Abstract
Separase has a critical role in dissolving the cohesion among sister chromatids during chromosome segregation 1–7. Separase is over-expressed in human tumors, making it a potential target for drug discovery 8. The protease activity of separase is strictly regulated by the inhibitor securin, which forms a tight complex with separase and may also stabilize this enzyme 9–16. Separases are large, 140–250 kD enzymes, with an N-terminal α-helical region and a caspase-like catalytic domain (CD) at the C-terminus. While crystal structures of the C-terminal two domains of separase 17 and low-resolution electron microscopy reconstructions of the separase-securin complex 18,19 have been reported, the atomic structures of full-length separase and especially the complex with securin are not known. Here we report crystal structures at up to 2.6 Å resolution of the yeast Saccharomyces cerevisiae separase-securin complex. The α-helical region of separase (also known as Esp1) contains four domains (I–IV), and a substrate-binding domain (SD) immediately precedes the CD and has tight associations with it. The separase-securin complex assumes a highly elongated structure. Residues 258–373 of securin (Pds1), named the separase interaction segment (SIS), is primarily in an extended conformation and traverses the entire length of separase, having interactions with all of its domains. Most importantly, residues 258–269 of securin are located in the separase active site, illuminating its mechanism of inhibition. Biochemical studies confirm the structural observations and indicate that contacts outside the separase active site are crucial for stabilizing the complex, thereby defining an important function for the helical region of separase.
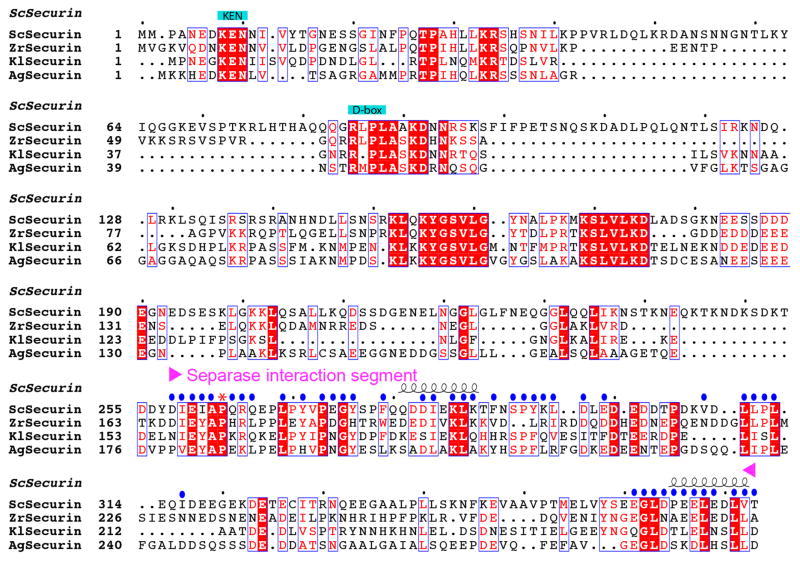
Saccharomyces cerevisiae separase contains 1630 amino acid residues and the N-terminal α-helical region covers ~1160 residues. We obtained good quality crystals for the complex of S. cerevisiae separase (residues 51–1630; Fig. 1a, Extended Data Figs. 1–3) and securin (residues 258–373; Extended Data Fig. 4). The N-terminal segment of securin contains the KEN- and D-box motifs, which are important for the destruction of securin but not for interaction with separase 20. The structure was determined and refined at 3.0 Å resolution (Extended Data Table 1). Under the same crystallization condition, we observed another crystal form and collected a data set to 3.7 Å resolution. We readily solved the structure of this crystal form and did not observe any significant differences.
