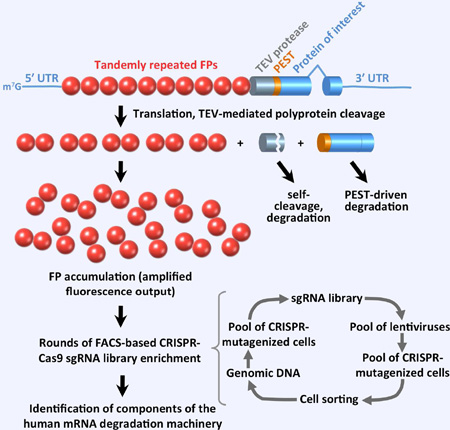
SUMMARY
Nonsense-mediated decay (NMD) degrades mRNAs containing a premature termination codon (PTC). PTCs are a frequent cause of human genetic diseases and the NMD pathway is known to modulate disease severity. Since partial NMD attenuation can potentially enhance nonsense suppression therapies, better definition of human-specific NMD is required. However, the majority of NMD factors were first discovered in model organisms and then subsequently identified by homology in human. Sensitivity and throughput limitations of existing approaches have hindered systematic forward genetic screening for NMD factors in human cells. We developed a method of in vivo amplification of NMD reporter fluorescence (Fireworks) that enables CRISPR-based forward genetic screening for NMD pathway defects in human cells. The Fireworks genetic screen identifies multiple known NMD factors and numerous human candidate genes, providing a platform for discovery of additional key factors in human mRNA degradation.
Graphical Abstract
INTRODUCTION
With nearly 20% of human disease-associated single base pair substitutions forming a premature termination codon (PTC) (Mort et al., 2008), these lesions often have drastic clinical consequences. PTC mutations can result in severe human genetic diseases such as variants of Duchenne and Becker muscular dystrophies (dystrophin), retinoblastoma (RB1), neurofibromatosis (NF1, NF2), ataxia-telangiectasia (ATM), Tay-Sachs disease (HEXA), cystic fibrosis (CFTR), Wilm's tumor (WT1), beta thalassemia (betaglobin), hemophilia A (factor VIII) and B (factor IX), von Willebrand disease (Willebrand factor), p53-associated cancers (p53), and numerous others (Holbrook et al., 2004). There are no cures for these genetic disorders.
Experimental therapies for PTC-induced disorders, aminoglycosides (Gunn et al., 2014; Keeling et al., 2014) and ataluren (PTC124) (Finkel et al., 2013; Kerem et al., 2014; Roy et al., 2016), as well as a number of small molecules, e.g. RTC13 and RTC14 (Du et al., 2009), GJ071 and GJ072 (Du et al., 2013), and amlexanox (Gonzalez-Hilarion et al., 2012), promote PTC readthrough. A growing body of evidence (Keeling et al., 2013; Keeling et al., 2014; Martin et al., 2014; Nomakuchi et al., 2016) suggests that successful therapies, in addition to promoting PTC readthrough, need to address degradation of PTC-containing mRNAs by the human nonsense-mediated decay (NMD) pathway (Kurosaki and Maquat, 2016). Because the NMD pathway reduces the levels of PTC-containing mRNA, inhibition of NMD increases the amount of PTC-containing mRNA available for readthrough, aiding in recovery of full-length protein product.
Not only does NMD play a prominent role in human disease, but it also serves as a major mRNA degradation pathway, regulating at least 10% of human mRNAs (Rehwinkel et al., 2005). NMD operates via the concerted action of multiple factors (Kurosaki and Maquat, 2016), many of which also function in other cellular processes. Knockdown of some of these factors (e.g. UPF1, UPF2, or SMG1) is embryonic lethal in mouse (McIlwain et al., 2010; Medghalchi et al., 2001; Weischenfeldt et al., 2008), whereas depletion of others, like SMG8, was found not to cause noticeable cell-growth defects (Usuki et al., 2013). Potential therapeutic inhibition of human NMD therefore must be carefully crafted to limit toxic/negative effects (Keeling et al., 2014), requiring better understanding of the precise cellular roles of human NMD components and mechanisms of human-specific and, prospectively, tissue-specific NMD.
However, the full scope of factors comprising the human NMD pathway is not known, largely because of the lack of comprehensive genetic screens. Comprehensive forward genetic screens for NMD factors have been accomplished only in model organisms such as S. cerevisiae, C. elegans, and D. melanogaster (Hodgkin et al., 1989; Leeds et al., 1991; Metzstein and Krasnow, 2006) leading to identification of a number of human NMD factors by homology search (Perlick et al., 1996). Yet, vertebrate NMD pathways appear to involve many vertebrate-specific factors (Lykke-Andersen and Jensen, 2015). For example, whereas the exon junction complex (EJC) is a major NMD player in mammals (Le Hir et al., 2000), the yeast S. cerevisiae has no known EJC; moreover, NMD in yeast does not depend on splicing, and known EJC components in D. melanogaster and C. elegans are not required for NMD (Gatfield et al., 2003; Longman et al., 2007). Therefore, screens in model (nonvertebrate) organisms are of only limited utility for identification of human-specific NMD components. Because, to our knowledge, no systematic forward genetic screen for human NMD factors has been successfully performed in human cells, the list of human NMD factors is undefined, with important factors, such as hCWC22 (Alexandrov et al., 2012; Barbosa et al., 2012; Steckelberg et al., 2012), MOV10 (Gregersen et al., 2014), GNL2, and SEC13 (Casadio et al., 2015), being only recently identified.
To increase understanding of human-specific NMD and expand the scope of human factors amenable for therapeutic NMD inhibition, we developed a forward genetic method involving in-vivo fluorescence amplification (Fireworks) that allows comprehensive screening for genes involved in NMD. As an example of successful forward genetic screening for NMD factors in human cells, we report identification of (i) known major NMD components (Kurosaki and Maquat, 2016): UPF1, UPF2, SMG5, SMG6, and SMG7; (ii) a known NMD regulator EIF2B4 (Gardner, 2008; Martin et al., 2010); as well as (iii) 11 candidate genes. Our method of fluorescence amplification coupled with sequential rounds of enrichment for functional sgRNAs provides a platform for forward genetic discovery of key components and modulators of the human mRNA degradation machinery. Our approach is also generally applicable to screening a variety of other human cellular pathways as long as their readout can be adjusted to result in fluorescent protein expression, providing a powerful forward genetic tool for multiple fields of human biology.
DESIGN
Lack of a sensitive FACS-compatible reporter system for forward genetic screening has impeded identification of components involved in human mRNA decay
Whereas numerous NMD reporter systems relying on mRNA detection by Northern blot, luminescence, or fluorescence are currently in use, they have difficultiy in attaining the sensitivity and throughput thresholds required for genome-wide genetic screening in human cells for the following reasons. (1) Although transient transfection with NMD reporters produces sufficient fluorescence signal intensity (Gurskaya et al., 2016; Pereverzev et al., 2015), the reporter’s expression can vary dramatically among individual cells, impeding analyses required to identify moderate signal differences (typical for CRISPR-based screens) on a highly variable expression background. (2) Existing chromosomally-integrated human NMD reporters do not yield fluorescent signal of sufficient intensity for meaningful forward genetic screening; the only published reporters (Paillusson et al., 2005) produce such weak signals that the fluorescence of the PTC-containing reporter completely overlaps the background signal of reporter-lacking control cells. The same study acknowledged that the signal of a chromosomally-integrated PTC-containing reporter was not observable – even by FACS or confocal microscopy – unless NMD was inhibited. Because of insufficient fluorescence intensity of these reporters, there are no reports of their use. Moreover, typically, chromosomally-integrated reporters in human cells are subject to transgene silencing, which produces random cell-to-cell signal variation that degrades the quality of a genetic screen. (3) Despite high sensitivity due to enzymatic amplification, the signal of luciferase-based NMD reporters (Nickless et al., 2014) is not directly compatible with FACS sorting, requiring plate-based well-to-well robotic processing that limits the throughput to tens of thousands, as opposed to a billion mutants per day, achievable with a FACS instrument. An NMD reporter system suitable for high-throughput FACS-based screening is therefore not readily available, precluding forward genetic identification of human-specific components and modulators of the mRNA decay machinery.
