Abstract
Normal skeletal development requires tight coordination of transcriptional networks, signaling pathways, and biomechanical cues, and many of these pathways are dysregulated in pathological conditions affecting cartilage and bone. Recently, a significant role has been identified for long noncoding RNAs (IncRNAs) in developing and maintaining cellular phenotypes, and improvements in sequencing technologies have led to the identification of thousands of IncRNAs across diverse cell types, including the cells within cartilage and bone. It is clear that IncRNAs play critical roles in regulating gene expression. For example, they can function as epigenetic regulators in the nucleus via chromatin modulation to control gene transcription, or in the cytoplasm, where they can function as scaffolds for protein-binding partners or modulate the activity of other coding and noncoding RNAs. In this review, we discuss the growing list of IncRNAs involved in normal development and/or homeostasis of the skeletal system, the potential mechanisms by which these IncRNAs might function, and recent improvements in the methodologies available to study IncRNA functions in vitro and in vivo. Finally, we address the likely utility of IncRNAs as biomarkers and therapeutic targets for diseases of the skeletal system, including osteoarthritis, osteoporosis, and in cancers of the skeletal system.
Keywords: Bone, cartilage, chondrocyte, long noncoding (Inc) RNAs, osteoarthritis, osteoblast, osteosarcoma
Introduction
The human genome project commenced in 1990, and its first complete assembly was announced in 2003. From this endeavor, it was found that while 70–90% of the human genome is actively transcribed, there is only 1–2% of the genome that contains protein-coding information (1,2). The remaining noncoding RNA (ncRNA) transcripts were found to lack conserved open reading frames (ORFs) and were predicted to have no biological functions in the cell. As such, these ncRNAs were usually initially referred to as “junk RNA.”
However, the past two decades of research on ncRNAs has overwhelmingly shown that these ncRNAs are in fact not “junk,” but rather play crucial and multifunctional roles in the regulation of cell phenotype. For example, classes of short ncRNAs [e.g., microRNAs (miRNAs) and Piwi-interacting RNAs (piRNAs)] have been shown to function as gene silencing agents (3–13). More recently, much interest has focused on another class of ncRNAs, known as long noncoding RNAs (IncRNAs). IncRNAs are defined as any ncRNA containing 200 or more nucleotides. The majority of IncRNAs are transcribed by RNA polymerase II (RNA Pol II), and they are often spliced (and even alternatively spliced) during RNA processing. From the most recent release of GENCODE, it is predicted that there are currently 15,941 human IncRNAs and 8793 mouse IncRNAs (14). In this article, we review the current literature on IncRNAs and their role in skeletal development and maintenance. A summary of the IncRNAs discussed here can be found in Table 1.
Table 1.
Summary of identified IncRNAs involved in skeletal biology and disease.
| Gene name | Gene symbol | ENSEMBL Gene ID | Expression pattern | Proposed function | Reference |
|---|---|---|---|---|---|
| Chondrogenesis associated transcript |
DA125942 Synonym: CISTR |
ENSG00000260492 | Genetic mutation results in brachydactyly type E (BDE) | Interacts with PTHLH in trans and with SOX9 in cis | (93) |
| Differentiation antagonizing non-protein coding RNA |
DANCR Synonym: ANCR |
ENSG00000226950 | Upregulated during chondrocyte differentiation | May promote cell proliferation and differentiation toward the chondrocyte lineage |
(90) |
| Downregulated during osteogenesis | Recruits EZH2 to the RUNX2 promoter and suppresses RUNX2 expression |
(165) | |||
| Inhibits osteogenesis via suppression of Wnt/ βcatenin pathway. |
(166) | ||||
| Upregulated in osteosarcoma | (193) | ||||
| F0XC2 antisense RNA 1 Synonym: Osteosarcoma doxorubicin-resistance upregulated IncRNA |
FOXC2-AS1 Synonym: ODRUL |
ENSG00000260944 | Expression correlates with poor response to doxorubicin chemotherapy |
(200) | |
| Growth arrest-specific 5 | GAS5 | ENSG00000234741 | Primarily expressed in the nucleus of healthy chondrocytes, and in the cytoplasm of OA chondrocytes |
Induces catabolic effect by regulating miR-21 | (142) |
| Elevated in OA chondrocytes | (138, 139) | ||||
| H19, imprinted maternally expressed transcript (Mus musculus) |
H19 | ENSMUSG00000000031 |
H19 is upregulated by overexpression of Hh and Yap1, Overexpression of Yap1 and H19 is responsible for the pathogenesis of osteoblastic osteosarcoma |
(173) | |
| H19, imprinted maternally expressed transcript (Homo sapiens) |
H19 | ENSG00000130600 | Highly expressed in mature chondrocytes | Functions downstream of SOX9. H19 is processed into miR-675 that subsequently upregulates COL2A1. |
(133) |
| Elevated in OA chondrocytes | (134, 138, 139) |
||||
| Upregulated in osteogenesis |
H19 is processed into miR-675, which targets TGFB1 for degradation. |
(136) | |||
|
H19 acts as a ceRNA, binding to miR-141 and miR- 22, thus preventing their binding to target mRNAs. |
(135) | ||||
| HIF1A antisense RNA | HIF1A-AS1 | ENSG00000258777 | Knockdown of HIF1a-AS1 decreases HOXD10 expression |
(168) | |
| Highly upregulated in liver cancer conserved region |
HULC | ENSG00000276019 | Upregulated in osteosarcoma | (189) | |
| HOX transcript antisense RNA (Mus musculus) | Hotair | ENSMUSG00000086903 | Hotair knockout mice exhibit patterning malformations in the limbs and lumbosacral junction |
Recruits EZH2 and SUZ12 to the HoxD cluster and silences the locus. |
(101) |
| HOX transcript antisense RNA (Homo sapiens) | HOTAIR | ENSG00000228630 | Elevated in OA chondrocytes | Recruits EZH2 and SUZ12 to the HoxD cluster and silences the locus. |
(138, 139) |
| Upregulated in osteosarcoma | (195) | ||||
| Decreased in AVICs by cyclic stretch, leading to calcification | β-catenin is a stretch responsive signaling pathway that represses HOTAIR |
(102) | |||
| HOXA transcript at the distal tip | HOTTIP | ENSG00000243766 | Elevated in OA chondrocytes | (139) | |
| Upregulated in osteosarcoma | (194) | ||||
| HOXA transcript induced by TGFβ |
IncRNA-HIT Synonym: 9530018H14RIK |
ENSMUSG00000102373 | Expressed in the developing mouse limb | Binds to p100/CBP complexes to maintain active chromatin mark H3K27ac at the HoxA locus |
(104) |
| KCNK15 antisense RNA 1 |
KCNK15-AS1 Synonym: RP11- 445H22.4 |
ENSG00000244558 | Elevated in OA chondrocytes | (138) | |
| Long intergenic non-protein coding RNA 707 |
LINC00707 | ENSG00000238266 | Downregulated during chondrogenesis | (87) | |
| Long intergenic non-protein coding RNA 1589 |
UNC01589 Synonym: CTA- 941F9.9 |
ENSG00000238120 | Upregulated during chondrogenesis | (87) | |
| Maternally expressed gene 3 | MEG3 | ENSG00000214548 | Downregulated in osteoarthritis chondrocytes | (144) | |
| Increased during osteogenic differentiation | Upregulates BMP4 by disrupting the interaction between the SOX2 and BMP4 promoter. |
(170) | |||
| Decreased expression of MEG3 is correlated with poor prognosis in osteosarcoma |
(198) | ||||
| Metastasis-associated lung adenocarcinoma transcript 1 |
MALAT1 | ENSG00000278217 | Upregulated in osteosarcoma | (183, 184) | |
| Downregulated in MG63 osteosarcoma cell line by high- dose estrogen |
Sequesters SFPQ and disrupts the SFPQ/PTBP2 complex, resulting in PTBP2-mediated increases in cellular proliferation and migration. |
(180, 185) | |||
| Not assigned | CTD2574D22.4 | ENSG00000260114 | Elevated in OA chondrocytes | (138) | |
| Not assigned | RP11–815M8.1 | ENSG00000238042 | Upregulated during chondrogenesis | (87) | |
| PMS1 homolog 2, mismatch repair system component pseudogene 2 |
PMS2L2 | ENSG00000278416 | Elevated in OA chondrocytes | (138) | |
| PTGS2 antisense NFKB1 complex-mediated expression regulator RNA Synonym: PS- associated COX2 extragenic RNA |
PACERR Synonym: |
ENSG00000273129 | Increased following 1L1-β treatment of primary OA chondrocytes |
(260) | |
|
PACER, PTGS2- AS1 |
Upregulated in osteosarcoma | Upregulates oncogene COX2 in an NF-kB dependent manner |
(196) | ||
| Small nucleolar RNA host gene 12 | SNHG12 | ENSG00000197989 | Upregulated in osteosarcoma | (187, 188) | |
| Taurine upregulated 1 | TUG1 | ENSG00000253352 | Increased expression is correlated with poor prognosis in osteosarcoma |
(199) | |
| Tumor-suppressor candidate 7 |
TUSC7 Synonym: LSAMP-AS3 LOC285194 |
ENSG00000243197 | Downregulated in osteosarcoma | (190, 191) | |
| Vimentin 2, pseudogene Synonym: Cartilage injury-related IncRNA |
VIM2P Synonym: IncRNA-CIR RP11- 162L10.1 |
ENSG00000220548 | Downregulated in OA chondrocytes | (141) | |
| ZBED3 antisense RNA 1 | ZBED3-AS1 | ENSG00000250802 | Upregulated during chondrogenesis | (87) |
IncRNAs: nomenclature and categorization
It is encouraged that IncRNAs should be named based on their functions (15), although at this point, many of their specific functions remain to be determined. For example, XIST is named for “X (Inactive)-Specific Transcript” (16,17), while DANCR stands for “Differentiation Antagonizing Non-Protein Coding RNA” (18–20). However, when IncRNAs are initially identified without a known function, they are named based on the associated genomic context or assigned an arbitrary name based on the GENCODE categorization, which divides IncRNAs into the following sub-categories (21,22):
Antisense: IncRNA whose sequence overlaps the exonic sequence of a protein-coding gene on the opposite strand (e.g., HIF1a-AS1)
lincRNA: IncRNA located within the intergenic regions of protein-coding genes (e.g., LINC00152)
Sense-overlapping: IncRNA whose intronic sequences encode a protein-coding or noncod-ing gene without overlap of exonic sequences (e.g., SOX2-OT)
Sense intronic: IncRNA residing within introns of a coding gene (e.g., SPRY-IT1)
Processed transcript: IncRNA that does not contain an ORF and does not fit in any of the above categories
IncRNAs: mechanisms of action
IncRNAs function in diverse ways by virtue of their primary sequence as well as their secondary and tertiary structures. In addition to interacting via base pair complementarity with DNA and RNA partners, they also fold into unique conformations that allow them to interact with DNA, RNA, and proteins. Importantly, these functions are not mutually exclusive, and the unique ability of IncRNAs to utilize these features simultaneously allows them to have far-reaching effects on cellular processes. While a more extensive discussion of the molecular mechanisms of IncRNAs can be found elsewhere (23), in this review we aim to highlight the general mechanisms by which IncRNAs execute their functions and to discuss the growing list of IncRNAs known to be involved in skeletal biology.
IncRNAs in the nucleus
IncRNAs appear to play a number of roles within the nucleus. For example, immunoprecipitation of various chromatin-modifying complexes reveals interactions with many nuclear IncRNAs, indicating a likely function for IncRNAs in epigenetic regulation (24,25). In a variety of vertebrate and invertebrate systems, IncRNAs have been identified that function as scaffolds for repressive chromatin modifiers such as: (i) the Polycomb repressive complexes 1 and 2 (PRC1, PRC2), which methylate his-tone 3 at lysine 27 (26), (ii) the repressive histone demethylase complex LSD1 (27), which demethylates histone 3 at lysine 4, and (iii) the heterochromatin-inducing EHMT2/G9a histone methyltransferase, which methylates histone H3 at lysine 9 (28). In contrast, others have been shown to interact with activating chromatin modifiers such as IncRNA-binding WDR5 subunit of the MLL (mixed-lineage leukemia) complex (29), which methylates histone 3 at lysine 4 (29–32). In some cases, these IncRNA-protein interactions are dictated by the structure of the IncRNA, as in the case of the stem-loop structures in some Polycomb-interacting IncRNAs (26,33). Notably, the presence of discrete structural “modules” along the length of the IncRNA can influence the binding of different protein complexes to the IncRNA, further diversifying its function [reviewed in (34)]. While it is not completely clear how IncRNAs navigate to their targets within the nucleus, it is apparent that IncRNAs can interact with transcription factors themselves (35), which could promote IncRNA targeting to discrete loci. In addition, a number of models have been proposed to describe how IncRNAs identify and interact with their targets, including sequenced-based or structure-based recognition of DNA loci (24,36).
A second layer of complexity in epigenetic regulation by IncRNAs is influenced by their ability to regulate nearby loci in cis (32), loci far from its site of transcription in trans (31), or a combination of both. Interestingly, knockdown of long intergenic ncRNAs (lincRNAs) demonstrated that the majority of their regulation occurred in trans (37), while IncRNAs near protein-coding genes often regulate these nearby loci in cis, as in the case of enhancer-associated RNAs [reviewed in (38)]. Notably, a number of IncRNAs are capable of regulating nearby and distant loci and may also bind chromatin-modifying complexes with opposing functions, suggesting that the regulatory capacity of a given IncRNA can be locus specific (39).
In addition to the well-established functions in regulating expression of discrete loci, IncRNAs also play important roles in nuclear organization and the establishment of chromosome territories. A special example of this occurs during X chromosome inactivation (XCI) in female mammalian cells, where one copy of the X chromosome is silenced to ensure proper gene dosage. Here, the Xist IncRNA interacts with the Polycomb repressive complex PRC2 to induce the formation of heterochromatin across the inactive X chromosome (26). Interestingly, Xist itself is regulated by other IncRNAs including Tsix and Jpx. Tsix is transcribed antisense to Xist and represses Xist expression on the active X chromosome via DNA methylation, while Jpx induces Xist transcription by sequestering a transcriptional repressor CTCF (40–42). A second well-documented example of IncRNA functioning in nuclear organization is the IncRNA NEAT1, which functions as an essential scaffold for the organization of RNA processing and editing “factories” called para-speckles (43). Knockdown of NEAT1 resulted in ablation of paraspeckles and alterations in export of mRNAs containing inverted repeats (44), suggesting an important role for this IncRNA in regulating this process.
IncRNAs in the cytoplasm
A number of IncRNAs have been demonstrated to be primarily localized within the cytoplasm or to shuttle between the nucleus and cytoplasm. These RNAs also function in diverse ways, interacting with a variety of protein-binding partners. IncRNAs have been shown to regulate protein levels (45) and function by base-pairing with the mRNA to upregulate protein translation, while some regulate mRNA degradation, recruiting proteins via the double-stranded RNA structures formed by extensive base-pairing between IncRNA and mRNA (46). Others regulate trafficking of proteins into the nucleus via their associations with the nuclear pore complex (47).
Furthermore, a growing body of evidence suggests that cytoplasmic IncRNAs can function as decoys for other ncRNAs such as miRNAs, and have been termed competing endogenous RNAs (ceRNAs) [reviewed in (48)]. While many ceRNAs function as a “sponge” to sequester miRNAs, others bind to mRNA targets and prevent miRNA-mediated downregulation (49). Interestingly, artificial ceRNAs have been utilized for more detailed characterization of miRNAs in mammalian biology [reviewed (50)].
IncRNAs regulating stem cell maintenance or differentiation
IncRNAs have also been shown to play a crucial role in maintaining pluripotency (35,37). One study (37) performed a systematic loss of function experiment in which 147 lincRNAs were knocked down in mouse embryonic stem cells (ESCs). While the differentiation capacity of these cells after lincRNA knockdown was not assessed, they observed that 93% of the lincRNAs targeted likely played an important role in maintenance of pluripotency, as their depletion resulted in gene expression profiles that were highly similar to those obtained when key pluripotency markers such as Nanog or Oct4 were depleted.
