Highlight
SgPAP7, SgPAP10, and SgPAP26 may differentially contribute to root-associated APase activities, and thus control extracellular dNTP utilization in stylo.
Key words: Acid phosphatase activities, dNTP, organic phosphorus, phosphate starvation, purple acid phosphatase, Stylosanthes.
Abstract
Stylo (Stylosanthes spp.) is a pasture legume predominant in tropical and subtropical areas, where low phosphorus (P) availability is a major constraint for plant growth. Therefore, stylo might exhibit superior utilization of the P pool on acid soils, particularly organic P. However, little is known about mechanisms of inorganic phosphate (Pi) acquisition employed by stylo. In this study, the utilization of extracellular deoxy-ribonucleotide triphosphate (dNTP) and the underlying physiological and molecular mechanisms were examined for two stylo genotypes with contrasting P efficiency. Results showed that the P-efficient genotype, TPRC2001-1, was superior to the P-inefficient genotype, Fine-stem, when using dNTP as the sole P source. This was reflected by a higher dry weight and total P content for TPRC2001-1 than for Fine-stem, which was correlated with higher root-associated acid phosphatase (APase) activities in TPRC2001-1 under low P conditions. Subsequently, three PAP members were cloned from TPRC2001-1: SgPAP7, SgPAP10, and SgPAP26. Expression levels of these three SgPAPs were up-regulated by Pi starvation in stylo roots. Furthermore, there was a higher abundance of transcripts of SgPAP7 and SgPAP10 in TPRC2001-1 than in Fine-stem. Subcellular localization analysis demonstrated that these three SgPAPs were localized on the plasma membrane. Overexpression of these three SgPAPs could result in significantly increased root-associated APase activities, and thus extracellular dNTP utilization in bean hairy roots. Taken together, the results herein suggest that SgPAP7, SgPAP10, and SgPAP26 may differentially contribute to root-associated APase activities, and thus control extracellular dNTP utilization in stylo.
Introduction
Phosphorus (P) is an essential macronutrient for plant growth and development. Inorganic phosphate (Pi) is the major form of P acquired by plants directly from soil (Marschner, 1995). However, Pi is easily fixed by soil particles and microorganisms, thus turning into unavailable forms (e.g. Fe/Al-P complexes and organic P), which cannot be directly utilized by plants (Lambers and Plaxton, 2015). It has been estimated that 30–65% of total P in soils is present as organic P, mainly in the form of phytate, nucleic acids, and phospholipids (Shen et al., 2011; Stutter et al., 2012). As a result, low Pi availability is a major constraint limiting plant growth and production on both natural and agricultural soils (Vance et al., 2003; López-Arredondo et al., 2014).
Plant adaptations to improve Pi acquisition and utilization under conditions of P deficiency lie within a wide-ranging set of available morphological, physiological, and molecular strategies, such as modification of root morphology and architecture (Lynch, 2015), increased organic acid and acid phosphatase (APase) secretion (Plaxton and Tran, 2011; Shen et al., 2011; Song and Liu, 2015; Tian and Liao, 2015), and enhanced high-affinity Pi transporter expression (Qin et al., 2012; Zhang et al., 2015). It has recently been established that the adaptive strategies listed above are tightly and coordinately mediated through a variety of regulators in complex P signalling networks (Chiou and Lin, 2011; Wu et al., 2013; Liang et al., 2014). Among the adaptive responses, increased APase activities and exudation is generally considered as a means for plants to mobilize and utilize organic P (Tran et al., 2010a; Tian and Liao, 2015).
Plant APases (EC 3.1.3.2) belong to the hydrolase enzymes, which catalyse phosphate from a group of phosphomonoesters and anhydrides with optimal activity pH below 7.0 (Duff et al., 1994). Among the APases, purple acid phosphatase (PAP) has distinctive biochemical and molecular characteristics, including a purple or pink colour in aqueous solution, activity that is insensitive to inhibition by L-tartrate, a bimetallic active site, and the presence of seven invariant residues in five conserved metal ligating motifs (Bozzo et al., 2004; Schenk et al., 2013; Matange et al., 2015; Tian and Liao, 2015). PAPs have been widely identified and characterized from a range of organisms, including mammals, plants, fungi, and bacteria (Schenk et al., 2000a, 2000b, 2013; Flanagan et al., 2006). Despite low homology between PAPs from different kingdoms, five conserved motifs have been identified, including DXG, GDXXY, GNH(D/E), VXXH, and GHXH (Oddie et al., 2000; Schenk et al., 2000a, 2000b, 2013; Flanagan et al., 2006; Mitić et al., 2006). Within the conserved motifs, these seven invariant residues shown in bold and italic have been suggested to participate in the formation of a binuclear centre by coordinating with Fe3+ and other divalent metal ions, such as Fe2+, Mn2+, and Zn2+ (Beck et al., 1986, 1988; Sträter et al., 1995; Schenk et al., 1999, 2005; Mitić et al., 2006). Unlike animal PAPs, the second divalent metal ion in non-recombinant plant PAP members varies among plant species. In red kidney bean (Phaseolus vulgaris), soybean (Glycine max), and sweet potato (Ipomoea batatas), PAPs containing a Fe3+–Zn2+ centre have been identified (Beck et al., 1986; Durmus, et al.,1999a; Schenk et al., 1999). Another PAP from sweet potato contains a Fe3+–Mn2+ centre (Schenk et al., 1999, 2001, 2005). Biochemical analysis of recombinant and non-recombinant PAPs has documented that most plant PAPs can catalyse the hydrolysis of a wide range of phosphorylated compounds both natural and synthetic, such as ρ-nitrophenyl phosphate, energetic compounds (e.g. ATP and inorganic pyrophosphate), phosphorylated sugars, and phosphorylated amino acids (Tran et al., 2010a; Tian and Liao, 2015). However, NtPAP from tobacco (Nicotiana tabacum) and AtPAP15 from Arabidopsis (Arabidopsis thaliana) exhibit relatively high rates of activities with phytate as the substrate (Lung et al., 2008; Zhang et al., 2008). Among plant pioneer PAP studies, one PAP from kidney bean has been well characterized from its protein structure to catalytic properties (Beck et al., 1986, 1988; Klabunde et al., 1994; Sträter et al., 1995). Subsequently, other plant PAPs have been purified, and their amino acid sequences elucidated in plant species including soybean, sweet potato, and Arabidopsis (Del Pozo et al., 1999; Durmus et al., 1999b; Schenk et al., 1999, 2000a). The conserved sequence patterns of PAPs facilitated the identification of a large number of novel PAP and PAP-like sequences in plants (Schenk et al., 2000a; Li et al., 2002). Furthermore, with the availability of genomic sequences for diverse plant species, PAP members have now been widely identified and characterized in plants such as Arabidopsis, rice (Oryza sativa), soybean, and maize (Zea mays). Phosphate starvation has been documented to increase PAP expression levels for 11 out of 29 members in Arabidopsis, 10 out of 26 in rice, 23 out of 35 in soybean, and 11 out of 33 in maize (Li et al., 2002; Zhang et al., 2011; Li et al., 2012a; Wang et al., 2014; González-Muñoz et al., 2015). These results suggest that many plant PAP members are involved in P scavenging and recycling under conditions of P deficiency. This hypothesis is supported by functional analysis results for a number of plant PAP members. For example, overexpression of AtPAP15 from Arabidopsis in soybean, MtPHY1 from Medicago truncatula in white clover (Trifolium repens), and OsPHY1 from rice in tobacco could significantly increase transgenic plant dry weight and total P content when phytate was supplied as the sole P source (Ma et al., 2009; Wang et al., 2009; Li et al., 2012b). In addition, several other PAP members appear to participate in the utilization of other forms of extracellular organic P, such as ATP, ADP, and dNTP (Liang et al., 2010, 2012; Wang et al., 2011; Robinson et al., 2012). These results strongly suggest that root-secreted PAP has a pivotal role in the scavenging and recycling of extracellular P. Still, comprehensive dissection of PAP functions remains elusive owing to the variation in gene and protein structures, regulation of expression at multiple levels (i.e. transcriptional and post-translational), and the localization in a wide range of subcellular and extracellular compartments (Tian and Liao, 2015). The ability of plant PAPs to serve in multiple and complex functions is evident in recent findings, such as participation of GmPAP3 in salt stress tolerance in soybean (Liao et al., 2003; Li et al., 2008), NtPAP12 in cell wall biosynthesis in tobacco (Kaida et al., 2009), and AtPAP2 in carbon metabolism and AtPAP5 in biotic stress resistance (Sun et al., 2012; Ravichandran et al., 2013; Law et al., 2015). These results suggest that integrating all available knowledge of PAP properties will be critical for further understanding the complex range of PAP functions related to P scavenging and recycling in plants (Tian and Liao, 2015).
Stylo (Stylosanthes spp.) is a dominant pasture legume that is widely grown in tropical and subtropical areas, where acid soils are widely distributed (Chandra, 2009). On acid soils, P deficiency, aluminium (Al) toxicity, and manganese (Mn) toxicity are considered as major constraints for crop growth and production (Kochian et al., 2004). Therefore, it is possible that stylo is capable of overcoming these constraints to grow on acid soils. Recently, it has been documented that malate synthesis and exudation in stylo allow it to tolerate both Al and Mn toxicity (Sun et al., 2014; Chen et al., 2015). Even so, wide variation in P efficiency has been observed among stylo genotypes (Du et al., 2009). To date, physiological and molecular mechanisms underlying stylo adaptation to P deficiency remain largely unknown. In this study, dNTP utilization was investigated between two stylo genotypes with contrasting P efficiency. Subsequently, three SgPAP members were cloned and characterized to elucidate molecular mechanisms of dNTP utilization in stylo.
