Key Points
Contrasting patterns of basal phosphorylation levels and α-BCR–induced signaling between CLL and MCL tumors.
Direct association between BCR-induced signaling strength and CD79B level, but inverse association with BTK and SYK inhibitor efficacy.
Abstract
Kinases downstream of B-cell antigen receptor (BCR) represent attractive targets for therapy in non-Hodgkin lymphoma (NHL). As clinical responses vary, improved knowledge regarding activation and regulation of BCR signaling in individual patients is needed. Here, using phosphospecific flow cytometry to obtain malignant B-cell signaling profiles from 95 patients representing 4 types of NHL revealed a striking contrast between chronic lymphocytic leukemia (CLL) and mantle cell lymphoma (MCL) tumors. Lymphoma cells from diffuse large B-cell lymphoma patients had high basal phosphorylation levels of most measured signaling nodes, whereas follicular lymphoma cells represented the opposite pattern with no or very low basal levels. MCL showed large interpatient variability in basal levels, and elevated levels for the phosphorylated forms of AKT, extracellular signal-regulated kinase, p38, STAT1, and STAT5 were associated with poor outcome. CLL tumors had elevated basal levels for the phosphorylated forms of BCR-signaling nodes (Src family tyrosine kinase, spleen tyrosine kinase [SYK], phospholipase Cγ), but had low α-BCR–induced signaling. This contrasted MCL tumors, where α-BCR–induced signaling was variable, but significantly potentiated as compared with the other types. Overexpression of CD79B, combined with a gating strategy whereby signaling output was directly quantified per cell as a function of CD79B levels, confirmed a direct relationship between surface CD79B, immunoglobulin M (IgM), and IgM-induced signaling levels. Furthermore, α-BCR–induced signaling strength was variable across patient samples and correlated with BCR subunit CD79B expression, but was inversely correlated with susceptibility to Bruton tyrosine kinase (BTK) and SYK inhibitors in MCL. These individual differences in BCR levels and signaling might relate to differences in therapy responses to BCR-pathway inhibitors.
Introduction
Non-Hodgkin lymphoma (NHL) is a diverse group of malignancies originating from mature B cells, most commonly germinal center (GC) B cells.1,2 Diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (FL) are the most frequent types, whereas mantle cell lymphoma (MCL) is less frequent, but remains more challenging to treat. The B-cell antigen receptor (BCR) is commonly maintained in malignant B cells,3 and its expression and downstream signaling is increasingly implicated in the pathogenesis of NHL. The BCR consists of the antigen-binding immunoglobulin heavy (IgH) and light (IgL) chains coupled to a heterodimer of the signaling subunits CD79A (Igα) and CD79B (Igβ).4,5 BCR signaling is thought to depend on ligand-induced aggregation. However, continuous BCR expression is needed for survival of healthy B cells,6,7 and BCR signal to maintain survival in the absence of receptor engagement.7,8
Crosslinking of BCR by antigen triggers the phosphorylation of tyrosines within the immunoreceptor tyrosine-based activation motifs (ITAMs) of CD79A and CD79B by Src family tyrosine kinases (SFKs) such as Lyn and by spleen tyrosine kinase (SYK), and provides a docking site for SYK. Activation of SYK is central in the propagation of BCR signaling, and initiates formation of the “signalosome” complex, composed of multiple tyrosine kinases and adaptor molecules including B-cell linker protein (BLNK), phospholipase Cγ2 (PLCγ2), and Bruton tyrosine kinase (BTK).9-11 The result of proximal BCR signaling is the activation of NF-κB, phosphatidylinositol 3-kinase, MAPK, nuclear factor of activated T cells, and RAS pathways, altering gene expression that directs fate decisions in normal and malignant B cells.12-14
Activation of BCR by autoantigen is thought to be an initial driving force for some NHLs, and several autoantigens have been identified in chronic lymphocytic leukemia (CLL),15 marginal zone lymphoma,16 FL,17-19 and DLBCL.20,21 In other lymphoma types, BCR signaling nodes are frequently altered by recurrent mutations. In the activated B-cell (ABC) subtype of DLBCL, mutations of CD79B, CARD11, and the negative regulator of NF-κB TNFAIP3/A20 occur in about 21%, 11%, and 30% of cases, respectively.22-24 The functional importance of BCR signaling in malignant B cells makes this pathway an attractive target for therapy with small-molecule inhibitors. In particular, the BTK inhibitor ibrutinib has shown overall response rates of 71% and durable responses in CLL and an overall response rate of 68% in MCL,25-28 whereas the response rates in FL and DLBCL have been lower.29 Therefore, BCR signaling differences in malignant B cells, caused by autoantigens, mutations, or other abnormalities, may shape treatment responses.
We previously used phosphospecific–flow cytometry to obtain clinically relevant signaling profiles of acute myeloid leukemia and lymphoma tumors30-33 and to explore patients’ individual intratumor T-cell signaling.34 Here, we investigate basal- and activation-induced phosphorylation levels in lymphoma cells across different types of NHL malignancies using the same approach, and explored the mechanisms behind variability in α-BCR–induced signaling capacity and relationship with BCR-pathway inhibitors.
Methods
Human samples
All specimens were obtained with informed consent in accordance with the Declaration of Helsinki from either Stanford University Medical Center or from the Norwegian Radium Hospital, Oslo, Norway.
Tonsils and autologous peripheral blood samples were obtained from children undergoing tonsillectomy at Stanford Hospital. All samples were processed to mononuclear cells by Ficoll gradient centrifugation (Ficoll-Paque PLUS; GE Healthcare), and cryopreserved in liquid nitrogen. An overview of the NHL patient samples is given in supplemental Table 1 (available on the Blood Web site). The FL cases were the same as in the test cohort previously described.33 Lymphoma cell line Granta 519 was from DSMZ (ACC 342), and was maintained in RPMI 1640 (PAA) supplemented with antibiotics penicillin and streptomycin (Sigma-Aldrich) and 10% fetal calf serum (HyClone).
Reagents
Signaling inputs included recombinant human (rh) interleukin-4 (IL-4), rhIL-7, rhIL-10, and rhIL-21 (Bioscience) at 20 ng/mL; interferon-γ (IFN-γ; R&D Systems) at 20 ng/mL; BCR crosslinking (α-BCR) by anti-IgM F(ab′2) and anti-IgG F(ab′2) from Life Technologies (Invitrogen-Biosource) at 10 μg/mL each; a soluble trimer of CD40L (Amgen-Immunex) at 100 ng/mL; a Toll-like receptor 9 (TLR9) agonist (oligodeoxynucleotides containing unmethylated cytosine/guanine [CpG] motifs; CpG 7909, Pfizer Inc.) at 10 μg/mL; phorbol 12-myristate 13-acetate (PMA) and ionomycin (Sigma-Aldrich) at 1.0 μg/mL each. The following small-molecule inhibitors were used: fostamatinib (R406; Selleckchem) used at 2.5 μM or as specified; ibrutinib (PCI-32765 from Selleckchem or from Pharmacyclics) used at 2.0 μM or as specified. Antibodies from Becton Dickinson were used to detect expression of CD5, CD3, CD20, BCL2, CD79B, phosphorylated STAT1 (p-STAT1; Y701), p-STAT3 (Y705), p-STAT5 (Y694), p-STAT6 (Y641), phosphorylated ζ-associated protein of 70 kDa (Zap70)/p-SYK (Y319/Y352), p-SFK/p-Lck (Y705), p-PLCγ (Y759), p-p65 NF-κB (S529), p-BTK (Y223), p-BLNK (Y84), phosphorylated extracellular signal-regulated kinase (p–extracellular signal-regulated kinase [ERK]; T202/Y204), whereas antibodies to detect p-S6 (S235/S236), p-AKT (S473) and total IκBα (L35A5) were from Cell Signaling Technologies (all antibodies are listed in supplemental Table 3). Anti-IgM, anti-IgG, anti-Ig λ, and anti-Ig κ were from Invitrogen. Pacific Blue and Pacific Orange used for fluorescent barcoding of cells were from Life Technologies/Molecular Probes.