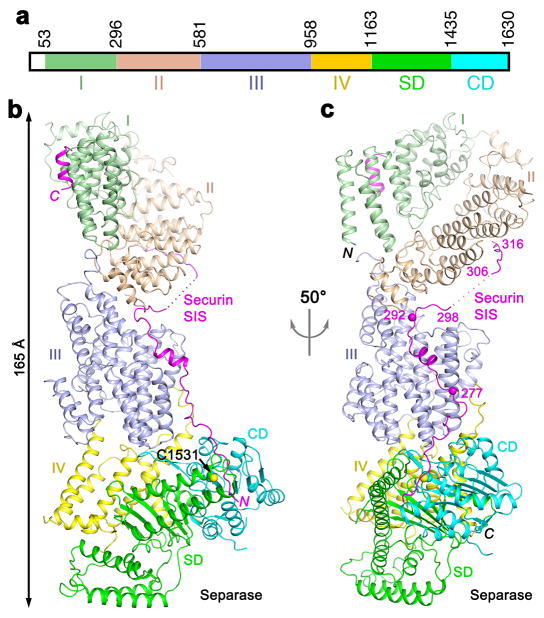
Figure 1.
Crystal structure of the yeast separase-securin complex. (a). Domain organization of S. cerevisiae separase. The domains are labeled and given different colors. (b). Overall structure of the yeast separase-securin complex. The domains in separase are colored according to panel a, and the securin SIS is in magenta. The side chain of the catalytic Cys1531 residue of separase is shown as a sphere model. (c). Overall structure of the complex viewed after a 50° rotation around the vertical axis. Two of the phosphorylation sites in the securin SIS (Ser277 and Ser292) 29,30 are indicated with the spheres and labeled. The structure figures were produced with PyMOL (www.pymol.org).
We produced crystals of another complex of S. cerevisiae separase (residues 71–1630) and securin (residues 258–373), and determined its structure at 2.6 Å resolution. The overall structure of this complex is essentially the same as that at 3.0 Å resolution (Extended Data Fig. 5). One difference is that residues 75–79 at the N-terminal end of separase form an anti-parallel β-sheet with the equivalent residues of another separase molecule in the crystal (Extended Data Fig. 5), which is an artifact due to the longer truncation of separase in this complex.
The overall structure of the separase-securin complex has a highly elongated shape, with dimensions of 65 Å × 70 Å × 165 Å (Figs. 1b, 1c). The CD is located at one end of the complex, far away from the N-terminus of separase at the other end. This elongated shape is generally consistent with that observed for the human separase-securin complex by EM 18, but distinct from a closed form reported for the C. elegans complex 19.
The structure shows that the N-terminal helical region (residues 53–1163) of separase can be divided into four domains (I–IV) (Figs. 1a–c). Their structures are generally like HEAT repeats, with extensions that connect each domain to the next one. Domain I is folded back onto domain II such that the loops at one end of each domain are facing each other (Fig. 2a), with a buried surface area of 1,100 Å2 for each domain, suggesting that this may be a stable association between them. Two consecutive helices (the fourth and fifth helices) in domain I are parallel to each other (Fig. 2a, Extended Data Fig. 6). The first 50 residues of separase are poorly conserved (Extended Data Fig. 1). Earlier studies showed that the first 155 residues of separase are important for function 12. These residues include helices α1 to α5 of domain I. Their deletion would disrupt the structure of this domain and its interaction with domain II (Fig. 2a), and helix α3 is in the binding site for the C-terminal helix of securin (see below), thereby explaining their functional importance. Human separase is stimulated by DNA of greater than 100bp 21, and this stimulation is likely also mediated by the helical region.