Materials and methods
Plant material and growth conditions
Two Stylosanthes guianensis Aubl. genotypes with contrasting P efficiency, TPRC2001-1 (P-efficient genotype) and Fine-stem (P-inefficient genotype), were used in this study. Seeds of stylo were soaked in 80°C water for 2min before being surface sterilized in 10% (v/v) sodium hypochlorite solution and sown in solidified Murashige and Skoog (MS) medium lacking KH2PO4. After germination, seedlings with 1 cm-long roots were transferred to 1.2mM KH2PO4 (+P), 0.4mM dNTP (dNTP), or no added P (−P) treatments on solid MS medium. For dNTP treatments, the medium was autoclaved and supplemented with 0.4mM filter sterilized dNTP, containing 0.1mM each of dATP, dGTP, dTTP, and dCTP. Plates were placed vertically in growth chambers with a 16/8h (light/dark) photoperiod (100 μmol m−2 s−1 photosynthetically active radiation) at 23°C. After 7 d growth, shoots and roots were separately harvested to analyse dry weight, soluble Pi concentration, APase activities, and total P content. Soluble Pi concentration and total P content were assessed using the phosphorus-molybdate blue colour reaction method according to Murphy and Riley (1962).
Cloning and characterization of SgPAP7, SgPAP10, and SgPAP26
To clone homologues of PvPAP3, AtPAP10, and AtPAP26 in stylo, primers (SgPAP7/10/26-EST-F and SgPAP7/10/26-EST-R, Supplementary Table S1) were designed separately according to the conserved motif sequences of PvPAP3 (NCBI accession FJ464333.1), AtPAP10 (NCBI accession NM_127196.3), AtPAP26 (NCBI accession NM_122874.3), and three GmPAPs (NCBI accessions NM_001254279.1, NM_001253997.1, NM_001249748.1). Three fragments were successfully amplified by PCR from the cDNA of TPRC2001-1 plants subjected to P deficiency, and then cloned into the pMD18-T vector (Takara, Japan). Subsequently, the 5′ and 3′ terminals of the three PAP fragments were amplified from a TPRC2001-1 RACE cDNA library constructed with the SMARTer™ RACE cDNA amplification kit (Clontech, USA) using specific primers paired with the universal primers RACE-UPM and RACE-NUP (Supplementary Table S1). After separately cloning 5′-terminal and 3′-terminal cDNA products into the pMD18-T vector, full-length sequences of SgPAP7, SgPAP10, and SgPAP26 were generated through the MEGA 5 program. The nucleotide sequences of these three SgPAPs were deposited in NCBI and assigned the accession numbers KU315544, KU315545, and KU315546 for SgPAP7, SgPAP10, and SgPAP26, respectively. Multiple sequence alignments and phylogenetic trees were constructed in ClusterX and MEGA 5, respectively. Signal peptides were predicted by SignalP 4.1 (http://www.cbs.dtu.dk/services). The transmembrane topology of the proteins was predicted by MEMSATSVM (http://bioinf.cs.ucl.ac.uk/web_servers/).
APase activity analysis
Acid phosphatase activity assays were performed as described by Liang et al. (2012). Briefly, to measure internal APase activities, protein extracted from plant tissues was mixed with 2mL of 45mM Na-acetate buffer (pH 5.0) containing 1mM ρ-nitrophenyl phosphate (ρ-NPP; Sigma, USA). After incubation at 37°C for 15min, the reaction was stopped by adding 1mL of 1M NaOH. APase activities were measured spectrophotometrically as absorbance at 405nm. Soluble protein concentration was analysed using the Coomassie Brilliant Blue method (Bradford, 1976). APase activities are presented as micromoles of ρ-NPP hydrolysed per milligram of soluble protein per minute.
For quantitative analysis of root-associated APase activities, roots of stylo or transgenic bean hairy roots were incubated in 45mM Na-acetate buffer containing 2mM ρ-NPP (pH 5.0). After incubation for 30min, the reaction was terminated by adding 1mL of 1M NaOH, and the absorbance of the reaction mix was measured at 405nm. Root-associated APase activities are expressed as micromoles of ρ-NPP hydrolysed per hour per centimetre of root.
For root-associated APase activity staining, transgenic bean hairy roots were placed on solid MS medium. These roots were then evenly overlaid with 0.5% (w/v) agar solution containing 0.02% (w/v) of the substrate 5-bromo-4-chloro-3-indolyl-phosphate (BCIP; Sigma, USA) for 2h at 25°C. Root-associated APase activities were visualized as the intensity of blue colour along root surfaces after BCIP hydrolysation (Lloyd et al., 2001; Wang et al., 2011). Images were captured using a single lens reflex camera (Canon, Japan). Root-associated APase activities were also investigated using ELF-97 phosphate as a substrate according to previous methods (Robinson et al., 2012). Briefly, transgenic bean hairy roots were rinsed three to five times with 45mM Na-acetate buffer (pH 5.0) and incubated in this buffer with 100 µM ELF-97 phosphate (Invitrogen, USA) for 30min at 25°C. After incubation, transgenic bean hairy roots were rinsed three times with 45mM Na-acetate buffer (pH 5.0) containing 25mM EDTA for 15min. The green fluorescent product of root-associated APase activities was imaged using a fluorescence microscope (Leica, Germany) with excitation at 345nm, and emission at 530nm. Images of root-associated APase activity staining were collected from three independent replicates, and representative results are shown.
Subcellular localization of SgPAPs
To analyse the subcellular localization of SgPAP7, SgPAP10, and SgPAP26, the open reading frame (ORF) sequences of SgPAP7, SgPAP10, and SgPAP26 without stop codons were amplified from cDNA stock of TPRC2001-1 with specific primers (Supplementary Table S1). Subsequently, these three SgPAP sequences were separately cloned into the binary vector pLGFP and fused with GFP at the C-terminus. The SgPAP-GFP fusion vectors and GFP empty constructs were separately introduced into Agrobacterium tumefaciens strain GV3101. Transformed GV3101 cells were grown overnight at 28°C in liquid yeast extract peptone (YEP) medium, and suspended to an absorbance of 0.5 at 600nm in agroinfiltration buffer (pH 5.6), containing 10mM MES, 10mM MgCl2, and 0.15mM acetosyringone. After incubating for 3h at 25°C in the dark, equal volumes of mixed suspensions were syringe-infiltrated into the abaxial side of near-fully expanded leaves of 5–6-week-old tobacco (Nicotiana benthamiana) plants. After 3 d, epidermal cells on the abaxial leaf side were imaged on a Zeiss LSM7 DUO confocal microscope (Zeiss, Germany). Co-localization experiments were performed using the mCherry-labelled plasma membrane marker AtPIP2A-mCherry.
For subcellular localization of SgPAPs in transgenic bean hairy roots, the SgPAP-GFP fusion and GFP empty constructs were separately transformed into Agrobacterium rhizogenes strain K599, which were then used to generate transgenic bean hairy roots with SgPAP-GFP or GFP overexpression. Transgenic bean hairy roots were generated as described by Liang et al. (2012). Confocal images were taken on a Zeiss LSM7 DUO confocal microscope (Zeiss, Germany). GFP fluorescence was stimulated at 488nm and detected with filter sets at 500–530nm. For western blot analysis, soluble proteins and membrane proteins were separately extracted using the FOCUS™ Global Fractionation kit (G-Biosciences, USA) from transgenic hairy roots, which were generated as above. Western blotting analysis was carried out as described by Liang et al. (2010). Briefly, extracted proteins were separated by 10% SDS-PAGE in a Mini-PROTEAN Tetra Cell (Bio-Rad, USA) and transferred onto polyvinylidene difluoride membranes (GE Healthcare Life Sciences, USA) using a Trans-Blot Cell (Bio-Rad, USA). Subsequently, the membranes were incubated overnight in 0.01M Tris buffer (pH 8.0) containing 5% (w/v) non-fat milk powder and 0.15M NaCl. Membranes were separately probed with a GFP antibody (HuaAn Biotechnology, China), a phosphoenolpyruvate carboxylase antibody (Agrisera, Sweden), and a plasma membrane proton ATPase antibody (Agrisera, Sweden). Antigenic polypeptides were visualized using alkaline-phosphatase-tagged secondary antibodies and BCIP/NBT substrates. For localization in Arabidopsis protoplasts, the SgPAP-GFP fusion or GFP empty constructs were transiently expressed in Arabidopsis mesophyll protoplasts following the methods of Wu et al. (2009). The GFP fusion constructs were co-transfected with the plasma membrane marker AtPIP2A-mCherry. Confocal images were taken on a Zeiss LSM7 DUO confocal microscope (Zeiss, Germany). GFP fluorescence was stimulated at 488nm and detected with filter sets at 500–530nm. The mCherry fluorescence was excited at 568nm and emission captured at 580–630nm.
Quantitative real-time PCR analysis
Total RNA was extracted using TRIzol reagent (Invitrogen, USA). DNase I-treated RNA (2 µg) was used for first-strand cDNA synthesis by M-MLV reverse transcriptase (Promega, USA) according to the manufacturer’s manual. The quantitative real-time PCR (qRT-PCR) analysis was carried out using SYBR Premix ExTaq II (Takara, Japan) on the Rotor-Gene 3000 qRT-PCR system (Corbett Research, Australia). The housekeeping genes SgEF-1a (accession number, JX164254) and PvEF-1a (PvTC3216 from the Dana-Farber Cancer Institute Computational Biology and Functional Genomics Laboratory) were used as internal controls to normalize gene expression for stylo and bean hairy roots, respectively. The primer pairs for amplification of SgPAPs and the housekeeping gene used for qRT-PCR analysis are listed in Supplementary Table S1. All of the gene expression analyses had four biological replicates.
Functional analysis of SgPAPs in bean hairy roots
To overexpress SgPAP7, SgPAP10, and SgPAP26 in bean hairy roots, the ORFs of SgPAP7, SgPAP10, and SgPAP26 were amplified from cDNA stock of TPRC2001-1 using gene-specific primers (see Supplementary Table S1), and then cloned into the pYLRNAi binary vector. The constructed vectors were transferred into Agrobacterium rhizogenes strain K599, which was used for bean hairy roots transformation and induction as described previously by Liang et al. (2012). Transgenic hairy roots were verified by qRT-PCR analysis.
In the dNTP utilization experiments, approximately 0.1g (fresh weight) of hairy roots was cultured in solid MS medium supplied with 0 µM KH2PO4 (−P), 1.2mM KH2PO4 (+P), or 0.4mM dNTP (i.e. 0.1mM each of dATP, dGTP, dTTP, and dCTP) as the P sources. After 14 d growth, transgenic bean hairy roots were photographed using a single lens reflex camera (Canon, Japan). Fresh weight and total P content of each transgenic line were determined as described above. Each treatment had four biological replicates.
Statistical analysis
All the data were analysed by ANOVA or t-test using SPSS software (version 18.0, SPSS Institute, USA).