Activation of signaling and phosphospecific fluorescent flow cytometry
Activation of signaling and detection of phosphoproteins were performed as previously described.31,33-35 Briefly, the samples were thawed, and 1 to 5 million cells were used for flow cytometry–based live/dead discrimination and immunophenotyping. Signaling was analyzed in the remaining samples which were allowed to rest for 30 minutes at 37°C in RPMI 1640, supplemented with 10% fetal calf serum, before redistribution at 200 µL per well into v-bottomed 96-well plates and being given another 20 minutes rest. In experiments using inhibitors, these were added 60 minutes prior to adding α-BCR. Signaling was activated by α-BCR for 4 minutes or 45 minutes, CD40L, IL-4, IL-7, IL-10, IL-21, or IFN-γ for 15 minutes, or CpG for 45 minutes. Paraformaldehyde at a final concentration of 1.6% was added to stop signaling and incubated for 5 minutes at room temperature, followed by permeabilization in >90% freezer-cold methanol. At this point, the samples could be stored at −80°C before further processing. After rehydrating the cells by washing by centrifugation 2 times in phosphate-buffered saline, the cells were stained with antibodies or the cells were “barcoded” prior to staining with antibodies as previously described34 (see supplemental Figure 1 for details). The samples were then collected on a LSR II flow cytometer (Becton Dickinson). Data were analyzed using Cytobank software, as previously described33 (see details in supplemental Methods). Hierarchical cluster analysis of arcsinh-transformed data (in rows, each column represented a patient sample) was performed by Cluster 3.0, and the rows were clustered using complete linkage for visual clarity. Java Treeview 3.0 was used for visualization.
Overexpression of CD79B by retroviral transduction
The CD79B coding sequence was isolated from tonsillar B cells using the primers CACCATGGCCAGGCTGGCGTTGTC and CTCTCTTGGCTCTCTCCTGGCCTGGGTGCTCAC. This amplicon was then linked to green fluorescent protein (GFP) via a picornavirus 2A sequence using the primers AGAGCCAAGAGAGGCAGCGGCG and CTCGAGTTACTTGTACAGCTCGTCCATGCCGAG to generate an equimolar amount of CD79B and GFP, as previously described for other genes.36 The fusion polymerase chain reaction was performed using the primers in italic. The product was subcloned into the pENTR vector, sequenced (Eurofins), and recombined into the modified retroviral vector pMP71.37 Granta 519 was transduced by spinoculation on Retronectin (Takara) and expanded for 10 days before use.
Classification of DLBCL
DLBCL cases were classified based on immunohistochemical staining of formalin-fixed paraffin-embedded tissue sections by the Hans algorithm and the Choi algorithm as previously described.38,39 Results were 100% concordant.
Results
Basal phosphorylation levels distinguish lymphoma types
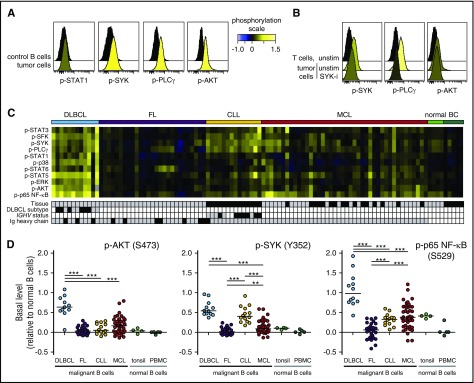
To investigate basal phosphorylation levels across B-NHL, we included patient samples from DLBCL, FL, MCL, and CLL (supplemental Table 1), as well as samples from healthy donors (tonsils and peripheral blood). All samples were acquired prior to therapy. The levels of 12 different phosphorylated kinases relevant for BCR, CD40, TLR, and cytokine signaling pathways were detected by phosphospecific flow cytometry (supplemental Figure 1). This revealed that MCL tumor cells had elevated levels of p-SYK, p-PLCγ, and p-AKT (Figure 1A). Our approach for detection of basal levels was sensitive and robust, as pretreatment with the SYK-inhibitor fostamatinib reduced the tumor cell levels of p-SYK and p-PLCγ (Figure 1B). A heatmap of phosphoprotein levels across NHL types revealed that DLBCL cells had elevated levels of many of the measured signaling nodes, and significantly higher levels of p-STAT5, p-AKT, and p-p65 NF-κB than the other NHL types (Figure 1C-D; supplemental Figure 2). In contrast, FL tumors had no or very low basal levels, similar to healthy donor B cells. A distinct feature of CLL cells was elevated levels of BCR-signaling nodes, including p-SFK, p-SYK, and p-PLCγ, but low levels of p-AKT and p-p65 NF-κB (Figure 1C-D; supplemental Figure 2). MCL tumors had variable expression levels across patient samples, with no clear pattern. Together, these data revealed not only lymphoma type-specific differences in basal phosphorylation levels but also great interpatient variability among MCL tumors.
Figure 1.
Elevated basal phosphorylation levels in subgroups of lymphoma patients. Single-cell suspensions from lymphoma patients or healthy donors were thawed and rested for 90 minutes before fixation, permeabilization, and staining with a combination of phosphospecific and lineage-marker–specific antibodies, followed by fluorescent flow cytometric measurements. Tumor cells were identified as CD20+BCL2+CD5−CD3− cells (FL, DLBCL) or CD20+BCL2+CD5+CD3− cells (MCL, CLL), normal B cells as CD20+CD5−CD3−, and T cells as CD20−CD5+CD3+. (A) Histogram overlays of basal (untreated) phosphorylation levels in MCL cells, relative to the basal level in healthy donor PBMC B cells. (B) Patient samples were left untreated or treated with the SYK inhibitor fostamatinib (SYK-i, 2.5 μM) for 1 hour. Shown are histogram overlays of phosphorylation levels in unstimulated (unstim) and fostamatinib-treated MCL cells, as compared with the level in unstimulated intratumor T cells. (C) Heatmap of basal phosphorylation levels across different types of NHLs. Each column represents a patient sample, and the rows were clustered for visual clarity (dendrogram not shown). Samples were annotated for tissue (gray, LN; black, peripheral blood), DLBCL subtype (gray, GCB; black, non-GCB; white, not determined [n.d.]), IGHV status (gray, unmutated: <2% change in IGHV gene sequence compared with germ line; black, mutated; white, n.d.) and Ig heavy chain (gray, IgM; black, IgG). (D) Shown are scatter plots of basal p-AKT, p-SYK, and p-p65 NF-κB in lymphoma cells, relative to healthy donor control PBMC B cells. Each dot represents 1 patient sample. In panels A, C, and D, phosphorylation levels are calculated relative to B cells in a healthy donor control sample, using arcsinh transformation. In panel B, phosphorylation is calculated relative to intratumor T cells. DLBCL (n = 12), FL (n = 27), CLL (n = 14), MCL (n = 42). Healthy donor controls: tonsillar B cells (n = 4) and PBMC B cells (n = 5). Statistical difference between NHL types was calculated using the Mann-Whitney nonparametric test (***P < .0001, **P < .005). BC, B cells.