Fireworks: a fluorescence amplification system to screen for factors that modulate NMD in human cells
Based on the above considerations, an ideal system would involve a chromosomally-integrated reporter that produces intense fluorescence signal. Fluorescence intensity is critical for a FACS-based screen because the higher the signal is over background, the more accurately it can be measured by FACS, defining sorting accuracy and eventually the quality of a genetic screen. Additionally, a well-balanced fluorescence-based NMD reporter system suitable for high-throughput screening must eliminate: (i) differential effects of the PTC(+) [PTC-containing] and PTC(−) [PTC-lacking] polypeptides [the PTC(+) peptide is always shorter] on localization, maturation, and half-life of the fused fluorescent protein and (ii) potential toxicity and mosaic-like silencing of genome-integrated transgenic reporters in stable cell lines.
We solved the above problems by developing the Fireworks system (Figures 1 and S1), which incorporates the following: (1) Translation of multiple (10 or 5) tandemly-repeated fluorescent proteins (FPs) into a long single polyprotein to amplify the fluorescence signal produced by a single transcription unit of a PTC-containing reporter without the need to increase the reporter copy number (Figures 1A and S1A). (2) In vivo proteolytic release of the multiple FPs from the polyprotein by tobacco etch virus (TEV) protease, expressed as part of the same polyprotein, to eliminate negative effects of the PTC-containing polypeptide (e.g. β-globin) on FP maturation and localization. (3) Multiple features to limit reporter silencing and toxicity, including use of reporter-flanking tDNA-based (Lee et al., 2013; Raab et al., 2012) chromatin insulators (Figure S3), introduction of a destabilizing N-terminal serine (Bachmair et al., 1986) into the TEV protease and the β-globin as a result of their proteolytic processing (Figure 1A and S1A), autocatalytic self-cleavage (Parks et al., 1995) of the TEV protease (Figures 1A and S1A), and destabilization of the β-globin via its fusion with the PEST (Rogers et al., 1986) peptide sequence (Figures 1A and S1A).
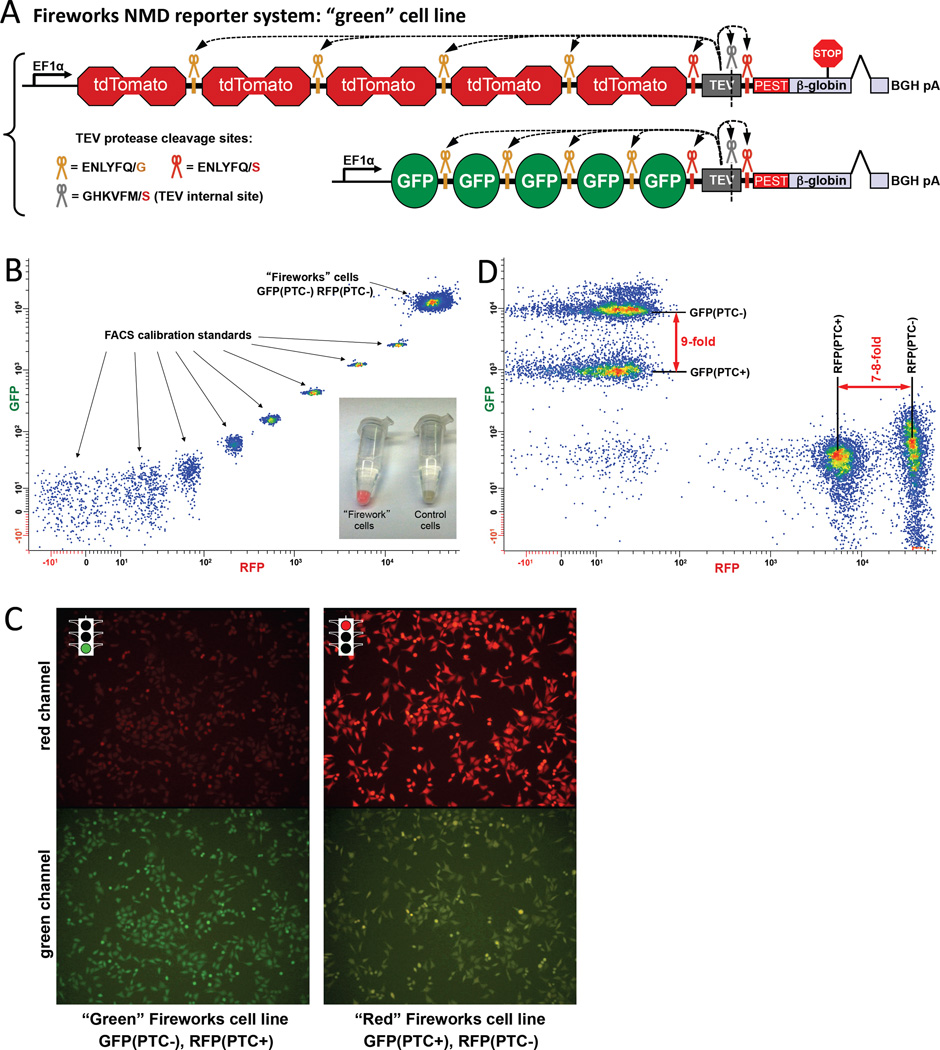
Figure 1. Fireworks in vivo NMD reporter system.
(A) Schematic of the Fireworks approach is shown for the “green” Fireworks cell line. Each Fireworks reporter expresses a single polyprotein consisting of (1) 10 or 5 FPs (RFP or GFP), (2) TEV protease, and (3) PEST-β-globin. The β-globin gene lacks the first intron and either carries or lacks a PTC (PTC39) upstream of its last (second) intron. The resulting polyprotein contains 7 TEV protease cleavage sites: (i) between each copy of fluorescent protein, (ii) between the last fluorescent protein and the protease, (iii) within the protease, and (iv) between the protease and the PEST-β-globin. As a result, in vivo translated FPs are separated from each other, the protease, and the PEST-β-globin through in vivo proteolytic cleavage. The PEST degradation sequence (Rogers et al., 1986) was added to shorten the β-globin protein half-life. (B) HeLa cells with genome-integrated Fireworks reporters produce strong fluorescence signals, which are above the calibration range of a FACS instrument. The fluorescence is stable, uniform, and virtually immune to silencing. Lower-right insert: Pellets of Fireworks cells at ambient lighting. (C) Fluorescence of stable orthogonal Fireworks cell lines observed with an epifluorescence microscope. Each (HeLa) cell line carries two FRT-integrated Fireworks reporters that differ by a single (PTC39) nucleotide: the cell line on the left carries PTC39 in the second exon of the β-globin sequence in the RFP-containing reporter; the cell line on the right carries the identical PTC39 in the GFP-containing reporter. mRNAs expressed by the PTC39-containing reporters are NMD substrates and yield lower fluorescence; mRNAs of the control Fireworks reporters are unaffected, producing the green (left lower panel) and red (right upper panel) signal. The brown color of the cells in the lower right panel results from the leakage of red color into the green channel. (D) Pools of HeLa cells with genome-integrated Fireworks reporters appropriately respond to the introduction of PTC39 by a 7- to 9-fold fluorescence drop (no attempt was made to fine-tune color compensation). See also Figure S1.