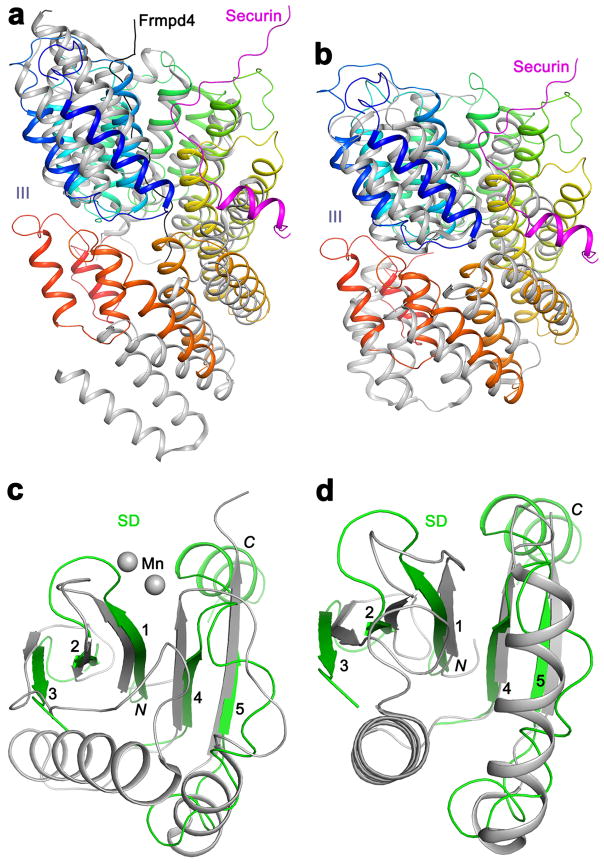
Figure 2.
Structures of the domains in separase. (a). Structure of domains I and II of separase. The directions of helices 4 and 5 in domain I are indicated with the red arrows. (b). Structure of domain III of separase. The helices are colored ramping from blue at the N-terminus to red at C-terminus. (c). Structure of domains IV, SD and CD of separase, and overlay of the structure of the SD-CD of C. thermophilum separase (gray) 17. The active site is indicated with the red asterisk. The red arrowhead indicates the region where the β4A-β4B hairpin has a different conformation and is partly disordered in C. thermophilum separase, and the purple box highlights the region where domain IV provides an extra strand (labeled 6) to the β-sheet of SD.
The helical hairpins of domain III are arranged in a tight, right-handed super-helix (Fig. 2b). This domain is located in the middle of the structure and has contacts with domain II at one end (buried surface area of ~900 Å2) and domain IV at the other (~1,100 Å2) (Fig. 1b). It also has a direct contact with a segment immediately following the catalytic Cys1531 residue in the CD (strands β4A, β4B and the connecting loop, Fig. 2c, Extended Data Figs. 3, 6). The overall arrangement of the helices in this domain has similarity to that in several other structures based on a DaliLite search 22, and the closest homologs include the TPR domain of the G-protein signaling modulator LGN 23–25 and others (Extended Data Fig. 7).
Domain IV has extensive interactions with the domain immediately prior to the CD (SD, Fig. 1a; see next), covering one face of its β-sheet (buried surface area of ~1,900 Å2 for each domain) and providing one extra strand to this β-sheet (Fig. 2c). It also interacts with two of the surface helices of the CD. Therefore, domain IV could be considered as forming a module together with the SD-CD.
An α+β domain is located just prior to the CD in separases 18,26. The backbone fold of this domain has no similarity to caspases 17 or other proteases. The domain is tightly associated with the CD and contributes to substrate binding. We will refer to it as the substrate-binding domain (SD) here (Fig. 1a). Other names for the SD and CD include PPD (pseudo-protease domain) and APD (active protease domain), respectively 17. The SD contains a five-stranded (β1-β5) mostly anti-parallel β-sheet, and there is a large insertion (residues 1200–1380) between the third and fourth strands, including a hairpin of two long helices (αA-αB) and a four-helical bundle (αC-αF) (Fig. 2c). The backbone fold of this β-sheet is RNase H-like, with close homologs including a part of the catalytic PIWI domain of Argonaute 27 and others (Extended Data Fig. 7).