Results
Utilization of exogenous dNTP in stylo
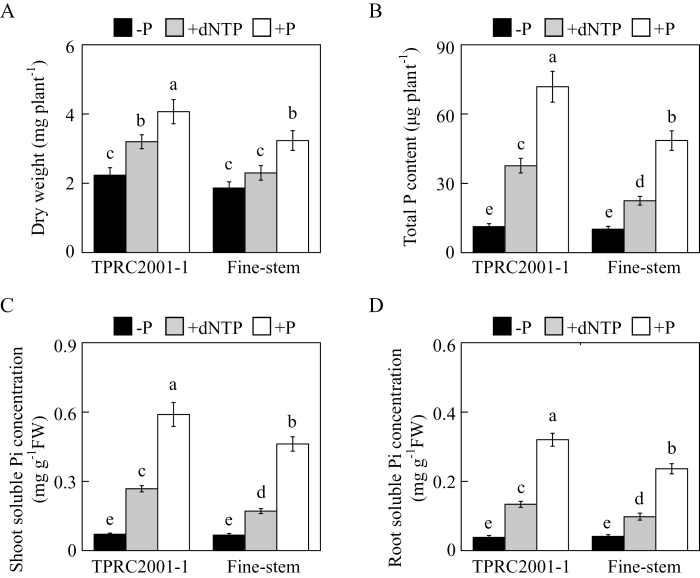
A P-efficient genotype, TPRC2001-1, and a P-inefficient genotype, Fine-stem, of stylo were used to investigate the capacity of genotypes to utilize extracellular dNTP. As expected, plant growth was significantly affected by P source (Fig. 1). Stylo growth was significantly inhibited, with the lowest plant dry weight and total P content found in the −P treatment (Fig. 1A, B). With application of dNTP or phosphate, both plant dry weight and total P content significantly increased, except for dry weight of Fine-stem in dNTP. With dNTP application, plant dry weight and total P content were higher in TPRC2001-1 than in Fine-stem by 39% and 68%, respectively (Fig. 1A, B). Consistent with these results, Pi concentration in both shoots and roots of both stylo genotypes was higher in dNTP than in −P. Again, genotypic variation was observed with dNTP application, with a higher soluble Pi concentration in TPRC2001-1 than Fine-stem for both shoots and roots, by 56% and 36%, respectively (Fig. 1C, D). Taken together, these results strongly suggest that TPRC2001-1 is more capable of utilizing dNTP as a P source than Fine-stem.
Fig. 1.
Effects of different P sources on plant growth and Pi acquisition in stylo. (A) Dry weight. (B) Total P content. (C) Soluble Pi concentration in shoots. (D) Soluble Pi concentration in roots. After seed germination, seedlings with 1cm root length were grown in MS medium containing 0 µM (−P), 1.2mM KH2PO4 (+P), or 0.4mM dNTP (+dNTP) as the sole P source. After 7 d, dry weight, total P content, and soluble Pi concentration were separately measured. Each bar shows the mean of four replicates with standard error. Significant differences (P < 0.05) are indicated by different letters. FW, fresh weight.
Root internal and root-associated APase activities at two P levels
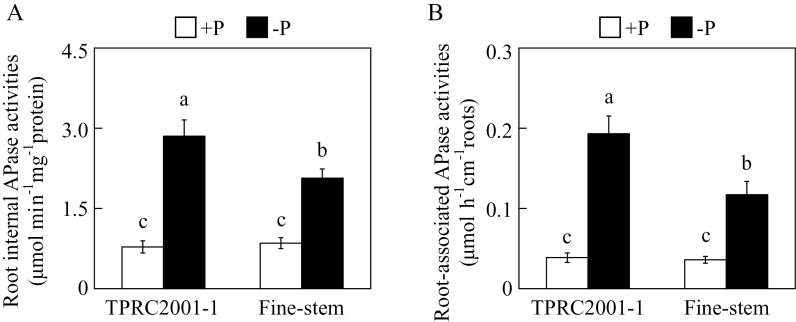
To examine whether APase participates in extracellular dNTP utilization, both root internal and root-associated APase activities were assayed in the two stylo genotypes. Both root internal and root-associated APase activities were significantly increased by P deficiency in both stylo genotypes (Fig. 2A, B). Furthermore, root internal and root-associated APase activities in TPRC2001-1 were 38% and 65% higher than those in Fine-stem under low P conditions, despite the fact that no differences in root APase activities were observed between two stylo genotypes under high P conditions (Fig. 2A, B). This strongly suggests that the different capabilities of the two stylo genotypes to utilize extracellular dNTP might be caused by them having different root APase activities.
Fig. 2.
APase activities in roots of two stylo genotypes at two P levels. (A) Root internal APase activities. (B) Root-associated APase activities. After seed germination, seedlings with 1cm root length were grown in MS medium containing 1.2mM KH2PO4 (+P) or 0 µM (−P). After 7 d, root internal and root-associated APase activities were separately measured. Each bar represents the mean of four independent replicates with standard error. Significant differences (P < 0.05) are indicated by different letters.
Isolation and bioinformatic analysis of three stylo SgPAPs
Because extracellular dNTP utilization has been documented for PvPAP3 in bean, as well as for AtPAP10 and AtPAP26 in Arabidopsis (Liang et al., 2010, 2012; Wang et al., 2011, 2014; Robinson et al., 2012), full-length cDNA of homologs of PvPAP3, AtPAP10, and AtPAP26 in stylo were cloned and named SgPAP7, SgPAP10, and SgPAP26, respectively. SgPAP7, SgPAP10, and SgPAP26 contain 993-bp, 1401-bp, and 1431-bp ORFs, which encode polypeptides with 330, 466, and 476 amino acid residues, respectively (see Supplementary Fig. S1). Bioinformatic analysis predicted that all three SgPAPs have an N-terminal signal peptide (Supplementary Fig. S1). Multiple sequence alignment of SgPAP7, SgPAP10, and SgPAP26 amino acid sequences with homologous plant proteins confirmed the presence of seven metal-binding residues in five conserved motifs [DXG/GDXXY/GNH(D/E)/VXXH/GHXH; bold italic letters represent invariant residues], which is characteristic of plant PAPs (Supplementary Fig. S1).
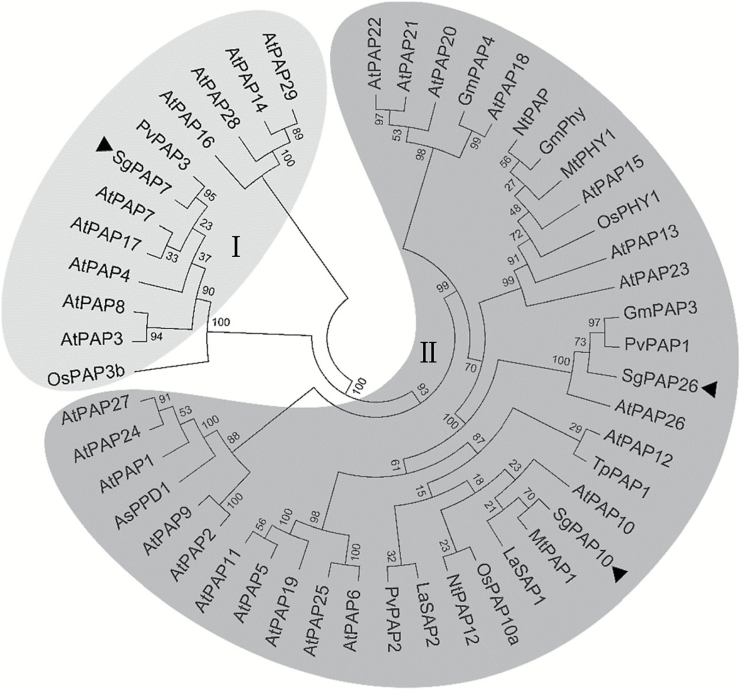
A neighbour-joining phylogenetic tree was constructed for plant PAPs, including SgPAP7, SgPAP10, and SgPAP26 from stylo; all PAP members in Arabidopsis, and other PAP members in rice, bean, tobacco, Medicago truncatula, soybean, astragalus (Astragalus sinicus), white lupin (Lupinus albus), and dwarf marigold (Tagetes patula). In the resulting phylogenetic tree, plant PAPs were widely classified into two groups, group I with low molecular mass PAPs and group II with high molecular mass PAPs (Fig. 3). Among stylo PAPs, SgPAP7 belonged to group I, with the highest similarity to PvPAP3, whereas SgPAP10 and SgPAP26 were classified into group II, including AtPAP10 and AtPAP26 (Fig. 3).
Fig. 3.
Phylogenetic analysis of plant PAP proteins. The phylogenetic tree was constructed using the neighbour-joining method with 1000 bootstrap replicates in the MEGA 5 program. The first two letters of each PAP protein label represent the abbreviated species name. As: Astragalus sinicus; At: Arabidopsis thaliana; Gm: Glycine max; La: Lupinus albus; Mt: Medicago truncatula; Nt: Nicotiana tabacum; Os: Oryza sativa; Pv: Phaseolus vulgaris; Sg: Stylosanthes guianensis; Tp: Tagetes patula. Roman numerals I and II designate the two groups of PAP proteins. Bootstrap values are indicated for major branches as percentages. The three SgPAPs are indicated by black triangles.