A subsequent study demonstrated that IncRNAs are not only important in maintaining pluripotency, but may also act as key regulators in promoting and preserving cell identity after differentiation (35). Using a microarray chip to quantify expression of 6671 IncRNA transcripts, Ng et al. identified 934 differentially expressed ncRNAs as human ESCs were differentiated into neuron progenitors in vitro. Their functional studies focused on two classes of IncRNAs: those that were much more abundant in ESCs than in neural progenitors (NPCs) and likely played a role in maintenance of pluripotency and those that were much more abundant in NPCs than in ESCs and likely played a role in neuronal differentiation. As expected, knockdown of several IncRNAs more abundant in ESCs led to spontaneous differentiation into a variety of cell types, while knockdown of several neurogenic IncRNAs resulted in inhibition of neuronal differentiation. Functional characterization of some of these neuronal IncRNAs (e.g., RMST, LINC01109, and CACNA2D1) showed that they were localized to the nucleus and interacted with either the Polycomb repressive complex PRC2 or the neuronal transcription factor REST. In contrast, the neurogenic IncRNA MIR100HG was localized to the cytoplasm. This IncRNA also serves as a precursor to the miRNAs miR- 125b and let-7 within its introns, which are known to function in neural development (51,52). Knockdown of MIR100HG substantially reduced the expression of these miRNAs, suggesting this IncRNA may also function as a reservoir for miRNAs important for differentiation.
Although the majority of IncRNA studies in stem cell differentiation have been carried out in vitro, comprehensive libraries of IncRNA expression during different stages of murine neurogenesis in vivo have also been described (53). In the study from Ramos et al., this library was developed by integrating multiple RNA-sequencing approaches (RNA-seq and RNA CaptureSeq (54)) with assays of chromatin state using chromatin immunoprecipitation and sequencing (ChlP-seq), which provides not only a comprehensive list of expressed transcripts and splice isoforms, but also provides some indication of their regulation in vivo. Interestingly, comparisons of murine ESCs and micro-dissected neural stem cells (NSCs) demonstrated that many IncRNAs exhibit substantial changes in chromatin state during differentiation that correspond with transitioning from a “poised” state to an active state. For example, lnc-pou3f2, which is encoded upstream of the well-established neurogenic transcription factor POU3F2, is bivalently marked with both activating (H3K4me3) and repressive (H3K27me3) histone modifications in ESCs, but transitions to a fully active state marked by only H3K4me3 in NSCs. However, a substantial number of IncRNAs remain bivalently marked in NSCs, suggesting that these IncRNAs may function later during neural lineage specification. Functional validation of several IncRNAs confirmed their important roles in NSC self-renewal or differentiation, demonstrating that well-constructed IncRNA expression libraries are extremely valuable resources for investigating the roles of IncRNAs in development and disease in vivo across various cell types and tissues.
Other studies have demonstrated the function of IncRNAs in the definition of other cell/tissue fates, including cardiac development. The IncRNA Fendrr was identified by differential expression in posterior mesoderm compared to other early somite-stage mouse tissues, and in vivo studies in which the transcript was prematurely terminated clearly demonstrated that loss of Fendrr was embryonic lethal (39). Defects in both cardiac development and in development of the ventral body wall suggested defects in differentiation of the lateral mesoderm, which was verified using gene expression analysis and chromatin profiling. Similarly, Klattenhoff and colleagues again utilized RNA-seq to identify IncRNAs that were most abundant in the murine heart compared to other tissues and identified the candidate Braveheart (Bvht), a IncRNA which has no obvious homologs in human or rat tissues (55). In vitro studies of Bvht in ESC differentiation showed that this IncRNA is not required for ESC maintenance or global differentiation but plays an important role in cardiac differentiation via interactions with the Polycomb repressive complex, PRC2. Notably, these studies were possible due to the natural propensity for ESC-derived embryoid bodies to generate cardiac tissue and well-established cardiomyocyte differentiation assays, highlighting the importance of developing well-defined in vitro differentiation assays for other cell types of interest.
LncRNA H19 in genomic imprinting
The highly coordinated process of genomic imprinting, in which one of two parental alleles is silenced, is a process involving IncRNAs that is of special importance to musculoskeletal biology and cancer development. One human disease associated with defective imprinting is Beckwith–Wiedemann syndrome, which results in an “overgrowth” phenotype and is caused by genetic or epigenetic changes within imprinting control regions (ICRs). These regions are regulatory loci defined by differential DNA methylation and are responsible for regulating a network of imprinted genes, including the IncRNA H19 and insulin-like growth factor, IGF2; however, mutations or epigenetic changes at many other loci involved in the imprinted gene network, such as the IncRNAs Kcnq1ot1 and Airn, can also confer similar phenotypes. As a result of the clear phenotypes seen with errors in imprinting and its conservation between mouse and human, the imprinted gene network involving H19 and IGF2 and the regulation of these loci is one of the best studied loci for genomic imprinting.
In normal circumstances, IGF2 is expressed from the paternal allele, and the downstream IncRNA H19 is expressed from the maternal allele. Generation of a series of genetic knockout (KO) models in mice (reviewed in (56)) revealed that genomic imprinting elements within the H19-IGF2 locus are essential for normal imprinting and thus properly regulated H19 and IGF2 expression. In order for normal imprinting to occur, the zinc finger protein CTCF binds to ICRs comprised of differentially methylated regions (DMRs) and the matrix attachment region (MAR) on the maternal allele to shield the IGF2 promoter from its enhancer. As a result, this enhancer is accessible to H19 and promotes H19 expression. Meanwhile on the paternal allele, methylation of the ICR blocks CTCF binding, allowing contact between the IGF2 promoter and enhancer to promote IGF2 expression. Simultaneously, the paternally inherited H19 promoter is also methylated, rendering it inaccessible by both enhancers and transcription factors. In Beckwith–Wiedemann syndrome, loss or epigenetic dysfunction of an ICR typically results in either overexpression of IGF2 or loss of the imprinted gene CDKN1C, which regulates cell proliferation; these phenotypes are thought to contribute to the overgrowth phenotypes observed in patients with this syndrome.
The proper regulation of H19 and IGF2 is critical for normal development and tissue homeostasis. During fetal development, H19 and IGF2 are highly expressed in endodermal and mesodermal tissues. More importantly, their expression is tightly coordinated with identical spatial and temporal patterns (57,58), and IGF2 utilizes fetal-specific promoters for its expression during development (59). Interestingly, in a KO mouse model in which the entire H19 sequence was deleted, normal imprinting patterns were observed on both parental alleles (60), suggesting that H19 does not function in cis to regulate IGF2 gene expression. However, other evidence proposes that H19 may regulate IGF2 expression post-transcriptionally by binding to insulinlike growth factor II mRNA-binding proteins (IMPs) (61,62). H19 can also be processed into miR-675, whose targets such as IGFR1 and Rb (Retinoblastoma) are involved in cell growth and regulation (63–65).
Given that H19 is highly abundant in the mesoderm during embryogenesis (66,67), it could be hypothesized that this IncRNA may play a role in regulating skeletal development. In the remaining sections of this review, we will focus on recent studies describing the expression and potential function of IncRNAs, including H19, in chondrogenesis and osteogenesis, as well as in cartilage and bone homeostasis and disease.
Cartilage Development
Cartilage is an important connective tissue that can be classified into three groups: hyaline (e.g., joint articular cartilage), elastic (e.g., ear), and fibrocartilage (e.g., meniscus and intervertebral discs in the spine). These cartilage types differ in extracellular matrix (ECM) composition and cell (chondrocyte) phenotype, thereby providing each tissue type with specific mechanical properties to carry out their function in the body.
Hyaline cartilage development is a complex process that begins with the condensation of mesenchymal stem cells (MSCs) that are derived from the mesoderm germ layer (68). In response to various signaling pathways, chondro-progenitor cells are formed. These cells proliferate and differentiate into chondrocytes that are responsible for generating the ECM which, in hyaline cartilage, consists primarily of type II collagen and the large aggregating proteoglycan, aggrecan. Two types of hyaline cartilage exist in the limb: articular cartilage and growth plate (epiphyseal) cartilage. Some controversy remains as to whether articular and growth plate cartilages are derived from common precursor cells or if the outer region of articular cartilage (i.e., the superficial zone) is derived from distinct precursor cells during synovial joint development (69,70). In any case, we know that chondrocytes of articular cartilage remain as permanent, differentiated chondrocytes that synthesize an ECM with appropriate mechanical properties to permit transmission of loads within the synovial joint during development and aging. Chondrocytes of the growth plate, however, are distinctly localized in columns where they continue to proliferate, thereby lengthening the limb during development. These cells then terminally differentiate to form large hypertrophic chondrocytes. The production of hypertrophic chondrocytes is essential to permit long bone formation via the process of endochondral ossification whereby the cartilage template is essentially replaced by bone (71). It was thought that the majority of hypertrophic chondrocytes become apoptotic during endochondral ossification, but recent studies have shown that these cells also contribute toward the osteoblast pool or that there are osteoprogenitor cells in hypertrophic cartilage that can differentiate toward bone-producing osteoblasts (72–75).
There is an abundance of knowledge with respect to the transcription factors and the signaling pathways/ mediators that regulate chondrocyte differentiation and ECM synthesis. In addition to protein-coding genes, many studies have been published on noncoding miRNAs and how their expression changes during chondrocyte differentiation (76–78) as well as how they may function in controlling chondrogenesis (79–82). In keeping with the epigenetic theme, studies are now emerging to investigate the expression and potential function of IncRNAs in cartilage development and disease.
IncRNAs regulating chondrocyte differentiation
A common method to study chondrogenesis in vitro involves three-dimensional culture of human mesenchymal stem/stromal cells (MSCs) isolated from bone marrow (83,84) or other sources such as adipose, synovium, and umbilical cord (85,86). A recent study utilized human bone marrow-derived MSCs (HBMSCs) to determine IncRNA expression signatures in cells 2 weeks following in vitro chondrogenesis compared to HBMSCs at day 0 (control) of the differentiation assay (87). Following microarray analysis (IncRNA + mRNA Human Gene Expression Microarray V4.0; CapitalBio Corp, Beijing, China), over 3000 IncRNAs were found to be significantly differentially expressed (±twofold) between control and chondro-induced cells. Among these, three IncRNAs were identified as upregulated following chondrogenesis (ZBED3-AS1, CTA-941F9.9, and ENST00000433576.1) and one was identified as downregulated (LINC00707; it should be noted that this IncRNA was reported as two individual genes in this report). Moreover, it is not known if these IncRNAs showed the most dramatic fold changes in this work since the entire array data was not reported. Another caveat with this study involves the relatively small increase in type II collagen expression (~twofold increase) after 2 weeks of chondrogenesis. Normally, COL2A1 levels are increased by several orders of magnitude during such MSC chondrogenesis assays (88,89). Also, no COL2A1 expression data was shown from their array study. It will be interesting to determine IncRNA expression by RNA-seq analysis in HBMSC-induced cells that produce higher levels of COL2A1 during chondrogenesis and investigate if the same IncRNAs found in this study by Wang et al. are also found to be differentially expressed.
Another recent study identified that the transcription factor, SOX4, can regulate the proliferation and chondrogenic differentiation of human synovium-derived MSCs (SMSCs) and that activation of the IncRNA, DANCR, is involved in this process (90). Here, SMSCs were extracted from the synovium of osteoarthritis (OA) patients and aggregate cultures were utilized for in vitro chondrogenesis. Apparently, the differentiation medium in these assays did not include TGF-β1/3, and so it is surprising that efficient chondrogenic differentiation was achieved. In any case, SOX4 overexpression was found to increase the expression of chondrocyte markers over 2 weeks in culture. To attempt to decipher SOX4 function in this scenario, promoter analysis of the IncRNA DANCR revealed a SOX4-binding site. Luciferase and ChIP assays showed that SOX4 can interact with its binding site on the DANCR promoter. Inhibition of DANCR by RNAi apparently suppressed the effects of SOX4 in promoting chondrogenesis, thereby suggesting that SOX4 functions upstream of DANCR. DANCR (Differentiation Antagonizing Non-protein Coding NA) was first described as a IncRNA (previously named ANCR), which functions in suppression of progenitor cell differentiation (18). Two recent studies have shown that DANCR can increase stem cell-like features in hepatocellular carcinoma cells (91) and that it can suppress odontoblast-like differentiation of human dental pulp cells by inhibiting the Wnt/β-catenin pathway (92). Given this “anti-differentiation” function, it is interesting that DANCR activation via SOX4 in SMSCs resulted in increased proliferation and differentiation toward the chondrocyte lineage in this study. Further research is required to thoroughly decipher the role of DANCR in chondrogenesis.
With respect to human skeletal pathology, a IncRNA was recently found to be associated with brachydactyly type E (BDE), a condition characterized by shortening of metacarpals and metatarsals. In this study (93), two families with autosomal dominant BDE were identified with translocations in chromosome 12 that resulted in genomic disruption of an important cis-regulatory element named CISTR-ACT, which also produces the IncRNA DA125942. Notably, the CISTR-ACT locus interacted in cis with the PTHLH locus, which encodes the chondrogenic regulator parathyroid hormone-like hormone (94), and in trans with the SOX9 locus, which encodes the master chondrogenic transcription factor SOX9 (95). Importantly, knockdown of DA125942 in a chondrogenic cell line resulted in downregulation of PTHLH and SOX9, as well as a plethora of other genes important in skeletal patterning (HOXB cluster, ETV4) or chondrogenesis (HIF1A), suggesting a potentially important role for this IncRNA in regulating chondrogenic differentiation.
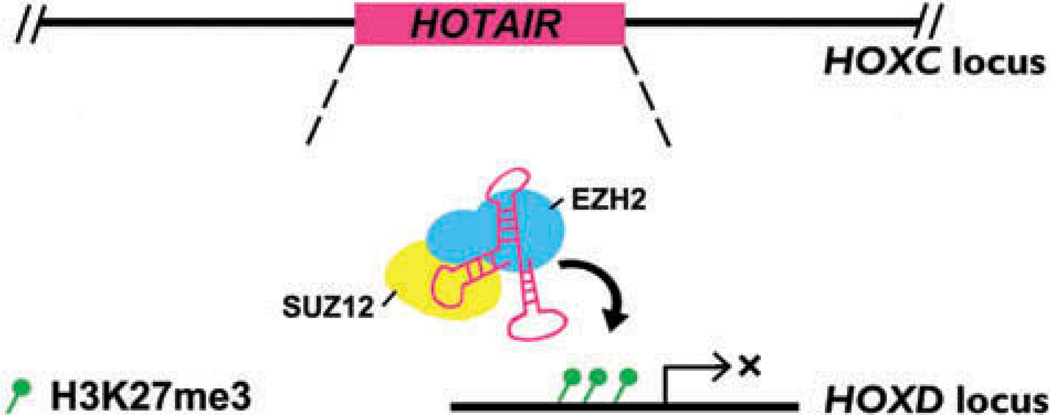
Studies on IncRNAs associated with HOX genes have provided insights into their functions in regulating skeletal development. HOX genes are a conserved family of developmental transcription factors that elicit specific developmental programs along the head-to-tail axis of animals (96). For example, IncRNA genes in the 5ˊ HOXA and HOXD clusters are known to regulate limb and spine growth and patterning (97–100). HOTAIR (HOX Transcript Antisense RNA) was the first vertebrate IncRNA described to regulate HOX function (31). Specifically, HOTAIR is expressed from the HOXC locus and functions to recruit EZH2 and SUZ12, part of the Polycomb repressive complex PRC2, to the HOXD cluster, thereby establishing a silent chromatin state using H3K27 methylation and resulting in repression of several 5ˊ HOXD genes (Figure 1). Targeted deletion of Hotair in mice resulted in patterning malformations in the lumbosacral junction and in the metacarpal and carpal bones in the limbs (101). Interestingly, cyclic stretch has been shown to decrease HOTAIR levels in human aortic interstitial cells (AVICs), and reducing HOTAIR levels via siRNA in AVICs results in increased expression of calcification genes (102). These findings suggest that HOTAIR is mechanoresponsive and may thus may play a potential role in mechanically regulated calcification, although additional work is needed to test this hypothesis in other cell types.
Figure 1.
