Abstract
Background
We performed non-targeted metabolomics analysis using liquid chromatography-mass spectrometry coupled technique to explore the biological mechanism of coronary artery disease (CAD) events for improved prediction.
Material/Methods
We studied the association of CAD events in 4092 individuals and observed the replication of sphingomyelin (28:1), lysophosphatidylcholine (18:2), lysophosphatidylcholine (18:1), and monoglyceride (18:2), which were independent of main CAD risk factors.
Results
We found that these 4 metabolites were responsible for traditional risk factors and also contributed to the modifications related to reclassification and discrimination. Monoglycerides (MonoGs) were positively associated with C-reactive proteins and body mass index, while lysophosphatidylcholines (LPPCs), which had less evidence of subclinical CAD in an additional 1010 participants, yielded a reverse pattern. An association between monoGs and CAD independence of triglycerides (triGs) were also observed. On the basis of Mendelian randomization analysis, we observed a positive but weak irregular effect (odds ratio per unit increase in standard deviation in monoG=1.11, P-value=0.05) on CAD.
Conclusions
Our work establishes the relationship of metabolome with coronary artery disease and explains the biological mechanism of CAD events, as we identified the above-mentioned metabolites along with the evidence supporting their clinical use.
MeSH Keywords: Biological Markers, Coronary Artery Disease, Gas Chromatography-Mass Spectrometry
Background
Metabolomics, an emerging technology, investigates the complex cellular and physiological processes through metabolic intermediates [1,2], and thus offer a broad assessment of human biochemical activities [3] through detection and quantification of low-weight molecules. We can see the importance of the metabolomics approach in the field of identification of new biomarkers, which could be applied further for early detection and prevention of coronary artery disease (CAD) and in the investigation of its biological mechanism. The metabolic pathways which play an irreplaceable role in the growth of atherosclerosis have been highlighted through the metabolomics approach, which was used before to investigate the associations of metabolites and CAD event risks [4,5]. Moreover, multivariate models calculating risk of CAD based on combinations of biomarkers have provided reasonable estimates and offer promising options for future risk estimation if based on large populations [6]. Recently, large-scale studies have been utilized to estimate the risk of CAD using several combinations of biomarkers [7–9].
On the basis of the above facts, we organized this work of non-targeted metabolomics profiling in 4092 individual participants not suffering from CAD at baseline from 3 cohort studies in order to identify novel CAD biomarkers and to depict the underlying biological mechanisms. Moreover, we also evaluated its clinical scope along with its potential irregular effects for strongly associated metabolites. We also investigated the associations with inflammation, integrated metabolomics along with their genetics data, subclinical CAD, and oxidative stress to perform the study.
Material and Methods
Ethics statement
The Ethics Review Board of Linyi People’s Hospital (Shandong, China) has approved this study (Approval number: 42082119861228). Linyi People’s Hospital has collected written consent from all the patients.
Study samples
Metabolomic profiling was performed for blood samples collected from 3 categories: Category 1 (Cat1M); Category 2 (Cat2M&W), and Category 3 (Cat3Twin). Cat1M: In this longitudinal cohort study, men born in Shandong, China were invited to participate at age 60 (N= 3127) and finally, plasma samples from 1272 individual participants (at 70 years of age) were included. Cat2M&W: In this study, both men and women (age 70 years) were chosen from a specific community, living in Shandong, China and plasma samples from 1010 individual fasting participants (at 70 years of age; 50% women) were included. Cat3Twin: In this longitudinal cohort study, twins born in Shandong, China were invited to participate (N=5534) and plasma samples from 1810 individuals were included. For metabolomics profiling, blood samples were frozen immediately after separation of plasma and stored (−80°C) in all 3 studies. Information related to anthropometric measurements, blood pressure, 24-h ambulatory blood pressure, and glucose tolerance test were collected for each subject. We chose healthy patients for all 3 categories on the basis of these medical lab results. We excluded non-fasting individuals and participants with previous CAD events in all the studies. We chose diagnosis of acute myocardial infarction or unstable angina stage of hospitalization or death as the parameter to define CAD cases.
Metabolomics profiling (UPLC-MS)
Metabolomics studies were performed as serum samples were thawed and 100 μL of serum was transferred to 400 μL methanol in 96-well plates to precipitate proteins, stored at −20°C overnight, and then subjected to centrifugation for 35 min at 4000 rpm (at 4°C) to pellet precipitated protein. The supernatant was aliquoted to 3 separate 96-well plates, sealed using a heat-seal foil, and stored at −20°C during the whole analysis. Duplicate injections were performed for all samples, with the second set of injections performed upon completion of the first set of injections for all samples. Then, 1 μL injections of protein-precipitated serum were subjected to liquid chromatography (Acquity UPLC) coupled to a mass spectrometer (Xevo G2 Q-TOFMS, Waters Corporation, Milford, USA). We used ESI (electrospray ionization) in positive ion mode (scanning range: 50–1200 and rate: 5 scans/s) onto a C8 analytical column (1.8 μM, 1.0×100 mm; Acquity UPLC). A gradient elution was used as: solvent A (0.1% formic acid, 5% methanol and 95% water) to solvent B (0.1% formic acid, 5% water and 95% methanol). The mobile phase A (90%) was used to make injections, which was held for 0.2 min, ramped to B (50%) in 0.8 min, to 80% B over 3 min, and to 90% B over 10 min.
We kept 100% B for 8 min as starting conditions over 0.2 min and then allowed it to re-equilibrate for 7.8 min. A constant flow rate at 150 μL/min was used. The temperature of 50°C was used to saturate the column and 10°C was used for the samples. A full scan mode was used for mass analysis, keeping m/z range as 50–1200. We used Waters DataBridge software for processing and converted the raw data files to cdf format.
