Highlight
Phs-A1 confers resistance to sprouting in wheat by delaying the rate of seed dormancy loss and is distinct from the previously proposed PM19 candidate genes.
Key words: After-ripening, dormancy, PM19, pre-harvest sprouting, seed, synteny, Triticum, aestivum.
Abstract
The precocious germination of cereal grains before harvest, also known as pre-harvest sprouting, is an important source of yield and quality loss in cereal production. Pre-harvest sprouting is a complex grain defect and is becoming an increasing challenge due to changing climate patterns. Resistance to sprouting is multi-genic, although a significant proportion of the sprouting variation in modern wheat cultivars is controlled by a few major quantitative trait loci, including Phs-A1 in chromosome arm 4AL. Despite its importance, little is known about the physiological basis and the gene(s) underlying this important locus. In this study, we characterized Phs-A1 and show that it confers resistance to sprouting damage by affecting the rate of dormancy loss during dry seed after-ripening. We show Phs-A1 to be effective even when seeds develop at low temperature (13 °C). Comparative analysis of syntenic Phs-A1 intervals in wheat and Brachypodium uncovered ten orthologous genes, including the Plasma Membrane 19 genes (PM19-A1 and PM19-A2) previously proposed as the main candidates for this locus. However, high-resolution fine-mapping in two bi-parental UK mapping populations delimited Phs-A1 to an interval 0.3 cM distal to the PM19 genes. This study suggests the possibility that more than one causal gene underlies this major pre-harvest sprouting locus. The information and resources reported in this study will help test this hypothesis across a wider set of germplasm and will be of importance for breeding more sprouting resilient wheat varieties.
Introduction
Pre- and post-harvest crop losses caused by biotic or abiotic stress factors are major drawbacks to attaining global food security. In addition to their detrimental effects on crop yield, their effects on quality are equally damaging. Pre-harvest sprouting (PHS) represents one such source of both yield and quality loss in global wheat production. PHS is characterized by the precocious germination of grains before harvest with consequent reductions in seed viability and end-use value, particularly for bread-making purposes. PHS is strongly influenced by the environment and is especially prevalent in wheat-growing regions with high rainfall during the period of grain maturation and ripening (Mares and Mrva, 2014). In addition, adverse environmental conditions like heat stress or water deficit during grain development have been generally associated with higher levels of seed germination upon grain maturation and this predisposes plants to incidences of PHS (Rodriguez et al, 2011). Given the current climate change projections of increased temperature and precipitation in parts of the world (Walther et al., 2002; Trenberth, 2011), the incidence of PHS is expected to increase and become a greater challenge in wheat production areas.
Breeding for PHS resistance is an effective and environmentally sustainable strategy to address this problem. This can be achieved by breeding for seed dormancy, which is the most dominant component of PHS resistance. In general, plants with higher seed dormancy and responsiveness to abscisic acid (ABA) – a key hormone regulating seed dormancy – display higher sprouting resistance (Walker-Simmons, 1987; Gerjets et al., 2010). On the other hand, prolonged seed dormancy is not desirable as it delays growing cycles and results in non-uniform germination upon sowing. The desired breeding objective is therefore an intermediate level of seed dormancy that confers resistance to sprouting during seed development and maturation, but declines shortly thereafter. However, breeding for PHS resistance is made difficult by the highly quantitative nature of this trait: numerous quantitative trait loci (QTL) accounting for varying degrees of resistance have been identified on all 21 wheat chromosomes (reviewed in Flintham et al., 2002; Gao et al., 2013b; Mares and Mrva, 2014), highlighting the complexity of its genetic control.
Despite this, a few QTL have been consistently identified across diverse germplasm which account for a large proportion of the sprouting variation observed in modern wheat cultivars. These loci include the Red grain colour genes (R-1) on the long arm of group 3 chromosomes, which encode a Myb transcription factor (TaMyb10) that regulates flavonoid biosynthesis (Himi and Noda 2005; Himi et al. 2011). Although not well understood, TaMyb10 also has pleiotropic effects on seed dormancy, with red-grained wheat being generally, but not always, more dormant than white-grained wheat (Mares et al. 2005; Kottearachchi et al., 2006; Mares and Mrva 2014). Besides the R-1 loci, two major QTL on chromosome arms 3AS and 4AL account for a considerable proportion of the variation in PHS resistance when independent of grain colour. The 3AS QTL was recently cloned and shown to encode for Mother of Flowering Time (TaMFT: Nakamura et al., 2011; Lei et al., 2013; Liu et al., 2013b), which promotes seed dormancy in wheat embryos. TaMFT is upregulated when seeds develop at temperatures below 13 °C, suggesting its involvement in the integration of environmental inputs into the regulation of seed dormancy (Nakamura et al., 2011).
The 4AL QTL for PHS resistance was first reported by Flintham (2000) and was originally named Phs, although here we refer to it as Phs-A1 in agreement with the current wheat gene nomenclature. This locus was subsequently identified in diverse germplasm (Imtiaz et al., 2008; Ogbonnaya et al., 2008; Torada et al., 2008). In these studies the QTL effect was rarely characterized, thus limiting the understanding of its physiological basis. Also, the gene(s) underlying Phs-A1 have recently been investigated. A number of candidate genes including those encoding for Aquaporin (Lohwasser et al., 2013) and GA20-Oxidase (Tyagi and Gupta, 2012; Cabral et al., 2014) have been suggested based on homology to model species. Recently a transcriptomics study of wheat seed dormancy by Barrero et al. (2015) identified two tandem genes, PM19-A1 and PM19-A2, encoding ABA-inducible Plasma Membrane 19 proteins, as the main candidates for Phs-A1. Furthermore, the PM19 genes were shown to be positive regulators of wheat seed dormancy as RNA interference (RNAi) lines with reduced expression of both genes show reduced level of seed dormancy (Barrero et al., 2015).
In the present study we identified, validated and characterized the effect of the 4AL Phs-A1 QTL. We show that this QTL exerts its effect by regulating the duration of seed dormancy loss after seed maturation. Furthermore, through multiple fine-mapping experiments in independent bi-parental mapping populations, we narrowed down the effective resistance locus to an interval less than 0.5 cM which, surprisingly, excludes the PM19 candidate genes reported by Barrero et al. (2015). We discuss the conflicting nature of these results and the implication of our finding on the effort to breed for more PHS resilient wheat varieties.
Materials and methods
Plant materials
The identification, characterization and high-resolution fine-mapping of Phs-A1 were done in two experimental populations made from the Option × Claire and Alchemy × Robigus crosses. These four cultivars are UK winter bread-wheat varieties with Alchemy and Option being PHS resistant while Claire and Robigus are PHS susceptible. The 4AL Phs-A1 QTL was originally detected in the doubled haploid (DH) populations made from these crosses using the wheat × maize technique from F1 plants (Laurie and Bennett, 1988). Forty-eight and 122 DH individuals were analysed in the Alchemy × Robigus and the Option × Claire population, respectively. Selected DH lines from these populations were subsequently used to develop the mapping population used in this study as detailed below.