HOTAIR function in regulating HOXD gene expression. Expressed from the HOXC locus, HOTAIR binds to SUZ12 and EZH2, part of the PRC2 complex. HOTAIR acts in trans by recruiting the PRC2 complex to the HOXD locus and silences the locus by establishing H3K27me3 marks (30).
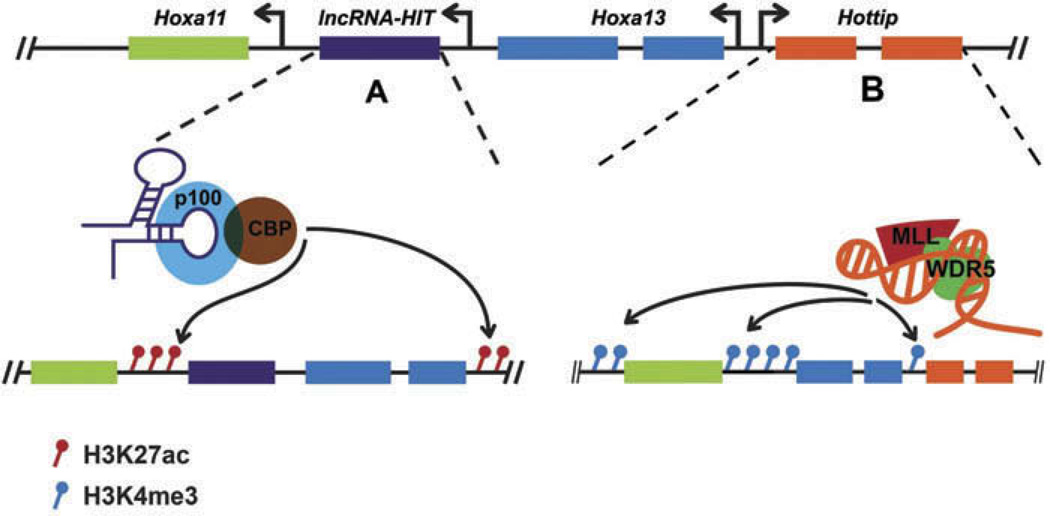
In contrast, HOTTIP (HOXA Transcript at the Distal Tip) is an enhancer IncRNA that regulates the expression of 5ˊ HOXA genes to control the growth and elongation of skeletal elements of the limb (32). HOTTIP functions by regulating chromosome looping to recruit the WDR5/ MLL histone methyltransferase complex to the 5ˊ HOXA genes, conferring an active chromatin state via H3K4 methylation (32,103) (Figure 2B). While HOTAIR has been shown to act in trans (located on the HOXC locus and regulates HOXD genes) (Figure 1), HOTTIP was noted to act in cis (Figure 2B) and its proximity was crucial for HOXA gene expression, as expression of HOXA genes was not changed when HOTTIP was ectopically expressed via lentivirus.
Figure 2.
IncRNA-HIT and Hottip function in regulating Hoxa gene expression. (A) IncRNA-HIT binds to the p100/CBP complex and maintains H3K27ac marks at HoxA genes, resulting in gene activation (103). (B) Hottip acts in cis by recruiting the WDR5/MLL histone methyltransferase complex to the Hoxa genes, resulting in gene activation by establishing H3K4me3 marks (31).
An elegant study published recently by Stadler’s group has identified another IncRNA located within the HOXA locus, named IncRNA-HIT, which appears to function as a critical epigenetic regulator of chondrogenesis (104). LncRNA-HIT (HOXA Transcript Induced by TGF-β) was initially characterized as a TGF-P-responsive IncRNA during epithelial-to-mesenchymal transition in mammary epithelia (105). LncRNA-HIT was previously mapped as a single exon in the mouse genome between Hoxa11 and Hoxa13 (106). Carlson et al. found that LncRNA-HIT was expressed in the developing mouse limb and hypothesized that it may play a role in chondrogenesis. RNA FISH analysis showed that LncRNA-HIT is localized to the nucleus of limb mesenchymal cells, while mass spectroscopy and immunoprecipitation experiments revealed that it associates with p100/CBP complexes (Figure 2A). Knockdown of IncRNA-HIT in micromass cultures of murine limb mesenchymal cells was found to inhibit chondrogenesis (i.e., cartilage nodule formation) in these cultures. Further experiments in this study strongly suggest that, mechanistically, IncRNA-HIT functions as an enhancer IncRNA via association with p100/CBP complexes to maintain H3K27ac at specific chromatin,sites thereby resulting in the activation of 5ˊ HoxA genes. In addition to affecting the expression of HoxA genes, IncRNA-HIT siRNA experiments also revealed decreased expression of a number of other genes, including Bmpr1b. This finding may also explain why knockdown of IncRNA-HIT reduces cartilage formation of murine limb mesenchyme in vitro since loss of Bmpr1b is known to negatively affect chondrogenesis in mice and humans (107–110). Overall, this study is the first to provide detailed mechanistic insights into the role of a specific IncRNA in chondrogenesis and reveals another level of complexity toward our understanding of how chondro-progenitor commitment and cartilage tissue development is controlled.
IncRNAs in cartilage homeostasis and osteoarthritis
OA is a painful and debilitating disease of the synovial joints that is characterized by progressive degenerative changes in the articular cartilage and other joint tissues such as the synovium and subchondral bone. Under normal circumstances, mature articular cartilage is maintained in a healthy balance of anabolic and catabolic processes by the activity of the chondrocytes. While many factors can contribute to the onset and progression of OA, disruption of cartilage homeostasis can occur if a joint is subjected to altered loading caused by malalignment or trauma (e.g., meniscal or ligament injury, articular cartilage fracture) (111–116). Obesity can also affect joint homeostasis due to increased loads as well as chronic inflammation (113,117–119). As a result, catabolism often predominates causing joint abnormalities including articular cartilage degradation, subchondral bone sclerosis, osteophyte formation, and synovitis which, collectively, can be defined as OA (120). OA is a challenging disease to treat, given that articular cartilage has little or no intrinsic regenerative capacity. Many of the cellular and molecular changes that occur in OA are known, such as alterations in growth factor and cytokine signaling, inflammation, oxidative stress, chemokine signaling, and metabolism (120–122). However, no effective therapies have yet been discovered to ameliorate or stop OA progression. It is now apparent that epigenetic modifications (e.g., DNA methylation, histone modifications) may confer susceptibility to OA (123–132) which could open up new avenues for alternative therapeutic approaches.
To date, a small number of studies have been published on IncRNA expression and potential function in mature chondrocytes within the context of OA. The IncRNA H19 was first reported by Dudek et al. to be highly expressed in mature primary articular chondrocytes and to be regulated by SOX9 (133). This study also showed that H19 knockdown in primary chondrocytes reduced COL2A1 levels; however, H19 also serves as the precursor for the miRNA miR-675, and they demonstrated that the effects of H19 knockdown could be rescued by overexpression of miR-675. Thus, they concluded that miR-675 is an important functional component of H19 to regulate the chondrocyte phenotype.
In addition, Steck et al. reported that H19 expression, as well as IGF2, was elevated in human OA cartilage compared to healthy, control tissue; however, H19 and IGF2 levels did not correlate significantly in cartilage (134). This study also showed that H19 levels are increased in chondrocytes under hypoxic conditions, while levels decreased in response to proinflammatory cytokines. From these findings, it was speculated that H19 may play a role in regulating chondrocyte metabolism in response to stress as well as induce chondrocyte anabolism. As will be discussed more later, H19 was recently shown to induce osteoblast differentiation via TGF-β1/Smad3/HDAC or β-catenin/Wnt signaling pathways (135,136). It remains to be discovered if H19 functions in a similar manner to induce chondrogenesis. Finally, a recent report revealed a new function for H19 in regulating DNMT3B-mediated DNA methylation (137). This is interesting from the standpoint that changes in DNA methylation patterns have been detected in human OA cartilage and that H19 levels have been reported to be higher in OA (134,138).
A study by Kim et al. showed that HOTTIP expression was increased (assessed by microarray analysis) in chondrocytes from OA patients compared to non-OA or normal control cells (139). They also detected a decrease in HOXA13 expression as a result of increased HOTTIP levels. However, it has been reported (and mentioned earlier in this review) that HOTTIP functions to facilitate H3K4 methylation and, therefore, activation of distal HOX genes. It is, therefore, curious that an increase in HOTTIP would lead to a reduced HOXA13 expression in this study. Further mechanistic studies are needed to clarify the functional role of HOTTIP in regulating cartilage homeostasis.
Other microarray-based studies have been published reporting IncRNA expression changes in OA chondrocytes compared to healthy control cells (138,140,141). One of these studies reported 121 differentially expressed IncRNAs, 73 of which were significantly upregulated including HOTAIR, GAS5, PMS2L2, RP11-445H22.4, H19, and CTD-2574D22.4 (138). Of these IncRNAs, HOTAIR and GAS5 were also found to be upregulated in OA chondrocytes in the Kim et al. study mentioned above (139). The study by Liu et al. (141) identified 152 differentially expressed IncRNAs and focused specifically on one IncRNA (IncRNA-CIR; Cartilage Injury Related) which was upregulated over tenfold in OA chondrocytes, as confirmed by qPCR, and was also shown to increase in chondrocytes following treatment with IL-1β and TNF-a. Attempts to decipher the function of IncRNA-CIR were done by knockdown (siRNA) and overexpression approaches; it was concluded from these experiments that IncRNA-CIR can induce chondrocyte catabolic events via increased expression of MMP13 and ADAMTS-5. The study by Fu et al. (140) provided expression data of differentially expressed IncRNAs followed by target predictions and pathway analyses of those IncRNAs found to be most highly up- or downregulated in OA. They utilized the same microarray (Human IncRNA Array v2.0; 8×60K; Arraystar) that was used by Liu et al. (141) and reported many more differentially expressed IncRNAs (4714 IncRNAs) based on the fact that they included IncRNAs whose levels changed by twofold or more (as opposed to the Liu et al. study that reported IncRNAs with an eightfold or more change in expression).
Another recent report also showed increased expression of GAS5 (Growth Arrest-Specific 5) in OA cartilage compared to healthy control tissue (142). RNA FISH analysis showed nuclear localization of GAS5 in healthy chondrocytes, while localization in OA chondrocytes also revealed cytoplasmic distribution. Overexpression of GAS5 in chondrocytes in vitro resulted in increased metalloproteinase expression, decreased expression of markers associated with autophagy, and decreased expression of miR-21. Manipulation of miR-21 levels in murine articular cartilage in vivo via lentivirus showed some degree of chondro-protection following OA-induced surgery by destabilization of the medial meniscus (DMM) (143). The overall conclusion from these findings is that GAS5 overexpression appears to contribute to a catabolic phenotype via regulation of miR-21 in chondrocytes. Another study was published recently reporting decreased expression of the IncRNA MEG3 (Maternally Expressed Gene 3) in OA and that its expression levels were inversely associated with VEGF levels (144). However, no functional data was included in this study to be able to make any conclusions on MEG3 regulation of VEGF in chondrocytes.
Finally, a study recently accepted for publication by Pearson et al. aimed to identify IncRNAs associated with the inflammatory response in human primary hip OA chondrocytes (145). These cells were treated for 4 h with IL-1β and differentially expressed IncRNAs between treated and untreated cells were identified following RNA-seq analysis (sequencing data is publically available via the GEO data repository: GSE74220). Of the 983 IncRNAs identified, 125 were found to be differentially expressed following IL-1β treatment including PACER (p5-Associated COX2-Extragenic RNA) and two novel chondrocyte inflammation-associated lincRNAs (CILinc01 and CILinc02). The increase in expression of these three lincRNAs was confirmed by additional in vitro assays using IL-1β-induced primary knee or hip chondrocytes followed by qPCR analysis. Interestingly, expression of these lincRNAs was found to be lower in OA cartilage compared to healthy control tissue. This finding may be due to the fact that the inflammatory status (and hence IncRNA expression) of chondrocytes embedded within late-stage OA cartilage tissue may not correlate with that in cultured chondrocytes isolated from cartilage following treatment with IL-1β. In any case, this report has provided in-depth information on a number of inflammatory-regulated IncRNAs that may be worth pursuing in greater detail with respect to determining their biological functions in regulating chondrocyte homeostasis.
Bone development
Unlike cartilage, bone is a rigid tissue made up mainly of collagen fibers, calcium carbonate, and hydroxyapatite. Three cell types reside within the bone tissue: osteoblasts that synthesize bone ECM, osteoclasts that resorb bone, and osteocytes that are the mechano-sensing cells embedded deep within the bone (146–148). During embryonic development, bone is formed from cells of three sources: the somites, the lateral plate mesoderm, and the cranial neural crest. Formation of bone can occur via two processes: endochondral ossification or intramembraneous ossification.
The vertebral, limb, and rib skeletons are formed by endochondral ossification that begins with the formation of a cartilage intermediate template (71). Under the control of multiple signaling pathways (e.g., BMP superfamily growth factors, FGFs, Wnts, Hedgehog), mesodermal stem cells become committed toward the chondrocyte lineage and undergo a process known as mesenchymal condensation (146). Here, the influence of cell adhesion molecules such as N-cadherin and N-CAM are noted to be highly critical in maintaining cell–cell contacts (149–151). While the master chondrocyte transcription factor Sox9 is one of the earliest markers expressed in the chondrocyte lineage, this transcription factor requires the co-activators L-Sox5 and Sox6 to fully induce the expression of other chondrogenic genes such as Col2a1 (type II collagen) and Acan (aggrecan). The cells later enlarge and become hypertrophic chondrocytes that express elevated levels of Runx2, Mmp13, Alp, Col10a1, and Vegf. This alteration in gene expression permits the generation of an ECM that can be mineralized and vascularized (152–154). At this stage, the hypertrophic chondrocytes can become apoptotic, although as mentioned earlier in this review, recent evidence suggests that these cells, or progenitor cells within hypertrophic cartilage, can also contribute to the osteoblast pool (72–75). Following vascularization of hypertrophic cartilage, pre-osteoblasts and pre-osteoclasts infiltrate this primary center of ossification. Mature osteoblasts then function to generate new bone matrix and, at the same time, a bone marrow cavity is created. Mature osteoclasts function to resorb the trabecular bone within the ossification center. Overall, there is a critical balance between osteoblasts and osteoclasts to maintain bone homeostasis.
In contrast, intramembraneous ossification occurs when the stem cells in mesenchymal tissue differentiate directly toward the osteoblast lineage without the requirement of a cartilage template. This can be seen perhaps most clearly by the construction of our facial skeleton. Under the influence of BMPs, the cranial neural crest cells differentiate to become pre-osteoblasts which express elevated levels of Runx2. These cells subsequently mature into osteoblasts that can produce an osteoid matrix rich in collagen, proteoglycan, and calcium (155). There are many signaling pathways involved in intramembraneous ossification. For example, BMP, IHH (Indian Hedgehog), and PTHrP (parathyroid hormone-related protein) signaling pathways function in the commitment of cranial neural crest cells toward an osteoblast identity (155). In addition, FGF and SHH (Sonic Hedgehog) are also involved in defining the shape and size of the facial midline (156–162).
IncRNAs regulating osteoblast differentiation
To date, studies on the role of IncRNAs in bone biology have been limited. However, one report showed that homozygous mice lacking the IncRNA, Hotair, resulted in lumbosacral transformation and fusion of the metacarpal and carpal bones (101). This skeletal phenotype was similar to transgenic mice with ectopic expression of HoxD resulting in elevation and anteriorization of Hoxd10 and Hoxd11 expression. It has been suggested earlier that Hotair acts in trans and regulates the expression of HoxD genes by recruiting the Polycomb repressive complex PRC2 to the HoxD locus (31). RNA-seq data of WT, Hotair +/−, and Hotair−/− confirms the earlier hypothesis: Hotair deletion leads to demethylation of H3K27 and methylation of H3K4 methylation at targeted genes, resulting in derepression of HoxD genes. Interestingly, several imprinted loci were also identified as Hotair targets, including Dlk1-Meg3 and Igf2-H19. Since Hotair and its well-known related gene Hottip regulate the Hox loci, and Hox genes are involved in body patterning and limb development, it was not surprising that mutation of this IncRNA led to such striking skeletal patterning defects. It will be interesting to investigate whether Hottip is also involved in the regulation of osteogenesis.