Metabolomics analysis
We used XCMS software [10] to process the raw data. The first step of the metabolomics workflow, including alignment, normalization, grouping, detection, and imputation, were performed separately for each study by using XCMS software. We detected 10742, 7668, and 9880 metabolic features in Cat1M, Cat2M&W, and Cat3Twin, respectively. As usual, the mass-to-charge ratios (m/z) of ions and their respective retention times were used to characterize each feature observed and we observed that more than 1 feature have represented a single metabolite. Common features between the 3 case studies were identified by matching of mass-to-charge ratio and retention time, followed by manual inspection of the spectra. As per a previous report [11] we selected the parameters used in XCMS to detect, align, and group peaks. To determine if the selected configuration was appropriate we performed manual inspection of the plots generated by the peak detection and grouping algorithms. The log-transformation and normalization were done for metabolic features intensities to take into account the factors of unwanted variation. As per a previous report [12], an ANOVA-type normalization approach was used, which has shown an increase in correlation between duplicates compared to other normalization methods in our data. We excluded those features with low Spearman correlation between duplicates and also manually excluded the samples with abnormal total feature intensity. The average correlation between duplicated features was 0.49 in Cat1M, 0.46 in Cat2M&W, and 0.41 in Cat3Twin.
For all the significant features, indiscriminant-mass spectrometry and indiscriminant-tandem mass spectrometric spectra were generated [13]. We knew that the spectra having high similarity, strong correlation, and similar retention time must belong to same metabolite, and thus used those spectra to decide that it belongs to a particular metabolite. We knew [14] that 4 annotation approaches, each with a different confidence level, were considered: MSI scale 1 (Metabolomics Standards Initiative: MSI; based on matching accurate mass, fragmentation pattern, and retention time with the in-house spectral library of authentic standards collected under the same experimental conditions); MSI scale 2 (based on spectrum and/or m/z similarities, but not retention time similarity); MSI scale 3 (based on a combination of spectral data, accurate mass, and retention time to assign the metabolite to a chemical class); and MSI scale 4 (annotated as “unknown” used when all the other approaches have failed in the annotation of the metabolite). Illumina Human Omni2.5M (≈2,500,000 SNPs) was used to genotype Cat1M participants and Illumina Human OmniExpress (≈700,000 SNPs) was used to genotype with Cat2M&W and Cat3Twin participants.
Statistical analysis
We used age-adjusted Cox proportional hazards model to test the association between each feature of Cat1M and CAD event for a 10-year follow-up because longer follow-up leads to a decrease in the association due to regression dilution bias. The Schoenfeld residual-based test was used to evaluate the P-value, which was further used to calculate the proportional hazard assumption. The results showed no significant deviation from the proportionality assumption. We have taken forward the CAD-associated features in Cat1M at 16% false discovery rate level to evaluate the replication in Cat3Twin. We utilized age- and sex-adjusted Cox models in Cat3Twin in order to explore main CAD risk factors associated with replicated features in the multivariable analysis. The main CAD risk factors in this study were sex, age, systolic blood pressure, body mass index, current smoking, antihypertensive treatment, low-density lipoprotein cholesterol, natural logarithm-transformed triGs, high-density lipoprotein cholesterol, and prevalent diabetes. In Cat2M&W association of metabolic features with markers of oxidative stress, inflammation and subclinical CAD was analyzed using age-, sex-, and aforementioned CAD risk factors-adjusted linear regression. As per a previous report [15] we calculated the individual 10-year risk of experiencing a CAD event to determine the Net Reclassification Index by selecting cut-offs of 10% and 20% thresholds. We estimate the false discovery rate with respect to validation as:
where is the false positive numbers and is declared a significant result in the validation study.
We used a t-mixture approach [16] to estimate these quantities. Moreover, we only used data from the discovery study (beta and standard errors) to determine these quantities and we assumed homogeneous discovery and validation samples. We observed the false discovery rate with respect to validation up to 0.21%.
Results
An overview of characteristics features of the 3 studies at baseline is shown in Table 1. At baseline, participants in Cat1M and Cat2M&W had the same approximate age (70.2 to 71.2 years), while participants in Cat3Twin had younger median age (65.1 years) and a wider range (59.4 to 69.8 years).
Table 1.
Baseline descriptive statistics for main CAD risk factors.