Subcellular localization of SgPAPs
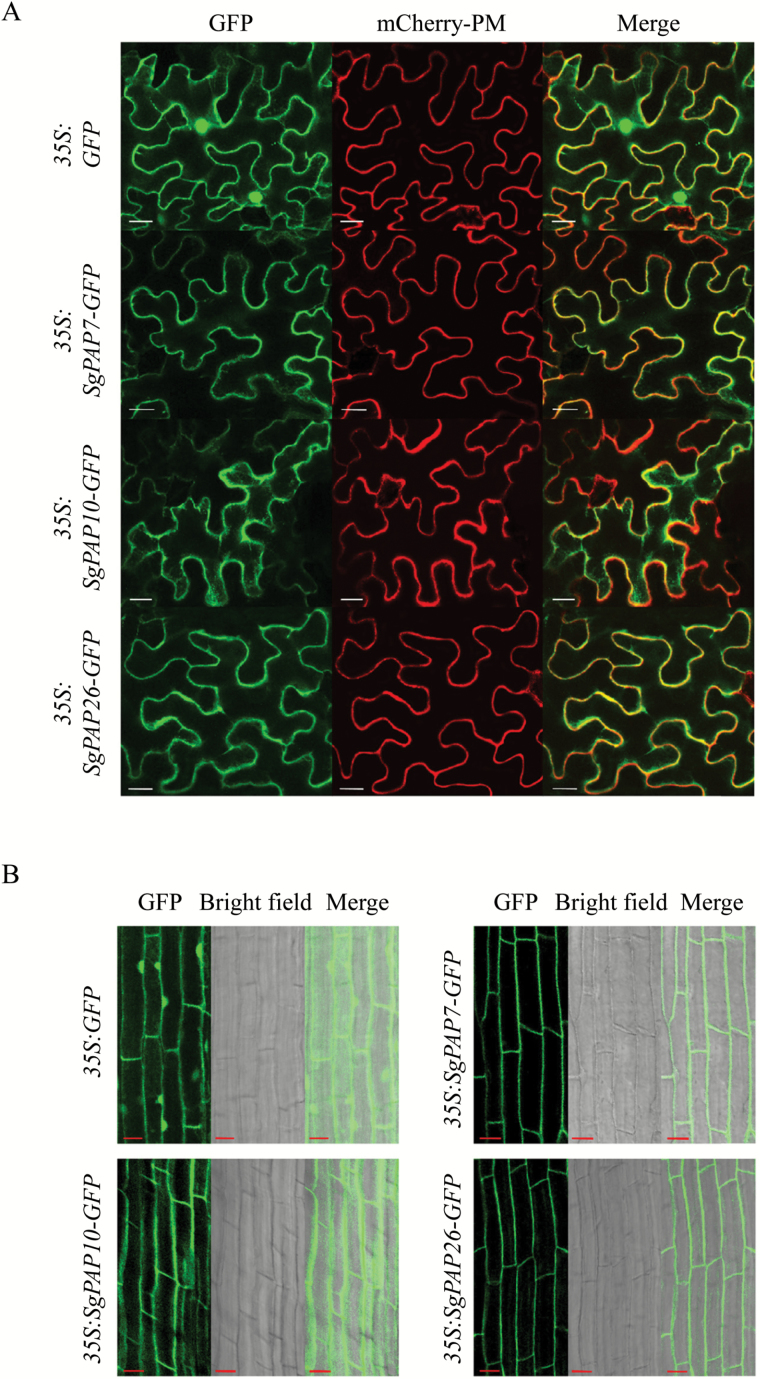
To investigate the subcellular localization of SgPAP7, SgPAP10, and SgPAP26, these encoding regions were separately fused to GFP at the C-terminus, and the constructs were transiently expressed in tobacco leaf epidermal cells and Arabidopsis mesophyll protoplasts. Localization was determined by the detection of GFP signals in confocal laser scanning microscopy analysis (Fig. 4A; Supplementary Fig. S2). The GFP signal of the empty vector control was detected in the plasma membrane, cytoplasm, and nucleus (Fig. 4A; Supplementary Fig. S2), whereas GFP signals from fusion with SgPAP7, SgPAP10, and SgPAP26 were mainly detected in the plasma membrane (Fig. 4A; Supplementary Fig. S2). GFP fluorescence from all the SgPAP fusions was also found in the cytoplasm (Fig. 4A; Supplementary Fig. S2).
Fig. 4.
Subcellular localization of SgPAPs. (A) Transient expression in tobacco leaf epidemic cells, with bars = 20 µm. (B) GFP fluorescence analysis of transgenic bean hairy roots, with bars =10 µm. The images are labelled to show the empty vector control and the 35S:SgPAP7-GFP, 35S:SgPAP10-GFP, and 35S:SgPAP26-GFP constructs. GFP fluorescence and mCherry fluorescence of the plasma membrane marker (mCherry-PM) were observed using confocal microscopy.
To further confirm the subcellular localization of SgPAP7, SgPAP10, and SgPAP26, the coding regions fused with the GFP were also overexpressed in transgenic bean hairy roots. As shown in Fig. 4B, the GFP signal was mainly localized to the cell periphery in transgenic bean hairy roots overexpressing SgPAP7-GFP, SgPAP10-GFP, or SgPAP26-GFP (Fig. 4B), while fluorescence was also observed in the cytoplasm (Fig. 4B). Furthermore, soluble protein and membrane protein were separately extracted from transgenic bean hairy roots overexpressing SgPAP-GFP, and analysed by western blot to verify their subcellular localization. Results showed that soluble proteins and membrane proteins were successfully separated because phosphoenolpyruvate carboxylase (PEPC), a cytoplasmic protein marker, and ATPase, a plasma membrane protein marker, were only detected in soluble protein and membrane protein fractions, respectively (see Supplementary Fig. S3A, B). However, SgPAP7-GFP, SgPAP10-GFP, and SgPAP26-GFP were detected in both soluble protein and membrane protein fractions (Supplementary Fig. S3C), suggesting that SgPAP7, SgPAP10, and SgPAP26 could be co-localized in the plasma membrane and cytoplasm.
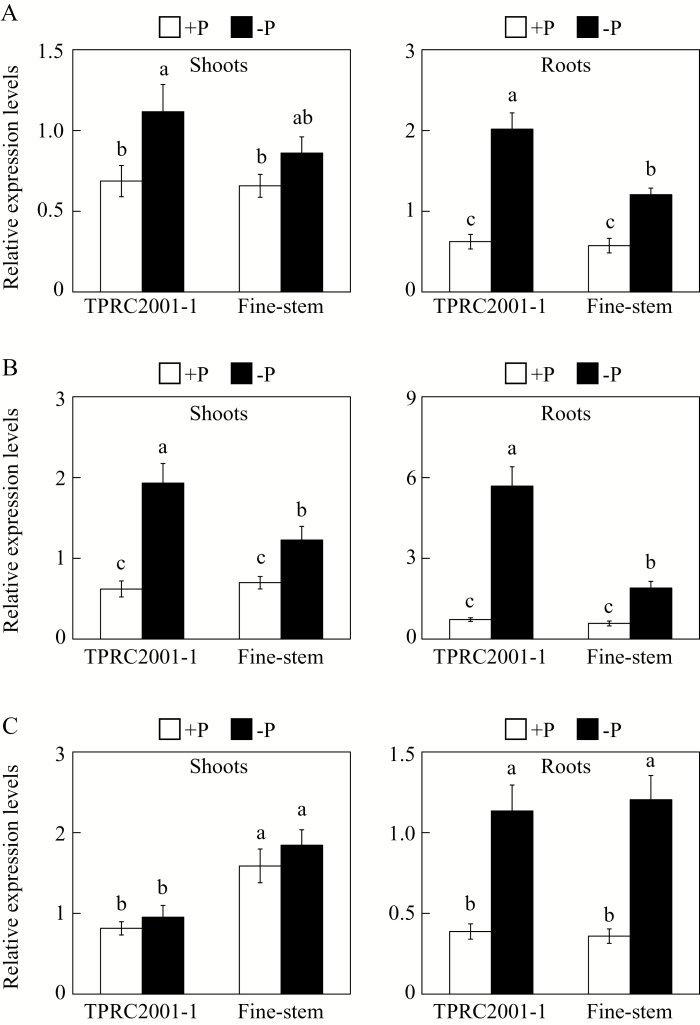
Transcriptional responses of SgPAPs to Pi starvation
To examine whether expression levels of SgPAP7, SgPAP10, and SgPAP26 were responsive to Pi starvation in stylo, their expression patterns were investigated in both shoots and roots of TPRC2001-1 and Fine-stem grown in +P or −P treatments. The results showed that, except for SgPAP7 expression in Fine-stem shoots, transcription levels of SgPAP7 and SgPAP10 were significantly increased by Pi starvation in both shoots and roots of these two stylo genotypes (Fig. 5A, B). In contrast, transcription levels of SgPAP26 were increased by Pi starvation in roots of both genotypes, but exhibited no response to Pi starvation in shoots of either genotype (Fig. 5C). Furthermore, expression levels of SgPAP7 and SgPAP10 were higher in TPRC2001-1 than in Fine-stem at low P levels, especially in roots, where expression of SgPAP7 and SgPAP10 in TPRC2001-1 was 67% and 200% higher than in Fine-stem, respectively (Fig. 5A, B).
Fig. 5.
Transcriptional responses of SgPAPs to Pi deprivation in two stylo genotypes. (A) Expression levels of SgPAP7. (B) Expression levels of SgPAP10. (C) Expression levels of SgPAP26. After seedlings were grown on +P (1.2mM KH2PO4) or −P (0 µM KH2PO4) MS medium for 7 d, total RNA isolated from shoots and roots was used for qRT-PCR analysis. Relative expression levels of the SgPAPs are shown relative to the housekeeping gene SgEF-1a, using arbitrary units. Each bar represents the mean with standard error of four replicates. Significant differences (P < 0.05) are indicated by different letters.