Alchemy × Robigus fine-mapping population
For subsequent characterization and fine mapping of Phs-A1 in the Alchemy × Robigus population, we developed near isogenic lines (NILs) and recombinant inbred lines (RILs). To accomplish this, five SSR markers including barc170, wmc420, wmc707, wmc760 and wmc313 were used to select DH lines homozygous for Alchemy in different, but overlapping, intervals across the 4AL chromosome arm. These were independently backcrossed to the recurrent parent Robigus and advanced to the BC3 generation by crossing heterozygous plants selected at each generation. Following self-pollination of selected BC3F1 lines, NILs homozygous for the Alchemy introgression found in the original DH lines were selected using the SSR markers flanking the introgressions. For the development of RILs used for high resolution fine-mapping, BC3F2 lines heterozygous for Phs-A1 interval (barc170-wmc420) were self-pollinated and BC3F3 lines with recombination events between the critical Phs-A1 interval were selected. These were advanced to the BC3F4 generation by self-pollination to obtain homozygous RILs.
Option × Claire fine-mapping population
We also developed F4 RILs from the Option × Claire cross. This was accomplished by crossing a DH line (OC69) homozygous for Option across the QTL interval with Claire. Following self-fertilization of F1 progeny, 2400 F2 plants were screened and 85 F2 recombinant lines with recombination events between markers barc170, wms894 and xhbe03 were recovered. Thirty of these lines were randomly selected, self-fertilized and lines with homozygous recombinant haplotype were extracted from the F3 population. In addition, lines with Claire or Option non-recombinant haplotype were also selected as controls. However, only 27 of these were initially phenotyped and advanced to the F4 generation for further phenotyping.
Growth conditions
Three germination index (GI) experiments and five sprouting experiments were conducted in the Alchemy × Robigus and the Option × Claire populations. All the GI experiments were conducted in the Alchemy × Robigus population, whereas sprouting experiments were conducted in both the Alchemy × Robigus (sprouting experiment-1 and 5) and Option × Claire populations (sprouting experiment-2, -3 and -4). In GI experiment-1 and sprouting experiment-1, -3 and -5, plants were grown in the glasshouse under long day conditions with 16h light (300 mmol) at 18 °C, 8h darkness at 15 °C and at relative humidity of 70%. In GI experiment-2 and sprouting experiment-4, plants were grown in controlled environment room (CER) under long day conditions with 16h light (250–400 mmol) at 20 °C, 8h darkness at 15 °C and at 70% relative humidity. GI experiment-3 was designed to test if Phs-A1 is still effective when grains are developed at low temperature. In this experiment, plants were transferred 1–7 d after anthesis into a CER and maintained at constant day and night temperature of 13 °C. Plant materials for sprouting experiment-2 were grown in the field at Thriplow, UK (52.1000° N, 0.1000° E) as single rows in 1 m2 plots using in a randomized complete block design with two replications per line.
SNP and SSR genotyping
Five SSR markers including wmc420, barc170, wmc707, wmc760 and wmc313 were used for the development of Alchemy × Robigus NILs. For the fine-mapping of Phs-A1, SSR wms894 and xhbe03 were used to genotype Option × Claire RILs while only xhbe03 was used for Alchemy × Robigus RILs as wms894 is not polymorphic in this cross. The primer sequences of SSR markers were obtained from the GrainGenes database (http://wheat.pw.usda.gov/GG3, last accessed 4 February 2016), except for wms894 which was obtained from RAGT Seed, UK. These were labelled with the FAM, VIC, NED or PET fluorescent dye (Applied Biosystems) for the multiplexing of assays. PCR were performed with the Qiagen Hotstart Master Mix (Qiagen, Cat No: 203443) and in volume of 6.25 µl containing 3.125 µl of Hotstart mix, 0.625 µl of primer mix and 2.5 µl of DNA. Thermal cycling conditions was as follow: Hotstart at 95 °C for 15min, 35 cycles of 95 °C for 1min; 50–60 °C (depending on annealing temperature of primers) for 1min, 72 °C for 1min and a final extension step of 72 °C for 10min. PCR amplicon were afterwards run on an Applied Biosystems 3730 DNA Analyzer using GeneScan 500 LIZ (Thermo Fisher Scientific; Cat. No:4322682) as size standard. Genotype data were analysed on the GeneScan® Analysis Software (Applied Biosystems). SSR markers barc170, wmc707, wmc760 and wmc313 gave Alchemy/Robigus band sizes of 170/180bp, 165/190bp, 110/100bp and 185320/340325bp, respectively, while wms894 and xhbe03 gave Option/Claire band sizes of 160/125bp and 140/138bp, respectively.
For the development of SNP markers, sequences of wheat genes orthologous to Brachypodium genes in the syntenic Phs-A1 interval were amplified and sequenced to identify SNPs between parental lines. Kompetitive Allelic Specific PCR assays (KASP: Smith and Maughan, 2015) were developed for each SNP. Assays were performed in 384 well plate format in a 5.07 µl volume containing 2.5 µl of DNA, 2.5 µl of KASP master mix (LGC, UK) and 0.07 µl of primer mix. PCR was performed on an Eppendorf Mastercycler pro 384 using the following protocol: Hotstart at 95 °C for 5min, ten touchdown cycles (95 °C for 20s; touchdown 65 °C, −1 °C per cycle, 25s) followed by 30–40 cycles of amplification (95 °C for 10s; 57 °C for 1min). No extension step is necessary as KASP amplicons are smaller than 100bp. Plates were read using the Tecan SAFIRE Fluorescent Scanner and genotype data was viewed graphically with the KlusterCaller™ software (LGC, UK).
Germination index (GI) assays
At four stages of grain maturation and after ripening including physiological maturity, harvest maturity (7 d after physiological maturity), as well as 14 and 28 d after harvest maturity, ears were harvested and gently threshed to obtain grains from the central portion of the ear. For plants grown under constant 13 °C in GI experiment-3, harvest maturity was reached 12 d after physiological maturity. Twenty grains were placed with the crease facing down in 90mm petri dishes containing two layers of Sartorius filter paper and were incubated in 5ml of sterile water for 7 d. After each day of incubation, germinated seeds (with ruptured seed coat) were counted and removed from the plate. The number of germinated seeds per day was used to calculate a weighted GI score using the formula described by Walker-Simmons (1987) with a slight modification: GI=(7×n1)+(6×n2)+(5×n3)+(4×n4)+(3×n5)+(2×n6)+(1×n7)/7×(N−M). Where n1, n2, … n7 are the number of germinated grains on the first, second, and nth days until the 7th day, respectively; N is the total number of grains per plate and M is the number of mouldy grains after the 7 d of incubation. GI test were conducted on ears from six independent plants per NIL group in GI experiment-1 and three to four independent plants per line in GI experiment-2 and -3. The average GI scores are presented.
Sprouting test
Plants were synchronized by days to flowering and maturity and ears were harvested at similar times post-anthesis and allowed to after-ripen at room temperature (20 °C) until GI differences were observed between the parental varieties. After-ripened ears (two to three per plant) were arranged standing upright on wire racks loaded on a revolving wheel inside a sprouting chamber. The ears were then misted for 5–7 d under 100% humidity. Misted ears were dried and ears from the same plant were gently threshed together to collect grains, which were examined for the symptoms of sprouting damage (breakage of the seed coat near the embryo). This was used to calculate the percentage of sprouting in each plant. The number of independent plants phenotyped per line ranged from 12 plants per NIL in sprouting experiment-1; two plants per RIL in sprouting experiment-2, 2–6 plants per RIL in sprouting experiment-3; 4–24 independent plants per RIL in sprouting experiment-4 and three plants per RIL in sprouting experiment-5. The total number of seeds across the experiments ranged from 50 to 200 seeds per genotype. The NILs and RILs were grouped according to their haplotype and the number of lines in each group are indicated in the figure legends. The average sprouting percentages of haplotype groups are presented in the main figures while the average sprouting percentages of individual RILs in experiments-3 to -5 are presented in the supplementary data.
