis we performed experiments to allow enumeration of Env-specific memory B cells and Ab-secreting plasma cells (62). By these methods, we determined the half-life of the plasma Ab response after boosting to be approximately 21 days, consistent with that the majority of the Abs measured after boost arise from short-lived plasma cells. Our B cell ELISpot results are consistent with the interpretation that what we detect derives from Env-specific IgG-producing cells that peak seven days after each boost. This B cell expansion is followed by a sharp decline, suggesting that they arise from the antigen-specific memory B cells, which expand and differentiate into short-lived plasmablasts. This is similar to the kinetics reported for other vaccines where boosting is assessed (76, 138, 139) and is consistent with early studies from human trials showing that the Ab response to gp120 immunization is poorly sustained following clearance of antigen (140, 141). We therefore investigated if HIV-1 Env is an unusual viral antigen in terms of the magnitude, kinetics or durability of the elicited B cell response. Macaques were inoculated with soluble HIV-1 Env and influenza virus HA proteins in adjuvant and the response was monitored over an eight-month period (142). After simultaneous immunization of an NHP with Env in one leg and HA in the other, we determined circulating Ab binding titers and frequencies of antigen-specific memory B cells and bone marrow plasma cells. We found that the two protein antigens behaved similarly in regards to all these parameters, suggesting that induction of short-lived Ab responses is not a unique property of HIV-1 Env, but is a general feature of soluble protein antigens. Finally, in regards to Env-specific plasma cells in the bone marrow, such cells are detected by B cell ELISpot analysis at frequencies of 1-10% of total Ab-secreting cells with some variation depending on the nature of the immunization protocol. Ongoing studies to trace Env-specific Ab lineages in the bone marrow will shed new light on the quality and the persistence of the Env-specific Ab repertoire present in this reclusive B cell compartment.
Summary
The chronicity of HIV-1 has generated “survival-fit” (battle-tested) variants of Env that have been molded by Darwinian selection so that the virus achieves successful persistence in the human population. Even though there are transmission bottlenecks during transmission of HIV-1, which may lower the range of transmitting/founder isolates, the number of HIV-1 variants that need to be neutralized by vaccine-induced antibody responses is orders of magnitude above the coverage afforded by other currently effective human vaccines. Even the effective tetra-valent HPV human vaccine, with a nine-valent product under development, does not generate breadth, but simply includes matches to the most clinically problematic papilloma virus strains. Therefore, the need to elicit broadly effective antibody responses against HIV-1, presumably against conserved neutralization targets, remains a critical objective for the design of immunogens and immunization regimens. The early soluble Env spike mimetics, although not structurally similar enough to mimic native Env, elicited tier 1 and, in some cases, tier 2 autologous neutralizing responses. These model immunogens provided an opportunity to obtain an initial understanding of host responses to different epitope regions of Env and promoted the development of new methodology for detailed B cell response studies. Methods for Ab specificity mapping, isolation of MAbs and germline Ab V gene discovery are invaluable in ongoing studies of new generation immunogens and are generally applicable to the evaluation of all human vaccines. This, together with developments of new well-ordered NFL, SOSIP and other soluble HIV-1 Env trimers, mark a new era in HIV-1 structure-directed vaccine design and B cell evaluation. In cases where new vaccine approaches are shown to activate effective B cell responses in relevant animal models, such protocols need to be rapidly translated into human clinical assessment (see FIG 4, lower right) to advance the development of a vaccine against this amazingly antibody- and neutralization-resistant global pathogen.
Acknowledgments
We would like to thank members of the Wyatt and Karlsson Hedestam laboratories for helpful discussions as well as by the other Principal Investigators within our HIVRAD P01 entitled “High resolution B cell analysis to HIV Env” that is based upon many of the principles described in this Review. We would like to thank Christiana Corbaci for graphical assistance with the figures. This work was supported by the NIH HIVRAD grant P01 AI104722, the Scripps CHAVI-ID AI100663, the Swedish Research Council and by IAVI and made possible by the support of its many donors, including: the Bill & Melinda Gates Foundation, the Ministry of Foreign Affairs of Denmark, Irish Aid, the Ministry of Finance of Japan in partnership with The World Bank, the Ministry of Foreign Affairs of the Netherlands, the Norwegian Agency for Development Cooperation (NORAD), the United Kingdom Department for International Development (DFID), and the United States Agency for International Development (USAID). The full list of IAVI donors is available at http://www.iavi.org. The contents of this manuscript are the responsibility of IAVI and do not necessarily reflect the views of USAID or the US Government.