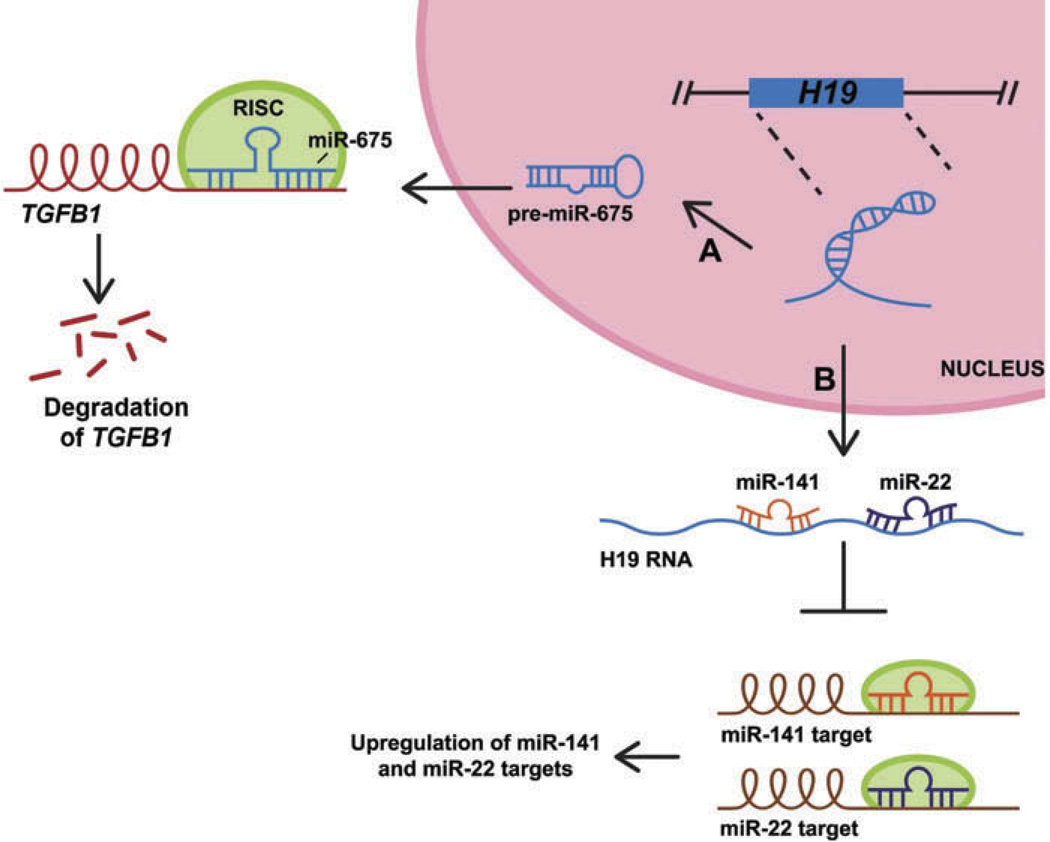
Recent studies have demonstrated that H19 is upregulated during osteogenesis (135,136). Different mechanisms, which may not be exclusive of one another, have been proposed to describe H19 function in this context. Huang and colleagues demonstrate that H19 can function to promote osteogenesis. They showed that H19 is processed into miR-675, which targets TGFB1 for degradation (136) (Figure 3A). Since TGFβ signaling has been previously shown to impede osteogenesis by inducing heterochromatin formation at several promoters such as RUNX2 and OCN (163), a drop in TGF-β1 would lead to derepression of these osteogenic markers and subsequently promote osteogenesis. Additionally, this study also showed that H19/miR-675 downregulated levels of HDAC4/5 which also resulted in increased expression of osteoblast genes, although it was not clear whether this effect is direct or indirect. Interestingly, a specific function for H19 outside of miR-675 biogenesis was not described by Huang et al. In contrast, the proposed mechanism described by Liang and colleagues demonstrated that H19 itself functions as a ceRNA, binding to miR-141 and miR-22 and sequestering them away from their mRNA targets (Figure 3B). Notably, these miRNAs were previously established to target and degrade CTNNB1 (β-catenin) (164), and H19 may relieve their repressive effect on the Wnt-signaling pathway by competitively binding to these miRNAs, thereby inducing osteoblast differentiation. As H19 had previously been shown to be enriched during chondrogenesis (133), it may be the case that H19 suppresses the stemness of MSCs in general instead of promoting specific differentiation programs in chondrogenesis and/or osteogenesis, although further evidence on the molecular mechanism of H19 in MSCs is required to prove this hypothesis.
Figure 3.
Different mechanisms of H19 in osteogenesis. (A) H19 is processed into miR-675, which targets TGFB1 for degradation (135). (B) H19 acts as a sponge to sequester miR-141 and miR-22 away from their targets, thus leading to an upregulation of targeted mRNAs that may be required for osteoblast differentiation (134).
There have been two reports of the IncRNA DANCR in osteogenesis. In one study, it was proposed that DANCR regulates expression of the osteogenic regulator RUNX2 by recruiting the Polycomb repressive complex component EZH2 to the RUNX2 promoter, thereby resulting in suppression of osteogenesis (165). While RNA pulldowns showed a direct association of DANCR and EZH2, there was no evidence that DANCR was directly binding or recruiting EZH2 to directly bind to RUNX2 promoter, which will be of interest for future investigation. A similar study manipulated DANCR expression in human periodontal ligament stem cells, and also showed that downregulation of DANCR was critical for osteogenesis (166). Here, DANCR knockdown upregulated β-catenin/Wnt signaling pathway in these cells, which has been well-documented to promote osteoblastogenesis. However, this study only suggests a correlation between DANCR and the osteogenic markers, and no direct interactions between the IncRNA and the genomic elements regulating osteogenic genes were investigated in the study. In addition, since DANCR was first proposed to function as an anti-differentiation player (18–20), it might be the case that reducing DANCR expression evokes a global effect on the epigenetic state of the cells. Instead of directly affecting their osteogenic capability, DANCR may be acting to make the cells more competent for differentiation into specific cell types given the appropriate medium conditions.
Additionally, a set of IncRNA microarray expression data was also generated from a mouse MSC cell line that was stimulated with BMP2 (167). The study identified 116 IncRNAs that were differentially expressed: 59 upregulated and 57 downregulated. However, several issues were noted in this study: (i) the cut-off values used for differential expression were arbitrary and changed between time points, (ii) the experimental methods were not well-described, and (iii) data showing appropriate osteogenic induction were relatively weak. Furthermore, no functional characterization was performed to test the requirements for these IncRNAs in osteogenesis. Therefore, additional validation of differentially expressed IncRNAs in this system is still needed.
The expression of the IncRNA HIF1A-AS1 in HBMSCs was also characterized, and the authors imply a potential role for this IncRNA in osteogenesis which is dependent on the histone deacetylase SIRT1, an important positive regulator of osteoblastogenesis and bone mass (168,169). Here, overexpression of SIRT1 downregulated HIF1α-AS1 expression, while knockdown and pharmacological inhibition of SIRT1 upregulated HIF1a-AS1 expression. Interestingly, HIF1a-AS1 knockdown decreased the expression of HOXD10, suggesting a potential role for this IncRNA in HOX gene regulation. However, the authors describe a pro-osteogenic role for TGF-β, which has been well documented to function in an anti-osteogenic capacity. In addition, the regulation of HIF1a-AS1 during in vitro osteogenic assays was not described, and it was not clear whether these experiments were performed under osteogenic conditions; so further work is needed to determine the function of this IncRNA during osteogenesis.
Last, the function of the IncRNA MEG3 was shown to play an important role in osteogenesis, especially in the context of multiple myeloma, a hematological cancer that also affects bone mass by inhibition of osteoblastogenesis of MSCs (170). Here, it was demonstrated that MEG3 expression increases during osteogenic differentiation of MSCs from normal patients, but its expression is reduced overall and is unchanged during osteogenesis. In normal cells, MEG3 knockdown reduced osteogenic differentiation and functioned by reducing transcription of BMP4, and ectopic MEG3 overexpression or treatment with exogenous BMP-4 rescued the osteogenic defect of MSCs from multiple myeloma patients. Mechanistically, it was shown that MEG3 regulates BMP4 transcription by disrupting the interaction between the SOX2 transcription factor and the BMP4 promoter. Interestingly, overexpression of MEG3 in MSCs from multiple myeloma patients also improved chondrogenic differentiation, suggesting a potential role for this IncRNA in chondrocyte differentiation as well.
IncRNAs in osteosarcoma
Osteosarcoma is a type of bone cancer that occurs at a rate of 5% in pediatric malignancies (171,172). In humans, osteoblastic osteosarcoma is characterized by highly mineralized tissues that are abundant in osteoids with multinucleated cells (173,174). From recent improvements in surgical, therapeutic, and radiation care, the survival rate of osteosarcoma patients has increased from less than 20–70% over the past 40 years (175–179). However, elucidation of the molecular basis of disease development and progression is required to establish better treatment options. Recently, several IncRNAs have been identified that show differential expression in osteosarcoma tissue.
One IncRNA that is dysregulated in osteosarcoma is MALAT-1 (Metastasis-Associated Lung Adenocarcinoma Transcript 1), a 6.8kb transcript that is conserved among 33 mammalian species (180). Unlike most IncRNA, MALAT-1 is not poly-adenylated at its 3ˊ end but instead folds into a triple helix (Figure 4), a unique structure that protects the RNA from degradation by nucleases (181). MALAT-1 is expressed in many types of cancers, where it often promotes proliferation [reviewed in (182)]. Therefore, it is not surprising that MALAT-1 was also observed to be upregulated in osteosarcoma (183,184). These independent studies established that knockdown of MALAT-1 in osteosarcoma cells not only decreased proliferation but also induced apoptosis, and knockdown reduced osteosarcoma growth and metastasis in vivo. In addition, a third study utilizing the MG63 osteosarcoma cell line demonstrated that MALAT-1 was downregulated by high-dose estrogen, and this downregulation reduced proliferation, migration, and invasion (185). MALAT-1 appeared to sequester the splicing complex factor SFPQ from its splicing co-regulator PTBP2 (Figure 4), and downregulation of MALAT-1 resulted in a higher accumulation of the SFPQ/PTBP2 complex. Interestingly, similar results have been observed in colon cancer cells, in which sequestration of SFPQ by MALAT-1 results in PTBP2-mediated increases in cellular proliferation and migration (186).
Figure 4.
MALAT-1 function in osteosarcoma. (A) MALAT-1 IncRNA is not polyadenylated, but instead contains a triple helical domain at its 3ˊ end that protects it from degradation (180). (B) MALAT-1 binds to SFPQ in nuclear speckles. This sequestration prevents the formation of the SFPQ/PTBP2 complex. Subsequently, unbound PTBP2 can induce tumor growth by increasing cellular proliferation and migration (185).
Many IncRNAs have been identified using similar methods, and in vitro studies show that they may play important roles in osteosarcoma. Much like MALAT-1, the IncRNA SNHG12 (Small Nucleolar RNA Host Gene 12) is upregulated in osteosarcoma patients (187,188). Both studies demonstrated that expression of SNHG12 correlated with expression of AMOT (angiomotin), a gene with unknown functions in osteosarcoma. While modulation of SNHG12 substantially altered expression of AMOT, the mechanism of regulation is not clear, and the function of this IncRNA and AMOThas not been clearly established (187). Similarly, the IncRNA HULC (Highly Upregulated in Liver Cancer) is upregulated in osteosarcoma tissues (189), and knockdown of HULC in osteosarcoma cell lines in vitro suppressed proliferation, migration, and invasion. In contrast, the IncRNA TUSC7 (Tumor Suppressor Candidate 7), also known as LOC285194, is substantially downregulated in osteosarcoma, and low expression of this IncRNA correlates with poor survival in osteosarcoma patients (190,191). Interestingly, expression changes in this IncRNA seemed to correlate with copy number alterations rather than an epigenetic silencing of the genes. In contrast to MALAT-1, knockdown of TUSC7 or LOC285194 led to increased proliferation and/or decreased apoptosis, and in vivo xenograft studies showed that TUSC7 silencing promoted tumor growth. Last, the promoter-upstream transcript HIF2PUT has been proposed to regulate expression of HIF2A in osteosarcoma (192). While the expression of HIF2PUT and HIF2A in osteosarcoma appeared to be relatively weakly correlated, knockdown of HIF2PUT increased cell proliferation and invasion in vitro in an osteosarcoma cell line, suggesting that this class of IncRNAs may also function in osteosarcoma.
In addition, IncRNAs that function in chondrocyte or osteoblast differentiation and homeostasis have also been found to be dysregulated in osteosarcoma, including DANCR, PACER, HOTAIR, HOTTIP, and H19. Much like MALAT-1, knockdown of DANCR in osteosarcoma cell lines reduced cell proliferation and resulted in cell cycle arrest (193). Similarly, the IncRNAs PACER, HOTAIR, and HOTTIP were shown to be upregulated in osteosarcoma, and knockdown of these IncRNAs inhibited proliferation and/or invasion of osteosarcoma cells (194–196). Mechanistic investigation of PACER function demonstrated that it upregulates the expression of the oncogene COX2 in an NF-κB-dependent manner. A role for H19 in osteosarcoma has been proposed by Chan and colleagues, who developed a model for spontaneous development of osteoblastic osteosarcoma in which p53-heterozygous mice containing an osteoblast-specific inactivation of Patch1 results in partial upregulation of Hedgehog signaling (173). In this model, the Hippo pathway transcriptional coactivator yes-associated protein 1 (Yap1) was upregulated by Hedgehog signaling, and in turn upregulates H19. Interestingly, loss of imprinting at the H19-IGF2 has previously been observed in osteosarcoma (197), suggesting a potential function for H19 and genomic imprinting in this setting.
Finally, other clinical investigations have revealed correlations between IncRNA expression levels and osteosarcoma patient survival. Decreased expression of MEG3 (198) and upregulation of TUG1 (Taurine-Upregulated Gene 1) (199) correlate strongly with poor prognosis in osteosarcoma, while the IncRNA ODRUL (Osteosarcoma Doxorubicin-Resistance Upregulated LncRNA) correlates strongly with poor response to doxorubicin chemotherapy (200). These preliminary data suggested that we may eventually be able to utilize IncRNAs as predictive biomarkers.
Characterization of IncRNAs
The field of IncRNA biology is relatively young by scientific standards, and significant investments are still being made to identify new IncRNAs across different tissues and among species. Many techniques that have been utilized to study well-characterized IncRNAs for some time will continue to be used to interrogate the functions of these newly identified IncRNAs. However, advances in other areas such as next-generation sequencing have improved our ability to characterize IncRNAs and have drastically improved assay throughput. Various approaches being utilized to determine IncRNA localization and function will be discussed in this section.
Identification of novel IncRNAs
While protein-coding genes can be predicted computationally from genomic sequences by virtue of their coding capacity and high levels of conservation, the identification of new IncRNAs cannot be performed in this manner, as noncoding transcripts lack these features. Notably, although comparisons of known IncRNAs demonstrated some degree of positive selection and conservation of secondary structure (201–203), predictive algorithms have a high false-positive rate for identification of new IncRNAs (204, 205). Furthermore, some well-characterized IncRNAs such as HOTAIR show highly divergent sequences and gene structures across species and are missed by these predictions, yet the function appears to be conserved (101).
As a result of this and significant advances in sequencing-based technologies and analysis pipelines, RNA sequencing (RNA-seq) experiments remain the gold standard for identification of IncRNAs and have been implemented to identify novel IncRNAs across a variety of tissues and species. While sufficient depth of sequencing (100–200 million reads) is a critical parameter for high-quality studies seeking to exhaustively identify novel low-abundance IncRNAs in a given tissue (206), lower-coverage experiments such as those utilized by Iyer and colleagues (207) can be pooled together to facilitate preliminary identification of novel transcripts. Identification of novel IncRNAs from RNA-seq datasets typically relies on analytical pipelines such as TopHat, which aligns reads that may contain splice junctions to the genome, and Cufflinks, which assembles transcripts de novo using these alignments (208,209).