Statistical analyses
Statistical significance was calculated using either one-way or two-way analyses of variance (ANOVA). Tukey’s Honestly Significant Difference (HSD) tests and Dunnett’s tests (using parental varieties as controls) were performed for multiple comparisons between NIL, RILs and parents. Data that did not meet the ANOVA assumption of homogeneity of variance were arcsin transformed and confirmed to meet the assumptions before being used for the ANOVA analysis. Statistical analyses were performed in Genstat (version15.2.0.8821) and Minitab (Version, 17.2.1).
Results
Validation of Phs-A1 in UK bi-parental populations
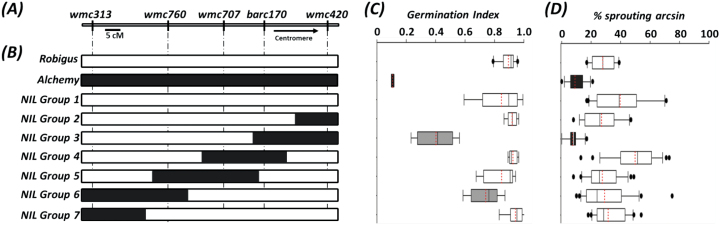
The 4AL QTL effect is known to segregate in UK wheat varieties (Flintham et al., 2011) and has been identified in DH populations derived from crosses between Alchemy × Robigus and Option × Claire (Alchemy and Option providing the resistance allele). The QTL is flanked by markers barc170 and wmc491 in the Alchemy × Robigus population (Supplementary Fig. S1 at JXB online) and as such collocated with the major Phs-A1 QTL identified across multiple studies (Flintham et al., 2002; Imtiaz et al., 2008; Torada et al., 2008; Barrero et al., 2015). To independently validate the effect of Phs-A1, we developed NILs from the Alchemy × Robigus cross through marker-assisted backcrossing (details in Methods). Five markers distributed across the 4AL chromosome arm were used for NIL development, including barc170 and wmc420 (1 cM proximal to the wmc491 flanking marker) as well as wmc707, wmc760 and wmc313 which are distal to barc170 (Fig. 1A). Seven overlapping recombination haplotypes (designated as NIL Groups 1–7) were developed (Fig. 1B) with only Group 3 NILs containing the Alchemy resistant haplotype across the complete QTL interval (barc170–wmc420).
Fig. 1.
Validation of Phs-A1 in Alchemy × Robigus NILs. (A) Genetic map of SSR markers across the 4AL chromosome arm used to develop the NILs. (B) Graphical genotypes of Alchemy × Robigus NILs. The NILs are grouped based on their recombination haplotype across the marker intervals, with each group comprising two independent NILs. The black filled portion in the graphical genotype represents the Alchemy alleles, whereas the white sections represent the Robigus alleles. (C) Mean germination index of each NIL group in GI experiment-1. (D) Sprouting phenotype of each NIL group in sprouting experiment-1. The left and right boundaries of the boxplot indicate the 25th and 75th percentile, respectively, while the error bars (whiskers) on either side of the boxplot indicate the 10th and 90th percentiles. The solid line within the boxplot marks the median (50th percentile) while the red line within the box marks the mean.
We assessed the seed dormancy and PHS resistance phenotype of these NILs through a GI test on threshed seeds (GI experiment-1) and an artificial sprouting test on whole spikes (sprouting experiment-1). In the GI test, highly significant differences were observed between the Robigus and Alchemy parental controls (P<0.001; Fig. 1C). NILs were classified as either resistant or susceptible based on a Dunnett’s test to the parental controls. NILs with higher or non-significant GI differences than Robigus were classified as susceptible, while NILs with lower or non-significant GI differences than Alchemy were classified as resistant (Supplementary Table S1). NIL Groups 1, 5 and 7 with the Robigus haplotype across the QTL interval, and NIL Groups 2 and 4 with recombinant haplotypes within the QTL interval, all showed the susceptible GI phenotype (Fig. 1C). Group 3 NILs showed significantly lower GI than Robigus (P<0.001) but also significantly higher GI than Alchemy (P<0.001). Likewise, Group 6 NILs also showed significant differences from both parents but the GI was only slightly lower than the susceptible Robigus parent.
In the sprouting test, all the NIL groups (except Group 3) were significantly different than Alchemy but not Robigus and were therefore classified as being susceptible to sprouting (sprouting experiment-1; Fig. 1D). Group 3 NILs showed comparable sprouting levels to the resistant variety Alchemy, and were significantly different from Robigus (P<0.001), consistent with the GI results. Taken together, the GI and sprouting results validate the resistance effect of Phs-A1 in NILs with the Alchemy haplotype across the complete barc170-wmc420 interval. NILs from Group 2 and 4, which have the Alchemy allele at either one or other – but not both – flanking markers, were susceptible suggesting that the Phs-A1 resistance locus is delimited by, but not linked to, these markers.
Phs-A1 reduces the rate of dormancy loss during dry seed after-ripening
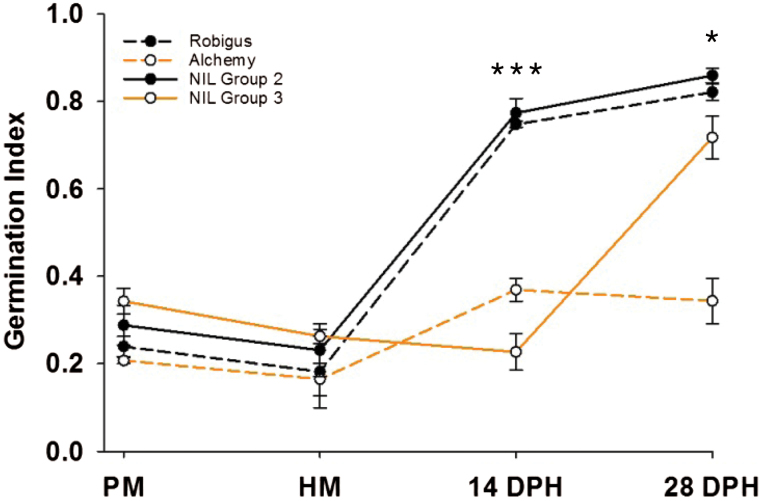
The validation experiments suggest that Phs-A1 confers PHS resistance by affecting seed dormancy since the effect was not only observed in whole spikes (sprouting test), but was also evident in the reduced germination potential of threshed seeds (GI test). To further understand this resistance mechanism, we examined the rate of dormancy loss in one of the resistant and susceptible NIL groups used in the previous experiment through the GI test (GI experiment-2). This was done across different stages of seed maturation and after-ripening including physiological maturity (PM, ~40% seed moisture content), harvest maturity (HM, ~20% seed moisture content) and two post-harvest time points (14 and 28 d post-harvest, DPH). The NILs used for the experiment (NIL Group 2 and 3) were genetically isogenic and only differed in the extent of the Alchemy introgression between barc170 and wmc420 (Fig. 1A). This allows for a precise characterization of this region without the confounding effect of other background loci.
At PM and HM, all lines showed similarly low GI with no difference between the contrasting alleles (P>0.51; Fig. 2). However, at 14 DPH there was a significant GI difference between the contrasting NILs and parents, with Robigus and the susceptible Group 2 NIL showing increased germination potential compared to Alchemy and the resistant Group 3 NIL (P<0.001). This difference in germination potential was maintained at 28 DPH, although the differences were reduced and less significant than at 14 DPH (P<0.05).