| Characteristics | Descriptive statistics | ||
|---|---|---|---|
| Cat1M (N=1272) | Cat2M&W (N=1010) | Cat3Twin (N=1810) | |
| Age in years, mean (SD) | 71.20 (0.58) | 70.20 (0.21) | 65.11 (6.62) |
| Female sex, % | 0 | 50 | 52 |
| Systolic blood pressure, mm Hg, mean (SD) | 149.11 (16.36) | 148.37 (21.09) | 137.55 (18.6) |
| Use of antihypertensive drugs, % | 35 | 33 | 16 |
| Prevalent type 2 diabetes mellitus, % | 18 | 11 | 16 |
| Glucose, mmol/l, mean (SD) | 5.51 (1.48) | 5.78 (1.59) | 5.52 (1.42) |
| Current smokers, % | 24 | 12 | 15 |
| Log-Triglycerides, mmol/l, mean (SD) | 0.21 (0.42) | 0.18 (0.33) | 0.16 (0.45) |
| BMI, kg/m2, mean (SD) | 26.39 (3.11) | 26.97 (4.98) | 26.43 (4.74) |
| Low-density lipoprotein-cholesterol, mmol/L, mean (SD) | 3.77 (0.69) | 3.31 (0.67) | 3.78 (0.87) |
| High-density lipoprotein-cholesterol, mmol/L, mean (SD) | 1.32 (0.43) | 1.56 (0.43) | 1.39 (0.57) |
| Log-C-reactive protein, mmol/L, mean (SD) | 0.71 (1.01) | 0.62 (0.91) | 0.59 (1.04) |
| N. events | 149 | – | 299 |
| Follow-up, y, median (max) | 10 (10) | – | 3.77 (6.54) |
Association of metabolic features with CAD: Discovery and validation
Out of 1272 Cat1M participants not suffering from CAD at baseline, a total 149 CAD events were detected (during follow-up of 10.0 years). Table 2 shows that at 16% false discovery rate 30 unique metabolites were associated with CAD events. On the other hand, 9, 11, 7, and 3 metabolites were annotated to MSI scale 1, MSI scale 2, MSI scale 3, and MSI scale 4 (as explained in “Material and Methods”), respectively. We observed 299 CAD events during the follow-up of 4.1 years when we replicated these 30 metabolites in the Cat3Twin study. Out of these 30, 25 metabolites were consistently detected in the Cat3Twin study (P-value <0.001). Table 3 shows that the other metabolites had a significant and consistent association (P-value < 0.05) [sphingomyelin (28:1; SM), lysophosphatidylcholine (18:2; LPPC), a cinnamic acid derivative, monoG (18:2; MonoG) and a monosaccharide]. A significant interaction of LPPC (18:2) and age (older than 70 years) was observed in individuals (Table 3, Figure 1A). Hence, this interaction was used further as a model to report the estimates for participants older than 70 years. We also found the association of 3 metabolites with main CAD risk factors and with meta-analysis-adjusted CAD with respect to Cat1M and Cat3Twin results (Table 1), as MonoG (18:2) (hazard ratio [HR] per unit increase in standard deviation=1.21; P-value=0.010) was positively associated while LPPC (18:2) (HR=0.87; P-value <0.001) and SM 28: 1 (HR=0.91; P-value=0.012) were negatively associated. Targeted MS/MS was explored further to confirm the chemical structures of these metabolites (spectra not shown).
Table 2.
Non-targeted LC/MS-based metabolomics for association with CAD in Cat1M and validation in Cat3Twin.
| Metabolite* | Metabolic class | ACL# | Cat1M& | Cat3Twin& | Meta-analysis@ | |||
|---|---|---|---|---|---|---|---|---|
| HR | P-value | HR | P-value | HR | P-value | |||
| SM (28:1) | Sphingolipids | 2 | 0.75 | 1.60E-03 | 0.88 | 3.70E-02 | 0.88 | 7.70E-03 |
| LPPC (18:2) | Glycerophospholipids | 1 | 0.88 | 3.50E-03 | 0.88 | 1.40E-02 | 0.86 | 1.80E-04 |
| – | Cinnamic acid derivatives | 3 | 0.82 | 1.20E-02 | 0.84 | 2.50E-02 | 0.88 | 8.20E-04 |
| LPPC (18:1) | Glycerophospholipids | 1 | 0.81 | 5.60E-05 | 0.83 | 5.40E-02 | 0.89 | 7.60E-04 |
| MG (18:2) | Lineolic acids and derivatives | 1 | 1.29 | 8.30E-03 | 1.35 | 2.20E-06 | 1.37 | 8.40E-08 |
| – | Monosaccharides | 3 | 1.22 | 2.10E-03 | 1.20 | 4.40E-04 | 1.22 | 2.80E-06 |
| Myristic acid | Fatty acids and conjugates | 1 | 0.72 | 4.80E-03 | 1.07 | 6.70E-01 | 0.93 | 4.40E-01 |
| PE (O–18:1/0:0) | Glycerophospholipids | 1 | 0.86 | 8.80E-03 | 0.92 | 6.10E-01 | 0.81 | 2.00E-01 |
| Hippuric acid | Amino acids and derivatives | 1 | 1.23 | 5.10E-03 | 0.99 | 4.70E-01 | 1.12 | 5.40E-01 |
| LPPC (20:0) | Glycerophospholipids | 1 | 0.74 | 5.30E-03 | 0.82 | 5.70E-02 | 0.87 | 5.40E-03 |
| PPC (34:1) | Glycerophospholipids | 1 | 0.89 | 1.40E-02 | 0.99 | 8.80E-02 | 0.82 | 4.50E-03 |
