Abstract
For structured RNAs that possess catalytic activity, this activity provides a powerful probe for measuring the progress of folding and the effects of RNA chaperone proteins on the folding rate. The crux of this approach is that only the natively folded RNA is able to perform the catalytic reaction. This method can provide a quantitative measure of the fraction of native RNA over time, and it can readily distinguish the native state from all misfolded conformations. Here we describe an activity-based method measuring native folding of ribozymes derived from self-splicing group I introns, and we show how the assay can be used to monitor acceleration of native folding by DEAD-box RNA helicase proteins that function as general RNA chaperones. By measuring the amount of substrate that is converted to product in a rapid first turnover, we describe how to determine the fraction of the ribozyme population that is present in the native state. Further, we describe how to perform a two-stage or discontinuous assay in which folding proceeds in stage one and then solution conditions are changed in stage two to permit catalytic activity and block further folding. This protocol allows folding to be followed under a broad range of solution conditions, including those that do not support catalytic activity, and facilitates studies of chaperone proteins.
Keywords: Catalysis, Ribozyme, Folding, Helicase, Chaperone
1 Introduction
Although it was known as early as the 1960s that certain RNAs folded to specific structures, interest in RNA structure and folding began to grow dramatically in the 1980s when it was discovered that some RNAs possess catalytic activity. Since then, the field of RNA folding has continued to grow because of continued interest in these and other functional RNAs, as well as a general interest in the fundamental properties of biological macromolecules. Although the conceptual challenges of folding are shared between RNA and the other major folded macromolecule, protein, the basic physical properties of RNA lead to important characteristics of its folding processes. RNA secondary structures (helices) and tertiary contacts can form rapidly, and local structure can be independently stable. With only four nucleotides in RNA, incorrect secondary and/or tertiary structure forms frequently, and the independent stability allows incorrect contacts to persist for long periods [1, 2]. This property can be restated as a folding free energy profile that is rugged, with deep valleys corresponding to stable misfolded forms, also termed kinetic traps [3]. Understanding how RNA folds correctly and avoids kinetic traps is universally important because structured RNAs are required in all cells.
DEAD-box proteins are ubiquitous in RNA-mediated processes and have been shown to resolve kinetic traps and promote folding to the native state [4, 5]. The mitochondrial DEAD-box proteins CYT-19 and Mss116p, from Neurospora crassa and Saccharomyces cerevisiae, respectively, have been shown to function as general RNA chaperones in vivo [6, 7]. Further, both of these proteins have been shown to facilitate native folding of group I and group II introns and their ribozyme derivatives in vitro [6, 8–11], primarily by resolving misfolded conformations [12–14]. Crystal structures and extensive biochemical analysis have indicated that DEAD-box proteins use cycles of ATP binding and hydrolysis to unwind RNA helices non-processively [15–19], and this unwinding is thought to underlie the ability of these proteins to function as RNA chaperones [13, 20].
A key method to probe native RNA folding is the catalytic activity assay [21–23]. In principle, this method can be applied to any RNA that possesses catalytic activity. Here we focus on ribozyme derivatives of group I introns, which lack exons and cleave an oligonucleotide substrate in a reaction that mimics the first step of the splicing reaction. The specific protocols shown were developed for use with the Tetrahymena ribozyme, which is derived from an intron found within the precursor rRNA that encodes the large ribosomal subunit RNAs. Folding of this ribozyme has been studied extensively by catalytic activity and other approaches [21, 24–29].