Fig. 2.
Phs-A1 delays the rate of seed dormancy loss during after-ripening. The germination index of seeds harvested from Robigus, Alchemy and NILs with either the recombinant haplotype (NIL Group 2) or Alchemy haplotype (NIL Group 3) between barc170 and wmc420 in GI experiment-2. Seeds were tested at physiological maturity (PM), harvest maturity (HM; 7 d after PM), 14 and 28 d post-harvest (DPH) and germinated at 16 °C. Error bars represent standard error of the means (SEM) of three biological replications for each time point. Significant differences between NILs at P<0.05 (*) and P<0.001 (***) are indicated.
The previously cloned seed-coat-independent PHS QTL regulated by TaMFT is effective when seeds develop under low temperature (Nakamura et al., 2011). We therefore examined if Phs-A1 was effective at low temperature by measuring the rate of seed germination in NILs grown at 13 °C post-anthesis (GI experiment-3). While the NILs displayed an overall higher depth of seed dormancy when plants were grown at 13 °C post anthesis, the same pattern of GI differences at the post-harvest time-points (but not at PM and HM) was observed (Supplementary Fig. S2). Together, these experiments suggest that Phs-A1 delays the rate of seed dormancy loss during after-ripening.
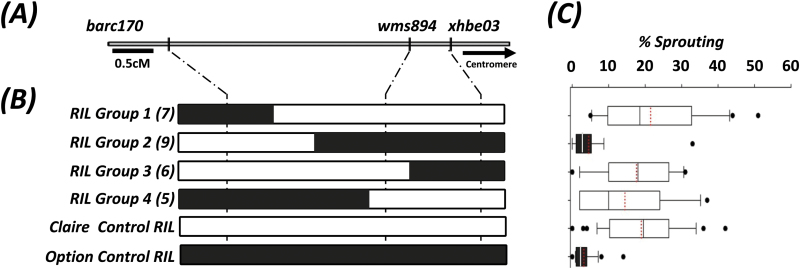
Phs-A1 maps to a 0.5 cM interval between wms894 and xhbe03
As previously stated, Phs-A1 is flanked by barc170 and wmc420 in the bi-parental populations used in this study. Torada et al. (2008) mapped Phs-A1 to a 2.6 cM interval between barc170 and xhbe03. We therefore used barc170, xhbe03, and another marker – wms894 – in the same physical bin (4AL_13-0.59–0.66), to characterize Option × Claire F4 RILs (Fig. 3A, B). We selected 27 homozygous recombinants across the interval, grouped these according to their haplotypes (Fig. 3B), and assessed the sprouting phenotype using the artificial sprouting test (sprouting experiment-2). Two significantly different sets were identified in this experiment: one was made up of RIL Group 2 and Option Control RILs with between 3 and 5% sprouting, whereas the second set contained RIL groups 1, 3, 4 and the Claire Control RILs with average sprouting between 15 and 22% (Fig. 3C). RIL Group 2 and the Option Control RILs were similar to the resistant Option parent and had the Option haplotype between wms894 and xhbe03. RIL Groups 1, 3, 4 and the Claire Control RILs were similar to the susceptible Claire parent and carried a homozygous Claire or recombinant haplotype across the wms894–xhbe03 interval. This suggests that the Phs-A1 resistance is only observed when RILs have the Option haplotype across the 0.5 cM wms894-xhbe03 interval.
Fig. 3.
Interval mapping of Phs-A1 in the Option × Claire RIL population. (A) Genetic map of the SSR markers flanking Phs-A1. (B) Graphical genotypes of RILs and controls are presented with the Option and Claire alleles represented in black and white, respectively. The RILs are grouped according to their fixed genotype across the Phs-A1 interval and the number of lines in each RIL group is indicated in parenthesis. (C) Sprouting phenotype of RIL groups and controls in sprouting experiment-3. The left and right boundaries of the boxplot indicate the 25th and 75th percentile, respectively, while the error bars (whiskers) on either side of the boxplot indicate the 10th and 90th percentiles. The solid line within the boxplot marks the median (50th percentile) while the red line within the box marks the mean. Boxes with the same colour are similar to each other based on pairwise comparisons.
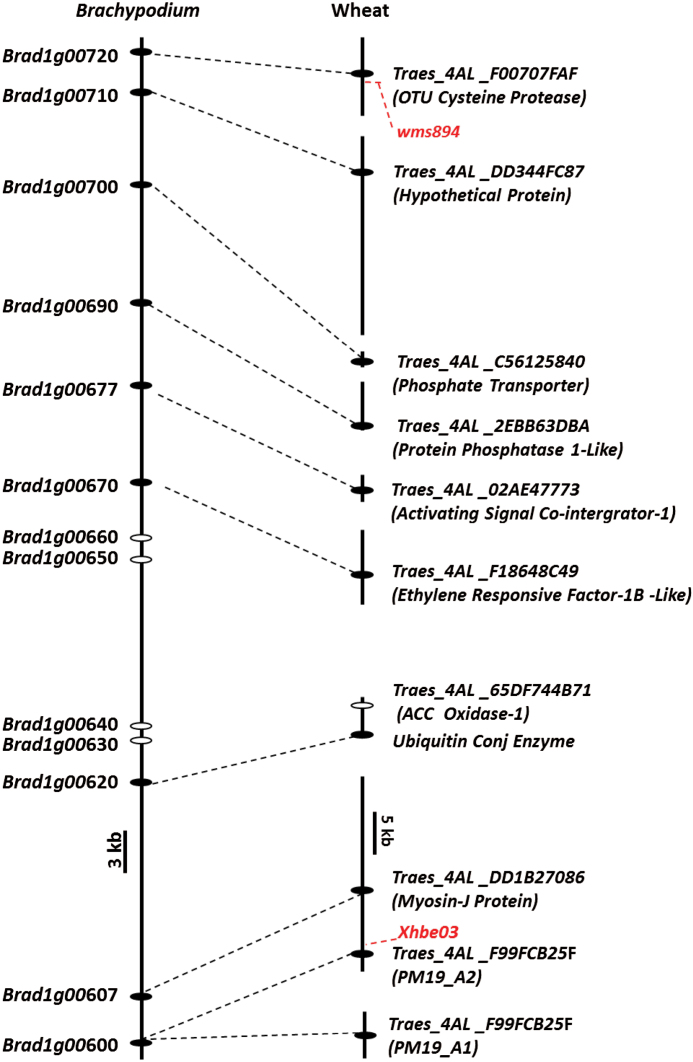
Synteny reveals the putative gene content of the Phs-A1 locus
Given the small genetic interval to which Phs-A1 mapped, we evaluated the gene content across this locus. We first identified genes containing the flanking markers (xhbe03 and wms894) in wheat: xhbe03 is designed from the 3ʹ UTR sequence of PM19-A2 (Traes_4AL _F99FCB25F) while the sequence of wms894 is located in the promoter region of an OTU Cysteine Protease gene (Traes_4AL _F00707FAF). We next examined the collinear region in Brachypodium: reciprocal BLASTs against the Brachypodium genome identified Bradi1g00600 and Bradi1g00720 as orthologues of PM19-A2 and OTU Cysteine Protease, respectively. This defined the collinear Phs-A1 interval in Brachypodium to a 75kb region which contains 11 genes (Bradi1g00607 to Bradi1g00710).