The Fireworks approach: implementation and experimental advantages
The fluorescence signal produced by HeLa cells with genome-integrated Fireworks NMD reporters is exceptionally bright, stable, and virtually immune to reporter silencing (Figures 1B, C and S1B, C). The signal is at least two orders of magnitude brighter than the signal of existing human chromosomally-integrated NMD reporters (Paillusson et al., 2005); it is even brighter than the brightest calibration standard of a FACS instrument (Rainbow Calibration Particles, 8 peaks, Spherotech) in both the RFP and GFP dimensions (Figure 1B). The remarkable brightness of the Fireworks reporters underscores their potential for use not only with human β-globin reporters but also with other disease-related PTC-containing mRNAs that are naturally much less stable. Additionally, because of careful selection of the FRT (Flp-recombinase target) (Sadowski, 1995) sites (Figure S3 and Experimental Procedures), Fireworks cell lines display low cell-to-cell signal variation (note the tight distribution in Figure 1B) and minimal reporter silencing even after long-term propagation.
RESULTS
The Fireworks approach provides the ability to perform forward genetic screens for human NMD factors at the speed of a FACS sorter
As designed, orthogonal Fireworks cell lines respond as expected to the introduction of a stop codon (PTC39) (Zhang et al., 1998) into the β-globin sequence by a 7- to 9-fold reduction in the reporter’s fluorescence (Figures 1C, D and S1C). PTC39, which is upstream of the last intron in the β-globin pre-mRNA, induces NMD of the Fireworks reporter mRNA. This, in turn, decreases the steady-state levels of all proteins (including FPs) produced from the Fireworks reporter. Each of the two orthogonal cell lines shown in Figures 1A and S1A (termed “green” and “red” cells, respectively) carries two genome-integrated Fireworks reporters. The “green” Fireworks cell line contains stably integrated PTC-containing RFP and PTC-lacking GFP reporters (Figures 1A and 1C, left panel). The PTC-containing RFP reporter allows rapid screening for PTC-dependent effects by FACS-isolating cells with increased RFP fluorescence; the fluorescence signal of the PTC-lacking GFP reporter serves as a same-cell control for variations in gene expression. This arrangement enables enrichment of NMD-defective cells at a rate approaching 50,000,000 cells/hour. In the “red” (Figures S1A and 1C, right panel) Fireworks cell line, mRNAs transcribed from the PTC-containing GFP reporter are destabilized by NMD, whereas the PTC-lacking RFP reporter serves as an expression control. These two orthogonal cell lines enable enrichment, discovery, and preliminary validation of candidate genes at a high rate, providing (i) a powerful platform to screen libraries of mutant cells for NMD defects and (ii) sound controls for cell line- and fluorescent protein-specific effects.
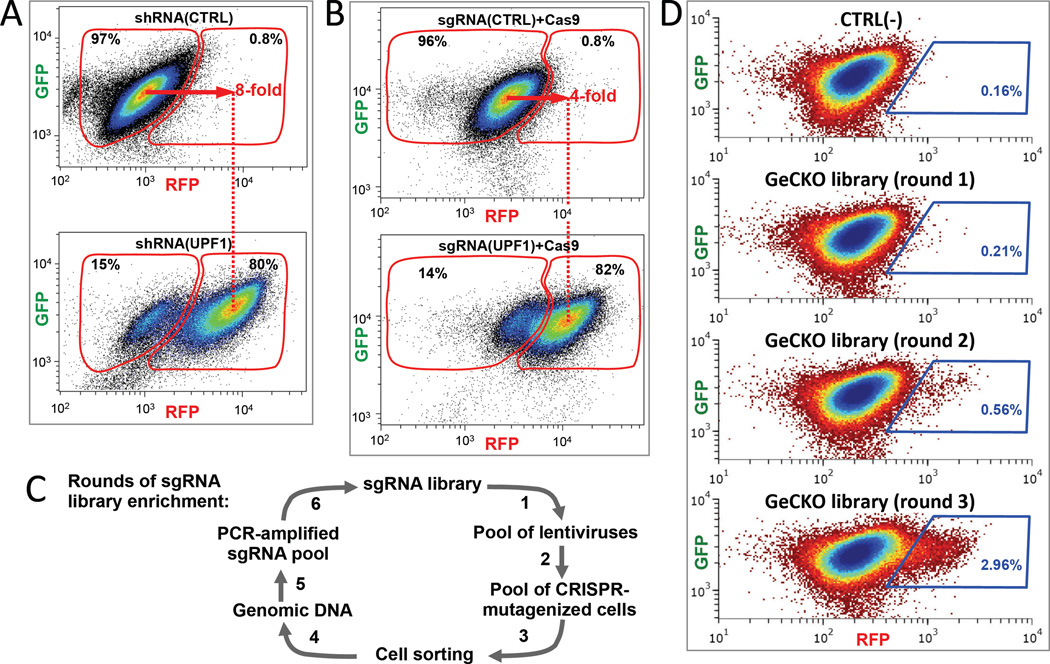
As proof of principle, we show that the fluorescence of the Fireworks cell lines faithfully increases with NMD inhibition, enabling FACS-based genetic screening for NMD factors in human cells. Each of the following modes of NMD inhibition results in 4- to 8-fold increase in fluorescence of the Fireworks cell lines: (1) inhibition via transient expression of a dominant-negative UPF1 mutant (versus UPF1(WT)) increases fluorescence 4-fold (not shown); (2) inhibition via lentiviral expression of a UPF1-targeting shRNA (versus shRNA(CTRL)) increases fluorescence 8-fold (Figure 2A); (3) inhibition via simultaneous lentiviral expression of wild-type Cas9 and an sgRNA targeting UPF1 (versus scrambled sgRNA) increases fluorescence 4-fold (Figure 2B). As expected, NMD inhibition increases fluorescence of only the PTC(+), not the PTC(−), reporter in each of the orthogonal Fireworks cell lines (Figures 2A, B, and S1D), ruling out non-specific effects. These predicted responses to NMD inhibition demonstrate that the Fireworks system can be applied to various methods of human genome interrogation, including (1) transposon-mediated dominant-negative (Landrette et al., 2011), (2) shRNA pLKO.1 knockdown library-based (Moffat et al., 2006), and (3) GeCKO-lentiCRISPR knockout library-based (Shalem et al., 2014) approaches.
Figure 2. Fireworks reporters permit rounds of efficient forward genetic screening for factors required for human NMD.