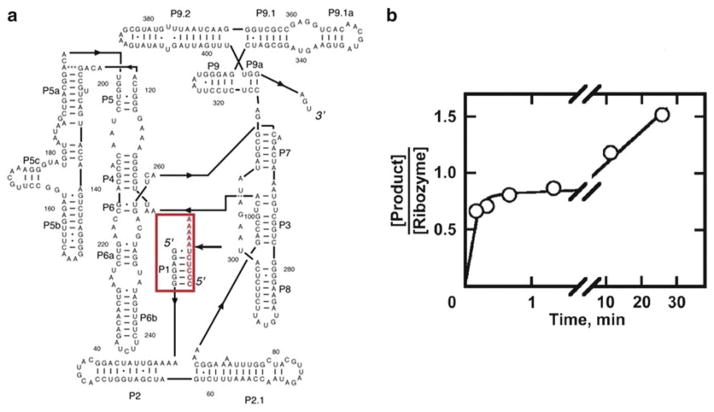
The operating principle of the catalytic activity assay is that only correctly folded, native RNA displays catalytic activity. Thus, the amount of native ribozyme can be determined from the amount of product produced rapidly when substrate is added in excess of the ribozyme. The “burst” of product formation is equal to the amount of native ribozyme if one of the products is released slowly, allowing the first round of cleavage to proceed to completion while preventing subsequent rounds. An example is shown in Fig. 1a, b for the Tetrahymena ribozyme. Upon cleavage of the substrate, the 3′ product is released fast while the 5′ product is released slowly because it forms a six base pair helix with the ribozyme that is additionally stabilized by tertiary contacts [30]. For some ribozyme constructs, both cleavage products are released slowly, allowing substrate cleavage and ligation to reach equilibrium [31]. In this case, the amount of product obtained in the burst is smaller than the amount of native ribozyme, requiring a correction based on the equilibrium constant for cleavage and ligation [22].
Fig. 1.
Tetrahymena ribozyme secondary structure and oligonucleotide substrate cleavage reaction. (a) The standard “L-21” Tetrahymena ribozyme, with the oligonucleotide substrate highlighted in red. The dark black arrow to the right of the substrate indicates the cleavage site. The 3′ product (A5) lacks base pairing with the ribozyme such that it dissociates fast after cleavage, while the 5′ product (CCCUCU) dissociates slowly, resulting in burst kinetics. (b) Cleavage reaction of pre-folded Tetrahymena ribozyme with excess substrate [22, 24]. The second slower phase is dominated by slow release of the 5′ product. It should be noted that an incomplete burst of 0.8–0.9 is characteristic of a homogenous population of native ribozyme, most likely due to a small fraction of damaged ribozyme which varies from preparation to preparation [24 ]. Reprinted from New pathways in folding of the Tetrahymena group I RNA enzyme, 291/5, Russell R. and Herschlag D., New pathways in folding of the Tetrahymena group I RNA enzyme, 1155–1167, Copyright (1999), with permission from Elsevier
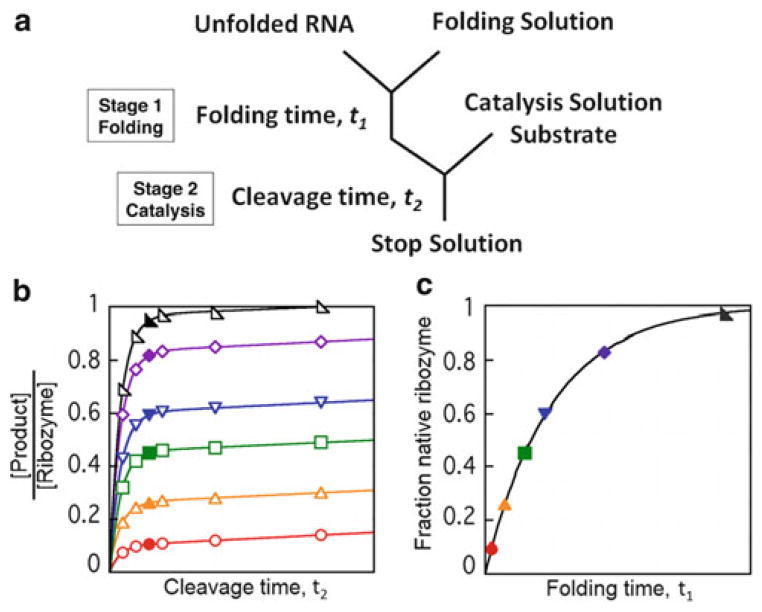
Although the simplest experimental design for catalytic activity examines catalysis and folding concurrently (termed a continuous assay), a more complex two-stage or “discontinuous” assay can be useful for many applications including studying the effects of RNA chaperone proteins [22, 23]. As shown in Fig. 2a, the reaction is separated into two stages: folding and catalysis. By arresting folding at different times and performing cleavage reactions, the accumulation of native ribozyme can be observed as the RNA folds in the folding stage. As shown in Fig. 2b, c, plotting the burst amplitudes from individual cleavage reactions over folding time reveals the progress of native folding.
Fig. 2.