Potentiated α-BCR–induced signaling in MCL cells
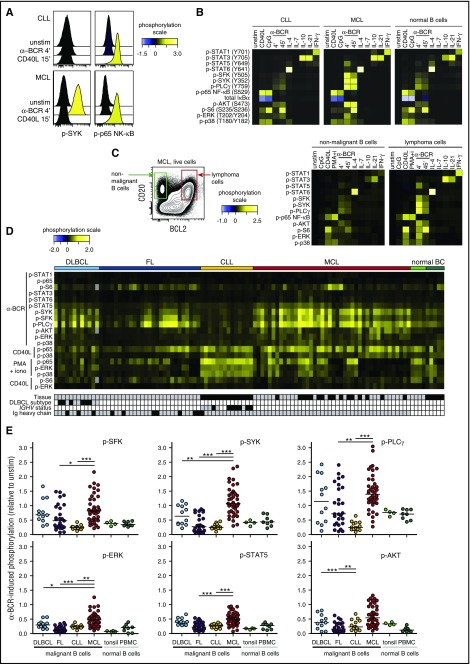
To obtain activation-induced signaling profiles, tumor samples were left unstimulated or were activated with CD40L, CpG, BCR engagement (α-BCR), IL-4, IL-7, IL-10, or IL-21, and the levels of phosphorylated kinases were determined by phosphospecific flow cytometry. Comparison of signaling profiles of intratumor T cells and lymphoma cells within the same sample provided distinct cell type–specific profiles as expected (supplemental Figure 3). Crosslinking of BCR (α-BCR) induced robust upregulation of p-SYK in MCL cells but not in CLL cells, whereas CD40L induced strong p-p65 NF-κB in both cases (Figure 2A-B). Furthermore, comparing signaling in lymphoma cells to healthy donor B cells revealed that the overall signaling pattern was similar; for example, the same kinases were phosphorylated upon specific stimulus, but the signaling strength varied (Figure 2B). In a MCL patient sample with detectable normal (CD20+CD5−BCL2−) and malignant (CD20+CD5+BCL2+) B cells, the malignant B cells demonstrated highly potentiated α-BCR–induced signaling (Figure 2C). Heatmap visualization of the major activation-induced phosphorylation events across B-NHL patient samples revealed that CLL tumors overall had very low α-BCR–induced p-SFK, p-SYK, and p-PLCγ levels (Figure 2D). In contrast, α-BCR–induced signaling was highly variable between patient samples in the FL, DLBCL, and MCL cohorts, but frequently higher than in CLL cells and healthy donor B cells (Figure 2D). Importantly, activation-induced signaling was not always low in CLL cells, as demonstrated by high IL-4–induced p-STAT6, IL-10–induced p-STAT3, and CD40L-induced p-p65 NF-κB in these tumors (supplemental Figure 4). BCR signaling in CLL is known to be heterogeneous, with CLL cells bearing unmutated IGVH (U-CLL) being more responsive than mutated CLL (M-CLL).40-42 However, our CLL cohort represented few cases and separation into U-CLL and M-CLL provided no significant differences (supplemental Figure 5A-C). Similarly, GCB was not significantly different from non-GCB (supplemental Figure 5D-E).
Figure 2.
Activation-induced signaling reveals contrasting differences between NHL subtypes, and potentiated α-BCR–induced signaling in MCL. Patient samples and control samples were thawed and rested for 20 minutes before activation with CD40L, IL-4, IL-7, IL-10, IL-21, or IFN-γ for 15 minutes, CpG for 45 minutes, BCR engagement with F(ab′)2 anti-IgM and anti-IgG (α-BCR) for 4 minutes or 45 minutes, or left untreated (unstim) (see supplemental Figure 1). Tumor cells were identified as described in Figure 1. (A) Histogram overlays of α-BCR– or CD40L-induced p-SYK and p-p65 NF-κB in CLL and MCL cells, shown relative to unstimulated cells. (B) Typical activation-induced signaling profiles shown as heatmaps of lymphoma cells from CLL and MCL patient samples as compared with healthy donor PBMC B cells. (C) Gating on lymphoma cells (CD20+BCL2+CD5+CD3−) and nonmalignant B cells (CD20+BCL2−CD5−CD3−) within the same patient sample revealed potentiated signaling in malignant B cells. (D) Heatmap of major activation-induced phosphorylation levels in lymphoma cells, across different types of NHLs. Each column represents a patient sample, and the rows were clustered for visual clarity (dendrogram not shown). Samples were annotated for tissue (gray, LN; black, peripheral blood), DLBCL subtype (gray, GCB; black, non-GCB; white, n.d.), IGHV status (gray, unmutated; black, mutated; white, n.d.), and Ig heavy chain (gray, IgM; black, IgG). (E) Scatter plots of α-BCR–induced phosphorylation levels in lymphoma cells, relative to unstimulated cells. Each dot represents a single patient sample. Phosphorylation levels are relative to unstimulated lymphoma cells from the same patient. DLBCL (n = 12), FL (n = 27), CLL (n = 14), MCL (n = 42). Healthy donor controls: tonsillar B cells (n = 4) and PBMC B cells (n = 8). Statistical difference was calculated using the Mann-Whitney nonparametric test (***P < .0001, **P < .005, *P < .05). iono, ionomycin.
Strikingly, MCL contrasted the other NHL types by overall high levels of both BCR proximal (p-SFK, p-SYK, p-PLCγ) and more distal signaling nodes (p-AKT, p-ERK, and p-STAT5), all significantly higher than in CLL and FL patients (P < .001; Figure 2E). Compared with DLBCL, only α-BCR–induced p-SYK and p-ERK were significantly higher in MCL. On average, α-BCR–induced p-SYK relative to unstimulated cells was 1.16 in MCL cells as compared with 0.67, 0.36, and 0.27 in DLBCL, FL, and CLL, respectively, and 0.40 and 0.48 in healthy donor tonsillar and peripheral blood mononuclear cell (PBMC) B cells (Figure 2E). The levels of BCR proximal readouts in MCL cells were strongly correlated, as demonstrated by α-BCR–induced p-SYK vs p-PLCγ (P < .0001, r = 0.96), and shown as a heatmap of 6 different signaling nodes across MCL cases (supplemental Figure 6). Low α-BCR–induced signaling in some MCL tumors could be due to higher basal phosphorylation levels; however, no relationship was found for basal vs α-BCR–induced phosphorylation levels of SFK, SYK, or PLCγ (supplemental Figure 7). For CLL, the tissue of origin is important, as demonstrated by stronger α-BCR–induced signaling and higher BCR gene signature score in the lymph node (LN) vs peripheral blood.43 As the MCL cohort contained LN and PBMC samples, central results from Figures 1 and 2 were reanalyzed with LN samples only and provided similar results (compare Figures 1, 2, and supplemental Figure 8). Overall, these data demonstrated heterogeneous α-BCR–induced signaling responses in lymphoma cells from DLBCL, FL, and MCL, which contrasted the low signaling responses in the malignant B cells from CLL patients.