The Brachypodium genes were used to search the wheat chromosome arm assemblies of the hexaploid wheat cultivar, Chinese Spring (IWGSC, 2014). Orthologous contigs and gene models to Bradi1g00600–Bradi1g00620 and Bradi1g00670–Bradi1g00720 were identified on chromosome arm 4AL. These included PM19-A2 and its paralogue PM19-A1, as well as genes encoding for Myosin J protein, Ubiquitin conjugating enzyme, Ethylene Responsive Factor-1B-Like (ERF-1B-Like), Activating Signal Co-integrator-1 (ASC1), Protein Phosphatase 1-Like (PP1-Like), a phosphate transporter, a hypothetical protein and the OTU Cysteine Protease (Fig. 4, Supplementary Table S2). No wheat orthologues were identified for Bradi1g00630 to Bradi1g00660. Within the wheat IWGSC contigs, a non-collinear gene encoding for an Aminocyclopropane Carboxylate Oxidase 1-Like protein (ACC Oxidase-1; Traes_4AL_65DF744B71) was also identified. All these genes/contigs were mapped within or linked to the critical wms894–xhbe03 interval using SNP-based KASP assays (Supplementary Table S3), except for Traes_4AL_C56125840 which we did not map due to the lack of a genetic marker. This confirmed the collinear gene order between wheat and Brachypodium and suggests possible candidate genes for Phs-A1.
Fig. 4.
Synteny reveals the putative gene content of the Phs-A1 locus. Sequences of genes containing the Phs-A1 flanking markers (wms894 and xhbe03; in red) were used to obtain genes in the orthologous Brachypodium interval (Brad1g00600–Brad1g00720). Collinear genes are represented by black ovals while non-collinear genes are represented by the white ovals. Orthologous wheat contigs (black lines) and gene models are connected to their corresponding Brachypodium genes. All the wheat genes were genetically mapped within or linked to the wms894–xhbe03 interval except for Traes_4AL _C56125840. Wheat orthologue could not be found for Brad1g00630–Brad1g00660.
Phs-A1 maps distal to the PM19 genes in two UK fine-mapping populations
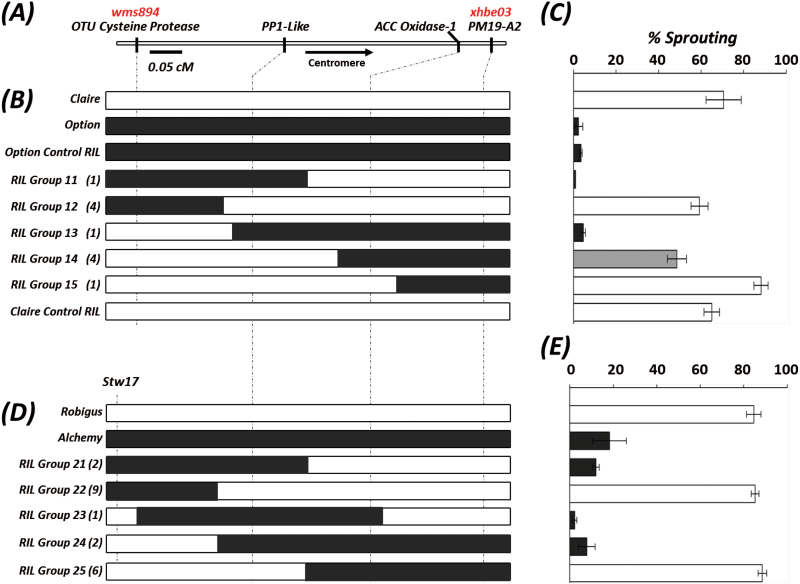
Barrero et al. (2015) identified PM19-A1 and PM19-A2 as the main candidates for a seed dormancy QTL on wheat 4AL chromosome arm in a multi-parental mapping population. To determine if these genes determined the allelic variation observed in the UK populations, we further fine-mapped Phs-A1 in the Option × Claire F4 RILs with homozygous recombinant and non-recombinant haplotypes in the Phs-A1 interval. We first defined the linkage between the gene-based KASP assays previously used to map the syntenic genes (Fig. 5A). The two PM19 genes were completely linked and so too were the PP1-like, ERF-1B-like and ASC1 genes. There were however recombination events between PP1-Like/ ERF-1B-Like/ ASC1 and OTU Cysteine Protease and between ACC Oxidase-1 and PM19-A2/PM19-A1. Given the genetic linkage between some of these markers, four SNP markers including OTU Cysteine Protease, PP1-Like, ACC Oxidase-1 and PM19-A2 (Supplementary Table S3) were used to define five distinct recombinant haplotypes (RIL Group 11–15; Fig. 5B).
Fig. 5.
High-resolution fine-mapping of Phs-A1 in Option × Claire and Alchemy × Robigus RIL populations. (A) Linkage map of SNP (black) and SSR (red) markers across the Phs-A1 interval. The graphical genotype of Option × Claire RILs (B) and Alchemy × Robigus RILs (D) are aligned against their sprouting phenotype (C and E, respectively). RILs are grouped based on their recombination haplotype across the marker interval and the number of lines in each group is indicated in parentheses. Resistant parent alleles (Option and Alchemy) are represented in black, whereas the susceptible parent alleles (Claire and Robigus) are shown in white. Marker stw17 (2 cM distal to wms894) was used in the Robigus × Alchemy population as wms894 and OTU Cysteine Protease are monomorphic. The sprouting phenotype of each RIL group is designated as susceptible (white), moderate (grey) or resistant (black) based on statistical comparison with the parental controls. Error bars represents SEM.
A subset of lines from each RIL group, in addition to the parental cultivars and non-recombinant Claire and Option control RILs were phenotyped using the artificial sprouting test (sprouting experiment-3; Fig. 5B). Variation in sprouting percentage was observed defining a bimodal distribution (Supplementary Fig. S3). To unequivocally assign sprouting phenotypes to these lines, the mean sprouting percentages of each RIL group (Fig. 5C) as well as the individual sprouting percentages of each RIL (Supplementary Table S4) were compared against those of Claire and Option using the Dunnett’s test. This showed that the sprouting phenotype is completely associated to the PP1-Like/ERF-1B-Like/ASC1 linkage in all the lines tested. Five independent recombination events (Group 12 and 13) map Phs-A1 proximal to the OTU Cysteine Protease (wms894) marker. Similarly, the six lines from RIL Groups 11, 14 and 15 map Phs-A1 distal to both the ACC Oxidase-1 and the PM19 genes. This was unexpected given the reported association of the PM19 genes with sprouting resistance. We confirmed this result in an independent sprouting experiment (sprouting experiment-4; Supplementary Table S5) using the critical Group 12–14 RILs. Although a higher level of sprouting was observed in this experiment, Phs-A1 still conferred a moderate level of resistance, which was associated with the PP1-Like/ERF-1B-Like/ASC1 linkage.
We also independently fined-mapped Phs-A1 in an Alchemy × Robigus RIL population, which contained similar recombination haplotypes (RIL Groups 21–25; Fig. 5D) as in the Option × Claire population. However, marker stw17 was used in place of the OTU Cysteine Protease marker as this was not polymorphic in the Alchemy × Robigus cross. We assessed the sprouting phenotype of these lines using the artificial sprouting test (sprouting experiment-5, Supplementary Table S6). Similar to the previous results, the mean sprouting percentages of each RIL group (Fig. 5E) confirmed the complete linkage of Phs-A1 to PP1-Like, ERF-1B-Like and ASC1 genes. Eleven independent recombination events in RIL Group 22 and 24 map Phs-A1 proximal to stw17, whereas nine independent RILs map Phs-A1 distal to the ACC Oxidase-1 and PM19 genes (RIL Groups 21, 23 and 25). This provides strong genetic evidence that in the two UK mapping populations Phs-A1 maps distal to the PM19 genes.