| PPC (O–36:5) | Glycerophospholipids | 1 | 0.87 | 5.90E-03 | 0.97 | 2.50E-01 | 0.81 | 5.30E-02 |
| SM (d38:2) | Sphingolipids | 2 | 0.80 | 4.10E-03 | 0.94 | 9.10E-01 | 0.92 | 3.10E-01 |
| SM (d32:1) | Sphingolipids | 2 | 0.88 | 1.20E-02 | 0.92 | 7.40E-01 | 0.98 | 2.30E-01 |
| SM (d18:2/14:0) | Sphingolipids | 2 | 0.81 | 5.80E-03 | 0.97 | 3.50E-01 | 0.83 | 8.40E-02 |
| SM (d18:2/15:0) | Sphingolipids | 2 | 0.72 | 2.20E-03 | 1.01 | 9.40E-01 | 0.83 | 3.30E-01 |
| LPPC (18:3) | Glycerophospholipids | 2 | 0.78 | 2.10E-05 | 0.94 | 1.00E-01 | 0.87 | 4.40E-02 |
| LPPC (20:2) | Glycerophospholipids | 2 | 0.83 | 6.00E-03 | 0.92 | 7.20E-01 | 0.80 | 2.30E-01 |
| LPPC (20:4) | Glycerophospholipids | 2 | 0.88 | 4.70E-03 | 1.03 | 7.60E-01 | 0.95 | 4.30E-01 |
| LPPC (20:5) | Glycerophospholipids | 2 | 0.79 | 2.20E-03 | 0.83 | 8.70E-02 | 0.87 | 1.50E-02 |
| LPPC (22:5) | Glycerophospholipids | 2 | 0.82 | 2.10E-03 | 0.84 | 8.10E-02 | 0.83 | 3.50E-03 |
| Eicosapentaenoic acid methyl ester | Fatty acid esters | 2 | 0.73 | 3.50E-03 | 0.97 | 2.60E-01 | 0.88 | 6.60E-02 |
| – | Fatty acids and conjugates | 3 | 0.74 | 3.40E-03 | 0.92 | 9.20E-01 | 0.83 | 3.30E-01 |
| PPC (31:1) or PE (34:1) | Glycerophospholipids | 3 | 0.79 | 1.00E-03 | 0.98 | 2.40E-01 | 0.85 | 1.10E-01 |
| PPC (32:2) or PE (35:2) | Glycerophospholipids | 3 | 0.87 | 1.10E-02 | 0.93 | 4.70E-01 | 0.82 | 1.40E-01 |
| PPC (36:1) or PE (39:1) | Glycerophospholipids | 3 | 0.72 | 1.70E-03 | 0.91 | 5.30E-01 | 0.83 | 2.10E-01 |
| PE (35:1) or PPC (32:1) | Glycerophospholipids | 3 | 0.81 | 3.10E-03 | 0.93 | 8.60E-01 | 0.87 | 2.70E-01 |
| – | Unknown class | 4 | 0.73 | 6.10E-03 | 1.05 | 9.10E-01 | 0.82 | 3.80E-01 |
| – | Unknown class | 4 | 0.72 | 1.80E-03 | 0.98 | 2.50E-01 | 0.81 | 7.80E-02 |
| – | Unknown class | 4 | 0.64 | 2.10E-05 | 0.95 | 1.50E-01 | 0.83 | 9.10E-02 |
For each metabolite, we reported the association of the precursor ion (M+H) or one of the main adducts (M+Na, M+H-H2O), if the precursor ion is not available;
Annotation Confidence Level;
Values are from Cox proportional hazards analyses per standard deviation increase of the metabolic feature adjusted by age and sex (only in Cat3Twin);
Values are from random effect meta-analysis. Metabolites in bold have false discovery rate < 0.15 in Cat1M and P-value < 0.05 in Cat3Twin.
Table 3.
Association between metabolites replicated in the univariable analysis and CHD in Cat1M and Cat3Twin, adjusted for established risk factors.
| Study | Strata | N. CAD Event/non-event | Metabolite* | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SM (28:1) | LPPC (18:2) | Cinnamic acid derivatives | MG (18:2) | Monosaccharides | ||||||||
| HR (95% CIs) | P-value | HR (95% CIs) | P-value | HR (95% CIs) | P-value | HR (95% CIs) | P-value | HR (95% CIs) | P-value | |||
| Cat1M | All | 132/988 | 0.74 (0.62–0.89) | 0.02 | 0.81 (0.75–0.96) | 0.04 | 0.88 (0.75–1.07) | 0.08 | 1.15 (0.90–1.30) | 0.29 | 1.16 (1.02–1.47) | 0.07 |
| Cat3Twin | All | 219/1479 | 0.8 (0.72–1.01) | 0.16 | – | – | 0.95 (0.84–1.08) | 0.28 | 1.21 (1.02–1.43) | 0.02 | 1.09 (0.95–1.22) | 0.43 |
| <65 yrs old | 60/462 | 0.95 (0.75–1.24) | 0.52 | 1.04 (0.82–1.33) | 0.71 | 0.95 (0.63–1.29) | 0.51 | 1.42 (1.07–1.90) | 0.02 | 1.19 (0.80–1.51) | 0.39 | |
| 65–70 yrs old | 56/479 | 0.88 (0.68–1.13) | 0.23 | 1.19 (0.88–1.60) | 0.22 | 0.73 (0.52–1.06) | 0.08 | 1.19 (0.88–1.58) | 0.36 | 1.09 (0.70–1.34) | 0.74 | |
| >70 yrs old | 147/598 | 0.97 (0.79–1.17) | 0.6 | 0.76 (0.63–0.93) | 0.009 | 1.04 (0.88–1.22) | 0.84 | 1.11 (0.99–1.47) | 0.15 | 1.03 (0.82–1.28) | 0.64 | |
| P-value for age* metabolic feature interaction | – | 0.51 | 0.02 | 0.28 | 0.36 | 0.41 | ||||||
Values are from Cox proportional hazards analyses per SD increment of the metabolic feature adjusted by age, sex (only in Cat3Twin), systolic blood pressure, body mass index, current smoking, antihypertensive treatment, low density lipoprotein cholesterol, high density lipoprotein cholesterol, log-triglycerides and diabetes at baseline. Associations in bold have P-value <0.05.
