Abstract
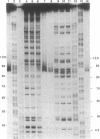
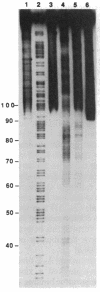
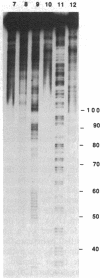
Metal complexes that bind to DNA on the basis of shape-selection have been used to map the conformational features of the DNA binding site for transcription factor IIIA. Conformationally distinct segments are detected on the 5S rRNA gene that correspond closely to the binding sites identified for the individual zinc finger domains of the protein. The local conformations are characterized by a major groove opened because of a change in base pair inclination and/or displacement at a central 5'-pyrimidine-purine-3' step, flanked by a widened minor groove, as would arise at the junctions between alternating B- and A-like DNA segments. Docking experiments with a consensus structure of a zinc finger reveal that the mixed A-B binding site accommodates the peptide domain better than either canonical B- or A-DNA helices. The close structural matching of the conformational variations in the 5S rDNA both to the proposed sites of zinc finger binding and to the shape of an individual zinc finger domain points to DNA structural polymorphism as providing an important determinant in recognition. In particular, shape selection in the 5' half of the internal control region may orient the multiple finger domains.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aboul-ela F., Varani G., Walker G. T., Tinoco I., Jr The TFIIIA recognition fragment d(GGATGGGAG).d(CTCCCATCC) is B-form in solution. Nucleic Acids Res. 1988 Apr 25;16(8):3559–3572. doi: 10.1093/nar/16.8.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. E., Ptashne M., Harrison S. C. Structure of the repressor-operator complex of bacteriophage 434. 1987 Apr 30-May 6Nature. 326(6116):846–852. doi: 10.1038/326846a0. [DOI] [PubMed] [Google Scholar]
- Barton J. K. Metals and DNA: molecular left-handed complements. Science. 1986 Aug 15;233(4765):727–734. doi: 10.1126/science.3016894. [DOI] [PubMed] [Google Scholar]
- Berg J. M. Proposed structure for the zinc-binding domains from transcription factor IIIA and related proteins. Proc Natl Acad Sci U S A. 1988 Jan;85(1):99–102. doi: 10.1073/pnas.85.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill M. E., Tullius T. D., Klug A. Mode of interaction of the zinc finger protein TFIIIA with a 5S RNA gene of Xenopus. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5528–5532. doi: 10.1073/pnas.87.14.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dervan P. B. Design of sequence-specific DNA-binding molecules. Science. 1986 Apr 25;232(4749):464–471. doi: 10.1126/science.2421408. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Fairall L., Martin S., Rhodes D. The DNA binding site of the Xenopus transcription factor IIIA has a non-B-form structure. EMBO J. 1989 Jun;8(6):1809–1817. doi: 10.1002/j.1460-2075.1989.tb03575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairall L., Rhodes D., Klug A. Mapping of the sites of protection on a 5 S RNA gene by the Xenopus transcription factor IIIA. A model for the interaction. J Mol Biol. 1986 Dec 5;192(3):577–591. doi: 10.1016/0022-2836(86)90278-0. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Blanco J., Tennant L. L. The 5S gene internal control region is B-form both free in solution and in a complex with TFIIIA. Nature. 1987 Oct 1;329(6138):460–462. doi: 10.1038/329460a0. [DOI] [PubMed] [Google Scholar]
- Huber P. W., Blobe G. C., Hartmann K. M. Conformational studies of the nucleic acid binding sites for Xenopus transcription factor IIIA. J Biol Chem. 1991 Feb 15;266(5):3278–3286. [PubMed] [Google Scholar]
- Kirshenbaum M. R., Tribolet R., Barton J. K. Rh(DIP)3(3+): a shape-selective metal complex which targets cruciforms. Nucleic Acids Res. 1988 Aug 25;16(16):7943–7960. doi: 10.1093/nar/16.16.7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S., Gippert G. P., Soman K. V., Case D. A., Wright P. E. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science. 1989 Aug 11;245(4918):635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- Majowski K., Mentzel H., Pieler T. A split binding site for TFIIIC on the Xenopus 5S gene. EMBO J. 1987 Oct;6(10):3057–3063. doi: 10.1002/j.1460-2075.1987.tb02612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCall M., Brown T., Hunter W. N., Kennard O. The crystal structure of d(GGATGGGAG): an essential part of the binding site for transcription factor IIIA. Nature. 1986 Aug 14;322(6080):661–664. doi: 10.1038/322661a0. [DOI] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- Mei H. Y., Barton J. K. Tris(tetramethylphenanthroline)ruthenium(II): a chiral probe that cleaves A-DNA conformations. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1339–1343. doi: 10.1073/pnas.85.5.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P. E., Møllegaard N. E., Jeppesen C. DNA conformational analysis in solution by uranyl mediated photocleavage. Nucleic Acids Res. 1990 Jul 11;18(13):3847–3851. doi: 10.1093/nar/18.13.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Sep 22;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Pieler T., Hamm J., Roeder R. G. The 5S gene internal control region is composed of three distinct sequence elements, organized as two functional domains with variable spacing. Cell. 1987 Jan 16;48(1):91–100. doi: 10.1016/0092-8674(87)90359-x. [DOI] [PubMed] [Google Scholar]
- Párraga G., Horvath S. J., Eisen A., Taylor W. E., Hood L., Young E. T., Klevit R. E. Zinc-dependent structure of a single-finger domain of yeast ADR1. Science. 1988 Sep 16;241(4872):1489–1492. doi: 10.1126/science.3047872. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. An underlying repeat in some transcriptional control sequences corresponding to half a double helical turn of DNA. Cell. 1986 Jul 4;46(1):123–132. doi: 10.1016/0092-8674(86)90866-4. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D. Contact points between a positive transcription factor and the Xenopus 5S RNA gene. Cell. 1982 Dec;31(2 Pt 1):395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D., Engelke D., Ng S. Y., Shastry B. S., Roeder R. G. The binding of a transcription factor to deletion mutants of a 5S ribosomal RNA gene. Cell. 1981 Mar;23(3):665–669. doi: 10.1016/0092-8674(81)90429-3. [DOI] [PubMed] [Google Scholar]
- Smith D. R., Jackson I. J., Brown D. D. Domains of the positive transcription factor specific for the Xenopus 5S RNA gene. Cell. 1984 Jun;37(2):645–652. doi: 10.1016/0092-8674(84)90396-9. [DOI] [PubMed] [Google Scholar]
- Vrana K. E., Churchill M. E., Tullius T. D., Brown D. D. Mapping functional regions of transcription factor TFIIIA. Mol Cell Biol. 1988 Apr;8(4):1684–1696. doi: 10.1128/mcb.8.4.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]