α-BCR–induced signaling strength in MCL cells is correlated to surface expression of BCR subunits
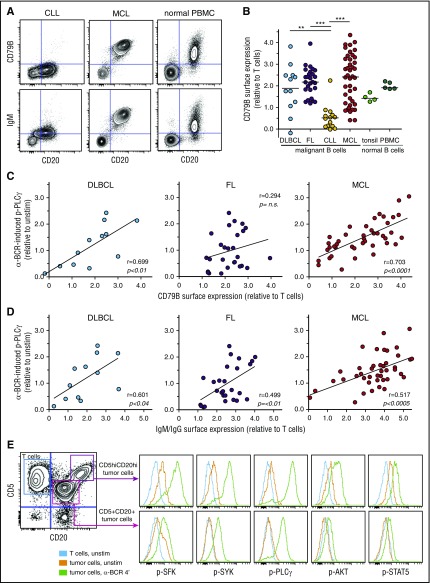
The interpatient variability in α-BCR–induced signaling strength for DLBCL, FL, and MCL could be due to variable surface expression levels of BCR subunits. To explore this relationship, we determined the surface expression of CD79B, IgG, IgM, Igκ, and Igλ in lymphoma cells. As expected from the signaling results, surface expression of CD79B was low in CLL cells, and highly variable in DLBCL, FL, and MCL cases (Figure 3A-B). Furthermore, there was a strong association between α-BCR–induced p-PLCγ and surface expression of CD79B for MCL patients (P < .0001, r = 0.70) and for DLBCL patients (P < .01, r = 0.70) but not for FL patients (Figure 3C). α-BCR–induced p-PLCγ was strongly correlated with surface expression of IgM in MCL patients (P < .0005, r = 0.52), but this association was less significant for DLBCL and FL (IgM or IgG heavy-chain expression, Figure 3D). Similar results were found for α-BCR–induced p-SYK vs CD79B or IgM/IgG expression, and for α-BCR–induced p-PLCγ vs IgM (supplemental Figure 9). In 1 of our MCL cases, we discovered 2 distinct lymphoma subclones based on different expression levels of CD5, CD20, Igκ, CD79B, and IgM (Figure 3E). The lymphoma subclone which possessed the highest expression of CD79B also had the strongest α-BCR–induced signaling response (Figure 3E; supplemental Figure 9). In ABC DLBCL, CD79B surface expression levels can be upregulated due to mutations.23 Sanger sequencing of CD79B was performed for 5 of the MCL cases, but no mutations were detected (not shown). Furthermore, exome sequencing of patient samples from our MCL cohort did not reveal mutations in CD79B. Hence, increased expression of CD79B in MCL cells is likely not due to mutations in the coding regions.
Figure 3.
α-BCR–induced signaling in MCL cells is associated with increased surface expression of BCR subunits CD79B and IgM. (A) Contour plots of CD20 expression vs CD79B or IgM expression in CLL, MCL, and healthy donor PBMC sample, respectively. (B) Expression of CD79B in lymphoma cells relative to T cells present in same sample in DLBCL, FL, CLL, MCL, and healthy donor tonsil and PBMC samples. (C) Association between α-BCR–induced p-PLCγ and CD79B surface expression in DLBCL, FL, and MCL. Each dot represents a patient sample. (D) Association between α-BCR–induced p-PLCγ and tumor heavy-chain expression (IgG or IgM for DLBCL, FL, and IgM for MCL). (E) The presence of 2 distinct lymphoma subclones in a MCL patient (MCL-R001), based on expression of CD5 and CD20, demonstrates potentiated α-BCR–induced phosphorylation events in the CD5hiCD20hi subclone. The CD20hiCD5hi cells are CD79Bhi, as compared with CD20+CD5+ lymphoma cells (see supplemental Figure 9E). DLBCL (n = 12), FL (n = 27), CLL (n = 14), MCL (n = 42). Healthy donor controls: tonsillar B cells (n = 4) and PBMC B cells (n = 5). Note that surface protein expression of CD79B and IgM were obtained as separate immunophenotypic staining, and not included in the signaling assay. Statistical difference was calculated using the Mann-Whitney nonparametric test (***P < .0001, **P < .005), and correlation was calculated by the Spearman rank test.
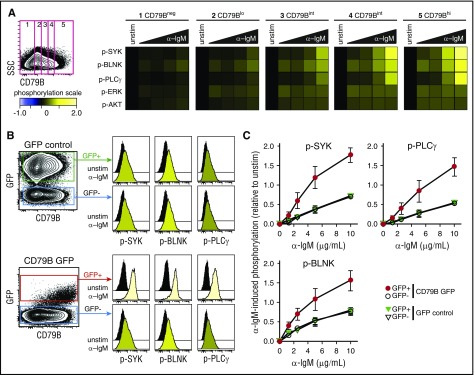
To test whether BCR–induced signaling per cell could be directly associated with surface expression of CD79B, we first measured α-IgM–induced signaling in an IgM+ MCL cell line, Granta 519, with highly heterogeneous expression of CD79B. A gating strategy to identify lymphoma cells with low, intermediate, and high levels of CD79B clearly demonstrated a direct relationship with the level of α-IgM–induced signaling (Figure 4A). To further study the impact of CD79B for regulating the strength of α-BCR–induced signaling, Granta 519 cells were transduced with bicistronic retroviral vector containing CD79B and GFP, or control vector containing GFP. The expression of GFP was then used to identify cells with an integrated vector. Introduction of CD79B led to higher and more homogeneous expression levels of CD79B in the GFP+ population (Figure 4B). Furthermore, cells overexpressing CD79B also exhibited higher levels of IgM. The introduction of GFP did not alter the α-IgM–induced signaling response, as this was equivalent in GFP+ vs GFP− cells for GFP control vector-transduced cells (Figure 4B-C). In contrast, the introduction of CD79B greatly enhanced α-IgM–induced p-SYK, p-PLCγ, and p-BLNK across a broad spectrum of α-IgM concentrations (Figure 4C). In these experiments, total CD79B protein was detected as the cells were stained postpermeabilization. However, the α-IgM–induced signaling pattern for distinct levels of CD79B gave similar results independent of method for CD79B detection (surface vs total; supplemental Figure 10). Together, these results demonstrate a direct relationship between the per-cell level of CD79B, IgM, and α-IgM–induced signaling strength in MCL cells.
Figure 4.
Expression of CD79B determines α-BCR–induced signaling strength. The MCL cell line Granta 519 was left unstimulated or was activated with different concentrations of F(ab′2) anti-IgM (α-IgM) for 4 minutes. (A) Cells were stained with anti-CD79B antibody (Ab), together with phosphospecific Abs. This strategy allowed gating on distinct levels of CD79B and mapping of α-BCR–induced signaling specifically to the distinct CD79B expression levels, from low to high. α-BCR–induced signaling is shown as heatmaps, where phosphorylation levels are relative to unstimulated cells within the same level of CD79B expression. (B) Overexpression of full length CD79B by retroviral transduction further enhances α-BCR–induced signaling. α-BCR–induced signaling assay was performed as in panel A. One representative experiment is shown, and in panel C, mean ± standard error of the mean (SEM) (n = 3). SSC, side scatter.