In addition to these first-pass strategies for identification of novel IncRNAs, additional methods are typically used to define the ends of the transcript and to validate the presence of additional splice isoforms. In many cases, these methods have been updated such that high-throughput sequencing-based methods may be used. While isoforms for a single gene can be identified from cDNA by PCR, detailed interrogation of many loci of interest can be accomplished utilizing protocols such as Capture-Seq, which enrich for sequences of interest using hybridization-based methods and significantly reduce the required depth of sequencing for identification of novel transcripts (54). In addition, identification of the 5ˊ end of the transcript, which is often underrepresented in sequencing libraries, using rapid amplification of cDNA ends (RACE) can be parallelized using methods like Deep-RACE (210).
Identifying clues to IncRNA function
Perhaps the most telling indicator of IncRNA function is its subcellular localization, as this provides insight into its potential binding partners and signals its capacity for re-wiring gene regulation at the transcriptional or post-transcriptional levels. A prominent method for IncRNA localization is single-molecule RNA in situ hybridization. When combined with fluorescence microscopy, these techniques can provide information relating to not only whether the IncRNA is nuclear or cytoplasmic, but whether it is present near putative mRNA or genomic targets within the cell (211). Furthermore, this methodology can also allow one to estimate the range of expression among single cells, and as a result, to more rigorously quantify the IncRNA of interest (212). Other methods relying on cellular fractionation can yield additional complementary information regarding IncRNA function. While the functions of ribosome-associated IncRNAs are not yet clear, ribosome profiling and ribosome fractionation experiments identified a number of ribosome-associated IncRNAs (213), and nuclear fractionation studies revealed a large number of IncRNAs associated to actively transcribed chromatin (214).
Identification of the DNA, RNA, and/or proteins that interact with a given IncRNA generates a second tier of information regarding its function (215). Unbiased approaches to identify these partners often rely on hybridization-based methods to isolate the IncRNA and its binding partners from the cell. Subsequently, next-generation sequencing can be used to identify interactions between other RNA molecules using RNA antisense purification (RAP-seq; (216)), chromatin domains using chromatin isolation by RNA purification (ChIRP; (217)) or capture hybridization analysis of RNA targets (CHART; (218)), or proteins using a wide range of RNA pulldown techniques coupled to mass spectrometry [RAP-MS; (219); Csy4 nuclease coupled by MS; (220)]. Notably, more direct interrogation of the functions of nuclear IncRNAs in regulation of individual loci is now possible with the development of the CRISPR-Display technique (221). This technique uses locus-specific guide RNAs to target a Cas9–lncRNA complex to the desired genomic location, which not only facilitates the detailed characterization of the IncRNA on the locus of interest, but also allows the identification of specific features within the IncRNA that are required for its function.
A third powerful predictor of IncRNA function is the secondary and tertiary structure of the transcript. For example, stem-loop structures are frequently present in ncRNAs that interact with the Polycomb complex protein PRC2, a key epigenetic regulator (33). As mentioned previously, a variety of methods are available to predict RNA secondary structure (202,203,222). However, more rigorous experimental analysis of RNA secondary and tertiary structure is frequently performed using chemical and enzymatic probes. Much like the previously described methods, these methods have been recently adapted to be compatible with next-generation sequencing technologies, yielding techniques like Structure-seq, SHAPE-seq, and FragSeq (223–225).
Finally, while RNA-seq is a powerful technique for identifying new IncRNAs, it also proves useful in predicting potential functions of IncRNAs in vivo through the identification of co-regulated gene networks. By identifying mRNAs that are regulated similarly to the IncRNAs of interest in response to developmental or environmental stimuli, a network of genes affecting specific pathways can be developed and tested using these techniques (226).
Validation of IncRNA function through reverse genetics
Probing the functions of IncRNAs using reverse genetics in vivo is complex, but a number of techniques can be used easily in vitro and in vivo. Antisense knockdown of IncRNAs has become commonplace, and techniques exist for robust knockdown of both nuclear and cytoplasmic IncRNAs (227,228). Furthermore, in vivo electroporation of short hairpin RNAs (shRNAs) to knockdown IncRNAs have also been used to target the Miat IncRNA, suggesting that this may be a relatively useful tool for characterizing IncRNA function. However, strategies relying on RNA interference may be limited by delivery of the siRNA or shRNA, low knockdown efficiency, and off-target effects, and the importance of including well-designed experimental controls can complicate the design of in vivo studies.
The utility of KO animal models for protein-coding genes has been essential for thorough investigations of their functions, but the design of similar models for IncRNAs is much more challenging. Exon-deletion strategies and the use of CRISPR-Cas technologies to introduce frameshifts in protein-coding genes have been very successful (229), but these are likely much less effective for IncRNAs, as there is no ORF to be altered and a IncRNA may retain its function even if significant regions are deleted (21,230–232). Furthermore, as the entire genomic region encoding the IncRNA needs to be removed in order to generate a true null mutant, it is especially important to ensure that regulatory regions that may be encoded within the locus are not perturbed.
Despite these challenges, a number of KO animal models have been developed for IncRNAs and have been reviewed elsewhere (233). In addition, a number of IncRNAs have been modeled successfully by a variety of genetic manipulations including gene disruptions [Xist; (234)], targeted promoter deletions [Kcnq1ot1; (235)], premature termination strategies [Kcnq1ot1; (28)], and the previously mentioned in vivo knockdown strategies [Miat; (236)]. Thus, as with other genetic models in cell culture and in vivo, recent advances in CRISPR-Cas technologies, especially those improving efficiency of targeted knock-in strategies, will likely improve the speed at which IncRNA targeting studies can be accomplished (237).
Conclusions and future studies
Although the half-lives of IncRNAs are, in general, less than those of mRNA (238), they are being recognized as potentially important biomarkers for a number of diseases, particularly cancers (239). For example, one of the most specific biomarkers for human prostate cancer identified to date is the IncRNA, PCA3 (prostate cancer antigen 3) (240,241). Circulating IncRNAs have been detected in whole blood, plasma, serum, and urine and have also recently been proposed as biomarkers for diseases such as gastric cancer and heart failure (242,243). LncRNAs have also been found in extracellular vesicles (i.e., exosomes), which likely increases their half-life in vivo, and thus their potential as a biomarker (244–246). In this regard, IncRNAs could potentially serve as a biomarker of OA or repair processes in the joint, although to date, little is known regarding the relationship between IncRNAs and OA severity. Similarly, future studies will inform us of whether or not IncRNAs could be used as predictive or prognostic biomarkers of bone diseases such as osteoporosis.
Given the size and cellular location of IncRNAs, challenges exist to attempt to knockdown or overexpress these ncRNAs as a means to treat disease. However, we can employ the knowledge acquired from manipulating protein-coding gene expression. In this regard, there are various ways to alter the expression of IncRNAs, including the use of viral or non-viral vectors, RNA-based or genome-editing approaches. While there have been approximately 1800 approved gene therapies in clinical trials from 1989 to 2012 (247), viral-based vectors face major limitations, most notably insertional mutagenesis and immunogenicity. For this reason, the use of non-viral vectors is an attractive alternative. While typical delivery systems (such as lipofectamine or polyethyleneimine) are associated with low efficiency and cytotoxicity (248,249), recent advances have presented the field with new improvements, such as chitosan for non-viral gene delivery that is both efficient and non-toxic (250). In addition, other methods to alter gene expression have also been explored. In the field of RNA therapeutics, it was demonstrated that higher potency and lower dose administration can be achieved with engineered antisense oligonucleotides, such as locked nucleic acid (LNA)-based gapmers (251), multimers, or multi-targeting oligonucleotides (252). Finally, lncRNA expression can potentially be controlled with modified CRISPR–Cas9 complexes. CRISPR (Clustered Regularly Interspaced Palindromic Repeats) and its associated nuclease Cas9 is a powerful and breakthrough genome editing tool. One of the many uses of CRISPR-Cas9 is knocking out protein-coding genes, mainly by introducing frame-shift mutations. However, the system is posed with difficulty when it comes to knocking out lncRNAs due to their lack of an ORF. Nevertheless, this can be achieved by establishing dual cuts to remove the entire gene fragment (237). Alternatively, modified CRISPR-dCas9 systems can also be exploited to enhance or reduce lncRNA expression. A derivative of Cas9, dCas9 is engineered to be nuclease deficient, and can be fused with various effectors to achieve desired transcriptional control. This includes, but not limited to, dCas9-KRAB (253,254) for gene repression and dCas9-VP48 (255), dCas9-VP64 (256) (257), dCas9-p300 (258), dCas9-VP64/p65 (259) for gene activation.
Clearly, lncRNAs play significant roles in regulating cellular fate and function, and in many cases, their dysregulation is associated with disease. With respect to chondrogenesis and osteogenesis, research on how lncRNAs regulate differentiation of stem/progenitor cells toward the chondrocyte or osteoblast lineage is still in its infancy. We predict that research interests in this area will significantly increase over the coming years and provide us with important knowledge on new players involved in the regulation of cartilage and bone development. Similarly, nothing is known yet on how lncRNAs could potentially control homeostasis of mature cartilage or bone; such discoveries will have a profound impact on understanding skeletal diseases such as OA and osteoporosis, and in determining new therapeutic strategies to treat these conditions. Given the size and cellular location of lncRNAs, challenges exist to attempt to knockdown or overexpress these ncRNAs as a means to treat disease. However, through advancing technologies, it is anticipated that novel, effective approaches to modulate lncRNA expression or function in vivo will soon be discovered.
References
- 1.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8(6):413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 2.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 3.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 4.Grishok A, Mello CC. RNAi (Nematodes: Caenorhabditis elegans) Adv Genet. 2002;46:339–360. doi: 10.1016/s0065-2660(02)46012-9. [DOI] [PubMed] [Google Scholar]
- 5.Montgomery MK, Fire A. Double-stranded RNA as a mediator in sequence-specific genetic silencing and cosuppression. Trends Genet. 1998;14(7):255–258. doi: 10.1016/s0168-9525(98)01510-8. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107(7):823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 7.Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19(7–8):454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 8.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442(7099):199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 9.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20(13):1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313(5785):363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 11.Lee HC, Chang SS, Choudhary S, Aalto AP, Maiti M, Bamford DH, Liu Y. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009;459(7244):274–277. doi: 10.1038/nature08041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313(5785):320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20(13):1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Girón CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kähäri AK, Keenan S, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Overduin B, Parker A, Patricio M, Perry E, Pignatelli M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Aken BL, Birney E, Harrow J, Kinsella R, Muffato M, Ruffier M, Searle SMJ, Spudich G, Trevanion SJ, Yates A, Zerbino DR, Flicek P. Ensembl 2015. Nucleic Acids Res. 2015;43(Database issue):D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright MW. A short guide to long non-coding RNA gene nomenclature. Hum Genomics. 2014;8:7. doi: 10.1186/1479-7364-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brockdorff N, Ashworth A, Kay GF, Cooper P, Smith S, McCabe VM, Norris DP, Penny GD, Patel D, Rastan S. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351(6324):329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 17.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 18.Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, Qu K, Zheng GX, Chow J, Kim GE, Rinn JL, Chang HY, Siprashvili Z, Khavari PA. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26(4):338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagase T, Miyajima N, Tanaka A, Sazuka T, Seki N, Sato S, Tabata S, Ishikawa K, Kawarabayasi Y, Kotani H, Nomura N. Prediction of the coding sequences of unidentified human genes III. The coding sequences of 40 new genes (KIAA0081-KIAA0120) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1995;2(1):37–43. doi: 10.1093/dnares/2.1.37. [DOI] [PubMed] [Google Scholar]