Discontinuous catalytic activity assay. (a ) Reaction scheme for the discontinuous assay. The assay is performed in two stages. Ribozyme folding occurs in the first stage and at different times (t1) aliquots are transferred to the catalysis stage, where the fraction of native ribozyme is determined by cleavage of substrate over time (t2). (b) Simulated data of cleavage reactions. The data follow substrate cleavage over time t2. Each curve represents an aliquot removed at a different time t1 during folding of the ribozyme, beginning with the red curve and ending with the black curve [23]. The solid curves simulate fits by an equation that includes an exponential term and a linear term [22 ]. The amplitudes of the exponential regions of the curves correspond approximately to the solid symbols. As indicated in the protocols and in the figure, at least six time points should be collected per cleavage reaction in initial experiments. However, after the substrate cleavage kinetics are well understood, it is possible to collect fewer time points. Even a single time point can be sufficient if it is collected at the time corresponding to the solid symbols. (c) The fraction of native ribozyme over folding time t1 taken from burst amplitudes in panel (b). Fitting the folding progress to a single exponential, in this case, results in determination of a single rate constant for folding to the native state. Reprinted from Catalytic activity as a probe of native RNA folding, 468, Wan Y., Mitchell D., and Russell R., Catalytic activity as a probe of native RNA folding, 195–218, Copyright (2009), with permission from Elsevier
The discontinuous assay has been used to study the effects of CYT-19 on native folding of the Tetrahymena ribozyme when starting from a long-lived misfolded conformation [8, 12]. Conditions that give accumulation of misfolded ribozyme have been previously established (10 min, 10 mM Mg2+, 25 °C). In experiments starting from misfolded ribozyme and using CYT-19 in the folding stage at different concentrations, CYT-19 has been shown to increase the observed rate constant for folding to the native state. In this chapter, we present a protocol used to measure this folding transition from misfolded to native ribozyme and the effects of CYT-19 [12]. It should be noted that variations of this protocol have also been used to monitor effects of CYT-19 and related DEAD-box proteins on the native ribozyme and effects of DEAD-box proteins on folding when starting from unfolded conformations. It should also be noted that while this method has been used principally to monitor effects of DEAD-box proteins—i.e., ATP-dependent RNA helicases—that function as chaperones, it is expected to be equally applicable to other groups of RNA chaperone proteins [32, 33].
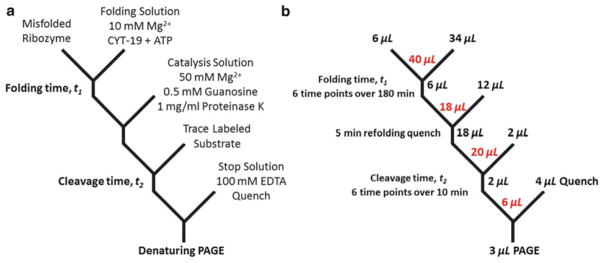
Figure 3a shows a detailed scheme for measurement of native ribozyme folding in the presence of CYT-19. A high concentration of magnesium (50 mM Mg2+) is used as a folding quench because it dramatically slows refolding transitions from the misfolded conformation, effectively blocking further folding, and it promotes fast catalysis, allowing measurement of the fraction of native ribozyme at different times during the folding reaction [34]. Proteinase K can also be present in the catalysis solution to ensure that the DEAD-box chaperone protein does not remain active in the catalysis stage and perturb the reaction by promoting rapid exchange of the substrate. In Fig. 3b, we present typical reaction volumes, a typical number of time points for each stage, and total reaction volumes for this particular experiment. The oligonucleotide substrate is typically radio-labeled using T4 polynucleotide kinase (PNK) and [γ-32P]-ATP, and time points from the cleavage reactions are analyzed using denaturing polyacrylamide gel electrophoresis (PAGE) to separate the substrate and product. Each cleavage reaction is expected to have a fast phase with a burst amplitude corresponding to the amount of native ribozyme and an observed rate constant for cleavage, and a slow phase limited by dissociation of one of the products from the ribozyme (the product that is released more slowly).
Fig. 3.