Efficacy of BCR-pathway drugs to suppress α-IgM–induced signaling depends on signaling strength
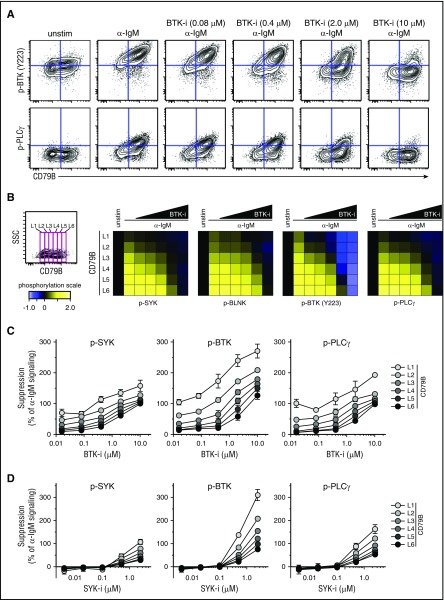
The strength of α-IgM–induced signaling in MCL could potentially affect the suppressing efficacy of BCR-signaling inhibitors. To test this, Granta 519 cells overexpressing CD79B were preincubated with fostamatinib or ibrutinib at different concentrations or were left untreated for 60 minutes before activation with α-IgM for 4 minutes. Visualization using dot plots for CD79B expression vs p-BTK or p-PLCγ showed a direct relationship between CD79B level and efficacy of ibrutinib to suppress α-IgM–induced phosphorylation (Figure 5A). Again, a gating strategy was used to measure signaling in cells expressing distinct levels of CD79B, from low to high (L1-L6), to test how different levels of CD79B affected the efficacy of the drugs. This approach revealed a direct dose-response relationship between the level of CD79B and efficacy of ibrutinib (Figure 5B). Here, high levels of CD79B expression (L5, L6) corresponding to strong α-IgM–induced signaling required much higher concentration of ibrutinib for efficient suppression as compared with cells with low CD79B expression and corresponding low IgM-induced signaling (Figure 5B-C). A similar relationship between the levels of CD79B/α-IgM–induced signaling and efficacy of fostamatinib was observed (Figure 5D).
Figure 5.
Higher concentrations of ibrutinib and fostamatinib are required to efficiently suppress α-BCR–induced signaling in CD79Bhicells as compared with CD79Blowcells. Granta 519 cells overexpressing CD79B were left untreated or treated with different concentrations of ibrutinib (BTK-i, 0.016-10.0 μM) or fostamatinib (SYK-i, 0.004-2.5 μM) for 1 hour, prior to α-IgM Ab stimulation for 4 minutes. The cells were stained with Abs as described in Figure 4. (A) Contour plots for CD79B expression vs p-BTK or p-PLCγ in cells treated with various concentrations of ibrutinib. (B) Same gating strategy was used to study α-BCR–induced signaling in cells with distinct expression levels of CD79B, from L1 to L6 (low-high). The effect of ibrutinib on α-IgM–induced phosphorylation is shown as heatmaps, relative to unstimulated cells. (C) Dose response for ibrutinib per CD79B expression level (mean ± SEM) (n = 3). (D) Dose response for fostamatinib per CD79B level (mean ± SEM) (n = 3).
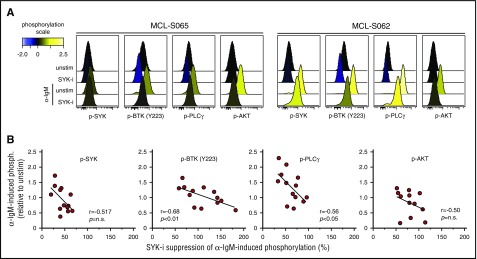
To test whether the relationship between α-IgM–signaling strength upon in vitro activation and efficacy of BCR-signaling inhibitors also was relevant for lymphoma cells from MCL patients, the efficacy of fostamatinib was tested in 13 MCL tumors. As shown for the MCL cell line, MCL cells from patients showed an inverse correlation between α-IgM–induced phosphorylation levels and drug–induced suppression (Figure 6). The correlation was strongest for phosphorylation of BTK (r = 0.68, P < .01) and PLCγ (r = 0.56, P < .05). A similar association was also found between basal phosphorylation levels and influence of fostamatinib (supplemental Figure 11). Hence, the BCR-signaling capacity might confound the efficacy of BCR-pathway inhibitory drugs.
Figure 6.
Efficacy of SYK-inhibitor fostamatinib to suppress α-BCR–induced signaling depends on BCR signaling strength in MCL tumors. Samples from 13 MCL patients were left untreated or were treated with the SYK-inhibitor fostamatinib (SYK-i, 2.5 μM) for 1 hour, prior to α-BCR stimulation for 4 minutes. The cells were stained with lineage-specific markers and phosphospecific Abs as earlier described. (A) Histogram overlays of p-SYK, p-BTK, p-PLCγ, and p-AKT for lymphoma cells treated with fostamatinib, α-IgM or α-IgM + fostamatinib, relative to unstimulated (untreated) cells. (B) Scatter plots of α-IgM–induced phosphorylation vs percentage of fostamatinib suppression of α-IgM–induced phosphorylation levels for SYK, PLCγ, BTK (Y223), and AKT. Each dot represents a MCL patient sample (n = 13). Correlations were calculated by the Spearman rank test.
Elevated basal levels are associated with shorter survival in MCL
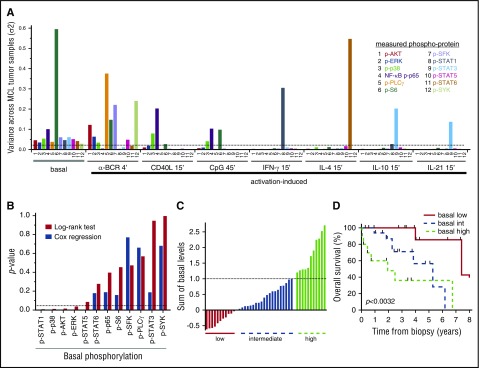
In FL, we previously demonstrated a pivotal role for α-BCR–induced signaling, as patients who harbored a large fraction of α-BCR–insensitive lymphoma cells were shown to have an inferior outcome.33 Therefore, basal- as well as activation-induced phosphorylation levels with highest variance across the MCL patients, which represented the largest cohort, were identified (Figure 7A; supplemental Table 2). Even though many of the activation-induced p-proteins showed high variance, including α-BCR–induced p-SYK and p-PLCγ, none of these reached statistical significance using the log-rank test. In contrast, elevated levels of basal p-STAT1, p-p38, p-AKT, and p-ERK were significantly associated with shorter survival (P < .05; Figure 7B). Cox proportional hazard regression confirmed that these, in addition to p-STAT5, were continuous variables (Figure 7B). Grouping of patients based on a single phosphoprotein value is likely to be less robust due to smaller differences than grouping of patients based on a combination of several p-proteins. Grouping the patients based on the combined sum of all significant basal levels from the Cox regression (p-AKT, p-ERK, p-p38, p-STAT1, and p-STAT5) into low, intermediate, and high levels translated into different overall survival (P < .0032), with median survival of 7.4, 5.3, and 1.9 years, respectively (Figure 7C-D). The Mantle Cell Lymphoma International Prognostic Index (MIPI) was available for a subset of patients, and was only weakly associated with the significant basal phosphoproteins (r from −0.01 to −0.25). Also, the statistical significance of the phosphoproteins was not changed upon inclusion of the MIPI in multiparameter Cox regression. These results will require validation with an independent data set, but suggest that measurements of basal phosphorylation levels are clinically relevant in MCL.