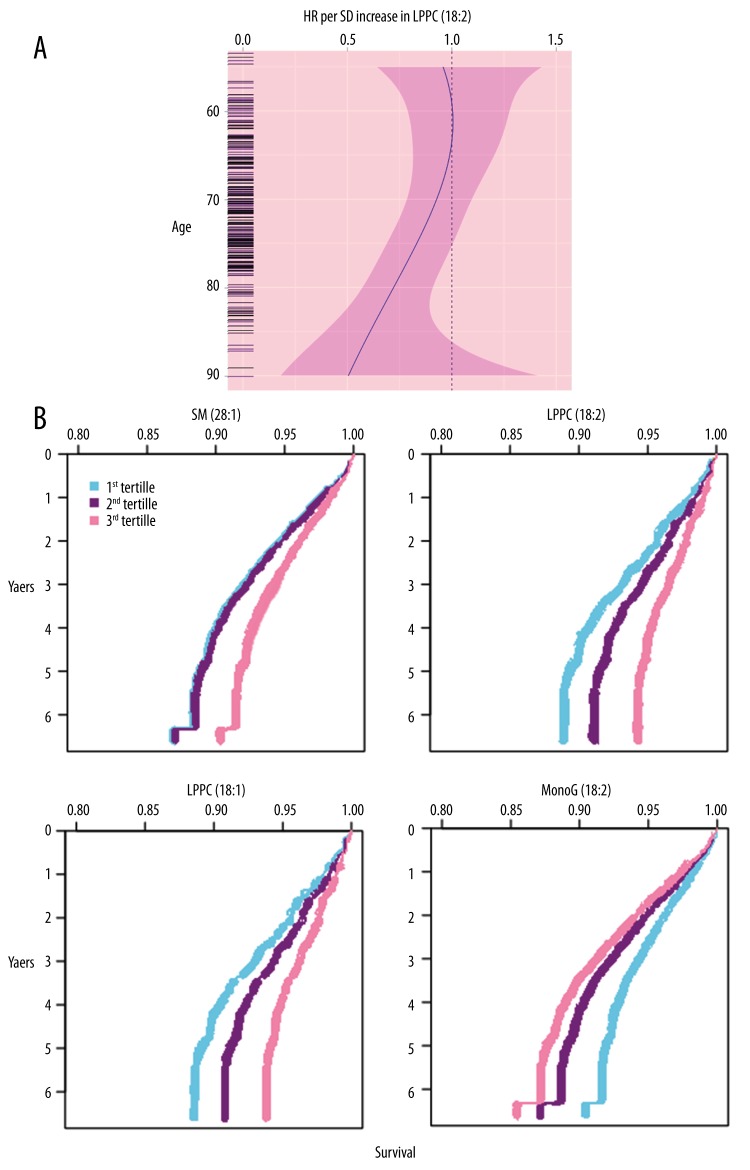
Figure 1.
(A) Splines modeled representation of association of LPPC (18:2) with CAD in terms of hazard ratio (HR) per unit increment in standard deviation with respect to function of age. (B) Representation of survival of a 75-year-old man who was a smoker but non-diabetic. His body mass index was 26 and he had 148-unit systolic blood pressure. Curves are tertiles of each of the 4 metabolites with respect to time vs. CAD.
Relation of lysophosphatidylcholines ratios with CAD and clinical utility of metabolites
We found the strongest associations between LPPC (18:2) metabolite and CAD in Cat1M and Cat3Twin studies and hence had a way to extend our work for another 4 LPPC species in order to explore the simplest patterns and simplest biological pathways. We knew that LPPCs are formed through the hydrolysis of phosphatidylcholines (PPC) [17]; hence, we detected the association of the most abundant LPPC/PPC ratios with CAD. After combining the adjustment for main CAD risk factors in Cat1M and Cat3Twin studies, we found that LPPC (18:1) is negatively associated with CAD (HR=0.80; P-value <0.001) (detailed data not shown). Thus, we found an increase in the number of CAD-associated (and independent increment in main CAD risk factors) metabolites up to 4. Figure 1B shows survival curves for each metabolite. We also found no significant association of the ratios between LPPCs and PPC with CAD (detailed data not shown). We found a negative correlation (r2=−0.18, P-value <0.001) between LPPC (18:1) and monoG (18:2), while a positive correlation (r2=0.79, P-value <0.001) was found between LPPC 18:1 and LPPC 18:2. In similar fashion, we found a negative correlation (r2=−0.15, P-value <0.001) between SM (28:1) and monoG (18:2), while a positive correlation (r2=0.46, P-value <0.001; r2=0.41, P-value <0.001, respectively) was found between SM (28:1) and LPPC (18:2) and LPPC (18:1).
After determining the association between 4 metabolites [SM (28:1), LysoPC (18:2), LysoPC (18:1) and monoG (18:2)] with main CAD risk factors-adjusted CAD, their utility as biomarkers for CAD prediction was assessed. For this purpose, we added these metabolites to the Framingham Heart Study risk score model [18] and achieved a slight enhancement in the Net Reclassification Index (10.1% [1.6; 21.4] for events and −0.9% [−8.0; 0.7] for non-events) and in C-index (0.761 vs. 0.757, P-value=0.028) (detailed data not shown).
Exploration of biological mechanisms: Association with subclinical CAD, with main CAD risk factors, with inflammation, and with oxidative stress-markers
Association of our 4 metabolites with main CAD risk factors (Table 4), with inflammation, and with oxidative stress-markers was explored and yielded similar patterns for 2 LPPC species as we found in the case of association of higher LPPC with 3 main CAD risk factors: lower body mass index, higher high-density lipoprotein cholesterol levels, and higher low-density lipoprotein cholesterol levels. We found a similar type of associations for SM (28:1). In all the 3 studies, we found that monoG (18:2) was positively associated with triGs and body mass index levels, but with high-density lipoprotein cholesterol it was reversely associated. Moreover, a strong correlation (r2 range: 0.29–0.61) was observed between monoG (18:2) and triGs. In Cat3Twin, triGs and monoG (18:2) was positively associated with CAD when we included them in same model adjusted for age and sex. Further, monoG (18:2) showed a better increase in the C-statistic (0.758 vs. 0.756) and likelihood ratio (206.2 vs. 198.4) compared with those models in which triGs were separately added along with all main CAD risk factors except triGs (detailed data not shown).