Detailed reaction scheme for using the discontinuous assay to probe CYT-19 effect on folding. (a) Solution conditions for each stage of the assay. Ribozyme is pre-folded to a population of predominantly misfolded RNA and enters the folding stage with the addition of CYT-19 and ATP at 10 mM Mg2+. Folding is quenched at different times t1 (typically ranging from 5 to 180 min) by raising the magnesium concentration to 50 mM and adding proteinase K with a short incubation between stages (typically 5 min, but this time can vary, provided that it is short enough to prevent additional accumulation of native ribozyme). Trace-labeled substrate is added to initiate the reaction and the fraction of native ribozyme is determined by the amplitude of the “burst” of product formation during time t2 (times for t2 typically range from 15 s to 10 min). (b ) Reaction volumes in this protocol. Red values indicate total reaction volumes after aliquots from each step are combined
We present two protocols below that are used commonly in our laboratory. The first protocol describes measurement of the rate constant of ribozyme folding to the native state when the folding process is begun from unfolded ribozyme by adding Mg2+, and the second protocol describes measurement of native folding starting from a population of predominantly misfolded ribozyme and including DEAD-box protein CYT-19 in the folding reaction.
2 Materials
To avoid RNA degradation, all solutions should be prepared using RNase-free water and conventional filter sterilization methods. Store all stock solutions at −20 °C (except the proteins, which should be stored at −80 °C or the temperature recommended by the manufacturer) and store all solutions on ice during the experiment. Folding and catalysis reactions are performed in siliconized or “slick” 500 μL microcentrifuge tubes (VWR), and solutions containing RNA are typically prepared in these tubes also. Ribozymes are transcribed using recombinant T7 RNA polymerase and run-off transcription, as defined by a restriction site, and purified using a QIAGEN RNeasy Mini or Midi kit as described [35].
2.1 Labeling 5′ Termini of Substrate Oligonucleotide
T4 polynucleotide kinase (PNK; 10 U/μL). Stored at −20 °C.
5× T4 PNK buffer: 350 mM Tris–HCl, 25 mM MgCl2, 12.5 mM dithiothreitol (DTT), pH 7.6. Stored at −20 °C.
[γ-32P]-adenosine triphosphate ([γ-32P]-ATP): 150 mCi/mL.
Oligonucleotide substrate: 5′ CCCUCUAAAAA 3′. Stored at −20 °C.
Tris–EDTA (TE) buffer: 1 mM EDTA and 10 mM Tris–HCl, pH 8.0.
Non-denaturing polyacrylamide gel: 20 % acrylamide/bisacryl-amide (29:1), 1× TBE buffer. Stored at 4 °C.
X-ray film.
Film processor (e.g., KODAK X-omat 2000A).
Microcentrifuge.
Scintillation counter.
2.2 Catalytic Activity Solutions
Working stock solutions: 0.5 M Na-MOPS, pH 7.0, 10 μM Tetrahymena ribozyme, 50 mM Mg(CH3COO)2 (magnesium acetate), 0.5 M MgCl2, 5 mM guanosine, 5 μM CYT-19, 20 mM ATP-Mg2+.
Folding solution 1: 40 μL solution of 0.6 μM ribozyme, 10 mM Mg(CH3COO)2, 50 mM Na-MOPS, pH 7.0. Do not add 8 μL of 50 mM Mg(CH3COO)2 until ready to initiate the folding (see Note 1).
Catalysis solution 1: 80 μL solution of 78.3 mM MgCl2, 833 μM guanosine, 50 mM Na-MOPS, pH 7.0 (see Notes 2 and 3).
Pre-folding solution: 20 μL solution of 0.6 μM ribozyme, 10 mM Mg(CH3COO)2, 50 mM Na-MOPS, pH 7.0. Do not add 4 μL of 50 mM Mg(CH3COO)2 until ready to initiate pre-folding (see Note 1).
20 mM ATP-Mg2+. Mix ATP with the same concentration of Mg(CH3COO)2 to achieve 20 mM ATP-Mg2+ (see Note 4).
Folding solution 2: 34 μL solution of 10 mM Mg(CH3COO)2, 0.25 μM CYT-19, 2 mM ATP-Mg2+, 50 mM Na-MOPS, pH 7.0. Wait to add 2 μL of 5 μM CYT-19 until ready to initiate the reaction (15 s–1 min) (see Note 5).
Catalysis solution 2: 80 μL solution of 78.3 mM MgCl2, 833 μM guanosine, 1 mg/mL proteinase K, 50 mM Na-MOPS, pH 7.0 (see Note 6).