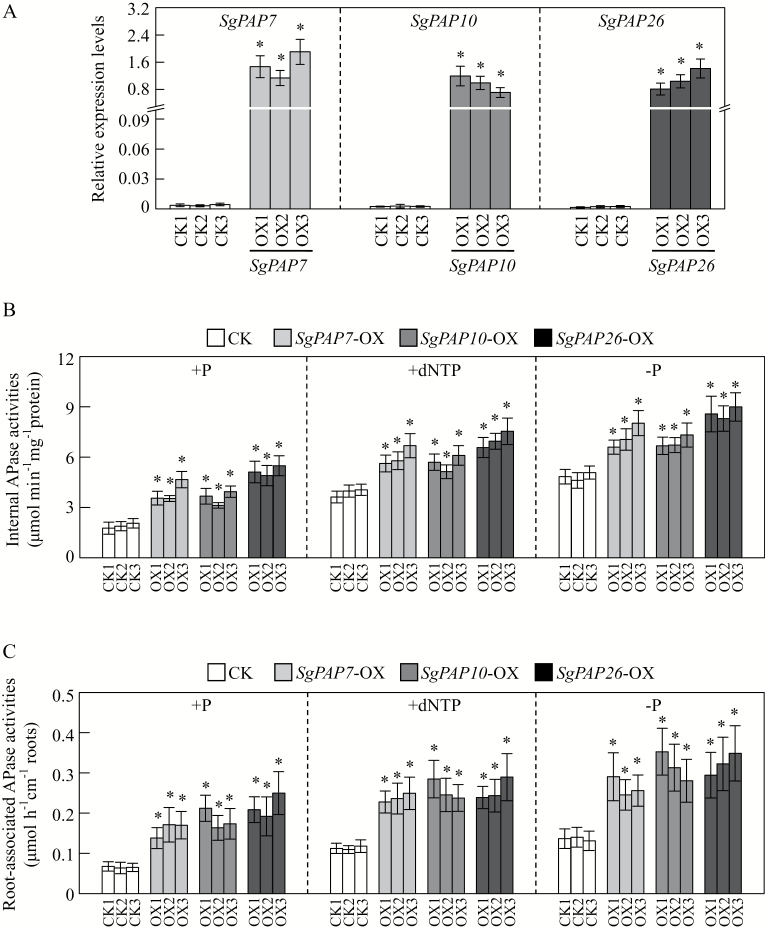
SgPAP overexpression results in increased root APase activities
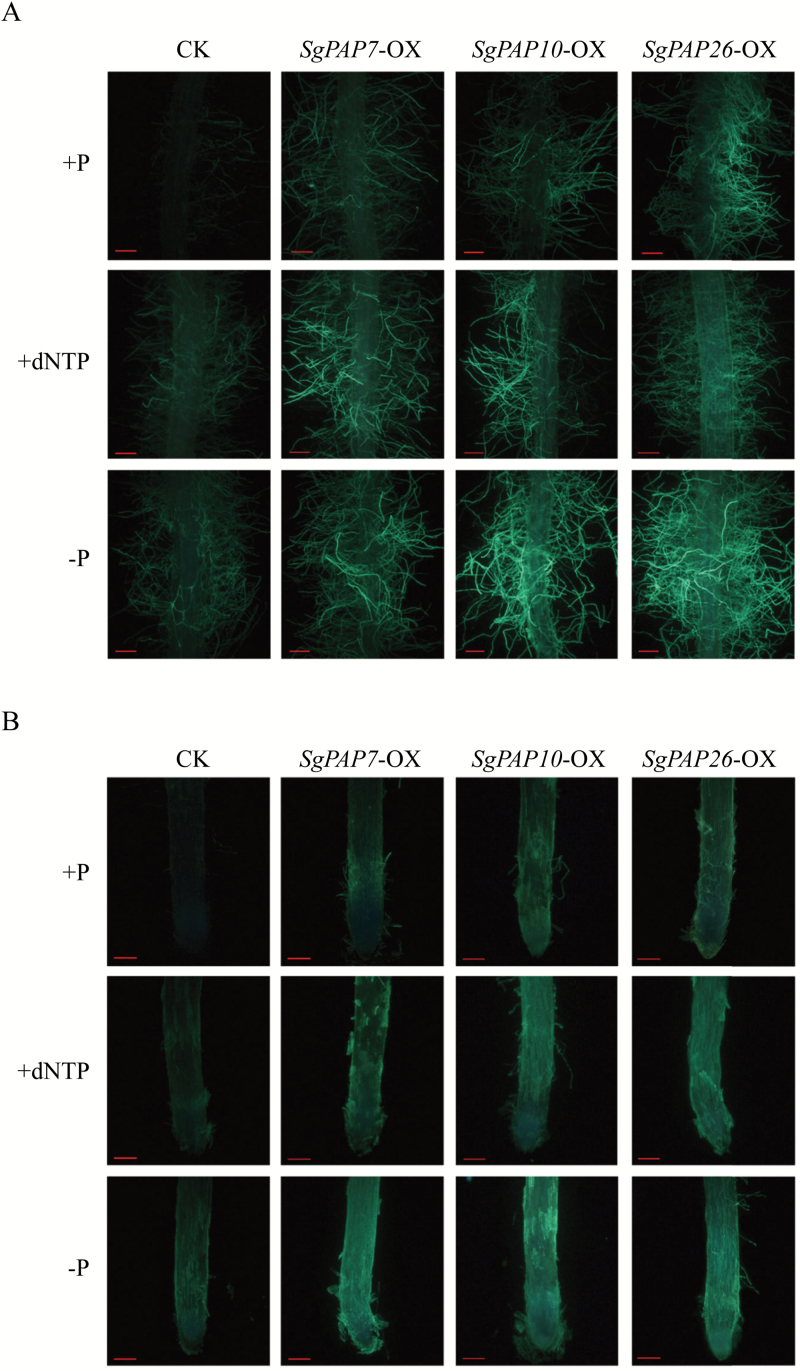
To elucidate the functions of SgPAP7, SgPAP10, and SgPAP26, the three SgPAPs were separately overexpressed (indicated by -OX) in transgenic bean hairy roots, with overexpression confirmed by qRT-PCR analysis (Fig. 6A). Overexpression of SgPAP7, SgPAP10, and SgPAP26 led to significant increases of internal APase activities in the transgenic bean hairy roots compared to those in the control lines with application of phosphate (+P) or dNTP (+dNTP) or without phosphate application (−P) (Fig. 6B). Furthermore, root-associated APase activities were also increased relative to those in controls by more than 75% in SgPAP7-OX lines, 100% in SgPAP10-OX lines, and 102% in SgPAP26-OX lines (Fig. 6C). To further validate root-associated APase activities, ELF-97 phosphate and BCIP were used as substrates to visualize root-associated APase activities in vivo (Fig. 7; Supplementary Fig. S4). When using ELF-97 phosphate as the substrate, in control bean hairy root lines grown in +P treatment, green fluorescence was weak and negligible in the root tip and in root hair (Fig. 7). In contrast, green fluorescence was more intense in both the root tip and root hairs of bean hairy roots overexpressing SgPAP7, SgPAP10, or SgPAP26 with phosphate application (Fig. 7). More intense green fluorescence was also observed in SgPAPs-OX lines than in control lines under −P and +dNTP conditions (Fig. 7). Similar results were observed when analysing root-associated APase activities in bean hairy roots using BCIP as the substrate (see Supplementary Fig. S4). A more intense blue colour was observed along root surfaces of SgPAP7-OX, SgPAP10-OX, and SgPAP26-OX lines when compared to staining in control lines, particularly for SgPAP10-OX and SgPAP26-OX lines (Supplementary Fig. S4), which strongly suggests that overexpression of SgPAP7, SgPAP10, or SgPAP26 can contribute to significant increases of root-associated APase activities in bean hairy roots.
Fig. 6.
Transcripts and APase activities in transgenic bean hairy roots overexpressing SgPAPs. (A) Relative expression levels of transcripts of SgPAPs in bean hairy roots grown under phosphate application conditions as determined by qRT-PCR analysis. (B) Internal APase activities. (C) Root-associated APase activities. CK1, CK2, and CK3 represent three transgenic hairy root lines transformed with the empty vector. OX1, OX2, and OX3 indicate transgenic hairy root lines overexpressing SgPAP7, SgPAP10, or SgPAP26, respectively. Transgenic bean hairy roots were grown on MS medium supplied with 1.2mM KH2PO4 (+P), 0.4mM dNTP (+dNTP), or 0 µM KH2PO4 (−P) as the sole P source for 14 d. Each bar represents the mean of four biological replicates with standard error. Asterisks indicate significant differences between SgPAP overexpression lines and CK lines (P < 0.05).
Fig. 7.
Histochemical staining of root-associated APase activities in bean hairy roots using ELF-97 phosphate as the substrate. (A) Detection of root-associated APase activities in the root hair zone. (B) Detection of root-associated APase activities in the root tip. Hairy roots were incubated in ELF-97 phosphate solution for 30min. Fluorescence was examined using fluorescence microscopy. Bars = 200 µm. Transgenic hairy roots cultivated on MS medium containing 1.2mM KH2PO4 (+P), 0.4mM dNTP (+dNTP), or 0 µM KH2PO4 (−P) for 14 d, before being used for staining of root-associated APase activities. CK indicates transgenic bean hairy roots transformed with the empty vector. SgPAP7-OX, SgPAP10-OX, and SgPAP26-OX indicate transgenic hairy roots overexpressing SgPAP7, SgPAP10, and SgPAP26, respectively.
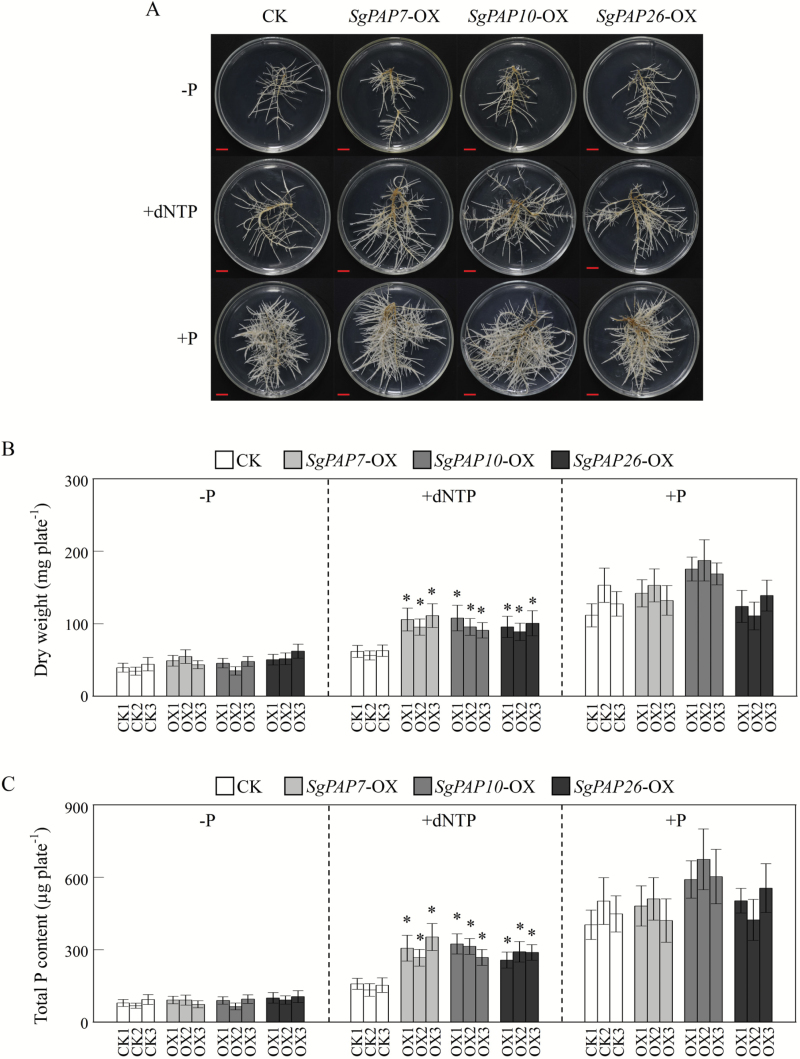
Overexpressing SgPAPs enhances extracellular dNTP utilization
To evaluate functions of SgPAP7, SgPAP10, and SgPAP26 involved in extracellular dNTP utilization, bean hairy roots were grown on MS medium with or without dNTP application. Without dNTP application, no significant difference was observed in appearance or dry weight among transgenic lines of bean hairy roots grown in either −P or +P treatments (Fig. 8A, B). However, with dNTP application, dry weight in SgPAP7, SgPAP10 and SgPAP26 overexpression bean hairy roots was more than 39% higher than in control lines (Fig. 8A, B). Consistent with changes of dry weight, when dNTP was used as the sole external P source in the growth medium, total P content increased by more than 68% in SgPAP7-OX lines, 69% in SgPAP10-OX lines, and 62% in SgPAP26-OX lines, compared with total P content in control lines (Fig. 8C). Yet, consistent with dry weight results, total P content among lines of bean hairy roots was similar in either the −P or +P treatments (Fig. 8C). This result suggests that SgPAP7, SgPAP10, and SgPAP26 can participate in extracellular dNTP utilization in stylo.