Table 4.
Association of four metabolites with main CAD risk factors in three case studies.
| Main CAD risk factors | Case study | Metabolite [HR (95% CIs)] | |||
|---|---|---|---|---|---|
| SM (28:1) | LPCC (18:2) | LPCC (18:1) | MonoG (18:1) | ||
| Systolic blood pressure | Cat1M | −0.06 (−0.09, −0.02) | −0.09 (−0.13, −0.05) | −0.08 (−0.13, −0.04) | 0.07 (0.02, 0.12) |
| Cat2M&W | −0.08 (−0.14, −0.02) | −0.02 (−0.06, 0.03) | 0.03 (−0.01, 0.07) | 0.02 (−0.03, 0.07) | |
| Cat3Twin | −0.05 (−0.10, 0.00) | 0.01 (−0.07, 0.06) | 0.04 (0.00, 0.07) | 0.04 (−0.01, 0.09) | |
| Body mass index | Cat1M | −0.10 (−0.14, −0.05) | −0.28 (−0.33, −0.22) | −0.28 (−0.35, −0.20) | 0.20 (0.16, 0.24) |
| Cat2M&W | −0.09 (−0.13, −0.06) | −0.14 (−0.19, −0.09) | −0.15 (−0.19, −0.09) | 0.15 (0.11, 0.19) | |
| Cat3Twin | −0.07 (−0.10, −0.05) | −0.26 (−0.33, −0.20) | −0.18 (−0.21, −0.15) | 0.18 (0.12, 0.22) | |
| Low density lipoprotein cholestrol | Cat1M | 0.38 (0.35, 0.41) | 0.17 (0.12, 0.22) | 0.17 (0.13, 0.22) | −0.08 (−0.13, −0.04) |
| Cat2M&W | 0.18 (0.14, 0.21) | 0.06 (0.04, 0.08) | 0.10 (0.08, 0.12) | −0.05 (−0.09, −0.01) | |
| Cat3Twin | 0.36 (0.32, 0.40) | 0.28 (0.25, 0.31) | 0.19 (0.16, 0.24) | −0.03 (−0.06, 0.03) | |
| Log TriGs | Cat1M | −0.18 (−0.21, −0.15) | −0.19 (−0.21, −0.14) | −0.06 (−0.10, −0.01) | 0.52 (0.48, 0.56) |
| Cat2M&W | −0.14 (−0.19, −0.09) | −0.06 (−0.11, −0.01) | 0.03 (0.00, 0.05) | 0.36 (0.34, 0.40) | |
| Cat3Twin | −0.13 (−0.18, −0.09) | −0.12 (−0.16, −0.09) | 0.06 (0.04, 0.09) | 0.54 (0.51, 0.58) | |
| High density lipoprotein cholestrol | Cat1M | 0.18 (0.14, 0.23) | 0.22 (0.18, 0.26) | 0.32 (0.29, 0.32) | −0.42 (−0.48, −0.36) |
| Cat2M&W | 0.15 (0.11, 0.21) | 0.16 (0.12, 0.20) | 0.22 (0.19, 0.24) | −0.32 (−0.38, −0.27) | |
| Cat3Twin | 0.17 (0.11, 0.22) | 0.28 (0.26, 0.31) | 0.31 (0.28, 0.36) | −0.40 (−0.44, −0.35) | |
In Cat2M&W, an association with less subclinical CVD and lower levels of inflammation markers was observed with the 2 LPPC species. We found that LPPC (18:1) was inversely associated with left ventricular mass index (P-value=2.6×10−7) and C-reactive protein (P-value=3.4×10−11). We found a similar association with plasminogen activator inhibitor-1 (P-value=2.0×10−12) and fibrinogen (P-value=7.8×10−9). On the other hand, we observed that monoG (18:2) was positively associated with subclinical CVD and oxidative stress-markers as: conjugated dienes (P-value=1.6×10−8), plasminogen activator inhibitor-1 (P-value=1.9×10−15), fibrogen (P-value=6.6×10−8), and tissue plasminogen activator (P-value=2.7×10−9). Interestingly, LPPC (18:1) showed a significant association with lower scales of fibrinogen and C-reactive protein and higher levels of monocyte chemotactic protein-1 (detailed data not shown).
Association between single-nucleotide polymorphisms and 4 metabolites
In all 4092 participants from Cat1M, Cat2M&W, and Cat3Twin studies, association of single-nucleotide polymorphisms (SNPs) with SM (28:1), LPPC (18:1), LPPC (18:2), and monoG (18:2) was tested with both metabolomics and genetic data. We found an associated signal near A4GALT on chromosome 22 (rs8141918; P-value=4.9×10−7) and a novel associated signal near C8orf87 on chromosome 8 (rs75729820; P-value=2.9×10−8), in analyses of LPPC (18:1) (detailed data not shown). For monoG (18:2), we detected a suggestive association with rs964184 (in the ZNF259/APOA5 region; P-value=1.4×10−7; detailed data not shown).
Association of metabolites with CAD-associated variants along with their biologically relevant pathways
We tested the association of 44 established CAD-associated SNPs [19] and biologically relevant pathways targeted 7 SNPs with SM (28:1), LPPC (18:1), LPPC (18:2), and monoG (18:2). We detected a significant association of CHD-associated SNPs with monoG (18:2) (P-values <0.05) and it remained intact (P-value=0.04) even after main CAD risk factors adjustment (detailed data not shown). We observed a significant enrichment of low p-values with respect to other metabolites. We also detected the association of LIPC with all 4 metabolites with respect to candidate SNPs-targeted relevant pathways; hence, we confirmed the role of hepatic lipase in regulation of LPPCs and monoG scales.