Stop solution: 100 mM EDTA, pH 8.0, 0.04 % bromophenol blue (BPB), 0.04 % xylene cyanol, 72 % (v/v) formamide, pH 8.0.
2.3 Denaturing Polyacrylamide Gel Electrophoresis (PAGE)
Running gel buffer (10× TBE): 890 mM Tris-base, 890 mM boric acid, 20 mM EDTA, pH 8.3.
Denaturing polyacrylamide mix: 20 % acrylamide/bisacrylamide (29:1), 7 M urea, 1× TBE buffer (see Note 7).
Two pairs of glass plates 20 cm × 20 cm × 4 mm with 0.35 mm thick spacers for each pair of plates and a comb that gives 5 μL wells with at least 20 wells per comb.
Two gel boxes for running gel electrophoresis and a power supply capable of generating 16 W per gel.
Whatman 3MM filter paper.
Gel dryer.
2.4 Autoradiography Equipment and Analysis Tools
Storm 820 phosphorimager (Amersham Pharmacia Biotech, Inc.—Molecular Dynamics Div.) or comparable equipment.
Phosphorimager exposure cassette and screen (Amersham Pharmacia Biotech, Inc.—Molecular Dynamics Div.) or comparable product.
ImageQuant TL v7.2 software (Molecular Dynamics) or comparable software: Used for densitometry analysis to determine fraction product produced over time.
3 Methods
We present typical reaction volumes for this assay in Fig. 3b and in the protocol, but reaction volumes can vary on an experiment by experiment basis (see Note 8). A minimum of six time points should be collected from each folding and cleavage reaction (but see Fig. 2 legend). Typical time points for cleavage reactions are 2 μL, such that the volume of solution in the catalysis stage must be at least 12 μL (typically 20 μL). Similarly, the folding stage requires enough volume to accommodate folding stage time points, with each folding time point requiring 6 μL.
3.1 Labeling 5′ Termini of Oligonucleotides with 32P
Prepare the kinase reaction solution by mixing 1 μL of 50 μM oligonucleotide substrate, 1 μL of 5× PNK buffer, 1 μL of [γ-32P]-ATP (150 mCi/mL), and 1 μL of T4 PNK (10 U/ μL). Note that this protocol produces a 4 μL reaction with a final buffer concentration of 1.25×.
Incubate the 4 μL reaction at 37 °C for 1 h in a water bath.
Prepare the 20 % non-denaturing gel for isolation of the 5′ labeled substrate.
Add 6 μL of stop solution to halt the labeling reaction.
Load 5 μL each into two adjacent wells on the gel and run the gel at 12 W for 60 min to ensure complete separation of substrate from unreacted [γ-32P]-ATP.
Separate the gel plates to expose the gel and cover the gel face with a layer of plastic wrap. Carefully press X-ray film sealed in envelope lightly to the plastic wrap for about 10–15 s while marking the gel and film for alignment by poking at least three holes with a needle. Develop the film in the film processor.
Align the exposed film with the gel and cut out the gel band of the labeled substrate using a flame-sterilized razor blade.
Elute the substrate from the gel piece by submerging the gel piece in ~150 μL of TE buffer overnight at 4 °C. The next morning, pellet the gel fragments by centrifugation using the microcentrifuge and transfer the supernatant containing the eluted oligonucleotide into a new tube. The specific activity of the labeled oligonucleotide is conveniently measured using Cerenkov detection by transferring 1 μL to a scintillation vial and detecting radioactive decay in the scintillation counter without adding scintillant. The efficiency of detection for 32P disintegrations is roughly 30 %. The labeled oligonucleotide should be stored at −20 °C and may be used for several weeks before it decays to background levels (t1/2 ≈ 2 weeks).
3.2 Rate of Folding to the Native State of Unfolded Tetrahymena Ribozyme
Aliquot 4 μL of stop solution into thirty-six tubes for six cleavage reactions with six time points each.
Heat folding solution 1 (without Mg2+) at 95 °C for 2 min and then place it on ice for 2 min. This step improves reproducibility by ensuring that the ribozyme molecules begin folding from the same ensemble of unfolded states in all experiments (see Note 1).
Aliquot 12 μL of catalysis solution 1 into six tubes. These will become the catalysis reactions when aliquots of the folding reaction are added to them in step 5 below.