The overall structure of the SD-CD of S. cerevisiae separase is similar to that of the C. thermophilum separase 17 (Fig. 2c), with rms distance of 1.5 Å for their equivalent Cα atoms and amino acid sequence identity of 39%, suggesting that binding of securin did not cause a large overall conformational change in these two domains. The β-sheets of the two domains are juxtaposed in the structure, although the two neighboring strands at the interface (β5 of SD and β6 of CD) are at an angle of nearly 90°. The αA-αB hairpin from SD covers a portion of the active site in CD and helps mediate substrate binding.
Securin is primarily in an extended conformation, with only two short helices, and runs along the entire length of separase (Figs. 1b, 3a). We will refer to this segment of securin, residues 258–373, as the separase interaction segment (SIS). The backbone direction of SIS is anti-parallel to that of separase, such that the N-terminal region of SIS interacts with the C-terminal region of separase. The securin SIS has contacts with every domain of separase, being positioned in prominent grooves in its surface (Fig. 3a). Approximately 4,600 Å2 of the surface area of securin is buried in the interface with separase (~3,900 Å2 surface area burial), indicating the tight association and the stability of this complex.
Figure 3.
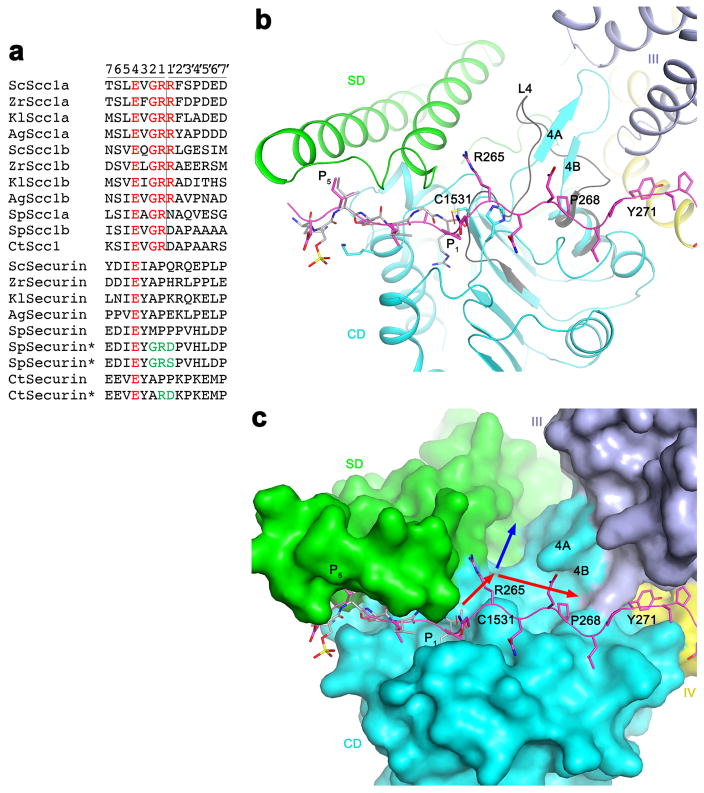
Interactions between separase and securin. (a). Molecular surface of separase, colored by the domains. The active site of separase is indicated with the red asterisk. The view is same as Fig. 1c. (b). Omit Fo–Fc electron density at 3.0 Å resolution for residues 258–269 of securin, contoured at 2σ. (c). Interactions between residues 258–265 of securin SIS (magenta) with the active site of separase. The side chains of residues in the interface are shown as stick models and labeled. The bound position of a substrate-mimic inhibitor to C. thermophilum separase is shown in gray 17. (d). Close-up of the active site region showing the differences between the bound position of securin (magenta) and that of the inhibitor to C. thermophilum separase (gray) 17. (e). Interactions between residues 271–288 of securin SIS (magenta) with domain III (light blue) of separase.