(A) NMD inhibition via expression of UPF1-targeting shRNA results in an 8- fold increase in red fluorescence of the Fireworks “green” cell line (shown in Figure 1A and the left panel of Figure 1C). (B) NMD inhibition via expression of Cas9 and UPF1-targeting sgRNA causes a 4- fold increase in red fluorescence of the Fireworks “green” cell line. (C) Schematic of sgRNA library enrichment using FACS sorting of Fireworks cells. Importantly, sgRNA-specific effects are amplified, whereas cell-specific effects are reset at the beginning of each enrichment round. (D) Enrichment of sgRNAs that affect human NMD is evident after FACS sorting of the GeCKO-Lt-CRISPR sgRNA library transduced into “green” Fireworks cells. An increase in the NMD-defective cell population is seen in the blue gates after sequential rounds of enrichment (round 1: 0.21%, round 2: 0.56%, and round 3: 2.96% of total cells).
CRISPR-based forward genetic screening for human NMD factors using Fireworks
We tested the Fireworks system with the CRIPR-based knockout screening using the lentiviral GeCKO-lentiCRISPR sgRNA library (Shalem et al., 2014), which contains 64,751 sgRNAs targeting 18,080 human genes. Ease of amplification of chromosomally-integrated sgRNAs by PCR allows multiple rounds of sgRNA library enrichment (Figure 2C) as sgRNAs are iteratively re-transduced into the original Fireworks cell line after the each round of FACS selection in a SELEX-like (Tuerk and Gold, 1990) manner. Fireworks-driven FACS throughput permits completion of one FACS cycle of genome-wide GeCKO library screening with nearly 800-fold library coverage in less than one hour. In our tests, leakage of negative control cells into the positive control cell population was below 1% (Figures 2A, B and S1D), demonstrating outstanding prospects for an unbiased genome-wide forward genetic screen. Importantly, sgRNA-specific effects are amplified, whereas cell-specific fluorescence noise is completely reset at the beginning of each sgRNA enrichment round (Figure 2C), effectively eliminating false-positives arising from spontaneous and lentiviral insertion-induced (and therefore sgRNA-independent) mutations affecting reporters or the NMD pathway. An increase in the population of cells with a defect in NMD becomes evident during FACS rounds 2 and 3 of sgRNA library enrichment, as shown in Figure 2D for the GeCKO-lentiCRISPR (Shalem et al., 2014) library in the “green” Fireworks cell line. Here, sorting gates were chosen so as to collect no more than 0.16% of the negative cell population.
CRISPR-based forward genetic screening in Fireworks cells identifies sgRNAs targeting a number of known NMD components and regulators, as well as candidate genes with possible roles in human NMD
The pool of sgRNAs isolated from round 2 (Figure 2C, D) of NMD-deficient population enrichment was deep sequenced to determine (i) the overall abundance of each sgRNA in the enriched pool and (ii) the enrichment factor for each surviving sgRNA over the starting sgRNA pool at this round. Here, the sgRNA enrichment factor serves as a measure of the increased fluorescence of the PTC(+) Fireworks reporter achieved by that sgRNA. The top 24 most abundant sgRNAs (of 64,751 sgRNAs originally present in the GeCKO library), which attained enrichment scores higher than 10, are listed in Table 1 along with their intended (Shalem et al., 2014) gene targets.
TABLE 1.
| sgRNA | sgRNA abundance1 (%) |
sgRNA enrichment2 (2nd round) |
Targeted gene | |
|---|---|---|---|---|
| 1 | TGATTACGTCCTCCACCTCG* | 1.081 | 47.0 | UPF1 |
| 2 | AGGTCCACTCACCATTGGAG | 0.888 | 113.9 | SMG7 |
| 3 | AGTGTCCTCCTTAGCTCTGC* | 0.872 | 118.7 | SMG7 |
| 4 | AGTGTCCTCCTTAGCTCTGC* | 0.440 | 33.8 | AVEN |
| 5 | GTTCTCTGATAATCAGAATG* | 0.439 | 14.7 | SMG6 |
| 6 | ACGCACCTGGTTGCTGGTAT* | 0.265 | 54.7 | SMG5 |
| 7 | GGATGACCAAGACGACATCA | 0.262 | 18.3 | SMG6 |
| 8 | CTATGAGGGGGTCAGTGACA | 0.232 | 18.3 | SMG5 |
| 9 | TGAGCACATACTCTTCACAC* | 0.180 | 102.2 | YAE1D1 |
| 10 | CCGTGACATCACAGACCCGT* | 0.176 | 21.2 | EIF2B4 |
| 11 | CGCATTGAAAACGTTTGCCG* | 0.153 | 18.4 | UPF1 |
| 12 | GGTACCTTCACTGGAGCCAC* | 0.132 | 16.1 | WDR55 |
| 13 | TATGTCTTACCAGAAGCTGC | 0.132 | 23.3 | UPF2 |
| 14 | CTCAACCGATTCCTTAGACG | 0.125 | 19.5 | SMG6 |
| 15 | TCACGCTTCACGCTGACAGT* | 0.083 | 80.0 | FARSB |
| 16 | TGCTTTGTGTTTCTTTACAC* | 0.083 | 17.0 | ZNHIT6 |
| 17 | TTAGCATCAGCTACTGCCAG* | 0.076 | 14.2 | NOP58 |
| 18 | GAAAGACTGAGGAGCTGCTG | 0.062 | 38.0 | SMG5 |
| 19 | TTCAAACTAGTAAGCAACAC* | 0.054 | 18.4 | SPATA5 |
| 20 | TAAGTTGTCAGAAAACATGA* | 0.054 | 17.1 | GAR1 |
| 21 | TCACATCGGCAGCCACCCGT* | 0.043 | 30.7 | SLMO2 |
| 22 | GCGCCCAGCTGCCAACACCA* | 0.036 | 53.9 | RPS8 |
| 23 | AGCCCATAATGTACCAGTGC | 0.033 | 14.4 | EIF2B4 |
| 24 | TACCTATGTGCGTCAGCAGC | 0.023 | 49.5 | SFI1 |
Bold type denotes known NMD factor or regulator.
sgRNAs marked with asterisks (*) were tested individually in the “green” Fireworks cell line as shown in Figures 3 and S2.
sgRNA abundance is the number of deep sequencing reads obtained for an sgRNA in the enriched (blue gates in Figure 2D) cell population divided by the total number of all reads for all sgRNAs in that population and multiplied by 100.
sgRNA enrichment is the number of deep sequencing reads obtained for an sgRNA in the enriched cell population divided by the number of deep sequencing reads obtained for this sgRNA in total cells at the beginning of the 2nd (Figure 2D) enrichment round.
Of these top 24 highly-enriched sgRNAs, 13 target well-established NMD factors (Kurosaki and Maquat, 2016): UPF1 (2 out of 4 sgRNAs present in the library [Table 1, rows 1 and 11]), SMG7 (2 out of 4 sgRNAs [rows 2 and 3]), SMG6 (3 out of 4 sgRNAs [rows 5, 7, and 14]), SMG5 (3 out of 6 sgRNAs [rows 6, 8, and 18]), UPF2 (1 out of 4 sgRNAs [row 13]), and a known human NMD regulator EIF2B4 (Gardner, 2008; Martin et al., 2010) (2 out of 5 sgRNAs [rows 10 and 23]) (Table 1). We therefore demonstrate successful unbiased identification of NMD factors and regulators in a forward genetic screen performed directly in human cells. Additionally, Table 1 contains 11 highly enriched sgRNAs targeting the following genes: AVEN, YAE1D1, WDR55, FARSB, ZNHIT6, NOP58, SPATA5, GAR1, SLMO2, RPS8, and SFI1; their potential NMD involvement is yet to be determined.