References
- 1.Karlsson Hedestam GB, Fouchier RA, Phogat S, Burton DR, Sodroski J, Wyatt RT. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat Rev Microbiol. 2008;6:143–155. doi: 10.1038/nrmicro1819. [DOI] [PubMed] [Google Scholar]
- 2.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 3.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 4.McLellan JS, Pancera M, Carrico C, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garces F, Lee JH, de Val N, et al. Affinity maturation of a potent family of HIV antibodies is primarily focused on accommodating or avoiding glycans. Immunity. 2015;43:1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ingale J, Tran K, Kong L, et al. Hyperglycosylated stable core immunogens designed to present the CD4 binding site are preferentially recognized by broadly neutralizing antibodies. J Virol. 2014;88:14002–14016. doi: 10.1128/JVI.02614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pejchal R, Doores KJ, Walker LM, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kepler TB, Liao HX, Alam SM, et al. Immunoglobulin gene insertions and deletions in the affinity maturation of HIV-1 broadly reactive neutralizing antibodies. Cell Host Microbe. 2014;16:304–313. doi: 10.1016/j.chom.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu X, Zhou T, Zhu J, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pejchal R, Walker LM, Stanfield RL, et al. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc Natl Acad Sci U S A. 2010;107:11483–11488. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garces F, Sok D, Kong L, et al. Structural evolution of glycan recognition by a family of potent HIV antibodies. Cell. 2014;159:69–79. doi: 10.1016/j.cell.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao HX, Lynch R, Zhou T, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simonich CA, Williams KL, Verkerke HP, et al. HIV-1 neutralizing antibodies with limited hypermutation from an infant. Cell. 2016;166:77–87. doi: 10.1016/j.cell.2016.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou T, Lynch RM, Chen L, et al. Structural repertoire of HIV-1-neutralizing antibodies targeting the CD4 supersite in 14 Donors. Cell. 2015;161:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheid JF, Mouquet H, Ueberheide B, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao HX, Lynch R, Zhou T, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou T, Xu L, Dey B, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker LM, Huber M, Doores KJ, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorman J, Soto C, Yang MM, et al. Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design. Nat Struct Mol Biol. 2016;23:81–90. doi: 10.1038/nsmb.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doria-Rose NA, Bhiman JN, Roark RS, et al. New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J Virol. 2016;90:76–91. doi: 10.1128/JVI.01791-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong L, Lee JH, Doores KJ, et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol. 2013;20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falkowska E, Le KM, Ramos A, et al. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blattner C, Lee JH, Sliepen K, et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doria-Rose NA, Schramm CA, Gorman J, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong R, Xu K, Zhou T, et al. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science. 2016;352:828–833. doi: 10.1126/science.aae0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kash N, Lee MA, Kollipara R, Downing C, Guidry J, Tyring SK. Safety and efficacy data on vaccines and immunization to human papillomavirus. J Clin Med. 2015;4:614–633. doi: 10.3390/jcm4040614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, Farzan M, Wyatt R, Sodroski J. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 2000;74:5716–5725. doi: 10.1128/jvi.74.12.5716-5725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X, Florin L, Farzan M, et al. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J Virol. 2000;74:4746–4754. doi: 10.1128/jvi.74.10.4746-4754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76:4634–4642. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen B, Cheng Y, Calder L, et al. A chimeric protein of simian immunodeficiency virus envelope glycoprotein gp140 and Escherichia coli aspartate transcarbamoylase. J Virol. 2004;78:4508–4516. doi: 10.1128/JVI.78.9.4508-4516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kovacs JM, Noeldeke E, Ha HJ, et al. Stable, uncleaved HIV-1 envelope glycoprotein gp140 forms a tightly folded trimer with a native-like structure. Proc Natl Acad Sci U S A. 2014;111:18542–18547. doi: 10.1073/pnas.1422269112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanders RW, Derking R, Cupo A, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guenaga J, de Val N, Tran K, et al. Well-ordered trimeric HIV-1 subtype B and C soluble spike mimetics generated by negative selection display native-like properties. PLoS Pathog. 2015;11:e1004570. doi: 10.1371/journal.ppat.1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma SK, de Val N, Bale S, et al. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 2015;11:539–550. doi: 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guenaga J, Dubrovskaya V, de Val N, et al. Structure-guided redesign increases the propensity of HIV Env to generate highly stable soluble trimers. J Virol. 2016;90:2806–2817. doi: 10.1128/JVI.02652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran K, Poulsen C, Guenaga J, et al. Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign. Proc Natl Acad Sci U S A. 2014;111:E738–747. doi: 10.1073/pnas.1319512111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundling C, Forsell MNE, O'Dell S, et al. Soluble HIV-1 Env trimers in adjuvant elicit potent and diverse functional B cell responses in primates. J Exp Med. 2010;207:2003–2017. doi: 10.1084/jem.20100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Douagi I, Forsell MN, Sundling C, et al. Influence of novel CD4 binding-defective HIV-1 envelope glycoprotein immunogens on neutralizing antibody and T-cell responses in nonhuman primates. J Virol. 2010;84:1683–1695. doi: 10.1128/JVI.01896-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navis M, Tran K, Bale S, et al. HIV-1 receptor binding site-directed antibodies using a VH1-2 gene segment orthologue are activated by Env trimer immunization. PLoS Pathog. 2014;10:e1004337. doi: 10.1371/journal.ppat.1004337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez P, Sundling C, O'Dell S, Mascola JR, Wyatt RT, Karlsson Hedestam GB. Primate immune responses to HIV-1 Env formulated in the saponin-based adjuvant AbISCO-100 in the presence or absence of TLR9 co-stimulation. Sci Rep. 2015;5:8925. doi: 10.1038/srep08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phad GE, Vazquez Bernat N, Feng Y, et al. Diverse antibody genetic and recognition properties revealed following HIV-1 envelope glycoprotein immunization. J Immunol. 2015;194:5903–5914. doi: 10.4049/jimmunol.1500122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai K, He L, Khan SN, et al. Rhesus macaque B-cell responses to an HIV-1 trimer vaccine revealed by unbiased longitudinal repertoire analysis. MBio. 2015;6 doi: 10.1128/mBio.01375-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dai K, Khan SN, Wang Y, et al. HIV-1 vaccine-elicited antibodies reverted to their inferred naive germline reveal associations between binding affinity and in vivo activation. Sci Rep. 2016;6:20987. doi: 10.1038/srep20987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Sundling C, Wilson R, et al. High-resolution longitudinal study of HIV-1 Env vaccine-elicited B cell responses to the virus primary receptor binding site reveals affinity maturation and clonal persistence. J Immunol. 2016;196:3729–3743. doi: 10.4049/jimmunol.1502543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez-Murillo P, Pramanik L, Sundling C, et al. CD138 and CD31 double-positive cells comprise the functional antibody-secreting plasma cell compartment in primate bone marrow. Front Immunol. 2016;7:242. doi: 10.3389/fimmu.2016.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seaman MS, Janes H, Hawkins N, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morner A, Douagi I, Forsell MN, et al. Human immunodeficiency virus type 1 env trimer immunization of macaques and impact of priming with viral vector or stabilized core protein. J Virol. 