Most importantly, residues 258–269 of the securin SIS, with good electron density (Fig. 3b), are located in the active site of separase, interacting with the SD and CD (Figs. 3a, 3c). Therefore, one mechanism for securin to inhibit the protease activity of separase is through blocking substrate access to the active site. Residues Ile259-Glu260-Ile261 of securin mimic the P5-P3 residues of the substrate 17 (Fig. 3c). Especially, the side chain of Ile259 has hydrophobic interactions with Trp1249 and Trp1250 located near the tip of the αA-αB helical hairpin of SD, and that of Glu260 is positioned near the N-terminus of helix αA in CD. However, the P1 Arg residue of the substrate is replaced with Pro263 in securin. It assumes a different conformation and its main-chain carbonyl is 6 Å away from the thiol group of the catalytic Cys1531 residue (Figs. 3c, 3d). The position of Pro263 also clashes with that of His2083 in the C. thermophilum CD 17, the second member of the catalytic machinery, and the conformation of the equivalent His1505 in the securin complex is probably an inactive one (Fig. 3d). These differences ensure that securin is not cleaved by separase even though it is located in the active site. Earlier studies showed that mutations of residues of securin equivalent to the P2, P1 and P1′ residues could turn it into a substrate (Extended Data Fig. 8) 14,17.
Residues 264–269 of securin have interactions with CD, and the P′ side of the substrate may be recognized by this region of separase as well. In contrast to the sequence conservation of the P side of the substrate, the P′ side is much less conserved (Extended Data Fig. 8). A P1′ Arg residue, found in several fungal species but not in S. pombe or C. thermophilum, may mediate the degradation of this fragment by the N-end rule pathway 28. The structure shows that a groove in the active site region can accommodate the P′ residues, and the β4A-β4B hairpin following the catalytic Cys1531 may contribute to substrate binding here (Extended Data Fig. 8; see below). Residues 264–269 of securin are in contact with the rim of this groove rather than being located at its bottom, possibly due to the shift in the position of the Pro263 residue relative to that of the P1 Arg of the substrate (Fig. 3c).
Following the active site region, residues 271–288 of the securin SIS are in close contact with domain III of separase (Fig. 3e), and residues 290–296 are located in a deep groove at the interface between domains III and II (Figs. 3a, Extended Data Fig. 6). These interactions are mostly hydrophobic in nature, and also involve several aromatic (Phe and Tyr) residues. Residues 306–316 of the securin SIS are placed on the side of domain II of separase (Fig. 1c). Finally, residues 365–373 at the C-terminus of securin form a short helix (Extended Data Fig. 4) and are located in a surface depression in domain I (Fig. 1b, Extended Data Fig. 6). Residues 299–305 and 317–362 of securin are disordered in the current structure.
The securin SIS contains three potential phosphorylation sites (Fig. 1c) 29,30, Ser277 (Fig. 3e), Ser292 (Extended Data Fig. 6) and Thr304. The S277A/S292A/T304A triple mutant had weaker interaction with separase and reduced nuclear location of separase, while the single and double mutants had no growth phenotype 30. pSer277 should have favorable interactions with Arg1130 in domain IV of separase (Fig. 3e). The side chain of Ser292 is hydrogen-bonded to the main-chain amides of residues 294 and 295 in the SIS and Thr304 is in a disordered region in the current structure, suggesting possible conformational rearrangements of Ser292 and Thr304 upon phosphorylation.
Separase residues in contact with securin are generally well conserved (Extended Data Figs. 1–3). The securin SIS is poorly conserved among species, but the residues in the major contact sites with separase are better conserved (Extended Data Fig. 4), suggesting that the securin homologs may share a common mechanism of interacting with and inhibiting separase. The observed binding mode of the SIS indicates that securin likely helps to stabilize the overall structure of separase, as suggested by prior biochemical studies 12–15. It might also be possible that once securin is removed by proteasome degradation the structure of separase alone could undergo a conformational change to assume a different arrangement of its domains, especially for domains I-III. The β4A-β4B segment is a loop (L4) in the structure of the C. thermophilum SD-CD (Extended Data Fig. 6), and residues in this loop are important for catalytic activity 17. This segment is stabilized by contacts with domain III (Extended Data Fig. 6), and therefore a change in the position of domain III might be necessary for catalysis. This would represent another, indirect mechanism for securin to inhibit the catalytic activity of separase.