Figure 7.
Associations of signaling nodes and overall survival in MCL. (A) Identification of the basal- and activation-induced signaling nodes with largest variance across MCL samples. (B) For all individual signaling nodes above a variance threshold set to 0.025, the patients were divided into 2 groups, with measurements greater than or less than the cohorts median value, and association with patient overall survival were tested with the log-rank test. Shown are histograms of P values using log-rank test or Cox regression analysis of the same variable. (C) Distribution of the sum of the 5 significant basal levels as determined by Cox regression (p-AKT, p-ERK, p-STAT1, p-STAT5, and p-p38) across MCL patients. (D) Kaplan-Meier analysis of overall survival, based on the 3 groups identified in panel C (n = 42; P < .0032).
Discussion
The BCR-signaling pathway represents an attractive target in therapy for several types of B-cell lymphoma, and there is intensive search for surrogate markers that can serve as predictors of response and understand the molecular basis for differential responses seen after treatment with BCR-pathway inhibitors. We performed single-cell signaling profiling of malignant B cells from MCL, FL, DLBCL, and CLL, with main focus on MCL which represented our largest cohort. The smaller size of DLBCL and CLL cohorts prevented meaningful comparison of the subcategories of these, but this has been well described in previous reports.22-24,40-42,44-46 Taking an overall averaged view of these categories, we discovered striking differences in basal phosphorylation levels as well as activation-induced signaling across the lymphoma types. MCL cells showed highly variable α-BCR–induced signaling responses but in general had highly potentiated signaling as compared with the other lymphoma types. The potentiated α-BCR–induced signaling in MCL was strongly associated with surface expression of BCR subunits (IgM and CD79B). In contrast, CLL tumors differed from the other NHL by having very low α-BCR–induced signaling, corresponding well with overall low surface expression of CD79B in this lymphoma type.
By comparing per cell level of α-IgM–induced signaling across distinct IgM expression levels in a MCL cell line that possessed highly heterogeneous expression of CD79B and IgM, a direct relationship was demonstrated between surface CD79B, IgM, and α-IgM–induced signaling levels. Similarly, overexpression of CD79B further enhanced the surface expression of IgM and potentiated α-IgM–induced signaling, confirming a direct relationship between surface CD79B, IgM, and α-IgM–induced signaling levels. Interestingly, there seems to be a stronger correlation between surface expression of CD79B and IgM as compared with CD79B and IgG. For IgM surface expression to occur, both CD79A and CD79B must be present.47 The cytoplasmic tail of IgM contains only 3 amino acids, and the heterodimeric complex of CD79A and CD79B is required to bring IgM to the plasma membrane. In contrast, IgG has a longer cytoplasmic tail and can pair with CD79A, and is thus less dependent on CD79B for proper surface expression.48 Furthermore, targeted mutation of CD79B in mice prevented expression of µ heavy chain, but not CD79A, and produced a complete block in B-cell development at the immature B-cell stage.49 The underlying cause for CD79B overexpression in MCL is not known, but the expression levels seen in MCL cells are within the range of healthy donor mature B cells. The CD79B gene is mutated in about 20% of the ABC subtype of DLBCL with hotspot mutation in the first ITAM tyrosine (Y196).23,45 Several of the mutated variants result in increased surface expression of CD79B/Ig and formation of microclustering of BCR,23 similar to BCRs in antigen-stimulated normal B cells; however, these mutations have not been found in MCL.50,51 In CLL, some early reports identified mutations in CD79B exons 5 and 6,52 but this has not been confirmed in recent exome-sequencing studies.53,54 However, CD79B surface expression in CLL is commonly downregulated,55,56 but can be upregulated upon IL-4 signaling.57,58 The overall higher surface expression of CD79B in MCL, but not in CLL, suggests it may be a better target for therapy in MCL. Indeed, a recent clinical trial using anti-CD79B antibody conjugate as a single agent or combined with rituximab had no clinical effect in CLL, but had an objective response in 14 of 24 DLBCL patients, and 6 of 7 patients with MCL.59
The difference between CLL and MCL for α-BCR–induced signaling is striking, given that MCL and CLL are the 2 types of B-cell malignancies that respond clinically to the BTK inhibitor ibrutinib.25-28 Ibrutinib might work in CLL by downregulating the constitutive BCR signaling that seems important for survival of CLL cells, whereas ibrutinib may work in MCL by inhibiting the potentiated BCR signaling characteristic of this malignancy. In MCL patients treated with ibrutinib, the BCR and NF-κB gene signatures were efficiently downregulated as early as 2 days after treatment initiation.60 A pathogenic role of BCR signaling in MCL was further demonstrated by poor clinical outcome for patients with high LN BCR signature scores.61 In addition to its central role in BCR signal propagation, BTK is activated upon ligation of crucial coreceptors such as CD40 and B-cell–activating factor receptor, as well as TLRs and chemokine receptors.62,63 Hence, BTK inhibitors can have a profound effect by targeting multiple signaling pathways. Another important facet of ibrutinib is its immune-modulating effects. Ibrutinib has been shown to inhibit the highly homologous IL-2–inducible T-cell kinase (ITK). ITK is expressed by T helper 2 (Th2) cells, and ibrutinib has been shown to shift the balance from Th2 cells toward Th1 cells which produce IFNγ, thereby enhancing antitumor responses.64,65
In FL, we previously identified a lymphoma subset that was unresponsive to α-BCR–induced signaling, of which high prevalence was associated with inferior outcome.33 A similar relationship was not found in our MCL cohort, despite large interpatient difference in α-BCR–induced signaling. Instead, elevated basal levels of p-AKT, p-ERK, p-STAT1, pSTAT5, and p-p38 were associated with shorter survival. These results will require validation with an independent data set, but suggest that measurements of basal phosphorylation levels are clinically relevant in MCL. Furthermore, our findings suggest that the lymphoma cells’ basal phosphorylation levels and BCR-signaling capacity might confound the efficacy of BCR pathway inhibitory drugs. Differences in the phosphorylation levels of signaling molecules between different NHL, as well as heterogeneity within each entity, might help us understand therapy responses to small-molecular inhibitors. As the lymphoma cell level of CD79B was strongly correlated with α-BCR–induced signaling, measurement of CD79B may turn out to be the biomarker of the future. Such measurements can easily be implemented in clinical trials with BCR-pathway inhibitory drugs and warrant further investigation. Going forward, combinations of high-dimensional single-cell approaches, such as mass cytometry, and machine-learning computational analysis tools are expected to systematically reveal and characterize clinically relevant alterations in cancer cell signaling.66-68
Acknowledgments
J.H.M. was supported by the Norwegian Cancer Society, the Research Council of Norway, South East-Regional Health Authorities, and the Norwegian Radium Hospital legacies. J.M.I. was supported as a Leukemia & Lymphoma Society Fellow and by the National Institutes of Health (NIH) National Cancer Institute (NCI) (K99/R00 CA143231). This work was supported by the NIH NCI (CA 34233 and CA 33399), the Leukemia & Lymphoma Society, and the Integrative Cancer Biology Program (U56 CA112973).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: J.H.M., J.M.I., J.B., R.H., and R.L. designed the research; J.H.M., J.M.I., D.K.C., M.R.G., G.T., and S.W. performed experiments; J.H.M., J.M.I., A.A.A., K.L., and R.L. analyzed data; H.E.K. provided tonsil specimens; J.D. revised histopathological analysis of patient samples; K.B. scored immunohistochemistry stainings for DLBCL subtype classification; A.K., A.A.A., and R.L. provided patient samples; J.B., H.E.K., A.A.A., and A.K. provided clinical data; J.H.M., J.B., J.M.I., and R.L. wrote the paper; all authors approved the final manuscript; and R.L. supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Holbrook E. Kohrt died on 24 February 2016.