- 20.Kikuchi K, Fukuda M, Ito T, Inoue M, Yokoi T, Chiku S, Mitsuyama T, Asai K, Hirose T, Aizawa Y. Transcripts of unknown function in multiple-signaling pathways involved in human stem cell differentiation. Nucleic Acids Res. 2009;37(15):4987–5000. doi: 10.1093/nar/gkp426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22(1):5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 23.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khalil AS, Ferrer JM, Brau RR, Kottmann ST, Noren CJ, Lang MJ, Belcher AM. Single M13 bacteriophage tethering and stretching. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(12):4892–4897. doi: 10.1073/pnas.0605727104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20(10):1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang YW, Flynn RA, Chen Y, Qu K, Wan B, Wang KC, Lei M, Chang HY. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. Elife. 2014;3:e02046. doi: 10.7554/eLife.02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152(4):743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, Pombo A, Fisher AG, Young RA, Jenner RG. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38(5):675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20(3):300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 35.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31(3):522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24(20):2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lam MT, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci. 2014;39(4):170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, Herrmann BG. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24(2):206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sado T, Hoki Y, Sasaki H. Tsix silences Xist through modification of chromatin structure. Dev Cell. 2005;9(1):159–165. doi: 10.1016/j.devcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 41.Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. Jpx RNA activates Xist by evicting CTCF. Cell. 2013;153(7):1537–1551. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143(3):390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35(4):467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, Forrest AR, Carninci P, Biffo S, Stupka E, Gustincich S. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491(7424):454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 46.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature. 2011;470(7333):284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309(5740):1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 48.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G, 3rd, Wahlestedt C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11(5):R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown BD, Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nature Rev Genet. 2009;10(8):578–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- 51.Le MT, Xie H, Zhou B, Chia PH, Rizk P, Um M, Udolph G, Yang H, Lim B, Lodish HF. MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol Cell Biol. 2009;29(19):5290–5305. doi: 10.1128/MCB.01694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10(8):987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 53.Ramos AD, Diaz A, Nellore A, Delgado RN, Park KY, Gonzales-Roybal G, Oldham MC, Song JS, Lim DA. Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell. 2013;12(5):616–628. doi: 10.1016/j.stem.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2012;30(1):99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, Abo R, Tabebordbar M, Lee RT, Burge CB, Boyer LA. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152(3):570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arney KL. H19 and Igf2-enhancing the confusion? Trends Genet. 2003;19(1):17–23. doi: 10.1016/s0168-9525(02)00004-5. [DOI] [PubMed] [Google Scholar]
- 57.Gabory A, Ripoche MA, Yoshimizu T, Dandolo L. The H19 gene: regulation and function of a non-coding RNA. Cytogenet Genome Res. 2006;113(1–4):188–193. doi: 10.1159/000090831. [DOI] [PubMed] [Google Scholar]
- 58.Ohlsson R, Hedborg F, Holmgren L, Walsh C, Ekstrom TJ. Overlapping patterns of IGF2 and H19 expression during human development: biallelic IGF2 expression correlates with a lack of H19 expression. Development. 1994;120(2):361–368. doi: 10.1242/dev.120.2.361. [DOI] [PubMed] [Google Scholar]
- 59.Taniguchi T, Schofield AE, Scarlett JL, Morison IM, Sullivan MJ, Reeve AE. Altered specificity of IGF2 promoter imprinting during fetal development and onset of Wilms tumour. Oncogene. 1995;11(4):751–756. [PubMed] [Google Scholar]
- 60.Ripoche MA, Kress C, Poirier F, Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997;11(12):1596–1604. doi: 10.1101/gad.11.12.1596. [DOI] [PubMed] [Google Scholar]
- 61.Hansen TV, Hammer NA, Nielsen J, Madsen M, Dalbaeck C, Wewer UM, Christiansen J, Nielsen FC. Dwarfism and impaired gut development in insulinlike growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol. 2004;24(10):4448–4464. doi: 10.1128/MCB.24.10.4448-4464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Runge S, Nielsen FC, Nielsen J, Lykke-Andersen J, Wewer UM, Christiansen J. H19 RNA binds four molecules of insulin-like growth factor II mRNA-binding protein. J Biol Chem. 2000;275(38):29562–29569. doi: 10.1074/jbc.M001156200. [DOI] [PubMed] [Google Scholar]
- 63.Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, Kwok TT. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31(3):350–358. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 64.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14(7):659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. Rna. 2007;13(3):313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lustig O, Ariel I, Ilan J, Lev-Lehman E, De-Groot N, Hochberg A. Expression of the imprinted gene H19 in the human fetus. Mol Reprod Dev. 1994;38(3):239–246. doi: 10.1002/mrd.1080380302. [DOI] [PubMed] [Google Scholar]
- 67.Poirier F, Chan CT, Timmons PM, Robertson EJ, Evans MJ, Rigby PW. The murine H19 gene is activated during embryonic stem cell differentiation in vitro and at the time of implantation in the developing embryo. Development. 1991;113(4):1105–1114. doi: 10.1242/dev.113.4.1105. [DOI] [PubMed] [Google Scholar]
- 68.Hall BK, Miyake T. The membranous skeleton: the role of cell condensations in vertebrate skeletogenesis. Anat Embryol (Berl) 1992;186(2):107–124. doi: 10.1007/BF00174948. [DOI] [PubMed] [Google Scholar]
- 69.Hyde G, Dover S, Aszodi A, Wallis GA, Boot-Handford RP. Lineage tracing using matrilin-1 gene expression reveals that articular chondrocytes exist as the joint interzone forms. Dev Biol. 2007;304(2):825–833. doi: 10.1016/j.ydbio.2007.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koyama E, Shibukawa Y, Nagayama M, Sugito H, Young B, Yuasa T, Okabe T, Ochiai T, Kamiya N, Rountree RB, Kingsley DM, Iwamoto M, Enomoto-Iwamoto M, Pacifici M. A distinct cohort of progenitor cells participates in synovial joint and articular cartilage formation during mouse limb skeletogenesis. Dev Biol. 2008;316(1):62–73. doi: 10.1016/j.ydbio.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 72.Park J, Gebhardt M, Golovchenko S, Perez-Branguli F, Hattori T, Hartmann C, Zhou X, deCrombrugghe B, Stock M, Schneider H, von der Mark K. Dual pathways to endochondral osteoblasts: a novel chondrocyte-derived osteoprogenitor cell identified in hypertrophic cartilage. Biol Open. 2015;4(5):608–621. doi: 10.1242/bio.201411031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jing Y, Zhou X, Han X, Jing J, von der Mark K, Wang J, de Crombrugghe B, Hinton RJ, Feng JQ. Chondrocytes directly transform into bone cells in mandibular condyle growth. J Dent Res. 2015;94(12):1668–1675. doi: 10.1177/0022034515598135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou X, von der Mark K, Henry S, Norton W, Adams H, de Crombrugghe B. Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet. 2014;10(12):e1004820. doi: 10.1371/journal.pgen.1004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang L, Tsang KY, Tang HC, Chan D, Cheah KS. Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc Natl Acad Sci USA. 2014;111(33):12097–12102. doi: 10.1073/pnas.1302703111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Swingler TE, Wheeler G, Carmont V, Elliott HR, Barter MJ, Abu-Elmagd M, Donell ST, Boot-Handford RP, Hajihosseini MK, Munsterberg A, Dalmay T, Young DA, Clark IM. The expression and function of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheumatism. 2012;64(6):1909–1919. doi: 10.1002/art.34314. [DOI] [PubMed] [Google Scholar]
- 77.Gabler J, Ruetze M, Kynast KL, Grossner T, Diederichs S, Richter W. Stage-specific mirs in chondrocyte maturation: differentiation-dependent and hypertrophy-related miR Clusters and the miR-181 family. Tissue Eng Part A. 2015;21(23–24):2840–2851. doi: 10.1089/ten.TEA.2015.0352. [DOI] [PubMed] [Google Scholar]
- 78.McAlinden A, Varghese N, Wirthlin L, Chang LW. Differentially expressed microRNAs in chondrocytes from distinct regions of developing human cartilage. PloS One. 2013;8(9):e75012. doi: 10.1371/journal.pone.0075012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gibson G, Asahara H. microRNAs and cartilage. J Orthopaedic Res. 2013;31(9):1333–1344. doi: 10.1002/jor.22397. [DOI] [PubMed] [Google Scholar]
- 80.Hong E, Reddi AH. MicroRNAs in chondrogenesis, articular cartilage, and osteoarthritis: implications for tissue engineering. Tissue Eng Part B. 2012;18(6):445–453. doi: 10.1089/ten.TEB.2012.0116. [DOI] [PubMed] [Google Scholar]
- 81.Le LT, Swingler TE, Clark IM. Review: the role of microRNAs in osteoarthritis and chondrogenesis. Arthritis Rheumatism. 2013;65(8):1963–1974. doi: 10.1002/art.37990. [DOI] [PubMed] [Google Scholar]
- 82.Shang J, Liu H, Zhou Y. Roles of microRNAs in prenatal chondrogenesis, postnatal chondrogenesis and cartilage-related diseases. J Cell Mol Med. 2013;17(12):1515–1524. doi: 10.1111/jcmm.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mackay AM, Beck SC, Murphy JM, Barry FP, Chichester CO, Pittenger MF. Chondrogenic differentiation of cultured human mesenchymal stem cells from marrow. Tissue Eng. 1998;4(4):415–428. doi: 10.1089/ten.1998.4.415. [DOI] [PubMed] [Google Scholar]
- 84.Johnstone B, Hering TM, Caplan AI, Goldberg VM, Yoo JU. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res. 1998;238(1):265–272. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- 85.Boeuf S, Richter W. Chondrogenesis of mesenchymal stem cells: role of tissue source and inducing factors. Stem Cell Res. Ther. 2010;1(4):31. doi: 10.1186/scrt31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Beane OS, Darling EM. Isolation, characterization, and differentiation of stem cells for cartilage regeneration. Ann Biomed Eng. 2012;40(10):2079–2097. doi: 10.1007/s10439-012-0639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang L, Li Z, Li Z, Yu B, Wang Y. Long noncoding RNAs expression signatures in chondrogenic differentiation of human bone marrow mesenchymal stem cells. Biochem Biophys Res Commun. 2015;456(1):459–464. doi: 10.1016/j.bbrc.2014.11.106. [DOI] [PubMed] [Google Scholar]
- 88.Barry F, Boynton RE, Liu B, Murphy JM. Chondrogenic differentiation of mesenchymal stem cells from bone marrow: differentiation-dependent gene expression of matrix components. Exp Cell Res. 2001;268(2):189–200. doi: 10.1006/excr.2001.5278. [DOI] [PubMed] [Google Scholar]
- 89.Hardingham TE, Oldershaw RA, Tew SR. Cartilage, SOX9 and Notch signals in chondrogenesis. J Anat. 2006;209(4):469–480. doi: 10.1111/j.1469-7580.2006.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang L, Chen S, Bao N, Yang C, Ti Y, Zhou L, Zhao J. Sox4 enhances chondrogenic differentiation and proliferation of human synovium-derived stem cell via activation of long noncoding RNA DANCR. J Mol Histol. 2015;46(6):467–473. doi: 10.1007/s10735-015-9638-z. [DOI] [PubMed] [Google Scholar]
- 91.Yuan SX, Wang J, Yang F, Tao QF, Zhang J, Wang LL, Yang Y, Liu H, Wang ZG, Xu QG, Fan J, Liu L, Sun SH, Zhou WP. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by dere-pression of CTNNB1. Hepatology. 2016;63(2):499–511. doi: 10.1002/hep.27893. [DOI] [PubMed] [Google Scholar]
- 92.Chen L, Song Z, Huang S, Wang R, Qin W, Guo J, Lin Z. lncRNA DANCR suppresses odontoblast-like differentiation of human dental pulp cells by inhibiting wnt/beta-catenin pathway. Cell Tissue Res. 2016;364(2):309–318. doi: 10.1007/s00441-015-2333-2. [DOI] [PubMed] [Google Scholar]
- 93.Maass PG, Rump A, Schulz H, Stricker S, Schulze L, Platzer K, Aydin A, Tinschert S, Goldring MB, Luft FC, Bahring S. A misplaced lncRNA causes brachydactyly in humans. J Clin Invest. 2012;122(11):3990–4002. doi: 10.1172/JCI65508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273(5275):613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 95.Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22(1):85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 96.Pineault KM, Wellik DM. Hox genes and limb musculoskeletal development. Curr Osteoporos Rep. 2014;12(4):420–427. doi: 10.1007/s11914-014-0241-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Favier B, Le Meur M, Chambon P, Dolle P. Axial skeleton homeosis and forelimb malformations in Hoxd-11 mutant mice. Proc Natl Acad Sci USA. 1995;92(1):310–314. doi: 10.1073/pnas.92.1.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gerard M, Chen JY, Gronemeyer H, Chambon P, Duboule D, Zakany J. In vivo targeted mutagenesis of a regulatory element required for positioning the Hoxd-11 and Hoxd-10 expression boundaries. Genes Dev. 1996;10(18):2326–2334. doi: 10.1101/gad.10.18.2326. [DOI] [PubMed] [Google Scholar]
- 99.Hashimoto K, Yokouchi Y, Yamamoto M, Kuroiwa A. Distinct signaling molecules control Hoxa-11 and Hoxa-13 expression in the muscle precursor and mesenchyme of the chick limb bud. Development. 1999;126(12):2771–2783. doi: 10.1242/dev.126.12.2771. [DOI] [PubMed] [Google Scholar]
- 100.Salsi V, Zappavigna V. Hoxd13 and Hoxa13 directly control the expression of the EphA7 Ephrin tyrosine kinase receptor in developing limbs. J Biol Chem. 2006;281(4):1992–1999. doi: 10.1074/jbc.M510900200. [DOI] [PubMed] [Google Scholar]
- 101.Li L, Liu B, Wapinski OL, Tsai MC, Qu K, Zhang J, Carlson JC, Lin M, Fang F, Gupta RA, Helms JA, Chang HY. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5(1):3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carrion K, Dyo J, Patel V, Sasik R, Mohamed SA, Hardiman G, Nigam V. The long non-coding HOTAIR is modulated by cyclic stretch and WNT/ beta-CATENIN in human aortic valve cells and is a novel repressor of calcification genes. PloS One. 2014;9(5):e96577. doi: 10.1371/journal.pone.0096577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128(4):735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 104.Carlson HL, Quinn JJ, Yang YW, Thornburg CK, Chang HY, Stadler HS. LncRNA-HIT Functions as an epigenetic regulator of chondrogenesis through its recruitment of p100/CBP complexes. PLoS Genet. 2015;11(12):e1005680. doi: 10.1371/journal.pgen.1005680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Richards EJ, Zhang G, Li ZP, Permuth-Wey J, Challa S, Li Y, Kong W, Dan S, Bui MM, Coppola D, Mao WM, Sellers TA, Cheng JQ. Long non-coding RNAs (LncRNA) regulated by transforming growth factor (TGF) beta: LncRNA-hit-mediated TGFbeta-induced epithelial to mesenchymal transition in mammary epithelia. J Biol Chem. 2015;290(11):6857–6867. doi: 10.1074/jbc.M114.610915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, Adachi J, Fukuda S, Aizawa K, Izawa M, Nishi K, Kiyosawa H, Kondo S, Yamanaka I, Saito T, Okazaki Y, Gojobori T, Bono H, Kasukawa T, Saito R, Kadota K, Matsuda H, Ashburner M, Batalov S, Casavant T, Fleischmann W, Gaasterland T, Gissi C, King B, Kochiwa H, Kuehl P, Lewis S, Matsuo Y, Nikaido I, Pesole G, Quackenbush J, Schriml LM, Staubli F, Suzuki R, Tomita M, Wagner L, Washio T, Sakai K, Okido T, Furuno M, Aono H, Baldarelli R, Barsh G, Blake J, Boffelli D, Bojunga N, Carninci P, de Bonaldo MF, Brownstein MJ, Bult C, Fletcher C, Fujita M, Gariboldi M, Gustincich S, Hill D, Hofmann M, Hume DA, Kamiya M, Lee NH, Lyons P, Marchionni L, Mashima J, Mazzarelli J, Mombaerts P, Nordone P, Ring B, Ringwald M, Rodriguez I, Sakamoto N, Sasaki H, Sato K, Schonbach C, Seya T, Shibata Y, Storch KF, Suzuki H, Toyo-oka K, Wang KH, Weitz C, Whittaker C, Wilming L, Wynshaw-Boris A, Yoshida K, Hasegawa Y, Kawaji H, Kohtsuki S, Hayashizaki Y Team RGERGPI, the FC. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409(6821):685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- 107.Suzuki T, Hasso SM, Fallon JF. Unique SMAD1/5/8 activity at the phalanx-forming region determines digit identity. Proc Natl Acad Sci US A. 2008;105(11):4185–4190. doi: 10.1073/pnas.0707899105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chimal-Monroy J, Rodriguez-Leon J, Montero JA, Ganan Y, Macias D, Merino R, Hurle JM. Analysis of the molecular cascade responsible for mesodermal limb chondrogenesis: Sox genes and BMP signaling. Dev Biol. 2003;257(2):292–301. doi: 10.1016/s0012-1606(03)00066-6. [DOI] [PubMed] [Google Scholar]
- 109.Lehmann K, Seemann P, Stricker S, Sammar M, Meyer B, Suring K, Majewski F, Tinschert S, Grzeschik KH, Muller D, Knaus P, Nurnberg P, Mundlos S. Mutations in bone morphogenetic protein receptor 1B cause brachydactyly type A2. Proc Natl Acad Sci USA. 2003;100(21):12277–12282. doi: 10.1073/pnas.2133476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Baur ST, Mai JJ, Dymecki SM. Combinatorial signaling through BMP receptor IB and GDF5: shaping of the distal mouse limb and the genetics of distal limb diversity. Development. 2000;127(3):605–619. doi: 10.1242/dev.127.3.605. [DOI] [PubMed] [Google Scholar]
- 111.Buckwalter JA. Osteoarthritis and articular cartilage use, disuse, and abuse: experimental studies. J Rheumatol Suppl. 1995;43:13–5. [PubMed] [Google Scholar]