Place the folding solution at the desired temperature for folding (typically 25 or 37 °C), allow at least one min for the temperature to equilibrate, and then add 8 μL of 50 mM Mg(CH3COO)2 (also equilibrated at the same temperature) to initiate folding (t1 =0) (see Note 9).
After 15 s, remove 6 μL of folding solution 1 and add it to one of the tubes prepared above in step 3 (containing 12 μL of catalysis solution 1) at 25 °C to quench folding (see Note 10).
Incubate the quenched folding reaction from step 5 for 5 min to allow formation of the native and long-lived misfolded conformations. Note that this protocol only measures the folding progress from the misfolded conformation to the native conformation. To follow the initial phase of folding of this ribozyme, which gives a small fraction of native ribozyme, the folding quench and substrate cleavage reaction are performed the same way but at 0 °C [36].
Repeat steps 5 and 6 for all six values of t1 to probe the fraction of native ribozyme at each time point as the RNA folds to the native state. Common times of t1 are 15 s, 1 min, 10 min, 30 min, 90 min, and 180 min (see Note 10).
Approximately 5 min after quenching folding in step 5, add 2 μL of excess substrate (twofold excess over the ribozyme concentration) mixed with trace labeled substrate (~20,000 dpm) to initiate the cleavage reaction (t2 =0). After 15 s, remove 2 μL of the cleavage reaction and add it to the previously aliquoted 4 μL of stop solution (step 1) for the 15 s time point (see Note 11).
Repeat step 8 for all six values of t2 to probe the fraction of product cleaved at each time point as the substrate is converted into product. Common times of t2 are 15 s, 30 s, 1 min, 2 min, 4 min, and 10 min (see Note 12).
Prepare the 20 % denaturing polyacrylamide gel by mixing 300 μL of 10 % APS and 30 μL of TEMED with 30 mL of denaturing polyacrylamide mix. Pour about 15 mL of polymerizing gel into the pre-assembled glass plates and set a comb in the gel with at least twenty 5 μL wells.
To facilitate quantitation of individual lanes, load alternate wells (wells 1, 3, 5, etc.) with 3 μL of quenched cleavage reaction from step 9. Run the gel for about 20 min at 16 W per gel to allow sample to enter the gel.
Once this time has elapsed, add 3 μL of the remaining time points into the remaining unused wells staggered in the same way (wells 2, 4, 6, etc.). Run the gel for 45 min at 16 W per gel to separate the shorter cleaved product from the longer substrate.
Separate the plates by leaving the gel attached to one side and cover the gel with plastic wrap before lifting it from the remaining plate. Place the gel on Whatman 3MM filter paper and dry it using the gel dryer. Expose the gel overnight to a phosphor screen and quantify it using a phosphorimager and ImageQuant or comparable software to determine the fraction of product at each time point for each substrate cleavage reaction.
Fit the fraction of material present as product against t2 to an equation that includes an exponential “burst” term and a linear phase for slower subsequent turnovers [22]. Plot the burst amplitude of each cleavage time course against the folding time t1 and fit the curve to a single exponential equation to determine the rate constant for native folding.
3.3 Rate of CYT-19-Mediated Refolding of Misfolded Tetrahymena Ribozyme
As in Subheading 3.2, aliquot 4 μL of stop solution into 36 tubes for six reactions with six time points each.
Initiate pre-folding of the ribozyme at 25 °C by adding 4 μL of 50 mM Mg(CH3COO)2 to the pre-folding solution and allow the RNA to fold for approximately 10 min.
While waiting, aliquot 12 μL of catalysis solution 2 to six tubes. These will become the catalysis reactions when aliquots of the folding reaction are added to them in step 5 below. Immediately prior to step 4 (15 s–1 min), complete the preparation of folding solution 2 by adding 2 μL of 5 μM CYT-19 (see Note 5).
When pre-folding of the ribozyme is complete, remove 6 μL of pre-folding solution and mix it with 34 μL of folding solution 2 at 25 °C to start the CYT-19-mediated folding reaction (see Note 13).
Repeat steps 5–14 from Subheading 3.2 using the CYT-19-mediated folding reaction generated in step 4 and catalysis solution 2 to determine the rate of native state formation.
Footnotes
To start from a homogeneously unfolded population, the ribozyme mixed with MOPS buffer may first be heated at 95 °C for 2 min and then incubated on ice for 2 min. When all solutions and tubes are prepared for the time course, Mg2+ is added to initiate folding. For the Tetrahymena ribozyme, this 95 °C step was found to give no detectable effect on folding (R.R., unpublished) and is now routinely omitted. However, when testing a new ribozyme it is necessary to compare the folding properties with and without this step.