Correspondence: Ronald Levy, Division of Oncology, Stanford University Medical Center, 269 Campus Dr, CCSR 1105, Stanford, CA 94305-5151; e-mail: levy@stanford.edu.
References
- 1.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8(1):22-33. [DOI] [PubMed] [Google Scholar]
- 2.Shaffer AL, Rosenwald A, Staudt LM. Lymphoid malignancies: the dark side of B-cell differentiation. Nat Rev Immunol. 2002;2(12):920-932. [DOI] [PubMed] [Google Scholar]
- 3.Harris NL, Jaffe ES, Stein H, et al. A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood. 1994;84(5):1361-1392. [PubMed] [Google Scholar]
- 4.Dal Porto JM, Gauld SB, Merrell KT, Mills D, Pugh-Bernard AE, Cambier J. B cell antigen receptor signaling 101. Mol Immunol. 2004;41(6-7):599-613. [DOI] [PubMed] [Google Scholar]
- 5.Harwood NE, Batista FD. Early events in B cell activation. Annu Rev Immunol. 2010;28(1):185-210. [DOI] [PubMed] [Google Scholar]
- 6.Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K. Survival of resting mature B lymphocytes depends on BCR signaling via the Igalpha/beta heterodimer. Cell. 2004;117(6):787-800. [DOI] [PubMed] [Google Scholar]
- 7.Lam KP, Kühn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90(6):1073-1083. [DOI] [PubMed] [Google Scholar]
- 8.Monroe JG. ITAM-mediated tonic signalling through pre-BCR and BCR complexes. Nat Rev Immunol. 2006;6(4):283-294. [DOI] [PubMed] [Google Scholar]
- 9.Baba Y, Hashimoto S, Matsushita M, et al. BLNK mediates Syk-dependent Btk activation. Proc Natl Acad Sci USA. 2001;98(5):2582-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geahlen RL. Syk and pTyr’d: signaling through the B cell antigen receptor. Biochim Biophys Acta. 2009;1793(7):1115-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurosaki T, Hikida M. Tyrosine kinases and their substrates in B lymphocytes. Immunol Rev. 2009;228(1):132-148. [DOI] [PubMed] [Google Scholar]
- 12.Niiro H, Clark EA. Regulation of B-cell fate by antigen-receptor signals. Nat Rev Immunol. 2002;2(12):945-956. [DOI] [PubMed] [Google Scholar]
- 13.Shinohara H, Kurosaki T. Comprehending the complex connection between PKCbeta, TAK1, and IKK in BCR signaling. Immunol Rev. 2009;232(1):300-318. [DOI] [PubMed] [Google Scholar]
- 14.Young RM, Staudt LM. Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat Rev Drug Discov. 2013;12(3):229-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dühren-von Minden M, Übelhart R, Schneider D, et al. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature. 2012;489(7415):309-312. [DOI] [PubMed] [Google Scholar]
- 16.Zhu D, Bhatt S, Lu X, et al. Chlamydophila psittaci-negative ocular adnexal marginal zone lymphomas express self polyreactive B-cell receptors. Leukemia. 2015;29(7):1587-1599. [DOI] [PubMed] [Google Scholar]
- 17.Cha SC, Qin H, Kannan S, et al. Nonstereotyped lymphoma B cell receptors recognize vimentin as a shared autoantigen. J Immunol. 2013;190(9):4887-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coelho V, Krysov S, Ghaemmaghami AM, et al. Glycosylation of surface Ig creates a functional bridge between human follicular lymphoma and microenvironmental lectins. Proc Natl Acad Sci USA. 2010;107(43):18587-18592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sachen KL, Strohman MJ, Singletary J, et al. Self-antigen recognition by follicular lymphoma B-cell receptors. Blood. 2012;120(20):4182-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montesinos-Rongen M, Purschke FG, Brunn A, et al. Primary central nervous system (CNS) lymphoma B cell receptors recognize CNS proteins. J Immunol. 2015;195(3):1312-1319. [DOI] [PubMed] [Google Scholar]
- 21.Young RM, Wu T, Schmitz R, et al. Survival of human lymphoma cells requires B-cell receptor engagement by self-antigens. Proc Natl Acad Sci USA. 2015;112(44):13447-13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Compagno M, Lim WK, Grunn A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459(7247):717-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis RE, Ngo VN, Lenz G, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lenz G, Davis RE, Ngo VN, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319(5870):1676-1679. [DOI] [PubMed] [Google Scholar]
- 25.Advani RH, Buggy JJ, Sharman JP, et al. Bruton tyrosine kinase inhibitor ibrutinib (PCI-32765) has significant activity in patients with relapsed/refractory B-cell malignancies. J Clin Oncol. 2013;31(1):88-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Byrd JC, Brown JR, O’Brien S, et al. ; RESONATE Investigators. Ibrutinib versus ofatumumab in previously treated chronic lymphoid leukemia. N Engl J Med. 2014;371(3):213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Byrd JC, Furman RR, Coutre SE, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369(1):32-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang ML, Rule S, Martin P, et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N Engl J Med. 2013;369(6):507-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maddocks K, Christian B, Jaglowski S, et al. A phase 1/1b study of rituximab, bendamustine, and ibrutinib in patients with untreated and relapsed/refractory non-Hodgkin lymphoma. Blood. 2015;125(2):242-248. [DOI] [PubMed] [Google Scholar]
- 30.Blix ES, Irish JM, Husebekk A, et al. Altered BCR and CD40 signalling are associated with clinical outcome in small lymphocytic lymphoma/chronic lymphocytic leukaemia and marginal zone lymphoma patients. Br J Haematol. 2012;159(5):604-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irish JM, Czerwinski DK, Nolan GP, Levy R. Altered B-cell receptor signaling kinetics distinguish human follicular lymphoma B cells from tumor-infiltrating nonmalignant B cells. Blood. 2006;108(9):3135-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Irish JM, Hovland R, Krutzik PO, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118(2):217-228. [DOI] [PubMed] [Google Scholar]
- 33.Irish JM, Myklebust JH, Alizadeh AA, et al. B-cell signaling networks reveal a negative prognostic human lymphoma cell subset that emerges during tumor progression. Proc Natl Acad Sci USA. 2010;107(29):12747-12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myklebust JH, Irish JM, Brody J, et al. High PD-1 expression and suppressed cytokine signaling distinguish T cells infiltrating follicular lymphoma tumors from peripheral T cells. Blood. 2013;121(8):1367-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irish JM, Czerwinski DK, Nolan GP, Levy R. Kinetics of B cell receptor signaling in human B cell subsets mapped by phosphospecific flow cytometry. J Immunol. 2006;177(3):1581-1589. [DOI] [PubMed] [Google Scholar]