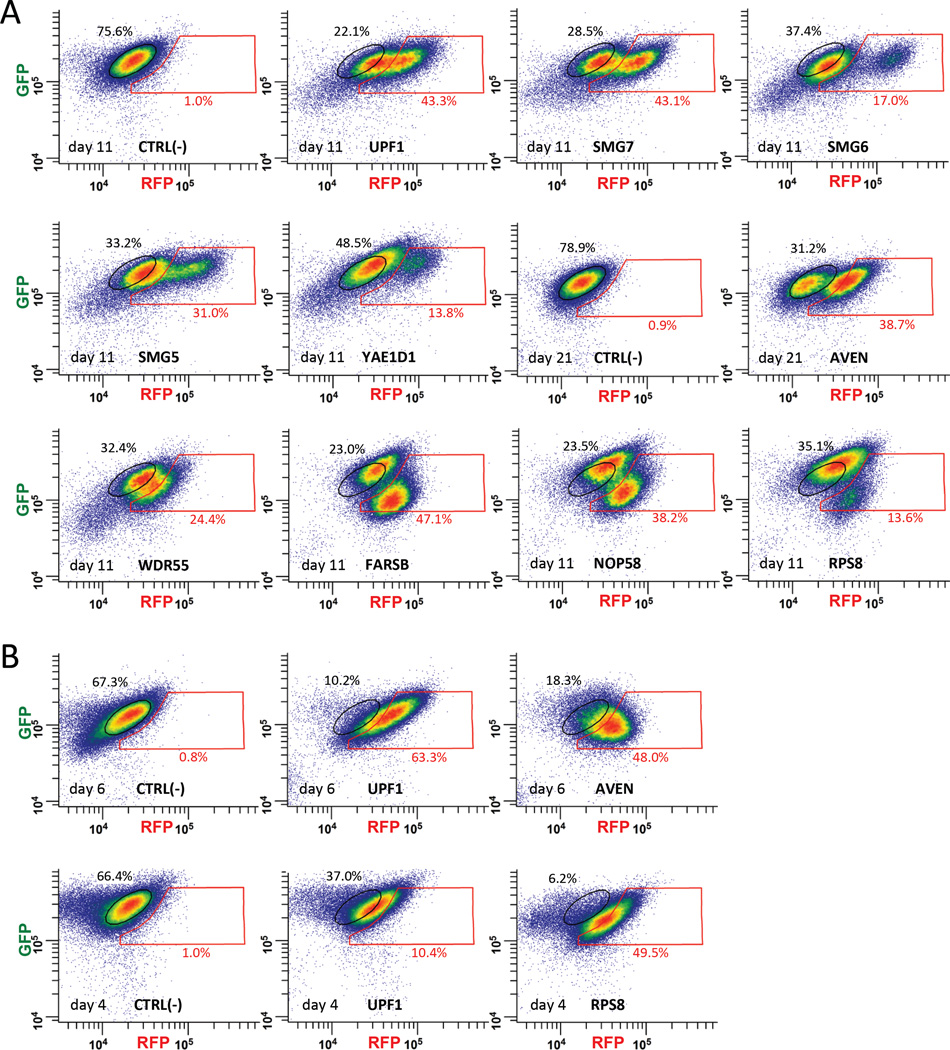
We reproduced the results of the genome-wide screen by individually transducing sgRNAs targeting 15 of the genes listed in Table 1 into the “green” Fireworks cell line and FACS-analyzing the fluorescence of the resulting cells. As shown in Figures 3A and S2A, cell sub-populations (albeit sometimes small) with increased fluorescence of the PTC-containing RFP reporter were observed for sgRNAs targeting each of the following genes: UPF1, SMG7, AVEN, SMG6, SMG5, YAE1D1, EIF2B4, WDR55, FARSB, ZNHIT6, NOP58, SPATA5, GAR1, SLMO2, and RPS8, demonstrating the high quality of our approach for FACS-based pooled sgRNA library enrichment. Whereas individually-tested sgRNAs produced varying degrees of increase in fluorescence of the PTC-containing RFP reporter, these sgRNAs can be broadly divided into two categories based on their effects on fluorescence of the PTC-lacking GFP control (Figures 3A and S2A). SgRNAs of the 1st category (like those targeting UPF1, SMG7, AVEN, SMG6, SMG5, YAE1D1, SPATA5, and ZNHIT6) shift the CRISPR-affected population of “green” Fireworks cells right and, in some cases, slightly up, reflecting increased fluorescence of the PTC-containing RFP reporter and largely unaffected fluorescence of the PTC-lacking internal GFP expression control. SgRNAs of the 2nd category (like those targeting EIF2B4, WDR55, FARSB, NOP58, GAR1, and RPS8) shift the CRISPR-affected cell population right and downward, reflecting an increase in fluorescence of the PTC-containing RFP reporter and simultaneous decrease in fluorescence of the PTC-lacking internal GFP expression control. Whereas both categories contain sgRNAs targeting well-established bona fide NMD factors or regulators, concurrent decreases in fluorescence of the PTC-lacking GFP control produced by sgRNAs of the 2nd category suggest complexity of their effects or, possibly, negative effects on general protein translation. We excluded potential off-target effects of sgRNAs on FP fluorescence for two of the genes, AVEN and RPS8, by targeting them with shRNAs. Unlike CRISPR-Cas9 guide RNAs (sgRNAs), which produce multiple diverse mutations at the targeted locus that result in widely varying degrees of NMD inhibition in individual cells, shRNAs do not produce mutations. Instead, shRNAs affect the level of the targeted transcript in transduced cells, resulting in a uniform fluorescence shift for the entire population of shRNA-transduced cells. FACS-analyzed fluorescence of the resulting Fireworks cell lines showed (Figures 3B and S2B) that shRNAs correctly reproduce the predicted fluorescence increase for PTC(+) but not PTC(−) reporters, eliminating the possibility of off-target sgRNA effects for these genes.
Figure 3. FACS analysis of the fluorescence shift produced by individual sgRNAs identified by Fireworks screening of the GeCKO-LtCRISPR sgRNA library for factors affecting human NMD.
(A) sgRNAs obtained from the genome-wide screen (Table 1) were individually transduced into the “green” Fireworks cell line (Figure 1A) and the resulting cells were FACS-analyzed for an increase in red fluorescence. Populations of cells with increased red fluorescence (nearly not observable [0.9–1.0%] in the corresponding negative controls) appear in the red gate. Fractions (percent) of cells in the original (black gates) and shifted (red gates) populations are indicated. (B) To exclude possible off-target effects of sgRNAs, two of the genes not previously implicated in NMD, AVEN and RPS8, were targeted by shRNAs. FACS-analyses of the shRNA-transduced “green” Fireworks cell line are shown; populations of cells with increased red fluorescence appear in the red gate (FACS-analysis of the orthogonal “red” Fireworks cell line transduced with shRNA targeting RPS8 is shown in Figure S2B). See also Figure S2.