2009;83:540–551. doi: 10.1128/JVI.01102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng Y, Tran K, Bale S, et al. Thermostability of well-ordered HIV spikes correlates with the elicitation of autologous tier 2 neutralizing antibodies. PLoS Pathog. 2016;12:e1005767. doi: 10.1371/journal.ppat.1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Svehla K, Mathy NL, Voss G, Mascola JR, Wyatt R. Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J Virol. 2006;80:1414–1426. doi: 10.1128/JVI.80.3.1414-1426.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dosenovic P, Chakrabarti B, Soldemo M, et al. Selective expansion of HIV-1 envelope glycoprotein-specific B cell subsets recognizing distinct structural elements following immunization. J Immunol. 2009;183:3373–3382. doi: 10.4049/jimmunol.0900407. [DOI] [PubMed] [Google Scholar]
- 52.Liao HX, Chen X, Munshaw S, et al. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. J Exp Med. 2011;208:2237–2249. doi: 10.1084/jem.20110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams WB, Liao HX, Moody MA, et al. HIV-1 VACCINES. Diversion of HIV-1 vaccine-induced immunity by gp41-microbiota cross-reactive antibodies. Science. 2015;349:aab1253. doi: 10.1126/science.aab1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, Migueles SA, Welcher B, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13:1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, Svehla K, Louder MK, et al. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J Virol. 2009;83:1045–1059. doi: 10.1128/JVI.01992-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li YX, Svehla K, Louder MK, et al. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J Virol. 2009;83:1045–1059. doi: 10.1128/JVI.01992-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng Y, McKee K, Tran K, et al. Biochemically defined HIV-1 envelope glycoprotein variant immunogens display differential binding and neutralizing specificities to the CD4-binding site. J Biol Chem. 2012;287:5673–5686. doi: 10.1074/jbc.M111.317776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chakrabarti BK, Feng Y, Sharma SK, et al. Robust neutralizing antibodies elicited by HIV-1 JRFL envelope glycoprotein trimers in nonhuman primates. J Virol. 2013;87:13239–13251. doi: 10.1128/JVI.01247-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanders RW, van Gils MJ, Derking R, et al. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng C, Pancera M, Bossert A, et al. Immunogenicity of a prefusion HIV-1 envelope trimer in complex with a quaternary-structure-specific antibody. J Virol. 2016;90:2740–2755. doi: 10.1128/JVI.02380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koch M, Pancera M, Kwong PD, et al. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology. 2003;313:387–400. doi: 10.1016/s0042-6822(03)00294-0. [DOI] [PubMed] [Google Scholar]
- 62.Sundling C, Li Y, Huynh N, et al. High-resolution definition of vaccine-elicited B cell responses against the HIV primary receptor binding site. Sci Transl Med. 2012;4:142r. doi: 10.1126/scitranslmed.3003752. a196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crooks ET, Tong T, Chakrabarti B, et al. Vaccine-elicited tier 2 HIV-1 neutralizing antibodies bind to quaternary epitopes involving glycan-deficient patches proximal to the CD4 binding site. PLoS Pathog. 2015;11:e1004932. doi: 10.1371/journal.ppat.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McCoy LE, van Gils MJ, Ozorowski G, et al. Holes in the glycan shield of the native HIV envelope are a target of trimer-elicited neutralizing antibodies. Cell Rep. 2016;16:2327–2338. doi: 10.1016/j.celrep.2016.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gibbs RA, Rogers J, Katze MG, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 66.Peruchon S, Chaoul N, Burelout C, et al. Tissue-specific B-cell dysfunction and generalized memory B-cell loss during acute SIV infection. PLoS ONE. 2009;4:e5966. doi: 10.1371/journal.pone.0005966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gujer C, Sundling C, Seder RA, Karlsson Hedestam GB, Lore K. Human and rhesus plasmacytoid dendritic cell and B-cell responses to Toll-like receptor stimulation. Immunology. 