Mendelian randomization analysis
In this analysis, we detected a positive but weak irregular effect (P-value=0.05) of monoG (18:2) with respect to CAD risk: odds ratio (1.06; 95% CI, 1.00–1.10) per unit increase in standard deviation. We also found the absence of irregular effect for SM (28:1), LPPC (18:2), and LPPC (18:1).
Discussion
The association between circulating metabolites and CAD was investigated by using liquid chromatography-mass spectrometry. There were a total of 4092 individuals from 3 studies, showing the association between 30 metabolites and CAD, 86% of which showed a directionally consistent association with CAD. However, we observed an association of 3 metabolites with main CAD risk factors-adjusted CAD multivariable analyses. Moreover, we found an additional significant association during targeted LPPC analysis, which showed an association between all 4 metabolites [SM (28:1); LPPC (18:2), LPPC (18:1) and monoG (18:2)] and main CAD risk factors-independent CAD. Interestingly, when we considered commonly used risk categories, we observed moderate improvement in risk reclassification by these biomarkers, even beyond traditional risk factors. Additionally, monoG (18:2) was associated positively with subclinical CAD, inflammation markers, and BMI, while LPPCs showed the reverse pattern. An irregular effect of monoG (18:2) on triGs levels-independent CAD was also detected. Several genome-wide significant SNPs were discovered along with discovery of their association with LPPC, which to the best of our knowledge, have not been previously reported.
MonoG (18:2) was strongly associated with CAD. We know that due to the effect of lipoprotein lipase and hepatic lipase and monoGs for tissue utilization by catalyzing the hydrolysis of triGs, the majority of circulating monoGs are released [20]. Due to the course of monoG lipase, monoGs further yield free fatty acids along with glycerol. MonoGs are used to resynthesize diGs and triGs within the intestinal wall through a monoacylglycerol pathway followed by lymph transportation. On the basis of our observations, we suggest that monoG (18:2) was involved in the pathogenesis of CAD. Recently, it is observed that in the triGs pathway and the irregular effect of plasma triG levels on CAD risk, monoG (18:2) plays the central role [21]. MonoG (18:2) and triGs were significantly associated with coronary heart disease when included in the same model, despite their high correlation. MonoG (18:2) was a better metabolite to forecast coronary heart disease than triGs when there was addition of monoG (18:2) to a main cardiovascular risk factors-independent model. Additionally, there was an association between monoG (18:2) and higher levels of oxidative stress, subclinical CAD-markers, and CAD risk factors. Mendelian randomization analysis also indicated a positive but weak irregular effect of monoG (18:2) on CAD risk. Even after adjustment for main CAD risk factors, we found an association between several SNPs associated with CAD and monoG (18:2).
However, LysoPC (18:2), LysoPC (18:1) were strongly age-dependently associated with CAD risk, but for older individuals we detected a strong inverse association. We also found that these LPPC species were further associated with subclinical CAD-markers, lower BMI, and higher total cholesterol levels and high-density lipoprotein cholesterol. Their high correlation with each other suggests shared biological mechanisms. PPCs are responsible for the derivation of LPPC, while several mechanisms are responsible for their formation. Glycoprotein lecithin cholesterol acyltransferase is responsible for the derivation of a large section of LPPC from PPC. Rozenberg et al. reported that LPPC is responsible for the inhibition of macrophage biosynthesis, and during the oxidative modification of low-density lipoprotein cholesterol, higher levels of LPPCs have been observed to lead to their conversion to atherogenic particles [22]. A recent study [23] has suggested LPPCs are responsible for protection from cardiovascular risk, before which LPPCs were known only as pro-atherogenic and pro-inflammatory metabolites. Recently, it was observed that several LPPC species are inversely associated with incidence of coronary heart disease [24]. Our results are quite consistent and extend the knowledge yielded by previous studies. The detected associations between LPPCs and CAD are not irregular, but rather are real and reasonable.
Our work is the widest and most unique study of its own kind to investigate metabolomics with respect to coronary artery disease. We used metabolomics approach using liquid chromatography-mass spectrometry, which is considered an extremely sensitive method to detect metabolites in comparison to other NMR-based methods [25]. Our results have been validated on the basis of an independent population and age range. We used a different blood collection method and serum blood partition rather than plasma blood partition. Our approach can increase the generalization of previous studies in this regard, which is another advantage of this work. Extensive characterization in depicting biological mechanisms, along with irregular effects and clinical usage of the metabolites, support the value of our work. However, out study has a few shortcomings. Since our work is a non-targeted study, we had to detect every ion by MS as a separate variable, thus creating a multiple-testing burden. Even so, our method is advantageous as it is independent of pre-elucidation and allows incorporation and subsequent identification of unidentified metabolites using targeted methods. Additionally, the use of the LC-MS method, which is a single analytical platform, might increase the number of detectable metabolites via integrating multiple analytical platforms.
Conclusions
To the best of our knowledge, this is the first study of the relationship of the metabolome with coronary artery disease. We identified sphingomyelin (28:1), lysophosphatidylcholine (18:2), lysophosphatidylcholine (18:1), and monoglyceride (18:2) as risk factor biomarkers for coronary artery disease and found an irregular effect for monoG (18:2) on coronary artery disease. This work furthers our understanding of the underlying biological mechanisms and also opens doors for future experiments to explore the mechanisms involved in the pathogenesis of coronary artery disease.