- 112.Gilbertson EM. Development of periarticular osteophytes in experimentally induced osteoarthritis in the dog. A study using microradiographic, microangio-graphic, and fluorescent bone-labelling techniques. Ann Rheum Dis. 1975;34(1):12–25. doi: 10.1136/ard.34.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guilak F. Biomechanical factors in osteoarthritis. Best Pract Res Clin Rheumatol. 2011;25(6):815–823. doi: 10.1016/j.berh.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hoch DH, Grodzinsky AJ, Koob TJ, Albert ML, Eyre DR. Early changes in material properties of rabbit articular cartilage after meniscectomy. J. Orthopaedic Res. 1983;1(1):4–12. doi: 10.1002/jor.1100010102. [DOI] [PubMed] [Google Scholar]
- 115.Moskowitz RW, Davis W, Sammarco J, Martens M, Baker J, Mayor M, Burstein AH, Frankel VH. Experimentally induced degenerative joint lesions following partial meniscectomy in the rabbit. Arthritis Rheumatism. 1973;16(3):397–405. doi: 10.1002/art.1780160317. [DOI] [PubMed] [Google Scholar]
- 116.Pond MJ, Nuki G. Experimentally-induced osteoarthritis in the dog. Ann Rheum Dis. 1973;32(4):387–388. doi: 10.1136/ard.32.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Griffin TM, Fermor B, Huebner JL, Kraus VB, Rodriguiz RM, Wetsel WC, Cao L, Setton LA, Guilak F. Diet-induced obesity differentially regulates behavioral, bio-mechanical, and molecular risk factors for osteoarthritis in mice. Arthritis Res Ther. 2010;12(4):R130. doi: 10.1186/ar3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wu CL, Diekman BO, Jain D, Guilak F. Diet-induced obesity alters the differentiation potential of stem cells isolated from bone marrow, adipose tissue and infrapatellar fat pad: the effects of free fatty acids. Int J Obes (Lond) 2013;37(8):1079–1087. doi: 10.1038/ijo.2012.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hugle T, Geurts J, Nuesch C, Muller-Gerbl M, Valderrabano V. Aging and osteoarthritis: an inevitable encounter? J Aging Res. 2012;2012:950192. doi: 10.1155/2012/950192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Goldring MB. Articular cartilage degradation in osteoarthritis. HSS J. 2012;8(1):7–9. doi: 10.1007/s11420-011-9250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sokolove J, Lepus CM. Role of inflammation in the pathogenesis of osteoarthritis: latest findings and interpretations. Ther Adv Musculoskelet Dis. 2013;5(2):77–94. doi: 10.1177/1759720X12467868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: a disease of the joint as an organ. Arthritis Rheumatism. 2012;64(6):1697–1707. doi: 10.1002/art.34453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Barter MJ, Bui C, Young DA. Epigenetic mechanisms in cartilage and osteoarthritis: DNA methylation, histone modifications and microRNAs. Osteoarthritis Cartilage. 2012;20(5):339–349. doi: 10.1016/j.joca.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 124.Barter MJ, Young DA. Epigenetic mechanisms and non-coding RNAs in osteoarthritis. Curr Rheumatol Rep. 2013;15(9):353. doi: 10.1007/s11926-013-0353-z. [DOI] [PubMed] [Google Scholar]
- 125.den Hollander W, Ramos YF, Bomer N, Elzinga S, van der Breggen R, Lakenberg N, de Dijcker WJ, Suchiman HE, Duijnisveld BJ, Houwing-Duistermaat JJ, Slagboom PE, Bos SD, Nelissen RG, Meulenbelt I. Transcriptional associations of osteoarthritis-mediated loss of epigenetic control in articular cartilage. Arthritis Rheumatol. 2015;67(8):2108–2116. doi: 10.1002/art.39162. [DOI] [PubMed] [Google Scholar]
- 126.Fernandez-Tajes J, Soto-Hermida A, Vazquez-Mosquera ME, Cortes-Pereira E, Mosquera A, Fernandez-Moreno M, Oreiro N, Fernandez-Lopez C, Fernandez JL, Rego-Perez I, Blanco FJ. Genome-wide DNA methylation analysis of articular chondrocytes reveals a cluster of osteoarthritic patients. Ann Rheum Dis. 2014;73(4):668–677. doi: 10.1136/annrheumdis-2012-202783. [DOI] [PubMed] [Google Scholar]
- 127.Gabay O, Sanchez C. Epigenetics, sirtuins and osteoarthritis. Joint Bone Spine. 2012;79(6):570–573. doi: 10.1016/j.jbspin.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 128.Jeffries MA, Donica M, Baker LW, Stevenson ME, Annan AC, Humphrey MB, James JA, Sawalha AH. Genome-wide DNA methylation study identifies significant epigenomic changes in osteoarthritic cartilage. Arthritis Rheumatol. 2014;66(10):2804–2815. doi: 10.1002/art.38762. [DOI] [PubMed] [Google Scholar]
- 129.Rushton MD, Young DA, Loughlin J, Reynard LN. Differential DNA methylation and expression of inflammatory and zinc transporter genes defines subgroups of osteoarthritic hip patients. Ann Rheum Dis. 2015;74(9):1778–1782. doi: 10.1136/annrheumdis-2014-206752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Taylor SE, Li YH, Wong WH, Bhutani N. Genome-wide mapping of DNA hydroxymethylation in osteoarthritic chondrocytes. Arthritis Rheumatol. 2015;67(8):2129–2140. doi: 10.1002/art.39179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Taylor SE, Smeriglio P, Dhulipala L, Rath M, Bhutani N. A global increase in 5-hydroxymethylcytosine levels marks osteoarthritic chondrocytes. Arthritis Rheumatol. 2014;66(1):90–100. doi: 10.1002/art.38200. [DOI] [PubMed] [Google Scholar]
- 132.Rushton MD, Reynard LN, Barter MJ, Refaie R, Rankin KS, Young DA, Loughlin J. Characterization of the cartilage DNA methylome in knee and hip osteoarthritis. Arthritis Rheumatol. 2014;66(9):2450–2460. doi: 10.1002/art.38713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dudek KA, Lafont JE, Martinez-Sanchez A, Murphy CL. Type II collagen expression is regulated by tissue-specific miR-675 in human articular chondrocytes. J Biol Chem. 2010;285(32):24381–24387. doi: 10.1074/jbc.M110.111328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Steck E, Boeuf S, Gabler J, Werth N, Schnatzer P, Diederichs S, Richter W. Regulation of H19 and its encoded microRNA-675 in osteoarthritis and under anabolic and catabolic in vitro conditions. J Mol Med. 2012;90(10):1185–1195. doi: 10.1007/s00109-012-0895-y. [DOI] [PubMed] [Google Scholar]
- 135.Liang WC, Fu WM, Wang YB, Sun YX, Xu LL, Wong CW, Chan KM, Li G, Waye MM, Zhang JF. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Sci Rep. 2016;6:20121. doi: 10.1038/srep20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huang Y, Zheng Y, Jia L, Li W. Long noncoding RNA H19 promotes osteoblast differentiation via TGF-beta1/Smad3/HDAC signaling pathway by deriving miR-675. Stem Cells. 2015;33(12):3481–3492. doi: 10.1002/stem.2225. [DOI] [PubMed] [Google Scholar]
- 137.Zhou J, Yang L, Zhong T, Mueller M, Men Y, Zhang N, Xie J, Giang K, Chung H, Sun X, Lu L, Carmichael GG, Taylor HS, Huang Y. H19 lncRNA alters DNA methyla-tion genome wide by regulating S-adenosylhomocysteine hydrolase. Nat Commun. 2015;6:10221. doi: 10.1038/ncomms10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xing D, Liang JQ, Li Y, Lu J, Jia HB, Xu LY, Ma XL. Identification of long noncoding RNA associated with osteoarthritis in humans. Orthop Surg. 2014;6(4):288–293. doi: 10.1111/os.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim D, Song J, Han J, Kim Y, Chun CH, Jin EJ. Two non-coding RNAs, MicroRNA-101 and HOTTIP contribute cartilage integrity by epigenetic and homeotic regulation of integrin-alpha1. Cell Signalling. 2013;25(12):2878–2887. doi: 10.1016/j.cellsig.2013.08.034. [DOI] [PubMed] [Google Scholar]
- 140.Fu M, Huang G, Zhang Z, Liu J, Zhang Z, Huang Z, Yu B, Meng F. Expression profile of long noncoding RNAs in cartilage from knee osteoarthritis patients. Osteoarthritis Cartilage. 2015;23(3):423–432. doi: 10.1016/j.joca.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 141.Liu Q, Zhang X, Dai L, Hu X, Zhu J, Li L, Zhou C, Ao Y. Long noncoding RNA related to cartilage injury promotes chondrocyte extracellular matrix degradation in osteoarthritis. Arthritis Rheumatol. 2014;66(4):969–978. doi: 10.1002/art.38309. [DOI] [PubMed] [Google Scholar]
- 142.Song J, Ahn C, Chun CH, Jin EJ. A long non-coding RNA, GAS5, plays a critical role in the regulation of miR-21 during osteoarthritis. J Orthopaedic Res. 2014;32(12):1628–1635. doi: 10.1002/jor.22718. [DOI] [PubMed] [Google Scholar]
- 143.Glasson SS, Blanchet TJ, Morris EA. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis Cartilage. 2007;15(9):1061–1069. doi: 10.1016/j.joca.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 144.Su W, Xie W, Shang Q, Su B. The Long Noncoding RNA MEG3 Is Downregulated and inversely associated with VEGF levels in osteoarthritis. Biomed Res Int. 2015;2015:356893. doi: 10.1155/2015/356893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pearson MJ, Philp AM, Heward JA, Roux BT, Walsh DA, Davis ET, Lindsay MA, Jones SW. Long intergenic noncoding RNAs mediate the human chondrocyte inflammatory response and are differentially expressed in osteoarthritis cartilage. Arthritis Rheumatol. 2016;68(4):845–856. doi: 10.1002/art.39520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gilbert SF. Developmental biology. 9th. Sunderland, MA: Sinauer Associates; 2010. p. 80. xxi, 711. [Google Scholar]
- 147.Ash P, Loutit JF, Townsend KM. Osteoclasts derived from haematopoietic stem cells. Nature. 1980;283(5748):669–670. doi: 10.1038/283669a0. [DOI] [PubMed] [Google Scholar]
- 148.Blair HC, Kahn AJ, Crouch EC, Jeffrey JJ, Teitelbaum SL. Isolated osteoclasts resorb the organic and inorganic components of bone. J Cell Biol. 1986;102(4):1164–1172. doi: 10.1083/jcb.102.4.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hall BK, Miyake T. Divide, accumulate, differentiate: cell condensation in skeletal development revisited. Int J Dev Biol. 1995;39(6):881–893. [PubMed] [Google Scholar]
- 150.Oberlender SA, Tuan RS. Expression and functional involvement of N-cadherin in embryonic limb chondrogenesis. Development. 1994;120(1):177–187. doi: 10.1242/dev.120.1.177. [DOI] [PubMed] [Google Scholar]
- 151.Oberlender SA, Tuan RS. Spatiotemporal profile of N-cadherin expression in the developing limb mesenchyme. Cell Adhes Commun. 1994;2(6):521–537. doi: 10.3109/15419069409014216. [DOI] [PubMed] [Google Scholar]
- 152.Gerber HP, Hillan KJ, Ryan AM, Kowalski J, Keller GA, Rangell L, Wright BD, Radtke F, Aguet M, Ferrara N. VEGF is required for growth and survival in neonatal mice. Development. 1999;126(6):1149–1159. doi: 10.1242/dev.126.6.1149. [DOI] [PubMed] [Google Scholar]
- 153.Haigh JJ, Gerber HP, Ferrara N, Wagner EF. Conditional inactivation of VEGF-A in areas of collagen2a1 expression results in embryonic lethality in the heterozygous state. Development. 2000;127(7):1445–1453. doi: 10.1242/dev.127.7.1445. [DOI] [PubMed] [Google Scholar]
- 154.Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, Karsenty G, Olson EN. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119(4):555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 155.Abzhanov A, Rodda SJ, McMahon AP, Tabin CJ. Regulation of skeletogenic differentiation in cranial dermal bone. Development. 2007;134(17):3133–3144. doi: 10.1242/dev.002709. [DOI] [PubMed] [Google Scholar]
- 156.Haworth KE, Healy C, Morgan P, Sharpe PT. Regionalisation of early head ectoderm is regulated by endoderm and prepatterns the orofacial epithelium. Development. 2004;131(19):4797–4806. doi: 10.1242/dev.01337. [DOI] [PubMed] [Google Scholar]
- 157.Lee SH, Fu KK, Hui JN, Richman JM. Noggin and retinoic acid transform the identity of avian facial prominences. Nature. 2001;414(6866):909–912. doi: 10.1038/414909a. [DOI] [PubMed] [Google Scholar]
- 158.Creuzet S, Couly G, Le Douarin NM. Patterning the neural crest derivatives during development of the vertebrate head: insights from avian studies. J Anat. 2005;207(5):447–459. doi: 10.1111/j.1469-7580.2005.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Trokovic N, Trokovic R, Mai P, Partanen J. Fgfr1 regulates patterning of the pharyngeal region. Genes Dev. 2003;17(1):141–153. doi: 10.1101/gad.250703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Trokovic N, Trokovic R, Partanen J. Fibroblast growth factor signalling and regional specification of the pharyngeal ectoderm. Int J Dev Biol. 2005;49(7):797–805. doi: 10.1387/ijdb.051976nt. [DOI] [PubMed] [Google Scholar]
- 161.Holleville N, Quilhac A, Bontoux M, Monsoro-Burq AH. BMP signals regulate Dlx5 during early avian skull development. Dev Biol. 2003;257(1):177–189. doi: 10.1016/s0012-1606(03)00059-9. [DOI] [PubMed] [Google Scholar]
- 162.Creuzet S, Schuler B, Couly G, Le Douarin NM. Reciprocal relationships between Fgf8 and neural crest cells in facial and forebrain development. Proc Natl Acad Sci USA. 2004;101(14):4843–4847. doi: 10.1073/pnas.0400869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kang JS, Alliston T, Delston R, Derynck R. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. EMBO J. 2005;24(14):2543–2555. doi: 10.1038/sj.emboj.7600729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Saydam O, Shen Y, Wurdinger T, Senol O, Boke E, James MF, Tannous BA, Stemmer-Rachamimov AO, Yi M, Stephens RM, Fraefel C, Gusella JF, Krichevsky AM, Breakefield XO. Downregulated microRNA-200a in meningiomas promotes tumor growth by reducing E-cadherin and activating the Wnt/beta-catenin signaling pathway. Mol Cell Biol. 2009;29(21):5923–5940. doi: 10.1128/MCB.00332-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhu L, Xu PC. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem Biophys Res Commun. 2013;432(4):612–617. doi: 10.1016/j.bbrc.2013.02.036. [DOI] [PubMed] [Google Scholar]
- 166.Jia Q, Jiang W, Ni L. Down-regulated non-coding RNA (lncRNA-ANCR) promotes osteogenic differentiation of periodontal ligament stem cells. Arch Oral Biol. 2015;60(2):234–241. doi: 10.1016/j.archoralbio.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 167.Zuo C, Wang Z, Lu H, Dai Z, Liu X, Cui L. Expression profiling of lncRNAs in C3H10T1/2 mesenchymal stem cells undergoing early osteoblast differentiation. Mol Med Rep. 2013;8(2):463–467. doi: 10.3892/mmr.2013.1540. [DOI] [PubMed] [Google Scholar]
- 168.Backesjo CM, Li Y, Lindgren U, Haldosen LA. Activation of Sirt1 decreases adipocyte formation during osteoblast differentiation of mesenchymal stem cells. J Bone Miner Res. 2006;21(7):993–1002. doi: 10.1359/jbmr.060415. [DOI] [PubMed] [Google Scholar]
- 169.Cohen-Kfir E, Artsi H, Levin A, Abramowitz E, Bajayo A, Gurt I, Zhong L, D’Urso A, Toiber D, Mostoslavsky R, Dresner-Pollak R. Sirt1 is a regulator of bone mass and a repressor of Sost encoding for sclerostin, a bone formation inhibitor. Endocrinology. 2011;152(12):4514–4524. doi: 10.1210/en.2011-1128. [DOI] [PubMed] [Google Scholar]
- 170.Zhuang W, Ge X, Yang S, Huang M, Zhuang W, Chen P, Zhang X, Fu J, Qu J, Li B. Upregulation of lncRNA MEG3 promotes osteogenic differentiation of mesenchymal stem cells from multiple myeloma patients by targeting BMP4 transcription. Stem Cells. 2015;33(6):1985–1997. doi: 10.1002/stem.1989. [DOI] [PubMed] [Google Scholar]
- 171.Haddox CL, Han G, Anijar L, Binitie O, Letson GD, Bui MM, Reed DR. Osteosarcoma in pediatric patients and young adults: a single institution retrospective review of presentation, therapy, and outcome. Sarcoma. 2014;2014:402509. doi: 10.1155/2014/402509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hagleitner MM, de Bont ES, Te Loo DM. Survival trends and long-term toxicity in pediatric patients with osteosarcoma. Sarcoma. 2012;2012:636405. doi: 10.1155/2012/636405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chan LH, Wang W, Yeung W, Deng Y, Yuan P, Mak KK. Hedgehog signaling induces osteosarcoma development through Yap1 and H19 overexpression. Oncogene. 2014;33(40):4857–4866. doi: 10.1038/onc.2013.433. [DOI] [PubMed] [Google Scholar]
- 174.Sathiyamoorthy S, Ali SZ. Osteoblastic osteosarcoma: cytomorphologic characteristics and differential diagnosis on fine-needle aspiration. Acta Cytol. 2012;56(5):481–486. doi: 10.1159/000339196. [DOI] [PubMed] [Google Scholar]
- 175.Bernthal NM, Federman N, Eilber FR, Nelson SD, Eckardt JJ, Eilber FC, Tap WD. Long-term results (>25 years) of a randomized, prospective clinical trial evaluating chemotherapy in patients with high-grade, operable osteosarcoma. Cancer. 2012;118(23):5888–5893. doi: 10.1002/cncr.27651. [DOI] [PubMed] [Google Scholar]
- 176.Eilber F, Giuliano A, Eckardt J, Patterson K, Moseley S, Goodnight J. Adjuvant chemotherapy for osteosarcoma: a randomized prospective trial. J Clin Oncol. 1987;5(1):21–26. doi: 10.1200/JCO.1987.5.1.21. [DOI] [PubMed] [Google Scholar]
- 177.Link MP, Goorin AM, Miser AW, Green AA, Pratt CB, Belasco JB, Pritchard J, Malpas JS, Baker AR, Kirkpatrick JA, Ayala AG, Shuster JJ, Abelson HT, Simone JV, Vietti TJ. The effect of adjuvant chemotherapy on relapse-free survival in patients with osteosarcoma of the extremity. N Engl J Med. 1986;314(25):1600–1606. doi: 10.1056/NEJM198606193142502. [DOI] [PubMed] [Google Scholar]