Because aliquots from the folding reaction contain magnesium and are diluted during the experiment, concentrations of magnesium and guanosine must be adjusted to achieve final desired concentrations.
Guanosine has a relatively low solubility. To maintain solubility of the stock solution of guanosine, heat it at 50 °C until it is fully dissolved before adding it to catalysis solutions.
The negatively charged triphosphate moiety of ATP binds magnesium ions. To keep the concentration of magnesium unchanged for RNA folding reactions, a stock solution of equimolar amount of magnesium and ATP is necessary.
To prevent loss of activity as a result of extended and premature interaction of protein with folding solution components, add CYT-19 immediately before starting the reaction (15 s–1 min).
With the solution conditions described and standard concentrations of CYT-19 (<1 μM), proteinase K is not necessary because the high MgCl2 concentration in stage 2 is sufficient to block activity of CYT-19. However, proteinase K addition may be necessary with different proteins or higher CYT-19 concentrations.
7 M urea is used to ensure complete RNA denaturation but is near the solubility limit. As such, the 1 L solution should be periodically mixed over 5–10 min at 37 °C to keep the urea in solution.
To determine the necessary reaction volumes, first determine how much catalysis reaction solution is needed in each cleavage reaction. Six time points of 2 μL each implies that at least 12 μL of volume is required in the catalysis stage (reaction volumes of 20 μL are typically used). Next, the volume needed in the folding reaction should be calculated. Aliquots of 6 μL of the folding reaction are typically added to 12 μL of folding quench solution (and 2 μL of substrate solution) for each folding time point (Fig. 3b). Thus, generating data at six different folding times will require at least 36 μL in the folding reaction.
Mixing the solutions well and rapidly is crucial. After each addition be sure to quickly flick the bottom of the tube gently or pipet several times in rapid succession to mix the solution.
Once the folding reaction starts, the fraction of native ribozyme needs to be determined at several different times during folding (i.e., several values of t1). Keep in mind while performing cleavage reactions from the first few folding time points that additional aliquots of the folding reaction may need to be removed, while these cleavage reactions are still in progress (step 5 in Subheading 3.2.).
Excess substrate relative to the ribozyme should be used to ensure that burst amplitudes are stoichiometrically correlated with the fraction of native ribozyme, i.e., twofold excess substrate over the ribozyme results in a burst amplitude of 50 % under conditions such that 100 % of the ribozyme is present in the native state. However, if the substrate binds with comparable rates to both misfolded and native RNA and remains bound on a time scale that is longer than the first turnover of substrate cleavage by the native ribozyme, then trace amounts of labeled substrate may be used. This alternative protocol can increase reproducibility of the burst amplitudes because the burst size is independent of the ratio of substrate to ribozyme in this regime.
Once the time course for folding and cleavage reactions are complete, quenched time points can be stored at 4 °C for up to several days before running gel electrophoresis if more time is needed.
After pre-folding for 10 min at 25 °C, misfolded Tetrahymena ribozyme is stable with a half-life on the order of hours. Because of this long lifetime, the folding reaction does not have to be initiated immediately at 10 min as long as the actual pre-folding time is recorded.