2011;134:257–269. doi: 10.1111/j.1365-2567.2011.03484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sundling C, Li Y, Huynh N, et al. High-resolution definition of vaccine-elicited B cell responses against the HIV primary receptor binding site. Sci Transl Med. 2012;4:142r. doi: 10.1126/scitranslmed.3003752. a196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Demberg T, Brocca-Cofano E, Xiao P, et al. Dynamics of memory B-cell populations in blood, lymph nodes, and bone marrow during antiretroviral therapy and envelope boosting in simian immunodeficiency virus SIVmac251-infected rhesus macaques. J Virol. 2012;86:12591–12604. doi: 10.1128/JVI.00298-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neumann B, Klippert A, Raue K, Sopper S, Stahl-Hennig C. Characterization of B and plasma cells in blood, bone marrow, and secondary lymphoid organs of rhesus macaques by multicolor flow cytometry. J Leukoc Biol. 2015;97:19–30. doi: 10.1189/jlb.1HI0514-243R. [DOI] [PubMed] [Google Scholar]
- 71.Demberg T, Mohanram V, Venzon D, Robert-Guroff M. Phenotypes and distribution of mucosal memory B-cell populations in the SIV/SHIV rhesus macaque model. Clin Immunol. 2014;153:264–276. doi: 10.1016/j.clim.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Silveira EL, Kasturi SP, Kovalenkov Y, et al. Vaccine-induced plasmablast responses in rhesus macaques: phenotypic characterization and a source for generating antigen-specific monoclonal antibodies. J Immunol Methods. 2015;416:69–83. doi: 10.1016/j.jim.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scheid JF, Mouquet H, Feldhahn N, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 74.Scheid JF, Mouquet H, Feldhahn N, et al. A method for identification of HIV gp140 binding memory B cells in human blood. J Immunol Methods. 2009;343:65–67. doi: 10.1016/j.jim.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sundling C, Zhang Z, Phad GE, et al. Single-cell and deep sequencing of IgG-switched macaque B cells reveal a diverse Ig repertoire following immunization. J Immunol. 2014 doi: 10.4049/jimmunol.1303334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wrammert J, Smith K, Miller J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poulsen TR, Meijer PJ, Jensen A, Nielsen LS, Andersen PS. Kinetic, affinity, and diversity limits of human polyclonal antibody responses against tetanus toxoid. J Immunol. 2007;179:3841–3850. doi: 10.4049/jimmunol.179.6.3841. [DOI] [PubMed] [Google Scholar]
- 78.Poulsen TR, Jensen A, Haurum JS, Andersen PS. Limits for antibody affinity maturation and repertoire diversification in hypervaccinated humans. J Immunol. 2011;187:4229–4235. doi: 10.4049/jimmunol.1000928. [DOI] [PubMed] [Google Scholar]
- 79.Foote J, Eisen HN. Kinetic and affinity limits on antibodies produced during immune responses. Proc Natl Acad Sci U S A. 1995;92:1254–1256. doi: 10.1073/pnas.92.5.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Klein F, Diskin R, Scheid JF, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Burton DR, Stanfield RL, Wilson IA. Antibody vs. HIV in a clash of evolutionary titans. Proc Natl Acad Sci U S A. 2005;102:14943–14948. doi: 10.1073/pnas.0505126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.MacLeod DT, Choi NM, Briney B, et al. Early antibody lineage diversification and independent limb maturation lead to broad HIV-1 neutralization targeting the Env high-mannose patch. Immunity. 2016;44:1215–1226. doi: 10.1016/j.immuni.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Breden F, Lepik C, Longo NS, Montero M, Lipsky PE, Scott JK. Comparison of antibody repertoires produced by HIV-1 infection, other chronic and acute infections, and systemic autoimmune disease. PLoS ONE. 2011;6:e16857. doi: 10.1371/journal.pone.0016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Scherer EM, Smith RA, Simonich CA, Niyonzima N, Carter JJ, Galloway DA. Characteristics of memory B cells elicited by a highly efficacious HPV vaccine in subjects with no pre-existing immunity. PLoS Pathog. 2014;10:e1004461. doi: 10.1371/journal.ppat.1004461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Georgiou G, Ippolito GC, Beausang J, Busse CE, Wardemann H, Quake SR. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat Biotechnol. 