- 36.Huse K, Bakkebø M, Wälchli S, et al. Role of Smad proteins in resistance to BMP-induced growth inhibition in B-cell lymphoma. PLoS One. 2012;7(10):e46117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wälchli S, Løset GÅ, Kumari S, et al. A practical approach to T-cell receptor cloning and expression. PLoS One. 2011;6(11):e27930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi WWL, Weisenburger DD, Greiner TC, et al. A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res. 2009;15(17):5494-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275-282. [DOI] [PubMed] [Google Scholar]
- 40.Lanham S, Hamblin T, Oscier D, Ibbotson R, Stevenson F, Packham G. Differential signaling via surface IgM is associated with VH gene mutational status and CD38 expression in chronic lymphocytic leukemia. Blood. 2003;101(3):1087-1093. [DOI] [PubMed] [Google Scholar]
- 41.Mockridge CI, Potter KN, Wheatley I, Neville LA, Packham G, Stevenson FK. Reversible anergy of sIgM-mediated signaling in the two subsets of CLL defined by VH-gene mutational status. Blood. 2007;109(10):4424-4431. [DOI] [PubMed] [Google Scholar]
- 42.Guarini A, Chiaretti S, Tavolaro S, et al. BCR ligation induced by IgM stimulation results in gene expression and functional changes only in IgV H unmutated chronic lymphocytic leukemia (CLL) cells. Blood. 2008;112(3):782-792. [DOI] [PubMed] [Google Scholar]
- 43.Herishanu Y, Pérez-Galán P, Liu D, et al. The lymph node microenvironment promotes B-cell receptor signaling, NF-kappaB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood. 2011;117(2):563-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503-511. [DOI] [PubMed] [Google Scholar]
- 45.Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevenson FK, Krysov S, Davies AJ, Steele AJ, Packham G. B-cell receptor signaling in chronic lymphocytic leukemia. Blood. 2011;118(16):4313-4320. [DOI] [PubMed] [Google Scholar]
- 47.Hombach J, Tsubata T, Leclercq L, Stappert H, Reth M. Molecular components of the B-cell antigen receptor complex of the IgM class. Nature. 1990;343(6260):760-762. [DOI] [PubMed] [Google Scholar]
- 48.Todo K, Koga O, Nishikawa M, Hikida M. IgG1 cytoplasmic tail is essential for cell surface expression in Igβ down-regulated cells. Biochem Biophys Res Commun. 2014;445(3):572-577. [DOI] [PubMed] [Google Scholar]
- 49.Gong S, Nussenzweig MC. Regulation of an early developmental checkpoint in the B cell pathway by Ig beta. Science. 1996;272(5260):411-414. [DOI] [PubMed] [Google Scholar]
- 50.Beà S, Valdés-Mas R, Navarro A, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci USA. 2013;110(45):18250-18255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J, Jima D, Moffitt AB, et al. The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood. 2014;123(19):2988-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson AA, Talley JA, Do HN, et al. Aberrations of the B-cell receptor B29 (CD79b) gene in chronic lymphocytic leukemia. Blood. 1997;90(4):1387-1394. [PubMed] [Google Scholar]
- 53.Landau DA, Tausch E, Taylor-Weiner AN, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526(7574):525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quesada V, Conde L, Villamor N, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2011;44(1):47-52. [DOI] [PubMed] [Google Scholar]
- 55.Cabezudo E, Carrara P, Morilla R, Matutes E. Quantitative analysis of CD79b, CD5 and CD19 in mature B-cell lymphoproliferative disorders. Haematologica. 1999;84(5):413-418. [PubMed] [Google Scholar]
- 56.Damle RN, Ghiotto F, Valetto A, et al. B-cell chronic lymphocytic leukemia cells express a surface membrane phenotype of activated, antigen-experienced B lymphocytes. Blood. 2002;99(11):4087-4093. [DOI] [PubMed] [Google Scholar]
- 57.Aguilar-Hernandez MM, Blunt MD, Dobson R, et al. IL-4 enhances expression and function of surface IgM in CLL cells. Blood. 2016;127(24):3015-3025. [DOI] [PubMed] [Google Scholar]
- 58.Guo B, Zhang L, Chiorazzi N, Rothstein TL. IL-4 rescues surface IgM expression in chronic lymphocytic leukemia. Blood. 2016;128(4):553-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palanca-Wessels MC, Czuczman M, Salles G, et al. Safety and activity of the anti-CD79B antibody-drug conjugate polatuzumab vedotin in relapsed or refractory B-cell non-Hodgkin lymphoma and chronic lymphocytic leukaemia: a phase 1 study. Lancet Oncol. 2015;16(6):704-715. [DOI] [PubMed] [Google Scholar]
- 60.Chang BY, Francesco M, De Rooij MFM, et al. Egress of CD19(+)CD5(+) cells into peripheral blood following treatment with the Bruton tyrosine kinase inhibitor ibrutinib in mantle cell lymphoma patients. Blood. 2013;122(14):2412-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saba NS, Liu D, Herman SEM, et al. Pathogenic role of B-cell receptor signaling and canonical NF-κB activation in mantle cell lymphoma. Blood. 2016;128(1):82-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hendriks RW, Yuvaraj S, Kil LP. Targeting Bruton’s tyrosine kinase in B cell malignancies. Nat Rev Cancer. 2014;14(4):219-232. [DOI] [PubMed] [Google Scholar]
- 63.Ponader S, Burger JA. Bruton’s tyrosine kinase: from X-linked agammaglobulinemia toward targeted therapy for B-cell malignancies. J Clin Oncol. 2014;32(17):1830-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dubovsky JA, Beckwith KA, Natarajan G, et al. Ibrutinib is an irreversible molecular inhibitor of ITK driving a Th1-selective pressure in T lymphocytes. Blood. 2013;122(15):2539-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sagiv-Barfi I, Kohrt HE, Czerwinski DK, Ng PP, Chang BY, Levy R. Therapeutic antitumor immunity by checkpoint blockade is enhanced by ibrutinib, an inhibitor of both BTK and ITK. Proc Natl Acad Sci USA. 2015;112(9):E966-E972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Diggins KE, Ferrell PB Jr, Irish JM. Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data. Methods. 2015;82:55-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Irish JM. Beyond the age of cellular discovery. Nat Immunol. 2014;15(12):1095-1097. [DOI] [PubMed] [Google Scholar]
- 68.Levine JH, Simonds EF, Bendall SC, et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162(1):184-197. [DOI] [PMC free article] [PubMed] [Google Scholar]