Abstract
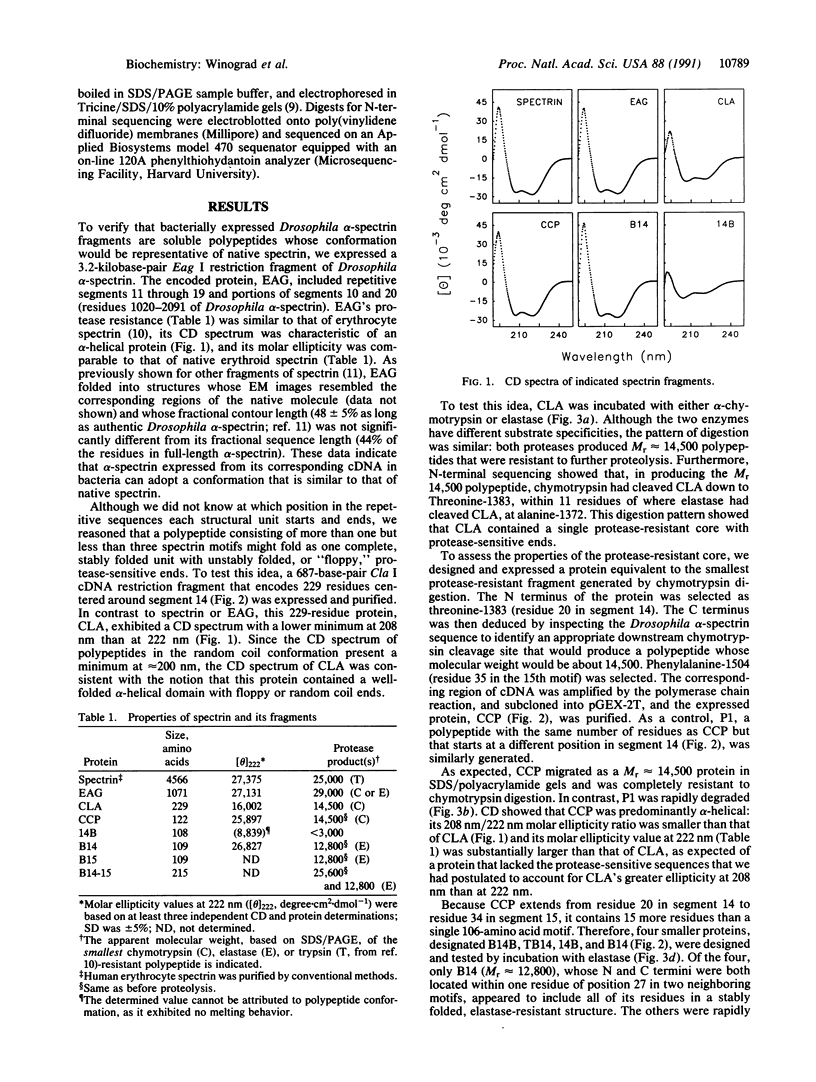
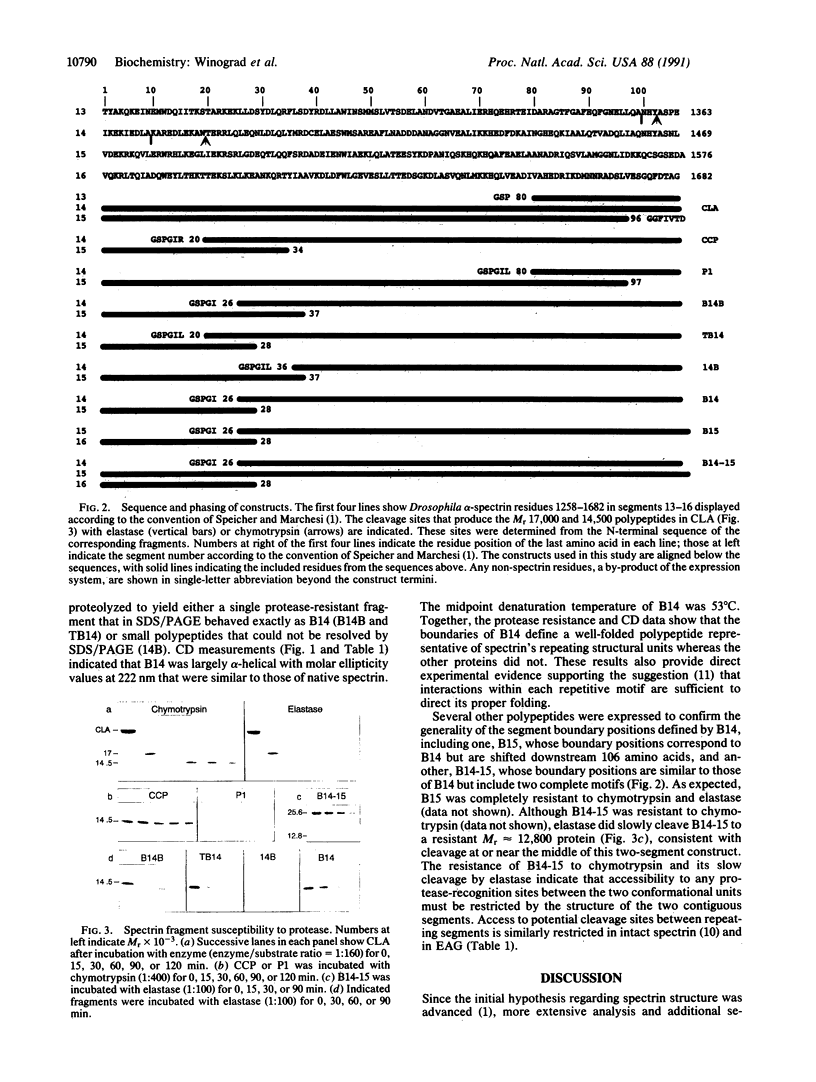
Many proteins contain a repetitive sequence motif, which implies that they contain a repetitive structural motif. Spectrin and the related proteins dystrophin and alpha-actinin consist largely of repeated motifs of 100-120 residues. But the repeating motif is degenerate and it has been difficult to define the boundaries of the repeating sequence unit or its corresponding structural unit. We have determined at which residues the structural units that correspond to spectrin's repeating 106-amino acid motifs begin and end. Drosophila alpha-spectrin cDNAs were expressed in bacteria to show that single segments (106 amino acids) and pairs of segments encoded by selected regions of spectrin cDNA can fold into stable conformations whose biophysical and biochemical properties are similar to those of native spectrin. Because such folding was critically dependent on the phasing of the expressed sequence with respect to the apparent boundaries of the repeating motifs, our data provide experimental evidence that relates the boundaries of the folded, conformational unit to the chemical sequence of repeating motifs.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Byers T. J., Husain-Chishti A., Dubreuil R. R., Branton D., Goldstein L. S. Sequence similarity of the amino-terminal domain of Drosophila beta spectrin to alpha actinin and dystrophin. J Cell Biol. 1989 Oct;109(4 Pt 1):1633–1641. doi: 10.1083/jcb.109.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen C., Parry D. A. Alpha-helical coiled coils and bundles: how to design an alpha-helical protein. Proteins. 1990;7(1):1–15. doi: 10.1002/prot.340070102. [DOI] [PubMed] [Google Scholar]
- Cross R. A., Stewart M., Kendrick-Jones J. Structural predictions for the central domain of dystrophin. FEBS Lett. 1990 Mar 12;262(1):87–92. doi: 10.1016/0014-5793(90)80160-k. [DOI] [PubMed] [Google Scholar]
- Davison M. D., Baron M. D., Critchley D. R., Wootton J. C. Structural analysis of homologous repeated domains in alpha-actinin and spectrin. Int J Biol Macromol. 1989 Apr;11(2):81–90. doi: 10.1016/0141-8130(89)90047-0. [DOI] [PubMed] [Google Scholar]
- Dubreuil R. R., Brandin E., Reisberg J. H., Goldstein L. S., Branton D. Structure, calmodulin-binding, and calcium-binding properties of recombinant alpha spectrin polypeptides. J Biol Chem. 1991 Apr 15;266(11):7189–7193. [PubMed] [Google Scholar]