DISCUSSION
Since the discovery of the destabilizing effect of certain β-thalassemia mutations on spliced β-globin mRNAs in the early 1980s (Maquat et al., 1981), the NMD pathway has been studied extensively, with more than 1,800 PubMed publications to date. However, the scope of factors comprising the human NMD pathway has not yet been determined because of the technological difficulties of performing forward genetic interrogation of mRNA degradation pathways in human cells.
Our Fireworks method successfully solves the problems that have hindered forward genetic identification of human mRNA degradation factors by providing a powerful experimental system to interrogate human NMD and mechanisms of its regulation. In just two iterative in vivo SELEX-like rounds of sgRNA library enrichment (Figure 2C, D), our method correctly identified 5 major known human NMD components (UPF1, UPF2, SMG5, SMG6, and SMG7), a known human NMD regulator (EIF2B4), and 11 candidate genes. Finding that more than half (13 out of 24) of the top-scoring sgRNAs (Table 1) in our genetic screen target known human NMD components and modulators provides excellent validation of the method. The fact that for each of the aforementioned known NMD components (except UPF2) the method identified two or more targeting sgRNAs (Table 1) – out of, on average, four sgRNAs per gene targeted by the original GeCKO-LentiCRISPR knockout library – adds additional credibility.
Since PTC recognition depends on ongoing translation, interference with protein synthesis readily suppresses NMD (Belgrader et al., 1993) predicting that numerous components/regulators of translation machinery might overwhelmingly dominate an NMD screen. However, this is not the case for Fireworks (Table 1) because, by design, Fireworks assesses NMD inhibition by measuring the absolute increase (Figure 2D) in fluorescence (i.e. absolute increase in protein levels) of the PTC(+) reporter. In contrast, inhibition of protein synthesis is expected to decrease protein levels, including those of PTC(+) and PTC(−) reporters. Inhibition of protein synthesis therefore results in diminished increase in PTC(+) reporter fluorescence and lower FACS enrichment scores as compared to those resulting from inactivation of bona fide NMD factors, reducing the number of translation components in the Fireworks screen.
Since potential interference with protein synthesis is readily reflected by decrease in fluorescence of the PTC(−) Fireworks reporter, a sizeable fraction of individually validated sgRNAs does not display noticeable defects in general protein translation (Figures 3A and S2A). For those sgRNAs that do decrease fluorescence of the control PTC(−) reporter, complex effects are possible, including NMD impairment in addition to or as a consequence of inhibition of general protein synthesis. Indeed, some of the genes obtained in the screen are known to act in translation: NOP58 and GAR1 are components of snoRNPs of the two different classes, C/D and H/ACA, involved in rRNA modifications; RPS8 is an essential constituent of the small (40S) subunit of the human ribosome; FARSB is a regulatory subunit of the only human phenylalanyl-tRNA synthetase; and EIF2B4 is one of the five subunits of a GTP exchange factor EIF2B (Eukaryotic Initiation Factor 2B), necessary for protein synthesis and known to be involved in regulation of the human NMD (Gardner, 2008; Martin et al., 2010). The candidate genes, obtained in the screen, must therefore be (i) tested to determine whether the increase in fluorescence of the PTC-containing reporter produced by an sgRNAs is solely due to a PTC-dependent decrease in mRNA stability, (ii) inspected for possible primary and secondary effects on stop codon recognition, and (iii) evaluated for possible roles in translation, considering potential effects of translation inhibition on NMD. Only then should prospective mechanistic roles in the NMD pathway be considered for these genes.
Undoubtedly, the list of human NMD factors identified in this screen is far from complete. In the currently employed GeCKO-LentiCRSPR sgRNA library (Shalem et al., 2014) certain known NMD components (for example, SMG1) are targeted by only one sgRNA, decreasing the likelihood of their identification. Additionally, since only established genes are targeted by this library, unknown, undefined human genes and genomic elements escape interrogation. One could, therefore, envisage more detailed genetic screening strategies based on larger, more comprehensive sgRNA libraries to identify additional human NMD components and regulators. Since the FACS-driven throughput of Fireworks permits screening for NMD-affecting mutations at a rate approaching 50,000,000 mutants per hour (up to a billion mutants per day), our Fireworks approach is not limited by screening rate. SgRNA libraries several orders (!) of magnitude larger than the one employed in this study could potentially be rapidly processed, if such libraries existed. Not only do sgRNAs vary in their efficiency of producing genomic mutations, but not all successfully produced genomic mutations result in bi-allelic gene inactivation (e.g., silent mutations or analogous amino acid substitutions may only partially, if at all, affect function of a gene’s product). Therefore, substantially larger libraries combined with more comprehensive screens containing higher numbers of sgRNA enrichment rounds hold significant potential for identification of novel human NMD components. Additionally, libraries can be designed to deliberately lack sgRNAs targeting known NMD factors. Screening such libraries should be more sensitive to new, previously unknown NMD components and regulators since their (presumably moderate) effects on NMD will not be masked by strong effects of knockouts of known NMD factors.
The remarkable brightness of the Fireworks reporters underscores their potential use with other disease-related PTC-containing mRNAs that are naturally much less abundant than β-globin mRNA. The system can be easily extended to genetic identification of mRNA-destabilizing factors in pathways other than NMD, such as those affecting mRNA stability via 5′- and 3′-UTR sequences. Since Fireworks-CRISPR screening does not require specialized haploid human cell lines (the current Fireworks HeLa cells are diploid/polyploid), forward genetic identification of tissue-specific mechanisms underlying mRNA stability is also possible. Though in this work we applied the Fireworks approach to screen for genes involved in human NMD, Firework’s main significance lies in its broad applicability to screening a variety of other human cellular pathways whose readout can be adjusted to result in fluorescent protein expression. Adaptations of the Fireworks approach are therefore poised to provide powerful forward genetic tools for a variety of different fields of human biology.
Limitations
Known Fireworks constraints mainly result from the currently employed methods of CRISPR-based genetic interrogation and FACS-driven screening as well as from the nature of the reporters and cell lines: (1) Since loss of function-based forward genetic approaches (including CRISPR-based approaches) have inherent difficulty in identifying components that possess counterparts acting redundantly in the same pathway, Fireworks exhibits this limitation. (2) The fact that CRISPR-based screening commonly requires cell lines that are easy to expand and inexpensive to propagate may limit the scope of new cell lines suitable for Fireworks-based genetic interrogation of NMD and other pathways. (3) Since our present Fireworks HeLa cell lines are diploid/polyploid, the probability of inactivation of all alleles of a targeted gene by CRISPR-Cas9-introduced mutations may be lower than that for haploid cell lines, resulting in lower (but, as we show here, entirely suitable for successful genetic screening) knockout efficiency. (4) For FACS-driven screening, cell lines should be able to form single-cell suspensions either in culture or upon trypsinization and withstand the stress of cell sorting. (5) Whereas we do not observe negative effects of transgenic expression of the TEV protease on HeLa cell growth or morphology, unanticipated effects cannot be ruled out. (6) Construction of new mammalian cell lines, including chromosomal insertion of FRT sites, FRT sites selection, and insertion of Fireworks reporters via FRT-Flp cassette exchange may require significant time and effort. (7) Finally, as for any new observation obtained using a reporter system in stable cell lines, results obtained in Fireworks cells must be independently validated using complementary experimental approaches.