Discussion
Phs-A1 confers resistance to sprouting in wheat
In this study we characterized and fine-mapped Phs-A1, a major PHS resistance and seed dormancy QTL in wheat. This QTL has been previously identified across diverse germplasm and agro-ecological zones, including Australia, Canada, China, Japan and Europe (Kato et al., 2001; Li et al., 2004; Mares et al., 2005; Torada et al., 2005, 2008; Ogbonnaya et al., 2007; Chen et al., 2008; Cabral et al., 2014; Albrecht et al., 2015). Given its widespread identification and the magnitude of its effect, Phs-A1 plays a crucial role in providing resilience to pre-harvest sprouting in wheat.
Despite its consistent identification, only Torada et al. (2008) had previously validated the effect of Phs-A1 in independent isogenic material. In this study, we also validated the effect in UK germplasm using isogenic lines. In both the GI and the artificial sprouting test, we show the QTL to be effective only in lines carrying the Alchemy (resistant) haplotype across the entire barc170–wmc420 QTL interval. However, the level of dormancy of these NILs in the GI test was only intermediate to that of Alchemy, unlike in the artificial sprouting test, where NILs showed similar sprouting resistance as Alchemy. This suggests the presence of additional loci controlling seed dormancy in the Alchemy × Robigus cross, which are independent of the 4AL region.
Phs-A1 delays the rate of dormancy loss during seed after-ripening
Understanding the mode and timing of expression of QTL is important to inform effective deployment strategies in breeding programmes and further define the underlying mechanisms. Our physiological characterization shows that Phs-A1 confers sprouting resistance by delaying the rate of dormancy loss during after-ripening (2–4 weeks after harvest ripeness). After-ripening could be described as a period of dry seed storage during which physiological changes within seeds ultimately lead to the release from dormancy (Gao and Ayele, 2014). Some of these physiological changes include non-enzymatic oxidation of mRNA and protein by Reactive Oxygen Species (ROS) giving rise to changes in protein levels, properties and function upon imbibition (Bykova et al., 2011; Gao et al., 2013a; Gao and Ayele, 2014). Also, changes in the transcript level of genes involved in the biosynthesis or signalling of several hormones, including ABA, indole acetic acid, brassinosteroid, ethylene, cytokinin and salicylic acid have been reported (Liu et al., 2013a; Chitnis et al., 2014). When and how these physiological and transcriptional changes are initiated in dry or imbibed seeds are not well understood. However, the strong effect of Phs-A1 on after-ripening provides an entry point to further understand the molecular pathway regulating seed after-ripening in wheat and possibly other cereals.
Phs-A1 maps 0.3 cM distal to the PM19 genes
Fine-mapping of Phs-A1 to an initial 0.5 cM interval revealed the presence of at least ten genes in the syntenic Brachypodium region with varied biological functions. However due to differential gene loss and duplication events between wheat and Brachypodium (Faris et al., 2008; Glover et al., 2015), it is possible that these do not represent the complete gene content in wheat. This is supported by the physical map surrounding the PM19 genes by Barrero et al. (2015), which revealed the presence of additional genes besides the ones defined solely by syntenic relationships. Importantly, the high-resolution fine-mapping conclusively excludes at least six of these genes as being causal for Phs-A1, including the OTU Cysteine Protease, ACC oxidase-1, Ubiquitin Conjugating Enzyme, Myosin J, PM19-A1 and PM19-A2 genes. These results are particularly surprising for the PM19 genes, but data from three independent experiments across two different mapping populations showed that Phs-A1 maps 0.3 cM distal to the PM19 locus in at least 16 recombinant lines (15 lines in Fig. 5 and one additional line in Supplementary Table S5). Based on these results, we argue that the PM19 genes are not the main cause of the Phs-A1 effect, at least in UK wheat varieties. In further support of this conclusion, Torada et al. (2008) mapped Phs-A1 0.5 cM distal to xhbe03 (located in the 3ʹ UTR of PM19-A2) in two independent populations derived from Japanese and Canadian germplasm. The results of Torada et al. (2008) are equivalent to those presented here and suggest that the two PM19 loci are not the causal genes defining the 4AL sprouting resistance in the UK and possibly other germplasm pools.
The discrepancy, with the results obtained by Barrero et al. (2015), could be explained by a number of factors. Firstly, although the PM19 genes affect seed dormancy in wheat, it is possible that they do not account for the natural variation in sprouting mapped to Phs-A1 but are instead closely linked to the causal locus. The transgenic data reported by Barrero et al. (2015) convincingly support the role of the PM19 genes in promoting primary seed dormancy in wheat. However, the genetic evidence presented by Barrero et al. (2015) is based on the phenotype of only one of five heterogeneous inbred families (F1038) developed from a four-parent MAGIC population. Given the diverse nature of this F7 MAGIC population, it is possible that the phenotype of the F1038 family might have been conditioned by other loci that are independent of Phs-A1, but were not accounted for in the study. Indeed, there is evidence of this occurring in this population: the phenotype of another heterogeneous inbred family presented in the study (F1516) did not completely support the PM19 genes as the causal genes.
Alternatively, it is possible that the 4AL PHS resistance originates from two independent natural variants in the closely linked PM19 and Phs-A1 loci. This scenario would be reminiscent of the VRN1 locus controlling vernalization requirement and flowering in wheat (Yan et al., 2003). The VRN1 locus contains three closely linked genes: APETALA1 (AP1), AGAMOUS – LIKE GENE 1 (AGLG1) and Phytochrome C (PHYC), which all function in transitioning to flowering in Arabidopsis (Irish and Sussex, 1990; Flanagan and Ma, 1994; Balasubramanian et al., 2006; Preston et al., 2009; Heijmans et al., 2012). Detailed genetic, expression and sequence analysis initially showed AP1 to be the main gene underlying VRN1. However, recent evidence suggests that PHYC is also important in accelerating flowering in wheat under long day conditions (Chen et al., 2014). Analogously, allelic variation at both PM19 genes and the Phs-A1 locus could determine resistance to sprouting, but distinct allelic variants could have been selected in Australian and UK germplasm. If true, this would account for the widespread identification of the wider Phs-A1 locus in diverse germplasms around the world.
In support of this multiple causal gene hypothesis, we identified similar but distinct polymorphisms in the PM19 genes in our experimental populations compared to those reported by Barrero et al. (2015). In the sequence of PM19-A1 (Supplementary Fig. S4), we found only six of the seven SNPs reported between the dormant (Yitpi) and one of the non-dormant (Chara) parents used by Barrero et al. (2015), with only one of these leading to amino acid change between our contrasting parents (Alchemy/Option and Robigus/Clare). We also found a 12-bp deletion close to the translation stop codon in our dormant parent which resulted in the loss of four amino acid residues. This deletion was not reported in the Australian germplasm. Similar haplotype differences were observed in the PM19-A2 sequence (Supplementary Fig. S5). A smaller deletion polymorphism (188bp) was found in the promoter sequence of PM19-A2 in UK germplasm compared to the 216bp deletion in Australian lines. Likewise, only one SNP was identified in the coding sequence of PM19-A2 by Barrero et al. (2015), while we identified nine SNPs of which three were non-synonymous. This clearly suggests differences in haplotype structure in these genes between Australian and UK germplasm. This might have functional implications on the regulation and role of PM19 in these different germplasm pools.