References
- 1.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 2.Russell R. RNA misfolding and the action of chaperones. Front Biosci. 2008;13:1–20. doi: 10.2741/2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Treiber DK, Williamson JR. Exposing the kinetic traps in RNA folding. Curr Opin Struct Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 4.Jarmoskaite I, Russell R. DEAD-box proteins as RNA helicases and chaperones. Wiley Interdiscip Rev RNA. 2011;2:135–152. doi: 10.1002/wrna.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 6.Mohr S, Stryker JM, Lambowitz AM. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- 7.Huang HR, Rowe CE, Mohr S, et al. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci USA. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tijerina P, Bhaskaran H, Russell R. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc Natl Acad Sci USA. 2006;103:16698–16703. doi: 10.1073/pnas.0603127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solem A, Zingler N, Pyle AM. A DEAD protein that activates intron self-splicing without unwinding RNA. Mol Cell. 2006;24:611–617. doi: 10.1016/j.molcel.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 10.Halls C, Mohr S, Del Campo M, et al. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohr S, Matsuura M, Perlman PS, et al. A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc Natl Acad Sci USA. 2006;103:3569–3574. doi: 10.1073/pnas.0600332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhaskaran H, Russell R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature. 2007;449:1014–1018. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Potratz JP, Del Campo M, Wolf RZ, et al. ATP-dependent roles of the DEAD-box protein Mss116p in group II intron splicing in vitro and in vivo. J Mol Biol. 2011;411:661–679. doi: 10.1016/j.jmb.2011.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Campo M, Tijerina P, Bhaskaran H, et al. Do DEAD-box proteins promote group II intron splicing without unwinding RNA? Mol Cell. 2007;28:159–166. doi: 10.1016/j.molcel.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Campo M, Mohr S, Jiang Y, et al. Unwinding by local strand separation is critical for the function of DEAD-box proteins as RNA chaperones. J Mol Biol. 2009;389:674–693. doi: 10.1016/j.jmb.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Potratz JP, Tijerina P, et al. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci USA. 2008;105:20203–20208. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sengoku T, Nureki O, Nakamura A, et al. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 18.Del Campo M, Lambowitz AM. Structure of the yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol Cell. 2009;35:598–609. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Q, Del Campo M, Lambowitz AM, et al. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 20.Russell R, Jarmoskaite I, Lambowitz AM. Toward a molecular understanding of RNA remodeling by DEAD-box proteins. RNA Biol. 2012;10:44–55. doi: 10.4161/rna.22210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russell R, Herschlag D. Probing the folding landscape of the Tetrahymena ribozyme: commitment to form the native conformation is late in the folding pathway. J Mol Biol. 2001;308:839–851. doi: 10.1006/jmbi.2001.4751. [DOI] [PubMed] [Google Scholar]
- 22.Wan Y, Mitchell D, 3rd, Russell R. Catalytic activity as a probe of native RNA folding. Methods Enzymol. 2009;468:195–218. doi: 10.1016/S0076-6879(09)68010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potratz JP, Russell R. RNA catalysis as a probe for chaperone activity of DEAD-box helicases. Methods Enzymol. 2012;511:111–130. doi: 10.1016/B978-0-12-396546-2.00005-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russell R, Herschlag D. New pathways in folding of the Tetrahymena group I RNA enzyme. J Mol Biol. 1999;291:1155–1167. doi: 10.1006/jmbi.1999.3026. [DOI] [PubMed] [Google Scholar]
- 25.Latham JA, Cech TR. Defining the inside and outside of a catalytic RNA molecule. Science. 1989;245:276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- 26.Sclavi B, Sullivan M, Chance MR, et al. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 27.Zarrinkar PP, Williamson JR. Kinetic intermediates in RNA folding. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- 28.Zhuang X, Bartley LE, Babcock HP, et al. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 29.Russell R, Millett IS, Tate MW, et al. Rapid compaction during RNA folding. Proc Natl Acad Sci USA. 2002;99:4266–4271. doi: 10.1073/pnas.072589599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narlikar GJ, Gopalakrishnan V, McConnell TS, et al. Use of binding energy by an RNA enzyme for catalysis by positioning and substrate destabilization. Proc Natl Acad Sci USA. 1995;92:3668–3672. doi: 10.1073/pnas.92.9.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinan S, Yuan X, Russell R. The Azoarcus group I intron ribozyme misfolds and is accelerated for refolding by ATP-dependent RNA chaperone proteins. J Biol Chem. 2011;286:37304–37312. doi: 10.1074/jbc.M111.287706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 33.Rajkowitsch L, Chen D, Stampfl S, et al. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 34.Russell R, Das R, Suh H, et al. The paradoxical behavior of a highly structured mis-folded intermediate in RNA folding. J Mol Biol. 2006;363:531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 35.Russell R, Herschlag D. Specificity from steric restrictions in the guanosine binding pocket of a group I ribozyme. RNA. 1999;5:158–166. doi: 10.1017/s1355838299981839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell R, Tijerina P, Chadee AB, et al. Deletion of the P5abc peripheral element accelerates early and late folding steps of the Tetrahymena group I ribozyme. Biochemistry. 2007;46:4951–4961. doi: 10.1021/bi0620149. [DOI] [PMC free article] [PubMed] [Google Scholar]