2014;32:158–168. doi: 10.1038/nbt.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bonsignori M, Zhou T, Sheng Z, et al. Maturation pathway from germline to broad HIV-1 neutralizer of a CD4-mimic antibody. Cell. 2016;165:449–463. doi: 10.1016/j.cell.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Soto C, Ofek G, Joyce MG, et al. Developmental pathway of the MPER-directed HIV-1-neutralizing antibody 10E8. PLoS ONE. 2016;11:e0157409. doi: 10.1371/journal.pone.0157409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Quail MA, Smith M, Jackson D, et al. SASI-Seq: sample assurance Spike-Ins, and highly differentiating 384 barcoding for Illumina sequencing. BMC Genomics. 2014;15:110. doi: 10.1186/1471-2164-15-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shugay M, Britanova OV, Merzlyak EM, et al. Towards error-free profiling of immune repertoires. Nat Methods. 2014;11:653–655. doi: 10.1038/nmeth.2960. [DOI] [PubMed] [Google Scholar]
- 90.Khan TA, Friedensohn S, Gorter de Vries AR, Straszewski J, Ruscheweyh HJ, Reddy ST. Accurate and predictive antibody repertoire profiling by molecular amplification fingerprinting. Sci Adv. 2016;2:e1501371. doi: 10.1126/sciadv.1501371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Collins AM, Wang Y, Roskin KM, Marquis CP, Jackson KJ. The mouse antibody heavy chain repertoire is germline-focused and highly variable between inbred strains. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2014.0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Watson CT, Breden F. The immunoglobulin heavy chain locus: genetic variation, missing data, and implications for human disease. Genes Immun. 2012;13:363–373. doi: 10.1038/gene.2012.12. [DOI] [PubMed] [Google Scholar]
- 93.Lavinder JJ, Hoi KH, Reddy ST, Wine Y, Georgiou G. Systematic characterization and comparative analysis of the rabbit immunoglobulin repertoire. PLoS ONE. 2014;9:e101322. doi: 10.1371/journal.pone.0101322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yu GY, Mate S, Garcia K, et al. Cynomolgus macaque (Macaca fascicularis) immunoglobulin heavy chain locus description. Immunogenetics. 2016;68:417–428. doi: 10.1007/s00251-016-0921-2. [DOI] [PubMed] [Google Scholar]
- 95.Gadala-Maria D, Yaari G, Uduman M, Kleinstein SH. Automated analysis of high-throughput B-cell sequencing data reveals a high frequency of novel immunoglobulin V gene segment alleles. Proc Natl Acad Sci U S A. 2015;112:E862–870. doi: 10.1073/pnas.1417683112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lyumkis D, Julien JP, de Val N, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Julien JP, Cupo A, Sok D, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.de Taeye SW, Ozorowski G, Torrents de la Pena A, et al. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dey B, Svehla K, Xu L, et al. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 2009;5:e1000445. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Decker JM, Bibollet-Ruche F, Wei X, et al. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med. 2005;201:1407–1419. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McLellan JS, Chen M, Joyce MG, et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Forsell MN, Dey B, Morner A, et al. B cell recognition of the conserved HIV-1 co-receptor binding site is altered by endogenous primate CD4. PLoS Pathog. 2008;4:e1000171. doi: 10.1371/journal.ppat.1000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Forsell MN, McKee K, Feng Y, Mascola JR, Wyatt RT. HIV-1 envelope glycoprotein trimer immunogenicity elicited in the presence of human CD4 alters the neutralization profile. AIDS Res Hum Retroviruses. 2014;30:1089–1098. doi: 10.1089/aid.2014.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guenaga J, Dosenovic P, Ofek G, et al. Heterologous epitope-scaffold prime:boosting immuno-focuses B cell responses to the HIV-1 gp41 2F5 neutralization determinant. PLoS ONE. 2011;6:e16074. doi: 10.1371/journal.pone.0016074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ofek G, Guenaga FJ, Schief WR, et al. Elicitation of structure-specific antibodies by epitope scaffolds. Proc Natl Acad Sci U S A. 2010;107:17880–17887. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lai RP, Hock M, Radzimanowski J, et al. A fusion intermediate gp41 immunogen elicits neutralizing antibodies to HIV-1. J Biol Chem. 2014;289:29912–29926. doi: 10.1074/jbc.M114.569566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bianchi E, Joyce JG, Miller MD, et al. Vaccination with peptide mimetics of the gp41 prehairpin fusion intermediate yields neutralizing antisera against HIV-1 isolates. Proc Natl Acad Sci U S A. 2010;107:10655–10660. doi: 10.1073/pnas.1004261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Binley JM, Sanders RW, Clas B, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion- associated structure. J Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sanders RW, Vesanen M, Schuelke N, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Georgiev IS, Joyce MG, Yang Y, et al. Single-chain soluble BG505.SOSIP gp140 trimers as structural and antigenic mimics of mature closed HIV-1 Env. J Virol. 2015;89:5318–5329. doi: 10.1128/JVI.03451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Julien JP, Lee JH, Ozorowski G, et al. Design and structure of two HIV-1 clade C SOSIP.664 trimers that increase the arsenal of native-like Env immunogens. Proc Natl Acad Sci U S A. 2015;112:11947–11952. doi: 10.1073/pnas.1507793112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li Y, Cleveland B, Klots I, et al. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J Virol. 2008;82:638–651. doi: 10.1128/JVI.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liang Y, Guttman M, Williams JA, et al. Changes in structure and antigenicity of HIV-1 Env trimers resulting from removal of a conserved CD4 binding site-proximal glycan. J Virol. 2016 doi: 10.1128/JVI.01116-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Crooks ET, Tong T, Osawa K, Binley JM. Enzyme digests eliminate nonfunctional Env from HIV-1 particle surfaces, leaving native Env trimers intact and viral infectivity unaffected. J Virol. 2011;85:5825–5839. doi: 10.1128/JVI.00154-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Grundner C, Mirzabekov T, Sodroski J, Wyatt R. Solid-phase proteoliposomes containing human immunodeficiency virus envelope glycoproteins. J Virol. 2002;76:3511–3521. doi: 10.1128/JVI.76.7.3511-3521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Leaman DP, Lee JH, Ward AB, Zwick MB. Immunogenic display of purified chemically cross-linked HIV-1 spikes. J Virol. 2015;89:6725–6745. doi: 10.1128/JVI.03738-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ingale J, Stano A, Guenaga J, et al. High-density array of well-ordered HIV-1 spikes on synthetic liposomal nanoparticles efficiently activate B cells. Cell Rep. 2016;15:1986–1999. doi: 10.1016/j.celrep.2016.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sliepen K, Ozorowski G, Burger JA, et al. Presenting native-like HIV-1 envelope trimers on ferritin nanoparticles improves their immunogenicity. Retrovirology. 2015;12:82. doi: 10.1186/s12977-015-0210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.He L, de Val N, Morris CD, et al. Presenting native-like trimeric HIV-1 antigens with self-assembling nanoparticles. Nat Commun. 2016;7:12041. doi: 10.1038/ncomms12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Maity PC, Blount A, Jumaa H, Ronneberger O, Lillemeier BF, Reth M. B cell antigen receptors of the IgM and IgD classes are clustered in different protein islands that are altered during B cell activation. Sci Signal. 2015;8:ra93. doi: 10.1126/scisignal.2005887. [DOI] [PubMed] [Google Scholar]
- 121.Liu M, Yang G, Wiehe K, et al. Polyreactivity and autoreactivity among HIV-1 antibodies. J Virol. 2015;89:784–798. doi: 10.1128/JVI.02378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang G, Holl TM, Liu Y, et al. Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. J Exp Med. 2013;210:241–256. doi: 10.1084/jem.20121977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Verkoczy L, Kelsoe G, Moody MA, Haynes BF. Role of immune mechanisms in induction of HIV-1 broadly neutralizing antibodies. Curr Opin Immunol. 2011;23:383–390. doi: 10.1016/j.coi.