Based on the present study, it is difficult to determine which of these hypotheses is true. We envisage that similar high-resolution fine-mapping of the Phs-A1 locus in different genetic backgrounds might help resolve this discrepancy. Importantly, the phenotypic characterization of the onset of PHS resistance, as well as the marker resources developed in this study, will facilitate comparative studies across other germplasm, helping clarify the association of the PM19 gene with the Phs-A1 natural allelic variation. If the multiple causal gene hypothesis is established, it would also be interesting to test the epistatic interaction between PM19 and Phs-A1 by examining lines with recombinant and non-recombinant Yipti and Alchemy/Option haplotypes between the two loci.
Towards the identification of Phs-A1 causal gene
In three independent fine-mapping experiments (sprouting experiment-3 to -5), Phs-A1 showed complete linkage to three genes: ERF-1B-Like, ASC1 and PP1-Like. None of the genes has previously been implicated in the control of dormancy and the induction of germination. ERF-1B-Like encodes for an ethylene responsive transcription factor orthologous to Arabidopsis AtERF1, which belongs to the AP2/ERF Group-IX family involved in defence response against pathogens (Licausi et al., 2013). ERF-1B-Like also shows good homology to AtERF15, another member of this AP2/ERF family, which was recently shown to be a positive regulator of ABA response (Lee et al., 2015). ABA is a critical regulator of seed dormancy, with higher seed responsiveness to ABA associated with the PHS resistance. ASC1 has not been characterized in plants, but sequence analysis of genes containing the ASC-homology domain suggest that they are involved in RNA metabolism as transcription co-activator, RNA processor and regulator of translation (Iyer et al., 2006). PP1-like is a member of the serine threonine phosphoprotein phosphatase (PPP), which are involved in a wide range of cellular processes. Since there are likely to be other non-syntenic genes within the interval, it would be premature to suggest these as unique candidate genes for Phs-A1. We are currently working to establish a physical map of this interval to reveal other possible candidate genes for Phs-A1.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. PHS resistance QTL on chromosome arm 4AL in the Alchemy × Robigus DH population.
Fig. S2. After-ripening effect of Phs-A1 in NILs grown at 13 °C post-anthesis.
Fig. S3. Distribution of the sprouting percentage of Option × Claire F4 RILs in sprouting experiment-3.
Fig. S4. Alignment of PM19-A1 coding sequences of UK and Australian germplasm.
Fig. S5. Alignment of PM19-A2 coding sequences of UK and Australian germplasm.
Table S1. Statistical comparison of the GI and sprouting phenotype of Alchemy × Robigus NILs.
Table S2. Information on genes found in the syntenic Phs-A1 intervals in wheat and Brachypodium.
Table S3. KASP SNP assays used to fine map Phs-A1.
Table S4. Statistical comparison of the sprouting scores of Option × Claire RILs in sprouting experiment-3.
Table S5. Statistical comparison of the sprouting score of Option × Claire RILs in sprouting experiment-4.
Table S6. Statistical comparison of the sprouting score of Alchemy × Robigus RILs in sprouting experiment-5.
Acknowledgements
We thank Peter Scott (JIC) for his help with plant husbandry and sample collection. This work was supported by the UK Biotechnology and Biological Sciences Research Council (BBSRC), the UK Agriculture and Horticulture Development Board (AHDB) and a consortium of four companies (KWS, Lantmännen, Limagrain and RAGT) through LINK grant BB/I01800X/1 and grants BB/J004588/1 and BB/J004596/1. TG, DS and MJH were supported by BBSRC grant BB/D007321/1 while BB and MV were supported by grant LO1204 from the National Program of Sustainability I, by the Czech Science Foundation (14-07164S). OS was supported by the John Innes Foundation.
References
- Albrecht T, Oberforster M, Kempf H, Ramgraber L, Schacht J, Kazman E, Zechner E, Neumayer A, Hartl L, Mohler V. 2015. Genome-wide association mapping of preharvest sprouting resistance in a diversity panel of European winter wheats. Journal of Applied Genetics 1–9. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Agrawal M, Michael TP, Wessinger C, Maloof JN, Clark R, Warthmann N, Chory J, Weigel D. 2006. The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana . Nature Genetics 38, 711–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero JM, Cavanagh C, Verbyla KL, et al. 2015. Transcriptomic analysis of wheat near-isogenic lines identifies PM19-A1 and A2 as candidates for a major dormancy QTL. Genome Biology 16, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykova NV, Hoehn B, Rampitsch C, Hu J, Stebbing JA, Knox R. 2011. Thiol redox-sensitive seed proteome in dormant and non-dormant hybrid genotypes of wheat. Phytochemistry 72, 1162–1172. [DOI] [PubMed] [Google Scholar]
- Cabral AL, Jordan MC, McCartney CA, You FM, Humphreys DG, MacLachlan R, Pozniak CJ. 2014. Identification of candidate genes, regions and markers for pre-harvest sprouting resistance in wheat (Triticum aestivum L.). BMC Plant Biology 14, 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Li C, Hu W, Lau MY, Lin H, Rockwell NC, Martin SS, Jernstedt JA, Lagarias JC, Dubcovsky J. 2014. PHYTOCHROME C plays a major role in the acceleration of wheat flowering under long-day photoperiod. Proceedings of the National Academy of Sciences, USA 111, 10037–10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CX, Cai SB, Bai GH. 2008. A major QTL controlling seed dormancy and pre-harvest sprouting resistance on chromosome 4A in a Chinese wheat landrace. Molecular Breeding 21, 351–358. [Google Scholar]
- Chitnis VR, Gao F, Yao Z, Jordan MC, Park SE, Ayele BT. 2014. After-ripening induced transcriptional changes of hormonal genes in wheat seeds: the cases of brassinosteroids, ethylene, cytokinin and salicylic acid. PLoS ONE 9, e87543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris JD, Zhang Z, Fellers JP, Gill BS. 2008. Micro-colinearity between rice, Brachypodium and T. monococcum at the wheat domestication locus Q. Functional & Integrative Genomics 8, 149–164. [DOI] [PubMed] [Google Scholar]
- Flanagan CA, Ma H. 1994. Spatially and temporally regulated expression of the MADS-box gene AGL2 in wild-type and mutant arabidopsis flowers. Plant Molecular Biology 26, 581–595. [DOI] [PubMed] [Google Scholar]
- Flintham J, Adlam R, Bassoi M, Holdsworth M, Gale MD. 2002. Mapping genes for resistance to sprouting damage in wheat. Euphytica 126, 39–45. [Google Scholar]
- Flintham J, Holdsworth M, Jack P, Kettlewell PS, Phillips A. 2011. An integrated approach to stabilising HFN in wheat: screens, genes and understanding. Project Report 480. HGCA, Agriculture and Horticulture Development Board.