EXPERIMENTAL PROCEDURES
Generation of Fireworks cell lines for the genetic screen
To obtain HeLa cell lines that minimally silence Fireworks reporters, we constructed a lentiviral FRT-flanked GFP cassette named Launchpad (AVA2590) and produced (using lentiviral transduction) a pool of HeLa cells carrying 1–2 copies of this cassette at various genomic locations. Propagated populations of these cells were FACS-selected for stable GFP fluorescence. The GFP-containing FRT cassette was then removed (using transient transfection of Flp recombinase) and exchanged for the PTC-lacking red Fireworks reporter (AVA2515) (Figure S3). Cells were subsequently FACS-selected for stable RFP expression and subjected to Flp recombinase-mediated cassette integration/exchange, as shown in Figure S3, resulting in “green” (RFP[PTC(+)], GFP[PTC(−)]) and “red” (RFP[PTC(−)], GFP[PTC(+)]) Fireworks cell lines.
Plasmid construction
Tandemly arranged DNA sequences of GFP and tdTomato were assembled using redundant DraIII restriction sites; the sequence of each individual FP within the repeat was verified by sequencing using DraIII sites as “bar codes” for annealing FP-specific sequencing primers. Plasmids with tandemly arranged FPs were routinely propagated in the XL1Blue strain of E. Coli; no deleterious effects of DNA recombination were observed for Fireworks reporters in this cell line. The first intron of the human β-globin gene was precisely removed using QuickChange to prevent skipping of β-globin exon 2, while the second intron was kept intact. Plasmids and plasmid sequences of the Launchpad (AVA2590) vector, PTC-lacking red (AVA2515) and green (AVA2598) Fireworks reporters, and PTC-containing red (AVA2626) and green (AVA2600) Fireworks reporters are deposited in and available from the Addgene plasmid repository (see also Supplemental Information: Supplemental vector sequences).
Forward genetic screen for NMD factors in Fireworks cells
1.8×108 (6 × 15 cm plates) “green” Fireworks cells were lentivirally transduced with the blasticidin-resistant GeCKO-LtCRISPR viral library (Shalem et al., 2014) using 4.0 µg/ml of polybrene and propagated for 8–9 days in DMEM media supplemented with 10% FBS and 120 µg/ml hygromycin, 0.3 µg/ml puromycin and 3.0 µg/ml blasticidin. NMD-deficient cell populations (2×105 cells) were isolated from 4×108 transduced cells using a Bio-Rad S3e cell sorter (100 µm nozzle) and their genomic DNA was phenol-extracted. sgRNAs pools were PCR-amplified using RandomF (5’-TAACTTGAAAGTATTTCGATTTCTTGGCTTTATATATCTTGTGGAAAGGACGAAACACCG-3’) and RandomR (5’-ACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3’) primers and Gibson-cloned into the BsmBI-inearized LentiCRISPR (Shalem et al., 2014) vector to obtain a lentiviral library (represented by 1.5×108 independent transformants) for the next enrichment round (Figure 2C). Three rounds of GeCKO Lt-CRISPR library enrichment were conducted; 1.4×108 and 1.2×108 cells were sorted in the 2nd and 3rd rounds, respectively. sgRNAs obtained from the 2nd round were deep-sequenced and ranked (Table 1) according to their (i) overall abundance in the enriched sgRNA pool and (ii) enrichment over the starting sgRNA population in this round (since successful enrichment increases both of these numbers).
16 of the most abundant individual sgRNAs (Table 1), whose enrichment scores in the 2nd round exceeded 10.0, were individually cloned into linearized Lt-CRISPR vector and lentivirally transduced into the “green” Fireworks cell line. Resulting cells were propagated for 11 days (or 21 days for AVEN sgRNA) and FACS-analyzed to observe an increase in red fluorescence of the PTC(+) Fireworks reporter. Indeed, the vast majority of sgRNAs reproduced the shift when tested individually. RPS8 and AVEN were targeted with shRNAs (sh22: 5’CCGGCCGTGCCCTGAGGTTGGACGTCTCGAGACGTCCAACCTCAGGGCACGGTTTTTTG3’ and sh01: 5’CCGGGACCTGAAATCCAAGGAAGATCTCGAGATCTTCCTTGGATTTCAGGTCTTTTTTG3’ for RPS8 and AVEN, respectively) cloned into the pLKO.1 vector (Moffat et al., 2006) that was used to transduce “green” and “red” Fireworks cells followed by FACS-analysis to observe effects of mRNA knockdown on fluorescence of the PTC(+) reporters.
Supplementary Material
Acknowledgments
K. Tycowski provided critical comments on the manuscript. This work was supported by grants GM026154 to J.A.S. and NS091637 to A.A. from the National Institutes of Health. J.A.S is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
A.A. and J.A.S. designed the studies and analyzed the data. A.A. performed and M-D.S assisted with performing the experiments. A.A and J.A.S. wrote the manuscript.
ACCESSION NUMBERS
Addgene (http://www.addgene.org) IDs of the Launchpad and Fireworks constructs reported in this paper are 85442–85446. All deep sequencing data have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) and can be found with the BioProject accession number PRJNA353310, study accession number SRP093318.
BIBLIOGRAPHY
- Alexandrov A, Colognori D, Shu MD, Steitz JA. Human spliceosomal protein CWC22 plays a role in coupling splicing to exon junction complex deposition and nonsense-mediated decay. Proc Natl Acad Sci U S A. 2012;109:21313–21318. doi: 10.1073/pnas.1219725110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Barbosa I, Haque N, Fiorini F, Barrandon C, Tomasetto C, Blanchette M, Le Hir H. Human CWC22 escorts the helicase eIF4AIII to spliceosomes and promotes exon junction complex assembly. Nat Struct Mol Biol. 2012;19:983–990. doi: 10.1038/nsmb.2380. [DOI] [PubMed] [Google Scholar]
- Belgrader P, Cheng J, Maquat LE. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc Natl Acad Sci U S A. 1993;90:482–486. doi: 10.1073/pnas.90.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadio A, Longman D, Hug N, Delavaine L, Vallejos Baier R, Alonso CR, Caceres JF. Identification and characterization of novel factors that act in the nonsense-mediated mRNA decay pathway in nematodes, flies and mammals. EMBO Rep. 2015;16:71–78. doi: 10.15252/embr.201439183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Damoiseaux R, Nahas S, Gao K, Hu H, Pollard JM, Goldstine J, Jung ME, Henning SM, Bertoni C, et al. Nonaminoglycoside compounds induce readthrough of nonsense mutations. J Exp Med. 2009;206:2285–2297. doi: 10.1084/jem.20081940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Jung ME, Damoiseaux R, Completo G, Fike F, Ku JM, Nahas S, Piao C, Hu H, Gatti RA. A new series of small molecular weight compounds induce read through of all three types of nonsense mutations in the ATM gene. Mol Ther. 2013;21:1653–1660. doi: 10.1038/mt.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel RS, Flanigan KM, Wong B, Bonnemann C, Sampson J, Sweeney HL, Reha A, Northcutt VJ, Elfring G, Barth J, et al. Phase 2a study of ataluren-mediated dystrophin production in patients with nonsense mutation Duchenne muscular dystrophy. PLoS One. 2013;8:e81302. doi: 10.1371/journal.pone.0081302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner LB. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol Cell Biol. 2008;28:3729–3741. doi: 10.1128/MCB.02284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Hilarion S, Beghyn T, Jia J, Debreuck N, Berte G, Mamchaoui K, Mouly V, Gruenert DC, Deprez B, Lejeune F. Rescue of nonsense mutations by amlexanox in human cells. Orphanet J Rare Dis. 2012;7:58. doi: 10.1186/1750-1172-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, Dieterich C, Landthaler M. MOV10 Is a 5' to 3' RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3' UTRs. Mol Cell. 2014;54:573–585. doi: 10.1016/j.molcel.2014.03.017. [DOI] [PubMed] [Google Scholar]