Fig. 8.
Growth and total P content of transgenic bean hairy roots supplied with different P sources. (A) Photograph of transgenic bean hairy roots supplied with different P sources. Bars = 1cm. (B) Dry weight in transgenic bean hairy roots. (C) Total P content in transgenic bean hairy roots. Transgenic bean hairy roots were grown on MS medium supplied with 0 µM KH2PO4 (−P), 0.4mM dNTP (+dNTP), or 1.2mM KH2PO4 (+P) as the sole P source for 14 d. Dry weight and total P content were separately measured. CK1, CK2, and CK3 represent three transgenic hairy root lines transformed with the empty vector. OX1, OX2, and OX3 indicate lines overexpressing SgPAP7, SgPAP10, and SgPAP26, respectively. Each bar is the mean of four biological replicates with standard error. Asterisks indicate significant differences between SgPAP overexpression lines and CK lines (P < 0.05).
Discussion
To cope with P deficiency, plants have developed a set of morphological and physiological strategies that are coordinately regulated by complex P signalling networks (Chiou and Lin, 2011; Liang et al., 2014; Sun et al., 2016). Although most adaptive strategies are well conserved in plants, significant variations have been observed across or within plant species for several adaptive responses, especially for alteration of root morphology and architecture (López-Arredondo et al., 2014). For example, it is generally observed that low P inhibits primary root growth in Arabidopsis, but not in other plant species, such as barley (Hordeum vulgare) and rice (Zhou et al., 2008; Péret et al., 2014). However, unlike various changes in root morphology and architecture, it seems that Pi-starvation-caused increases in APase activities are well conserved in a wide range of plant species, such as Arabidopsis, bean, soybean, wheat (Triticum aestivum), barley, and rice. It is therefore thought to be an important adaptive strategy in plants responsive to P deficiency (Tadano and Sakai, 1991; Asmar et al., 1995; Yan et al., 2001; George et al., 2008; Tran et al., 2010a). Consistent with this hypothesis, both root internal and root-associated APase activities were significantly increased by Pi starvation in two stylo genotypes (Fig. 2), suggesting that stylo, like other plant species, can adapt to P deficiency via increased APase activities.
It has been documented through forward genetic, biochemical, and protein MS analyses that increased APase activities are mainly due to increased transcripts or post-transcriptional modification of PAP members in plants under low P conditions. In Arabidopsis, knockout of AtPAP12 and AtPAP26 led to nearly 50% and 30% decreases, respectively, in intracellular APase activities under P-deficient conditions (Wang et al., 2014). Furthermore, atpap12/atpap26 double mutants displayed a 60% reduction in root-secreted APase activities under low P conditions (Robinson et al., 2012; Wang et al., 2014), suggesting that enzymatic activities of PAPs contribute to APase activities in plants.
Consistent with this conclusion, biochemical and MS analyses of purified APase isoforms also revealed that increased APase activities are mainly caused by increased enzymatic activities of plant PAPs, such as AtPAP12 and AtPAP26 in Arabidopsis, LeSAP1 and LeSAP2 in tomato (Lycopersicon esculentum), PvPAP3 in bean, LaSAP in white lupin, and NtPAP in tobacco (Miller et al., 2001; Bozzo et al., 2004; Lung et al., 2008; Liang et al., 2010; Tran et al., 2010b). For example, a Pi-starvation-induced APase isoform was purified from bean roots, and identified as PvPAP3 by sequence analysis (Liang et al., 2010). It has been suggested that increased PvPAP3 transcripts lead to PvPAP3 accumulation, and thus increase APase activities in both bean roots and leaves under low P conditions (Liang et al., 2010). In this study, SgPAP7, SgPAP10, and SgPAP26 were cloned from stylo, and found to be highly homologous with PvPAP3 in bean, and AtPAP10 and AtPAP26 in Arabidopsis, respectively (Fig. 3). It was observed that root internal and root-associated APase activities increased with Pi starvation, which was accompanied by increased transcripts of SgPAP7, SgPAP10, and SgPAP26 (Figs 2 and 5). Furthermore, overexpression of SgPAP7, SgPAP10, or SgPAP26 in bean hairy roots led to increases in internal and root-associated APase activities of more than 27% and 75%, respectively (Fig. 6B, C). These results strongly suggest that increased transcripts of SgPAP7, SgPAP10, or SgPAP26 can contribute to increased APase activities in stylo under low P conditions.
Functions of plant PAPs are partially determined by their subcellular localization. It has been well documented that plant PAPs can localize in many subcellular regions, including the cell wall, vacuole, nucleus, plastids, mitochondria, apoplast, and secretome, which implies complex and diverse functions for plant PAPs (Li et al., 2008; Kaida et al., 2009; Hurley et al., 2010; Liang et al., 2010, 2012; Sun et al., 2012; Del Vecchio et al., 2014). However, plasma membranes have been found to harbour only a few plant PAPs, such as PvPAP1 and PvPAP3 in bean, and AsPPD1 in Astragalus sinicus (Liang et al., 2010, 2012; Wang et al., 2015).
Through bioinformatic analysis, a transmembrane helix was found in the topology of SgPAP7, SgPAP10, and SgPAP26 (see Supplementary Fig. S5), suggesting that these three SgPAPs might localize on the plasma membrane. That these three SgPAPs were localized on the plasma membrane was consistently observed through transient expression analysis of SgPAP-GFP in tobacco leaf epidermal cells and Arabidopsis mesophyll protoplasts, as well as overexpression analysis of SgPAP-GFP in transgenic bean hairy roots (Fig. 4A, B; Supplementary Fig. S2). Like SgPAP7, its ortholog PvPAP3 has been found to be localized on the plasma membrane through transient expression of PvPAP3-GFP in onion epidermal cells (Liang et al., 2010), which suggests, along with sequence similarity, that PvPAP3 and SgPAP7 perform similar functions in plants. Although SgPAP26 exhibited similar subcellular localization to PvPAP1, its ortholog in bean (Liang et al., 2012), variation in subcellular localization from other orthologs has been found, including dual-targeting to both the cell wall and vacuole for AtPAP26 in Arabidopsis, and to the mitochondria for GmPAP3 in soybean (Li et al., 2008; Hurley et al., 2010; Robinson et al., 2012). This suggests that the diverse functions of PAPs in plants are attributable to the variation in their subcellular localization.
It has been demonstrated that soil microorganisms play a key role in soil–plant P cycling, with activities including immobilization of Pi from the soil solution, mineralization of soil organic P, and incorporation of Pi into microbial biomass (Richardson et al., 2011). It is estimated that microbial P levels can reach 30–40% of the total soil P, which mainly consists of nucleic acids (Achat et al., 2010). Therefore, plants might be capable of utilizing extracellular nucleic acids (e.g. DNA and dNTP). Recently, utilization of nucleic acid P and other phosphomonoesters has been reported for a variety of plant PAPs, such as PvPAP3 in bean, OsPAP10a in rice, and AtPAP12 and AtPAP26 in Arabidopsis (Liang et al., 2010; Robinson et al., 2012; Tian et al., 2012; Tian and Liao, 2015). In Arabidopsis, the biomass of atpap12/atpap26 mutant plants was significantly lower than that of wild-type plants when DNA was supplied as the sole P source in growth medium, suggesting that both AtPAP12 and AtPAP26 might be involved in the utilization of extracellular DNA (Robinson et al., 2012). PvPAP1 and PvPAP3 in bean were also demonstrated to augment extracellular dNTP utilization (Liang et al., 2012). In the current study, when dNTP was supplied as the sole extracellular P source, significant increases of both total P content and soluble Pi concentration were observed in two stylo genotypes (Fig. 1), suggesting that stylo is able to utilize exogenous dNTP. To elucidate molecular mechanisms underlying stylo utilization of extracellular dNTP, homologues of PvPAP3, AtPAP10, and AtPAP26 in stylo were cloned and named SgPAP7, SgPAP10, and SgPAP26. The significant increases in root-associated APase activities as well as the higher dry weight and total P content in transgenic bean hairy roots overexpressing SgPAP7, SgPAP10, or SgPAP26 when dNTP was supplied as the sole extracellular P source (Figs 6, 7, and 8) strongly suggest that these genes participate in extracellular dNTP utilization in stylo. Furthermore, low-P-enhanced higher expression levels of SgPAP7 and SgPAP10 in TPRC2001-1 than in Fine-stem (Fig. 5) might facilitate the utilization of more extracellular dNTP for TPRC2001-1 than for Fine-stem (Fig. 1).