- Dubreuil R. R., Byers T. J., Sillman A. L., Bar-Zvi D., Goldstein L. S., Branton D. The complete sequence of Drosophila alpha-spectrin: conservation of structural domains between alpha-spectrins and alpha-actinin. J Cell Biol. 1989 Nov;109(5):2197–2205. doi: 10.1083/jcb.109.5.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenicke L. A rapid micromethod for the determination of nitrogen and phosphate in biological material. Anal Biochem. 1974 Oct;61(2):623–627. doi: 10.1016/0003-2697(74)90429-1. [DOI] [PubMed] [Google Scholar]
- Koenig M., Kunkel L. M. Detailed analysis of the repeat domain of dystrophin reveals four potential hinge segments that may confer flexibility. J Biol Chem. 1990 Mar 15;265(8):4560–4566. [PubMed] [Google Scholar]
- Koenig M., Monaco A. P., Kunkel L. M. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988 Apr 22;53(2):219–228. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- Marchesi S. L., Letsinger J. T., Speicher D. W., Marchesi V. T., Agre P., Hyun B., Gulati G. Mutant forms of spectrin alpha-subunits in hereditary elliptocytosis. J Clin Invest. 1987 Jul;80(1):191–198. doi: 10.1172/JCI113047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Speicher D. W., Marchesi V. T. Erythrocyte spectrin is comprised of many homologous triple helical segments. Nature. 1984 Sep 13;311(5982):177–180. doi: 10.1038/311177a0. [DOI] [PubMed] [Google Scholar]
- Speicher D. W., Morrow J. S., Knowles W. J., Marchesi V. T. Identification of proteolytically resistant domains of human erythrocyte spectrin. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5673–5677. doi: 10.1073/pnas.77.10.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speicher D. W. The present status of erythrocyte spectrin structure: the 106-residue repetitive structure is a basic feature of an entire class of proteins. J Cell Biochem. 1986;30(3):245–258. doi: 10.1002/jcb.240300306. [DOI] [PubMed] [Google Scholar]
- Tse W. T., Lecomte M. C., Costa F. F., Garbarz M., Feo C., Boivin P., Dhermy D., Forget B. G. Point mutation in the beta-spectrin gene associated with alpha I/74 hereditary elliptocytosis. Implications for the mechanism of spectrin dimer self-association. J Clin Invest. 1990 Sep;86(3):909–916. doi: 10.1172/JCI114792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Prabhakaran M., Johnson M. E., Fung L. W. Secondary structure prediction for the spectrin 106-amino acid segment, and a proposed model for tertiary structure. J Biomol Struct Dyn. 1990 Aug;8(1):55–62. doi: 10.1080/07391102.1990.10507789. [DOI] [PubMed] [Google Scholar]