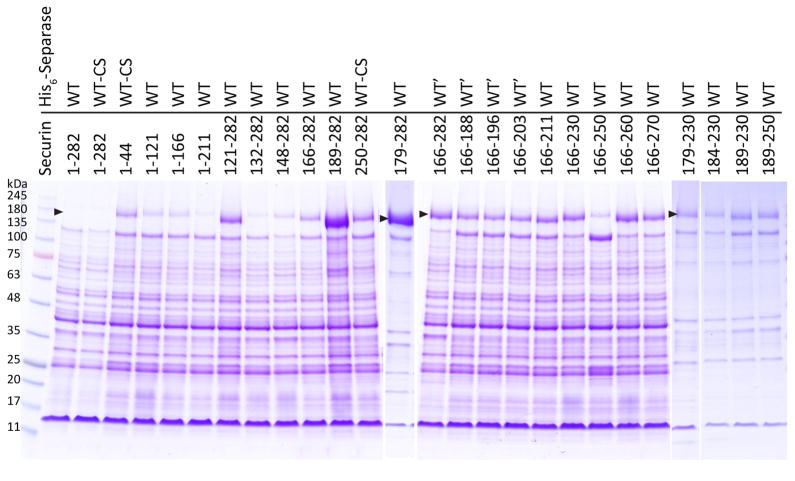
The structure of the separase-securin complex provides a molecular basis for the large amount of experimental data on this complex. This structure is also supported by our biochemical observations. The complex from the fungus Zygosaccharomyces rouxii was used for these studies, and its separase and the SIS of securin are closely related to those from S. cerevisiae (Extended Data Figs. 1–4). Z. rouxii securin contains only 282 residues due to a shorter N-terminal segment (Fig. 4a). We found that separase was produced in the insoluble fraction in insect cells unless an appropriate securin was co-expressed with it (Fig. 4b; Extended Data Fig. 9), which was likely due to the instability/misfolding of separase. Based on this observation, we assessed the importance of various segments of securin for producing soluble separase, which would reflect both its binding and chaperone functions. The N-terminal segment of securin, up to residue 166 (equivalent to residue 258 of S. cerevisiae securin, Extended Data Fig. 4), could not help produce soluble separase, and deleting these 165 N-terminal residues had little effect on the production of soluble separase (Figs. 4b, 4c). Additional experiments showed that deleting residue 231 to the C-terminus of securin led to a relatively small reduction in the solubility of separase (Figs. 4b, 4c). These data indicate that residues 166–230 of Z. rouxii securin, equivalent to residues 258–316 of S. cerevisiae securin, are required for interactions with separase, in agreement with our structural information.
Figure 4.
Biochemical characterizations of the interactions between separase and securin. (a). Domain organizations of S. cerevisiae and Z. rouxii securin. The KEN motif and the destruction box (D-box) are also indicated. (b). Z. rouxii separase (with an N-terminal His tag) was co-expressed with various segments of Z. rouxii securin. The eluates from nickel columns were separated by SDS gel electrophoresis. The positions of separase and securin are indicated with the black and red arrowheads, respectively. WT: full-length Z. rouxii separase; WT-CS: full-length Z. rouxii separase C1497S mutant; WT′: Z. rouxii separase with an internal deletion of residues 952–1010, corresponding to a poorly conserved, disordered loop in domain IV of the S. cerevisiae separase structure. For gel source data, see Supplementary Figure 1. (c). Summary of the expression results in panel b. The solubility levels of separase for various securin segments are indicated.