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Malaspina A, Moir S, Ho J, et al. Appearance of immature/transitional B cells in HIV-infected individuals with advanced disease: correlation with increased IL-7. Proc Natl Acad Sci U S A. 2006;103:2262–2267. doi: 10.1073/pnas.0511094103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moir S, Malaspina A, Ogwaro KM, et al. HIV-1 induces phenotypic and functional perturbations of B cells in chronically infected individuals. Proc Natl Acad Sci U S A. 2001;98:10362–10367. doi: 10.1073/pnas.181347898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Doria-Rose NA, Klein RM, Manion MM, et al. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J Virol. 2009;83:188–199. doi: 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kardava L, Moir S, Shah N, et al. Abnormal B cell memory subsets dominate HIV-specific responses in infected individuals. J Clin Invest. 2014;124:3252–3262. doi: 10.1172/JCI74351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dosenovic P, Soldemo M, Scholz JL, et al. BLyS-mediated modulation of naive B cell subsets impacts HIV Env-induced antibody responses. J Immunol. 2012;188:6018–6026. doi: 10.4049/jimmunol.1200466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dosenovic P, von Boehmer L, Escolano A, et al. Immunization for HIV-1 broadly neutralizing antibodies in human Ig knockin mice. Cell. 2015;161:1505–1515. doi: 10.1016/j.cell.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McGuire AT, Gray MD, Dosenovic P, et al. Specifically modified Env immunogens activate B-cell precursors of broadly neutralizing HIV-1 antibodies in transgenic mice. Nat Commun. 2016;7:10618. doi: 10.1038/ncomms10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jardine J, Julien JP, Menis S, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nutt SL, Hodgkin PD, Tarlinton DM, Corcoran LM. The generation of antibody-secreting plasma cells. Nat Rev Immunol. 2015;15:160–171. doi: 10.1038/nri3795. [DOI] [PubMed] [Google Scholar]
- 133.Hessell AJ, Rakasz EG, Poignard P, et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 2009;5:e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Barouch DH, Whitney JB, Moldt B, et al. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013;503:224–228. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hessell AJ, Rakasz EG, Tehrani DM, et al. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. J Virol. 2010;84:1302–1313. doi: 10.1128/JVI.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shingai M, Donau OK, Plishka RJ, et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med. 2014;211:2061–2074. doi: 10.1084/jem.20132494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gautam R, Nishimura Y, Pegu A, et al. A single injection of anti-HIV-1 antibodies protects against repeated SHIV challenges. Nature. 2016;533:105–109. doi: 10.1038/nature17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Blanchard-Rohner G, Pulickal AS, Jol-van der Zijde CM, Snape MD, Pollard AJ. Appearance of peripheral blood plasma cells and memory B-cells in a primary and secondary immune response in humans. Blood. 2009 doi: 10.1182/blood-2009-03-211052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Blink EJ, Light A, Kallies A, Nutt SL, Hodgkin PD, Tarlinton DM. Early appearance of germinal center-derived memory B cells and plasma cells in blood after primary immunization. J Exp Med. 2005;201:545–554. doi: 10.1084/jem.20042060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Anderson KP, Lucas C, Hanson CV, et al. Effect of dose and immunization schedule on immune response of baboons to recombinant glycoprotein 120 of HIV-1. J Infect Dis. 1989;160:960–969. doi: 10.1093/infdis/160.6.960. [DOI] [PubMed] [Google Scholar]
- 141.Graham BS, Keefer MC, McElrath MJ, et al. Safety and immunogenicity of a candidate HIV-1 vaccine in healthy adults: recombinant glycoprotein (rgp) 120. A randomized, double-blind trial. NIAID AIDS Vaccine Evaluation Group. Ann Intern Med. 1996;125:270–279. doi: 10.7326/0003-4819-125-4-199608150-00003. [DOI] [PubMed] [Google Scholar]
- 142.Sundling C, Martinez P, Soldemo M, et al. Immunization of macaques with soluble HIV type 1 and influenza virus envelope glycoproteins results in a similarly rapid contraction of peripheral B-cell responses after boosting. J Infect Dis. 2013;207:426–431. doi: 10.1093/infdis/jis696. [DOI] [PubMed] [Google Scholar]