Taken together, the results herein strongly suggest that Pi starvation can increase root-associated APase activities in stylo, which might be caused by enhanced expression levels of SgPAP7, SgPAP10, and SgPAP26 in roots of both stylo genotypes. Furthermore, higher expression levels of SgPAP7 and SgPAP10 in TPRC2001-1 roots contribute to its higher root-associated APase activities, and thus facilitate greater utilization of extracellular dNTP by TPRC2001-1 than by Fine-stem.
Supplementary material
Table S1. A list of primers used in the study.
Figure S1. Multiple sequence alignment of three SgPAPs and orthologous plant PAPs.
Figure S2. Subcellular localization of SgPAPs in Arabidopsis mesophyll protoplasts.
Figure S3. Western-blot analysis of soluble protein and membrane protein extracts from transgenic bean hairy roots.
Figure S4. Histochemical staining of root-associated APase activities in bean hairy roots using BCIP as the substrate.
Figure S5. Prediction of SgPAP transmembrane topology.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China [31422046], the National Key Basic Research Program of China [2014CB138706], the Earmarked Fund for Modern Agro-Industry Technology Research System [CARS-35], and Comprehensive Strength Promotion Plan of College in the Western Regions [M8K3124001001003002]. The authors thank Dr Thomas Walk and Cuiyue Liang for critical reading. We also thank Dr Li-Juan Luo, Mr Hong Qiu and Miss Ze-Fei Cai for their helpful subcellular localization analysis in Arabidopsis protoplasts and bean hairy roots.
References
- Achat DL, Morel C, Bakker MR, Augusto L, Pellerin S, Gallet-Budynek A, Gonzalez M. 2010. Assessing turnover of microbial biomass phosphorus: combination of an isotopic dilution method with a mass balance model. Soil Biology and Biochemistry 42, 2231–2240. [Google Scholar]
- Asmar F, Gahoonia T, Nielsen NE. 1995. Barley genotypes differ in activity of soluble extracellular phosphatase and depletion of organic phosphorus in the rhizosphere soil. Plant and Soil 172, 117–122. [Google Scholar]
- Beck JL, de Jersey J, Zerner B, Hendrich MP, Debrunner PG. 1988. Properties of the Fe(II)-Fe(III) derivative of red kidney bean purple phosphatase. Evidence for a binuclear Zn-Fe center in the native enzyme. Journal of the American Chemical Society 110, 3317–3318. [Google Scholar]
- Beck JL, McConachie LA, Summors AC, Arnold WN, de Jersey J, Zerner B. 1986. Properties of a purple phosphatase from red kidney bean: a zinc-iron metalloenzyme. Biochimica et Biophysica Acta 869, 61–68. [Google Scholar]
- Bozzo GG, Raghothama KG, Plaxton WC. 2004. Structural and kinetic properties of a novel purple acid phosphatase from phosphate-starved tomato (Lycopersicon esculentum) cell cultures. Biochemical Journal 377, 419–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Chandra A. 2009. Diversity among Stylosanthes species: habitat, edaphic and agro-climatic affinities leading to cultivar development. Journal of Environmental Biology 30, 471–478. [PubMed] [Google Scholar]
- Chen Z, Sun L, Liu P, Liu G, Tian J, Liao H. 2015. Malate synthesis and secretion mediated by a manganese-enhanced malate dehydrogenase confers superior manganese tolerance in Stylosanthes guianensis . Plant Physiology 167, 176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI. 2011. Signaling network in sensing phosphate availability in plants. Annual Review of Plant Biology 62, 185–206. [DOI] [PubMed] [Google Scholar]
- Del Pozo JC, Allona I, Rubio V, Leyva A, de la Peña A, Aragoncillo C, Paz-Ares J. 1999. A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. The Plant Journal 19, 579–589. [DOI] [PubMed] [Google Scholar]
- Del Vecchio HA, Ying S, Park J, Knowles VL, Kanno S, Tanoi K, She YM, Plaxton WC. 2014. The cell wall-targeted purple acid phosphatase AtPAP25 is critical for acclimation of Arabidopsis thaliana to nutritional phosphorus deprivation. The Plant Journal 80, 569–581. [DOI] [PubMed] [Google Scholar]
- Du YM, Tian J, Liao H, Bai CJ, Yan XL, Liu GD. 2009. Aluminium tolerance and high phosphorus efficiency helps Stylosanthes better adapt to low-P acid soils. Annals of Botany 103, 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff SMG, Sarath G, Plaxton WC. 1994. The role of acid phosphatases in plant phosphorus metabolism. Physiologia Plantarum 90, 791–800. [Google Scholar]
- Durmus A, Eicken C, Sift BH, Kratel A, Kappl R, Hüttermann J, Krebs B. 1999. a The active site of purple acid phosphatase from sweet potatoes (Ipomoea batatas): metal content and spectroscopic characterization. European Journal of Biochemistry 260, 709–716. [DOI] [PubMed] [Google Scholar]
- Durmus A, Eicken C, Spener F, Krebs B. 1999. b Cloning and comparative protein modeling of two purple acid phosphatase isozymes from sweet potatoes (Ipomoea batatas). Biochimica et Biophysica Acta 1434, 202–209. [DOI] [PubMed] [Google Scholar]
- Flanagan JU, Cassady AI, Schenk G, Guddat LW, Hume DA. 2006. Identification and molecular modeling of a novel, plant-like, human purple acid phosphatase. Gene 377, 12–20. [DOI] [PubMed] [Google Scholar]
- George TS, Gregory PJ, Hocking P, Richardson AE. 2008. Variation in root-associated phosphatase activities in wheat contributes to the utilization of organic P substrates in vitro, but does not explain differences in the P-nutrition of plants when grown in soils. Environmental and Experimental Botany 64, 239–249. [Google Scholar]
- González-Muñoz E, Avendaño-Vázquez AO, Montes RA, de Folter S, Andrés-Hernández L, Abreu-Goodger C, Sawers RJH. 2015. The maize (Zea mays ssp. mays var. B73) genome encodes 33 members of the purple acid phosphatase gene family. Frontiers in Plant Science 6, 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley BA, Tran HT, Marty NJ, Park J, Snedden WA, Mullen RT, Plaxton WC. 2010. The dual-targeted purple acid phosphatase isozyme AtPAP26 is essential for efficient acclimation of Arabidopsis to nutritional phosphate deprivation. Plant Physiology 153, 1112–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida R, Satoh Y, Bulone V, Yamada Y, Kaku T, Hayashi T, Kaneko TS. 2009. Activation of β-glucan synthases by wall-bound purple acid phosphatase in tobacco cells. Plant Physiology 150, 1822–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klabunde T, Stahl B, Suerbaum H, Hahner S, Karas M, Hillenkamp F, Krebs B, Witzel H. 1994. The amino acid sequence of the red kidney bean Fe(III)-Zn(II) purple acid phosphatase: determination of the amino acid sequence by a combination of matrix-assisted laser desorption/ionization mass spectrometry and automated Edman sequencing. European Journal of Biochemistry 226, 369–375. [DOI] [PubMed] [Google Scholar]
- Kochian LV, Hoekenga OA, Pineros MA. 2004. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annual Review of Plant Biology 55, 459–493. [DOI] [PubMed] [Google Scholar]
- Lambers H, Plaxton WC. 2015. Phosphorus: back to the roots. In: Plaxton WC, Lambers H, eds. Annual Plant Reviews, Volume 48, Phosphorus Metabolism in Plants.John Wiley & Sons, Inc: Hoboken, NJ, 3–22. [Google Scholar]
- Law YS, Zhang R, Guan X, Cheng S, Sun F, Duncan O, Murcha MW, Whelan J, Lim BL. 2015. Phosphorylation and dephosphorylation of the presequence of precursor MULTIPLE ORGANELLAR RNA EDITING FACTOR3 during import into mitochondria from Arabidopsis. Plant Physiology 169, 1344–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Gui S, Yang T, Walk T, Wang X, Liao H. 2012. a Identification of soybean purple acid phosphatase genes and their expression responses to phosphorus availability and symbiosis. Annals of Botany 109, 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhu H, Liu K, Liu X, Leggewie G, Udvardi M, Wang D. 2002. Purple acid phosphatases of Arabidopsis thaliana: comparative analysis and differential regulation by phosphate deprivation. Journal of Biological Chemistry 277, 27772–27781. [DOI] [PubMed] [Google Scholar]
- Li R, Lu W, Guo C, Li X, Gu J, Xiao K. 2012. b. Molecular characterization and functional analysis of OsPHY1, a purple acid phosphatase (PAP)-type phytase gene in rice (Oryza sativa L.). Journal of Integrative Agriculture 11, 1217–1226. [Google Scholar]
- Li WF, Shao G, Lam HM. 2008. Ectopic expression of GmPAP3 alleviates oxidative damage caused by salinity and osmotic stresses. New Phytologist 178, 80–91. [DOI] [PubMed] [Google Scholar]
- Liang C, Sun L, Yao Z, Liao H, Tian J. 2012. Comparative analysis of PvPAP gene family and their functions in response to phosphorus deficiency in common bean. PLoS One 7, 65–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Tian J, Lam HM, Lim BL, Yan X, Liao H. 2010. Biochemical and molecular characterization of PvPAP3, a novel purple acid phosphatase isolated from common bean enhancing extracellular ATP utilization. Plant Physiology 152, 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Wang J, Zhao J, Tian J, Liao H. 2014. Control of phosphate homeostasis through gene regulation in crops. Current Opinion in Plant Biology 21, 59–66. [DOI] [PubMed] [Google Scholar]
- Liao H, Wong FL, Phang TH, Cheung MY, Li WY, Shao G, Yan X, Lam HM. 2003. GmPAP3, a novel purple acid phosphatase-like gene in soybean induced by NaCl stress but not phosphorus deficiency. Gene 318, 103–111. [DOI] [PubMed] [Google Scholar]
- Lloyd JC, Zakhleniuk OV, Raines CA. 2001. Identification of mutants in phosphorus metabolism. Annals of Applied Biology 138, 111–115. [Google Scholar]
- López-Arredondo DL, Leyva-González MA, González-Morales SI, López-Bucio J, Herrera-Estrella L. 2014. Phosphate nutrition: improving low-phosphate tolerance in crops. Annual Review of Plant Biology 65, 95–123. [DOI] [PubMed] [Google Scholar]