- Gunn G, Dai Y, Du M, Belakhov V, Kandasamy J, Schoeb TR, Baasov T, Bedwell DM, Keeling KM. Long-term nonsense suppression therapy moderates MPS I-H disease progression. Mol Genet Metab. 2014;111:374–381. doi: 10.1016/j.ymgme.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurskaya NG, Pereverzev AP, Staroverov DB, Markina NM, Lukyanov KA. Analysis of nonsense-mediated mRNA decay at the single-cell level using two fluorescent proteins. Methods Enzymol. 2016;572:291–314. doi: 10.1016/bs.mie.2016.02.008. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004;36:801–808. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- Keeling KM, Wang D, Dai Y, Murugesan S, Chenna B, Clark J, Belakhov V, Kandasamy J, Velu SE, Baasov T, et al. Attenuation of nonsense-mediated mRNA decay enhances in vivo nonsense suppression. PLoS One. 2013;8:e60478. doi: 10.1371/journal.pone.0060478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling KM, Xue X, Gunn G, Bedwell DM. Therapeutics based on stop codon readthrough. Annu Rev Genomics Hum Genet. 2014;15:371–394. doi: 10.1146/annurev-genom-091212-153527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerem E, Konstan MW, De Boeck K, Accurso FJ, Sermet-Gaudelus I, Wilschanski M, Elborn JS, Melotti P, Bronsveld I, Fajac I, et al. Ataluren for the treatment of nonsense-mutation cystic fibrosis: a randomised, double-blind, placebo-controlled phase 3 trial. Lancet Respir Med. 2014;2:539–547. doi: 10.1016/S2213-2600(14)70100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Maquat LE. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci. 2016;129:461–467. doi: 10.1242/jcs.181008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrette SF, Cornett JC, Ni TK, Bosenberg MW, Xu T. piggyBac transposon somatic mutagenesis with an activated reporter and tracker (PB-SMART) for genetic screens in mice. PLoS One. 2011;6:e26650. doi: 10.1371/journal.pone.0026650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Lee NC, Kononenko AV, Lee HS, Tolkunova EN, Liskovykh MA, Masumoto H, Earnshaw WC, Tomilin AN, Larionov V, Kouprina N. Protecting a transgene expression from the HAC-based vector by different chromatin insulators. Cell Mol Life Sci. 2013;70:3723–3737. doi: 10.1007/s00018-013-1362-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Longman D, Plasterk RH, Johnstone IL, Caceres JF. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev. 2007;21:1075–1085. doi: 10.1101/gad.417707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16:665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- Maquat LE, Kinniburgh AJ, Rachmilewitz EA, Ross J. Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell. 1981;27:543–553. doi: 10.1016/0092-8674(81)90396-2. [DOI] [PubMed] [Google Scholar]
- Martin L, Grigoryan A, Wang D, Wang J, Breda L, Rivella S, Cardozo T, Gardner LB. Identification and characterization of small molecules that inhibit nonsense-mediated RNA decay and suppress nonsense p53 mutations. Cancer Res. 2014;74:3104–3113. doi: 10.1158/0008-5472.CAN-13-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L, Kimball SR, Gardner LB. Regulation of the unfolded protein response by eif2Bdelta isoforms. J Biol Chem. 2010;285:31944–31953. doi: 10.1074/jbc.M110.153148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain DR, Pan Q, Reilly PT, Elia AJ, McCracken S, Wakeham AC, Itie-Youten A, Blencowe BJ, Mak TW. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc Natl Acad Sci U S A. 2010;107:12186–12191. doi: 10.1073/pnas.1007336107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- Metzstein MM, Krasnow MA. Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet. 2006;2:e180. doi: 10.1371/journal.pgen.0020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Mort M, Ivanov D, Cooper DN, Chuzhanova NA. A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat. 2008;29:1037–1047. doi: 10.1002/humu.20763. [DOI] [PubMed] [Google Scholar]
- Nickless A, Jackson E, Marasa J, Nugent P, Mercer RW, Piwnica-Worms D, You Z. Intracellular calcium regulates nonsense-mediated mRNA decay. Nat Med. 2014;20:961–966. doi: 10.1038/nm.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomakuchi TT, Rigo F, Aznarez I, Krainer AR. Antisense oligonucleotide-directed inhibition of nonsense-mediated mRNA decay. Nat Biotechnol. 2016;34:164–166. doi: 10.1038/nbt.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillusson A, Hirschi N, Vallan C, Azzalin CM, Muhlemann O. A GFP-based reporter system to monitor nonsense-mediated mRNA decay. Nucleic Acids Res. 2005;33:e54. doi: 10.1093/nar/gni052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks TD, Howard ED, Wolpert TJ, Arp DJ, Dougherty WG. Expression and purification of a recombinant tobacco etch virus NIa proteinase: biochemical analyses of the full-length and a naturally occurring truncated proteinase form. Virology. 1995;210:194–201. doi: 10.1006/viro.1995.1331. [DOI] [PubMed] [Google Scholar]
- Pereverzev AP, Gurskaya NG, Ermakova GV, Kudryavtseva EI, Markina NM, Kotlobay AA, Lukyanov SA, Zaraisky AG, Lukyanov KA. Method for quantitative analysis of nonsense-mediated mRNA decay at the single cell level. Sci Rep. 2015;5:7729. doi: 10.1038/srep07729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlick HA, Medghalchi SM, Spencer FA, Kendzior RJ, Jr, Dietz HC. Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proc Natl Acad Sci U S A. 1996;93:10928–10932. doi: 10.1073/pnas.93.20.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab JR, Chiu J, Zhu J, Katzman S, Kurukuti S, Wade PA, Haussler D, Kamakaka RT. Human tRNA genes function as chromatin insulators. EMBO J. 2012;31:330–350. doi: 10.1038/emboj.2011.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Roy B, Friesen WJ, Tomizawa Y, Leszyk JD, Zhuo J, Johnson B, Dakka J, Trotta CR, Xue X, Mutyam V, et al. Ataluren stimulates ribosomal selection of near-cognate tRNAs to promote nonsense suppression. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1605336113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski PD. The Flp recombinase of the 2-microns plasmid of Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol. 1995;51:53–91. [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckelberg AL, Boehm V, Gromadzka AM, Gehring NH. CWC22 connects pre-mRNA splicing and exon junction complex assembly. Cell Rep. 2012;2:454–461. doi: 10.1016/j.celrep.2012.08.017. [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Usuki F, Yamashita A, Shiraishi T, Shiga A, Onodera O, Higuchi I, Ohno S. Inhibition of SMG-8, a subunit of SMG-1 kinase, ameliorates nonsense-mediated mRNA decay-exacerbated mutant phenotypes without cytotoxicity. Proc Natl Acad Sci U S A. 2013;110:15037–15042. doi: 10.1073/pnas.1300654110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sun X, Qian Y, Maquat LE. Intron function in the nonsense-mediated decay of beta-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.