- Flintham JE. 2000. Different genetic components control coat-imposed and embryo-imposed dormancy in wheat. Seed Science Research 10, 43–50. [Google Scholar]
- Gao F, Ayele BT. 2014. Functional genomics of seed dormancy in wheat: advances and prospects. Frontiers in Plant Science 5, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Rampitsch C, Chitnis VR, Humphreys GD, Jordan MC, Ayele BT. 2013. a Integrated analysis of seed proteome and mRNA oxidation reveals distinct post-transcriptional features regulating dormancy in wheat (Triticum aestivum L.). Plant Biotechnology Journal 11, 921–932. [DOI] [PubMed] [Google Scholar]
- Gao X, Hu CH, Li HZ, Yao YJ, Meng M, Dong L, Zhao WC, Chen QJ, Li XY. 2013. b Factors affecting pre-harvest sprouting resistance in wheat (Triticum aestivum L.): a review. The Journal of Animal and Plant Sciences 23, 556–565. [Google Scholar]
- Gerjets T, Scholefield D, Foulkes MJ, Lenton JR, Holdsworth MJ. 2010. An analysis of dormancy, ABA responsiveness, after-ripening and pre-harvest sprouting in hexaploid wheat (Triticum aestivum L.) caryopses. Journal of Experimental Botany 61, 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover NM, Daron J, Pingault L, Vandepoele K, Paux E, Feuillet C, Choulet F. 2015. Small-scale gene duplications played a major role in the recent evolution of wheat chromosome 3B. Genome Biology 16, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans K, Morel P, Vandenbussche M. 2012. MADS-box genes and floral development: the dark side. Journal of Experimental Botany 63, 5397–5404. [DOI] [PubMed] [Google Scholar]
- Himi E, Maekawa M, Miura H, Noda K. 2011. Development of PCR markers for Tamyb10 related to R-1, red grain color gene in wheat. Theoretical and Applied Genetics 122, 1561–1576. [DOI] [PubMed] [Google Scholar]
- Himi E, Noda K. 2005. Red grain colour gene (R) of wheat is a Myb-type transcription factor. Euphytica 143, 239–242. [Google Scholar]
- Imtiaz M, Ogbonnaya FC, Oman J, van Ginkel M. 2008. Characterization of quantitative trait loci controlling genetic variation for preharvest sprouting in synthetic backcross-derived wheat lines. Genetics 178, 1725–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium (IWGSC) 2014. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788. [DOI] [PubMed] [Google Scholar]
- Irish VF, Sussex IM. 1990. Function of the apetala-1 gene during Arabidopsis floral development. The Plant Cell 2, 741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Burroughs AM, Aravind L. 2006. The ASCH superfamily: novel domains with a fold related to the PUA domain and a potential role in RNA metabolism. Bioinformatics 22, 257–263. [DOI] [PubMed] [Google Scholar]
- Kato K, Nakamura W, Tabiki T, Miura H, Sawada S. 2001. Detection of loci controlling seed dormancy on group 4 chromosomes of wheat and comparative mapping with rice and barley genomes. Theoretical and Applied Genetics 102, 980–985. [Google Scholar]
- Kottearachchi NS, Uchino N, Kato K, Miura H. 2006. Increased grain dormancy in white-grained wheat by introgression of preharvest sprouting tolerance QTLs. Euphytica 152, 421–428. [Google Scholar]
- Laurie DA, Bennett MD. 1988. The production of haploid wheat plants from wheat × maize crosses. Theoretical and Applied Genetics 76, 393–397. [DOI] [PubMed] [Google Scholar]
- Lee SB, Lee SJ, Kim SY. 2015. AtERF15 is a positive regulator of ABA response. Plant Cell Reports 34, 71–81. [DOI] [PubMed] [Google Scholar]
- Lei L, Zhu X, Wang S, Zhu M, Carver BF, Yan L. 2013. TaMFT-A1 is associated with seed germination sensitive to temperature in winter wheat. PLoS ONE 8, e73330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Ni P, Francki M, Hunter A, et al. 2004. Genes controlling seed dormancy and pre-harvest sprouting in a rice-wheat-barley comparison. Functional & Integrated Genomics 4, 84–93. [DOI] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P. 2013. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytologist 199, 639–649. [DOI] [PubMed] [Google Scholar]
- Liu A, Gao F, Kanno Y, Jordan MC, Kamiya Y, Seo M, Ayele BT. 2013. a Regulation of wheat seed dormancy by after-ripening is mediated by specific transcriptional switches that induce changes in seed hormone metabolism and signaling. PLoS ONE 8, e56570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Sehgal SK, Li J, Lin M, Trick HN, Yu J, Gill BS, Bai G. 2013. b Cloning and characterization of a critical regulator for preharvest sprouting in wheat. Genetics 195, 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohwasser U, Rehman, Arif MA, Börner A. 2013. Discovery of loci determining pre-harvest sprouting and dormancy in wheat and barley applying segregation and association mapping. Biologia Plantarum 57, 663–674. [Google Scholar]
- Mares D, Mrva K. 2014. Wheat grain preharvest sprouting and late maturity alpha-amylase. Planta 240, 1167–1178. [DOI] [PubMed] [Google Scholar]
- Mares D, Mrva K, Cheong J, Williams K, Watson B, Storlie E, Sutherland M, Zou Y. 2005. A QTL located on chromosome 4A associated with dormancy in white- and red-grained wheats of diverse origin. Theoretical and Applied Genetics 111, 1357–1364. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Abe F, Kawahigashi H, et al. 2011. A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. The Plant Cell 23, 3215–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbonnaya FC, Imtiaz M, DePauw RM. 2007. Haplotype diversity of preharvest sprouting QTLs in wheat. Genome 50, 107–118. [DOI] [PubMed] [Google Scholar]
- Ogbonnaya FC, Imtiaz M, Ye G, Hearnden PR, Hernandez E, Eastwood RF, van, Ginkel M, Shorter SC, Winchester JM. 2008. Genetic and QTL analyses of seed dormancy and preharvest sprouting resistance in the wheat germplasm CN10955. Theoretical and Applied Genetics 116, 891–902. [DOI] [PubMed] [Google Scholar]
- Rodriguez MV, Toorop PE, Benech-Arnold RL. 2011. Challenges facing seed banks in relations to seed quality. In: Kermode AR, ed. Seed dormancy: methods and protocols, method in molecular biology, Vol 773 New York: Humana Press, 17–40. [DOI] [PubMed] [Google Scholar]
- Preston JC, Christensen A, Malcomber ST, Kellogg EA. 2009. MADS-box gene expression and implications for developmental origins of the grass spikelet. American Journal of Botany 96, 1419–1429. [DOI] [PubMed] [Google Scholar]
- Smith SM, Maughan PJ. 2015. SNP genotyping using KASPar assays. Methods in Molecular Biology 1245, 243–256. [DOI] [PubMed] [Google Scholar]
- Torada A, Ikeguchi S, Koike M. 2005. Mapping and validation of PCR-based markers associated with a major QTL for seed dormancy in wheat. Euphytica 143, 251–255. [Google Scholar]
- Torada A, Koike M, Ikeguchi S, Tsutsui I. 2008. Mapping of a major locus controlling seed dormancy using backcrossed progenies in wheat (Triticum aestivum L.). Genome 51, 426–432. [DOI] [PubMed] [Google Scholar]
- Trenberth KE. 2011. Changes in precipitation with climate change. Climate Research 47, 123–138. [Google Scholar]
- Tyagi S, Gupta PK. 2012. Meta-analysis of QTLs involved in pre-harvest sprouting tolerance and dormancy in bread wheat. Triticeae Genomics and Genetic 3, 9–24. [Google Scholar]
- Walker-Simmons M. 1987. ABA levels and sensitivity in developing wheat embryos of sprouting resistant and susceptible cultivars. Plant Physiology 84, 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther GR, Post E, Convey P, Menzel A, Parmesan C, Beebee TJC, Fromentin JM, Hoegh-Guldberg O, Bairlein F. 2002. Ecological responses to recent climate change. Nature 416, 389–395. [DOI] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. 2003. Positional cloning of the wheat vernalization gene VRN1. Proceedings of the National Academy of Sciences, USA 100, 6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.