Footnotes
Source of support: Departmental sources
References
- 1.Mercuro G, Bassareo PP, Deidda M, et al. Metabolomics: A new era in cardiology? J Cardiovasc Med (Hagerstown) 2011;12(11):800–5. doi: 10.2459/JCM.0b013e32834a658f. [DOI] [PubMed] [Google Scholar]
- 2.Suhre K, Shin SY, Petersen AK, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watkins SM, German JB. Toward the implementation of metabolomic assessments of human health and nutrition. Curr Opin Biotechnol. 2002;13(5):512–16. doi: 10.1016/s0958-1669(02)00363-4. [DOI] [PubMed] [Google Scholar]
- 4.Shah SH, Bain JR, Muehlbauer MJ, et al. Association of a peripheral blood metabolic profile with coronary artery disease and risk of subsequent cardiovascular events. Circ Cardiovasc Genet. 2010;3:207–14. doi: 10.1161/CIRCGENETICS.109.852814. [DOI] [PubMed] [Google Scholar]
- 5.Magnusson M, Lewis GD, Ericson U, et al. A diabetes-predictive amino acid score and future cardiovascular disease. Eur Heart J. 2013;34:1982–89. doi: 10.1093/eurheartj/ehs424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koenig W. Cardiovascular biomarkers: Added value with an integrated approach? Circulation. 2007;116(1):3–5. doi: 10.1161/CIRCULATIONAHA.107.707984. [DOI] [PubMed] [Google Scholar]
- 7.Wilson PW, D’Agostino RB, Levy D, et al. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97(18):1837–47. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 8.Assmann G, Cullen P, Schulte H. Simple scoring scheme for calculating the risk of acute coronary events based on the 10-yr follow-up of the prospective cardiovascular Munster (PROCAM) study. Circulation. 2002;105(3):310–15. doi: 10.1161/hc0302.102575. [DOI] [PubMed] [Google Scholar]
- 9.Conroy RM, Pyorala K, Fitzgerald AP, et al. Estimation of ten-yr risk of fatal cardiovascular disease in Europe: the SCORE project. Eur Heart J. 2003;24(11):987–1003. doi: 10.1016/s0195-668x(03)00114-3. [DOI] [PubMed] [Google Scholar]
- 10.Smith CA, Want EJ, O’Maille G, et al. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–87. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 11.Patti GJ, Tautenhahn R, Siuzdak G. Meta-analysis of untargeted metabolomic data from multiple profiling experiments. Nat Protoc. 2012;7:508–16. doi: 10.1038/nprot.2011.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jauhiainen A, Madhu B, Narita M, et al. Normalization of metabolomics data with applications to correlation maps. Bioinformatics. 2014;30(15):2155–61. doi: 10.1093/bioinformatics/btu175. [DOI] [PubMed] [Google Scholar]
- 13.Broeckling CD, Heuberger AL, Prenni JE. Large scale non-targeted metabolomic profiling of serum by ultra performance liquid chromatography-mass spectrometry (UPLC-MS) J Vis Exp. 2013;(73):e50242. doi: 10.3791/50242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sumner LW, Amberg A, Barrett D, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–21. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leening MJ, Vedder MM, Witteman JC, et al. Net reclassification improvement: computation, interpretation, and controversies: A literature review and clinician’s guide. Ann Intern Med. 2014;160:122–31. doi: 10.7326/M13-1522. [DOI] [PubMed] [Google Scholar]
- 16.Pawitan Y, Murthy KR, Michiels S, Ploner A. Bias in the estimation of false discovery rate in microarray studies. Bioinformatics. 2005;21:3865–72. doi: 10.1093/bioinformatics/bti626. [DOI] [PubMed] [Google Scholar]
- 17.Boyanovsky BB, Webb NR. Biology of secretory phospholipase A2. Cardiovasc Drugs Ther. 2009;23:61–72. doi: 10.1007/s10557-008-6134-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson PW, D’Agostino RB, Levy D, et al. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–47. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 19.CARDIoGRAMplusC4D Consortium. Deloukas P, Kanoni S, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller M, Stone NJ, Ballantyne C, et al. Triglycerides and cardiovascular disease: A scientific statement from the American Heart Association. Circulation. 2011;123:2292–333. doi: 10.1161/CIR.0b013e3182160726. [DOI] [PubMed] [Google Scholar]
- 21.Do R, Willer CJ, Schmidt EM, Sengupta S, et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet. 2013;45:1345–52. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rozenberg O, Shih DM, Aviram M. Human serum paraoxonase 1 decreases macrophage cholesterol biosynthesis: possible role for its phospholipase-A2-like activity and lysophosphatidylcholine formation. Arterioscler Thromb Vasc Biol. 2003;23:461–67. doi: 10.1161/01.ATV.0000060462.35946.B3. [DOI] [PubMed] [Google Scholar]
- 23.Schmitz G, Ruebsaamen K. Metabolism and atherogenic disease association of lysophosphatidylcholine. Atherosclerosis. 2010;208:10–18. doi: 10.1016/j.atherosclerosis.2009.05.029. [DOI] [PubMed] [Google Scholar]
- 24.Stegemann C, Pechlaner R, Willeit P, et al. Lipidomics profiling and risk of cardiovascular disease in the prospective population-based Bruneck study. Circulation. 2014;129(18):1821–31. doi: 10.1161/CIRCULATIONAHA.113.002500. [DOI] [PubMed] [Google Scholar]
- 25.Tzoulaki I, Ebbels TM, Valdes A, et al. Design and analysis of metabolomics studies in epidemiologic research: A primer on-omic technologies. Am J Epidemiol. 2014;180:129–39. doi: 10.1093/aje/kwu143. [DOI] [PubMed] [Google Scholar]