- Lung SC, Leung A, Kuang R, Wang Y, Leung P, Lim BL. 2008. Phytase activity in tobacco (Nicotiana tabacum) root exudates is exhibited by a purple acid phosphatase. Phytochemistry 69, 365–373. [DOI] [PubMed] [Google Scholar]
- Lynch JP. 2015. Root phenes that reduce the metabolic costs of soil exploration: opportunities for 21st century agriculture. Plant Cell and Environment 38, 1775–1784. [DOI] [PubMed] [Google Scholar]
- Ma XF, Wright E, Ge Y, Bell J, Xi Y, Bouton JH, Wang ZY. 2009. Improving phosphorus acquisition of white clover (Trifolium repens L.) by transgenic expression of plant-derived phytase and acid phosphatase genes. Plant Science 176, 479–488. [DOI] [PubMed] [Google Scholar]
- Marschner H. 1995. Function of mineral nutrients: micronutrients. In: Marschner H, ed. Mineral Nutrition of Higher Plants, 2nd edn Academic Press: London, UK, 379–396. [Google Scholar]
- Matange N, Podobnik M, Visweswariah SS. 2015. Metallophosphoesterases: structural fidelity with functional promiscuity. Biochemical Journal 467, 201–216. [DOI] [PubMed] [Google Scholar]
- Miller SS, Liu J, Allan DL, Menzhuber CJ, Fedorova M, Vance CP. 2001. Molecular control of acid phosphatase secretion into the rhizosphere of proteoid roots from phosphorus-stressed white lupin. Plant Physiology 127, 594–606. [PMC free article] [PubMed] [Google Scholar]
- Mitić N, Smith SJ, Neves A, Guddat LW, Gahan LR, Schenk G. 2006. The catalytic mechanisms of binuclear metallohydrolases. Chemical Reviews 106, 3338–3363. [DOI] [PubMed] [Google Scholar]
- Murphy J, Riley JP. 1962. A modified single solution method for the determination of phosphate in natural waters. Analytica Chimica Acta 27, 31–36. [Google Scholar]
- Oddie GW, Schenk G, Angel NZ, Walsh N, Guddat LW, de Jersey J, Cassady AI, Hamilton SE, Hume DA. 2000. Structure, function, and regulation of tartrate-resistant acid phosphatase. Bone 27, 575–584. [DOI] [PubMed] [Google Scholar]
- Péret B, Desnos T, Jost R, Kanno S, Berkowitz O, Nussaume L. 2014. Root architecture responses: in search of phosphate. Plant Physiology 166, 1713–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC, Tran HT. 2011. Metabolic adaptations of phosphate-starved plants. Plant Physiology 156, 1006–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Zhao J, Tian J, Chen L, Sun Z, Guo Y, Lu X, Gu M, Xu G, Liao H. 2012. The high-affinity phosphate transporter GmPT5 regulates phosphate transport to nodules and nodulation in soybean. Plant Physiology 159, 1634–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran S, Stone SL, Benkel B, Prithiviraj B. 2013. Purple Acid Phosphatase 5 is required for maintaining basal resistance against Pseudomonas syringae in Arabidopsis. BMC Plant Biology 13, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AE, Lynch JP, Ryan PR, et al. 2011. Plant and microbial strategies to improve the phosphorus efficiency of agriculture. Plant and Soil 349, 121–156. [Google Scholar]
- Robinson WD, Park J, Tran HT, Del Vecchio HA, Ying S, Zins JL, Patel K, McKnight TD, Plaxton WC. 2012. The secreted purple acid phosphatase isozymes AtPAP12 and AtPAP26 play a pivotal role in extracellular phosphate-scavenging by Arabidopsis thaliana . Journal of Experimental Botany 63, 6531–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk G, Boutchard CL, Carrington LE, Noble CJ, Moubaraki B, Murray KS, de Jersey J, Hanson GR, Hamilton S. 2001. A purple acid phosphatase from sweet potato contains an antiferromagnetically coupled binuclear Fe-Mn center. Journal of Biological Chemistry 276, 19084–19088. [DOI] [PubMed] [Google Scholar]
- Schenk G, Gahan LR, Carrington LE, Mitić N, Valizadeh M, Hamilton SE, de Jersey J, Guddat LW. 2005. Phosphate forms an unusual tripodal complex with the Fe-Mn center of sweet potato purple acid phosphatase. Proceedings of the National Academy of Sciences of the United States of America 102, 273–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk G, Ge Y, Carrington LE, Wynne CJ, Searle IR, Carroll BJ, Hamilton S, de Jersey J. 1999. Binuclear metal centers in plant purple acid phosphatases: Fe-Mn in sweet potato and Fe-Zn in soybean. Archives of Biochemistry and Biophysics 370, 183–189. [DOI] [PubMed] [Google Scholar]
- Schenk G, Guddat LW, Ge Y, Carrington LE, Hume DA, Hamilton S, de Jersey J. 2000. a Identification of mammalian-like purple acid phosphatases in a wide range of plants. Gene 250, 117–125. [DOI] [PubMed] [Google Scholar]
- Schenk G, Korsinczky MLJ, Hume DA, Hamilton S, de Jersey J. 2000. b Purple acid phosphatases from bacteria: similarities to mammalian and plant enzymes. Gene 255, 419–424. [DOI] [PubMed] [Google Scholar]
- Schenk G, Mitić N, Hanson GR, Comba P. 2013. Purple acid phosphatase: a journey into the function and mechanism of a colorful enzyme. Coordination Chemistry Reviews 257, 473–482. [Google Scholar]
- Shen J, Yuan L, Zhang J, Li H, Bai Z, Chen X, Zhang W, Zhang F. 2011. Phosphorus dynamics: from soil to plant. Plant Physiology 156, 997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Liu D. 2015. Ethylene and plant responses to phosphate deficiency. Frontiers in Plant Science 6, 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträter N, Klabunde T, Tucker P, Witzel H, Krebs B. 1995. Crystal structure of a purple acid phosphatase containing a dinuclear Fe(III)-Zn(II) active site. Science 268, 1489–1492. [DOI] [PubMed] [Google Scholar]
- Stutter MI, Shand CA, George TS, Blackwell MS, Bol R, MacKay RL, Richardson AE, Condron LM, Turner BL, Haygarth PM. 2012. Recovering phosphorus from soil: a root solution? Environmental Science and Technology 46, 1977–1978. [DOI] [PubMed] [Google Scholar]
- Sun F, Suen PK, Zhang Y, Liang C, Carrie C, Whelan J, Ward JL, Hawkins ND, Jiang L, Lim BL. 2012. A dual-targeted purple acid phosphatase in Arabidopsis thaliana moderates carbon metabolism and its overexpression leads to faster plant growth and higher seed yield. New Phytologist 194, 206–219. [DOI] [PubMed] [Google Scholar]
- Sun L, Liang C, Chen Z, Liu P, Tian J, Liu G, Liao H. 2014. Superior aluminium (Al) tolerance of Stylosanthes is achieved mainly by malate synthesis through an Al-enhanced malic enzyme, SgME1. New Phytologist 202, 209–219. [DOI] [PubMed] [Google Scholar]
- Sun L, Song L, Zhang Y, Zheng Z, Liu D. 2016. Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiology 170, 499–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadano T, Sakai H. 1991. Secretion of acid phosphatase by the roots of several crop species under phosphorus-deficient conditions. Soil Science and Plant Nutrition 37, 129–140. [Google Scholar]
- Tian J, Liao H. 2015. The role of intracellular and secreted purple acid phosphatases in plant phosphorus scavenging and recycling. In: Plaxton WC, Lambers H, eds. Annual Plant Reviews, Volume 48, Phosphorus Metabolism in Plants.John Wiley & Sons, Inc: Hoboken, NJ, 265–287. [Google Scholar]
- Tian J, Wang C, Zhang Q, He X, Whelan J, Shou H. 2012. Overexpression of OsPAP10a, a root-associated acid phosphatase, increased extracellular organic phosphorus utilization in rice. Journal of Integrative Plant Biology 54, 631–639. [DOI] [PubMed] [Google Scholar]
- Tran HT, Hurley BA, Plaxton WC. 2010. a Feeding hungry plants: the role of purple acid phosphatases in phosphate nutrition. Plant Science 179, 14–27. [Google Scholar]
- Tran HT, Qian W, Hurley BA, She YM, Wang D, Plaxton WC. 2010. b Biochemical and molecular characterization of AtPAP12 and AtPAP26: the predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana . Plant, Cell and Environment 33, 1789–1803. [DOI] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. 2003. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytologist 157, 423–447. [DOI] [PubMed] [Google Scholar]
- Wang J, Si Z, Li F, Xiong X, Lei L, Xie F, Chen D, Li Y, Li Y. 2015. A purple acid phosphatase plays a role in nodule formation and nitrogen fixation in Astragalus sinicus . Plant Molecular Biology 88, 515–529. [DOI] [PubMed] [Google Scholar]
- Wang L, Li Z, Qian W, et al. 2011. The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiology 157, 1283–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lu S, Zhang Y, Li Z, Du X, Liu D. 2014. Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10, AtPAP12, and AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. Journal of Integrative Plant Biology 56, 299–314. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang Y, Tian J, Lim BL, Yan X, Liao H. 2009. Overexpressing AtPAP15 enhances phosphorus efficiency in soybean. Plant Physiology 151, 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS. 2009. Tape-Arabidopsis sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5, 8567–8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Shou H, Xu G, Lian X. 2013. Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Current Opinion in Plant Biology 16, 205–212. [DOI] [PubMed] [Google Scholar]
- Yan X, Liao H, Trull MC, Beebe SE, Lynch JP. 2001. Induction of a major leaf acid phosphatase does not confer adaptation to low phosphorus availability in common bean. Plant Physiology 125, 1901–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Sun Y, Pei W, et al. 2015. Involvement of OsPht1;4 in phosphate acquisition and mobilization facilitates embryo development in rice. The Plant Journal 82, 556–569. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang C, Tian J, Li K, Shou H. 2011. Identification of rice purple acid phosphatases related to phosphate starvation signalling. Plant Biology 13, 7–15. [DOI] [PubMed] [Google Scholar]
- Zhang W, Gruszewski HA, Chevone BI, Nessler CL. 2008. An Arabidopsis purple acid phosphatase with phytase activity increases foliar ascorbate. Plant Physiology 146, 431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P. 2008. OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiology 146, 1673–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.