- 178.Meyers PA, Gorlick R, Heller G, Casper E, Lane J, Huvos AG, Healey JH. Intensification of preoperative chemotherapy for osteogenic sarcoma: results of the Memorial Sloan-Kettering (T12) protocol. J Clin Oncol. 1998;16(7):2452–2458. doi: 10.1200/JCO.1998.16.7.2452. [DOI] [PubMed] [Google Scholar]
- 179.Rosen G, Marcove RC, Huvos AG, Caparros BI, Lane JM, Nirenberg A, Cacavio A, Groshen S. Primary osteogenic sarcoma: eight-year experience with adjuvant chemotherapy. J Cancer Res Clin Oncol. 1983;106(Suppl):55–67. doi: 10.1007/BF00625054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Muller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 181.Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3’ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26(21):2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yoshimoto R, Mayeda A, Yoshida M, Nakagawa S. MALAT1 long non-coding RNA in cancer. Biochim Biophys Acta. 2016;1859(1):192–199. doi: 10.1016/j.bbagrm.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 183.Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015;36(3):1477–1486. doi: 10.1007/s13277-014-2631-4. [DOI] [PubMed] [Google Scholar]
- 184.Cai X, Liu Y, Yang W, Xia Y, Yang C, Yang S, Liu X. Long noncoding RNA MALAT1 as a potential therapeutic target in osteosarcoma. J Orthopaedic Res. 2016;34(6):932–941. doi: 10.1002/jor.23105. [DOI] [PubMed] [Google Scholar]
- 185.Fang D, Yang H, Lin J, Teng Y, Jiang Y, Chen J, Li Y. 17beta-estradiol regulates cell proliferation, colony formation, migration, invasion and promotes apoptosis by upregulating miR-9 and thus degrades MALAT-1 in osteosarcoma cell MG-63 in an estrogen receptor-independent manner. Biochem Biophys Res Commun. 2015;457(4):500–506. doi: 10.1016/j.bbrc.2014.12.114. [DOI] [PubMed] [Google Scholar]
- 186.Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, Sui H, Tang Y, Wang Y, Liu N, Ren J, Hou F, Li Q. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/ PTBP2 complex. Br J Cancer. 2014;111(4):736–748. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ruan W, Wang P, Feng S, Xue Y, Li Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes cell proliferation and migration by upregulating angiomotin gene expression in human osteosarcoma cells. Tumour Biol. 2016;37(3):4065–4073. doi: 10.1007/s13277-015-4256-7. [DOI] [PubMed] [Google Scholar]
- 188.Li JP, Liu LH, Li J, Chen Y, Jiang XW, Ouyang YR, Liu YQ, Zhong H, Li H, Xiao T. Microarray expression profile of long noncoding RNAs in human osteosarcoma. Biochem. Biophys Res Commun. 2013;433(2):200–206. doi: 10.1016/j.bbrc.2013.02.083. [DOI] [PubMed] [Google Scholar]
- 189.Sun XH, Yang LB, Geng XL, Wang R, Zhang ZC. Increased expression of lncRNA HULC indicates a poor prognosis and promotes cell metastasis in osteosarcoma. Int J Clin Exp Pathol. 2015;8(3):2994–3000. [PMC free article] [PubMed] [Google Scholar]
- 190.Pasic I, Shlien A, Durbin AD, Stavropoulos DJ, Baskin B, Ray PN, Novokmet A, Malkin D. Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res. 2010;70(1):160–171. doi: 10.1158/0008-5472.CAN-09-1902. [DOI] [PubMed] [Google Scholar]
- 191.Cong M, Li J, Jing R, Li Z. Long non-coding RNA tumor suppressor candidate 7 functions as a tumor suppressor and inhibits proliferation in osteosarcoma. Tumour Biol. 2016 doi: 10.1007/s13277-015-4414-y. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 192.Wang Y, Yao J, Meng H, Yu Z, Wang Z, Yuan X, Chen H, Wang A. A novel long non-coding RNA, hypoxia-inducible factor-2alpha promoter upstream transcript, functions as an inhibitor of osteosarcoma stem cells in vitro. Mol Med Rep. 2015;11(4):2534–2540. doi: 10.3892/mmr.2014.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Min L, Hong S, Duan H, Zhou Y, Zhang W, Luo Y, Shi R, Tu C. Antidifferentiation noncoding RNA regulates the proliferation of osteosarcoma cells. Cancer Biother Radiopharm. 2016;31(2):52–57. doi: 10.1089/cbr.2015.1888. [DOI] [PubMed] [Google Scholar]
- 194.Li F, Cao L, Hang D, Wang F, Wang Q. Long non-coding RNA HOTTIP is up-regulated and associated with poor prognosis in patients with osteosarcoma. Int J Clin Exp Pathol. 2015;8(9):11414–11420. [PMC free article] [PubMed] [Google Scholar]
- 195.Wang B, Su Y, Yang Q, Lv D, Zhang W, Tang K, Wang H, Zhang R, Liu Y. Overexpression of long non-coding RNA HOTAIR promotes tumor growth and metastasis in human osteosarcoma. Mol Cells. 2015;38(5):432–440. doi: 10.14348/molcells.2015.2327. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 196.Qian M, Yang X, Li Z, Jiang C, Song D, Yan W, Liu T, Wu Z, Kong J, Wei H, Xiao J. P50-associated COX-2 extragenic RNA (PACER) overexpression promotes proliferation and metastasis of osteosarcoma cells by activating COX-2 gene. Tumour Biol. 2016;37(3):3879–3886. doi: 10.1007/s13277-015-3838-8. [DOI] [PubMed] [Google Scholar]
- 197.Ulaner GA, Vu TH, Li T, Hu JF, Yao XM, Yang Y, Gorlick R, Meyers P, Healey J, Ladanyi M, Hoffman AR. Loss of imprinting of IGF2 and H19 in osteosarcoma is accompanied by reciprocal methylation changes of a CTCF-binding site. Hum Mol Genet. 2003;12(5):535–549. doi: 10.1093/hmg/ddg034. [DOI] [PubMed] [Google Scholar]
- 198.Tian ZZ, Guo XJ, Zhao YM, Fang Y. Decreased expression of long non-coding RNA MEG3 acts as a potential predictor biomarker in progression and poor prognosis of osteosarcoma. Int J Clin Exp Pathol. 2015;8(11):15138–15142. [PMC free article] [PubMed] [Google Scholar]
- 199.Ma B, Li M, Zhang L, Huang M, Lei JB, Fu GH, Liu CX, Lai QW, Chen QQ, Wang YL. Upregulation of long non-coding RNA TUG1 correlates with poor prognosis and disease status in osteosarcoma. Tumour Biol. 2016;37(4):4445–4455. doi: 10.1007/s13277-015-4301-6. [DOI] [PubMed] [Google Scholar]
- 200.Zhang CL, Zhu KP, Shen GQ, Zhu ZS. A long non-coding RNA contributes to doxorubicin resistance of osteosarcoma. Tumour Biol. 2016;37(2):3737–3748. doi: 10.1007/s13277-015-4130-7. [DOI] [PubMed] [Google Scholar]
- 201.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long non-coding RNAs. Genome Res. 2007;17(5):556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Smith MA, Gesell T, Stadler PF, Mattick JS. Widespread purifying selection on RNA structure in mammals. Nucleic Acids Res. 2013;41(17):8220–8236. doi: 10.1093/nar/gkt596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Washietl S, Hofacker IL, Lukasser M, Huttenhofer A, Stadler PF. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat. Biotechnol. 2005;23(11):1383–1390. doi: 10.1038/nbt1144. [DOI] [PubMed] [Google Scholar]
- 204.Nielsen MM, Tehler D, Vang S, Sudzina F, Hedegaard J, Nordentoft I, Orntoft TF, Lund AH, Pedersen JS. Identification of expressed and conserved human non-coding RNAs. Rna. 2014;20(2):236–251. doi: 10.1261/rna.038927.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Torarinsson E, Yao Z, Wiklund ED, Bramsen JB, Hansen C, Kjems J, Tommerup N, Ruzzo WL, Gorodkin J. Comparative genomics beyond sequence-based alignments: RNA structures in the ENCODE regions. Genome Res. 2008;18(2):242–251. doi: 10.1101/gr.6887408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tarazona S, Garcia-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21(12):2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Olivarius S, Plessy C, Carninci P. High-throughput verification of transcriptional starting sites by Deep-RACE. BioTechniques. 2009;46(2):130–132. doi: 10.2144/000113066. [DOI] [PubMed] [Google Scholar]
- 211.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5(10):877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, Rinn JL, Raj A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015;16:20. doi: 10.1186/s13059-015-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.van Heesch S, van Iterson M, Jacobi J, Boymans S, Essers PB, de Bruijn E, Hao W, MacInnes AW, Cuppen E, Simonis M. Extensive localization of long noncoding RNAs to the cytosol and mono- and poly-ribosomal complexes. Genome Biol. 2014;15(1):R6. doi: 10.1186/gb-2014-15-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Werner MS, Ruthenburg AJ. Nuclear fractionation reveals thousands of chromatin-tethered noncoding RNAs adjacent to active genes. Cell Rep. 2015;12(7):1089–1098. doi: 10.1016/j.celrep.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.McHugh CA, Russell P, Guttman M. Methods for comprehensive experimental identification of RNA-protein interactions. Genome Biol. 2014;15(1):203. doi: 10.1186/gb4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159(1):188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP) J Visualized Exp. 2012;(61):e3912. doi: 10.3791/3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci USA. 2011;108(51):20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, Sweredoski MJ, Shishkin AA, Su J, Lander ES, Hess S, Plath K, Guttman M. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Lee HY, Haurwitz RE, Apffel A, Zhou K, Smart B, Wenger CD, Laderman S, Bruhn L, Doudna JA. RNA-protein analysis using a conditional CRISPR nuclease. Proc Natl Acad Sci USA. 2013;110(14):5416–5421. doi: 10.1073/pnas.1302807110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Shechner DM, Hacisuleyman E, Younger ST, Rinn JL. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods. 2015;12(7):664–670. doi: 10.1038/nmeth.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Pedersen JS, Bejerano G, Siepel A, Rosenbloom K, Lindblad-Toh K, Lander ES, Kent J, Miller W, Haussler D. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput Biol. 2006;2(4):e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Fang R, Moss WN, Rutenberg-Schoenberg M, Simon MD. Probing xist RNA structure in cells using targeted structure-seq. PLoS Genet. 2015;11(12):e1005668. doi: 10.1371/journal.pgen.1005668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Loughrey D, Watters KE, Settle AH, Lucks JB. SHAPE-Seq 2.0: systematic optimization and extension of high-throughput chemical probing of RNA secondary structure with next generation sequencing. Nucleic Acids Res. 2014;42(21):e165. doi: 10.1093/nar/gku909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods. 2010;7(12):995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H, Zhao G, Luo H, Bu D, Zhao H, Skogerbo G, Wu Z, Zhao Y. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011;39(9):3864–3878. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016;44(2):863–877. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Goff LA, Rinn JL. Linking RNA biology to lncRNAs. Genome Res. 2015;25(10):1456–1465. doi: 10.1101/gr.191122.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grutzner F, Kaessmann H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505(7485):635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 231.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, Rinn JL, Lander ES, Regev A. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28(5):503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Li L, Chang HY. Physiological roles of long noncoding RNAs: insight from knockout mice. Trends Cell Biol. 2014;24(10):594–602. doi: 10.1016/j.tcb.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379(6561):131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 235.Fitzpatrick GV, Soloway PD, Higgins MJ. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet. 2002;32(3):426–431. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- 236.Rapicavoli NA, Poth EM, Blackshaw S. The long non-coding RNA RNCR2 directs mouse retinal cell specification. BMC Dev Biol. 2010;10:49. doi: 10.1186/1471-213X-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Ho TT, Zhou N, Huang J, Koirala P, Xu M, Fung R, Wu F, Mo YY. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res. 2015;43(3):e17. doi: 10.1093/nar/gku1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22(5):885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Silva A, Bullock M, Calin G. The clinical relevance of long non-coding RNAs in cancer. Cancers (Basel) 2015;7(4):2169–2182. doi: 10.3390/cancers7040884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Mouraviev V, Lee B, Patel V, Albala D, Johansen TE, Partin A, Ross A, Perera RJ. Clinical prospects of long noncoding RNAs as novel biomarkers and therapeutic targets in prostate cancer. Prostate Cancer Prostatic Dis. 2016;19(1):14–20. doi: 10.1038/pcan.2015.48. [DOI] [PubMed] [Google Scholar]
- 241.Salman JW, Schoots IG, Carlsson SV, Jenster G, Roobol MJ. Prostate specific antigen as a tumor marker in prostate cancer: biochemical and clinical aspects. Adv Exp Med Biol. 2015;867:93–114. doi: 10.1007/978-94-017-7215-0_7. [DOI] [PubMed] [Google Scholar]
- 242.Crea F, Watahiki A, Quagliata L, Xue H, Pikor L, Parolia A, Wang Y, Lin D, Lam WL, Farrar WL, Isogai T, Morant R, Castori-Eppenberger S, Chi KN, Wang Y, Helgason CD. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 2014;5(3):764–774. doi: 10.18632/oncotarget.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, Lemesle G, de Groote P, Pinet F, Thum T. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 2014;114(10):1569–1575. doi: 10.1161/CIRCRESAHA.114.303915. [DOI] [PubMed] [Google Scholar]
- 244.Mohankumar S, Patel T. Extracellular vesicle long non-coding RNA as potential biomarkers of liver cancer. Brief Funct Genomics. 2016;15(3):249–256. doi: 10.1093/bfgp/elv058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Takahashi K, Yan IK, Kogure T, Haga H, Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4:458–467. doi: 10.1016/j.fob.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Gezer U, Ozgur E, Cetinkaya M, Isin M, Dalay N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int. 2014;38(9):1076–1079. doi: 10.1002/cbin.10301. [DOI] [PubMed] [Google Scholar]
- 247.Edelstein ML, Abedi MR, Wixon J. Gene therapy clinical trials worldwide to 2007-an update. J Gene Med. 2007;9(10):833–842. doi: 10.1002/jgm.1100. [DOI] [PubMed] [Google Scholar]
- 248.Corsi K, Chellat F, Yahia L, Fernandes JC. Mesenchymal stem cells, MG63 and HEK293 transfection using chitosan-DNA nanoparticles. Biomaterials. 2003;24(7):1255–1264. doi: 10.1016/s0142-9612(02)00507-0. [DOI] [PubMed] [Google Scholar]
- 249.Regnstrom K, Ragnarsson EG, Fryknas M, Koping-Hoggard M, Artursson P. Gene expression profiles in mouse lung tissue after administration of two cationic polymers used for nonviral gene delivery. Pharm Res. 2006;23(3):475–482. doi: 10.1007/s11095-006-9563-7. [DOI] [PubMed] [Google Scholar]
- 250.Raftery RM, Tierney EG, Curtin CM, Cryan SA, O’Brien FJ. Development of a gene-activated scaffold platform for tissue engineering applications using chitosan-pDNA nanoparticles on collagen-based scaffolds. J Control Release. 2015;210:84–94. doi: 10.1016/j.jconrel.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 251.Kurreck J, Wyszko E, Gillen C, Erdmann VA. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30(9):1911–1918. doi: 10.1093/nar/30.9.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Subramanian RR, Wysk MA, Ogilvie KM, Bhat A, Kuang B, Rockel TD, Weber M, Uhlmann E, Krieg AM. Enhancing antisense efficacy with multimers and multi-targeting oligonucleotides (MTOs) using cleavable linkers. Nucleic Acids Res. 2015;43(19):9123–9132. doi: 10.1093/nar/gkv992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Thakore PI, D’Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, Gersbach CA. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015;12(12):1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23(10):1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, Gersbach CA. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10(10):973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10(10):977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33(5):510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517(7536):583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Pearson MJ, Philp AM, Heward JA, Roux BT, Walsh DA, Davis ET, Lindsay MA, Jones SW. Long intergenic non-coding RNAs mediate the human chondrocyte inflammatory response and are differentially expressed in osteoarthritis cartilage. Arthritis Rheumatol. 2016;68(4):845–856. doi: 10.1002/art.39520. [DOI] [PMC free article] [PubMed] [Google Scholar]