We also observed that the segment covering residues 189 to the C-terminus of Z. rouxii securin was able to produce a small amount of soluble separase (Figs. 4b, 4c). This segment would be missing the residues located in the active site of separase, suggesting that binding to the active site is not essential for complex formation with separase. On the other hand, gel-filtration studies of this complex showed that it migrated in the void volume, indicating that it was mostly aggregated. Therefore, even though this segment of securin could bind separase, it might not be sufficient for its chaperone function to promote the correct separase folding and/or conformation.
The segment covering residues 166–188 of Z. rouxii securin was not able to help produce soluble separase, indicating that binding to the active site region of separase alone is not sufficient to produce a stable complex and that the N-terminal helical region of separase has a crucial role in the regulation by securin. In addition, residues 189–250 of securin produced a much lower amount of soluble separase compared to that for residues 189–282 of securin, indicating an appreciable role for the C-terminal residues of securin.
Overall, our studies have produced the first crystal structure information on the separase-securin complex and defined the molecular basis for the tight regulation of separase by securin. The structure provides a foundation for understanding the biochemical and biological functions of this complex from S. cerevisiae. While the N-terminal helical region is poorly conserved among separases at the amino acid sequence level, they are better conserved at the secondary structure level (Extended Data Figs. 1–2). Therefore, it might be possible that the separase-securin complexes from other organisms could assume a similar overall architecture as observed here, which is supported by the similar shape for the human separase-securin complex 18. The information obtained from this structure may also be useful for deciphering the molecular mechanisms of other separase-securin complexes.
Online Methods
Protein expression and purification
Separase and securin were co-expressed in insect cell system using Multibac technology (Geneva Biotech) 31. Separase and securin gene fragments were cloned into the pFL dual plasmid through its PH and p10 promoters, respectively. The construct used for crystallization contained residues 51–1630 or 71–1630 of Saccharomyces cerevisiae separase and residues 258–373 of securin. A hexa-histidine tag was added to the N terminus of separase while securin did not carry a tag. Bacmids were generated by transforming constructs to DH10EMBacY competent cells (Geneva Biotech) and isolated by Bac-to-Bac protocol (ThermoFisher). Bacmids were further transfected into Sf9 cells using Cellfectin II (ThermoFisher) following the manufacturer’s instructions to produce P1 baculoviruses. P2 viruses were obtained by amplifying P1 viruses in Sf9 once and were used for large-scale protein expression.
High5 cells were grown in Fernbach flasks (PYREX) in ESF-921 medium (Expression System) by shaking at 120 rpm at 27 °C until the density reaches 2 × 106 cells/ml. Cells were infected by P2 virus and harvested 48 hours after infection by centrifugation at 500×g. The harvested cells were resuspended in lysis buffer containing 50 mM phosphate (pH 7.6), 500 mM NaCl, 5% (v/v) glycerol, 20 mM imidazole and protease inhibitor cocktail (Sigma) and lysed by sonication. Cell lysates were centrifuged at 24,000×g for 40 min at 4 °C before incubating with nickel beads (Qiagen). After 1 h, beads were transferred to a gravity flow column (Biorad) and washed extensively with lysis buffer. Protein was eluted with a buffer containing 50 mM phosphate (pH 7.6), 500 mM NaCl, 5% (v/v) glycerol and 500 mM imidazole. Protein eluate was further purified by gel filtration using Sephacryl S-300 column (GE Healthcare) equilibrated in a buffer containing 5 mM HEPES (pH 7.6), 400 mM NaCl and 2 mM DTT. The protein samples were concentrated to 10 mg/ml and stored at −80 °C.
Protein crystallization
Crystals were grown using the sitting-drop method at 20 °C by mixing 1.0 μl protein solution at 3 mg/ml concentration with 1.0 μl well solution containing 0.25 M Na/K-phosphate (pH 6.0) and 14% (w/v) polyethylene glycol 3,350. Crystals appeared in 3 days and were transferred to the well solution plus 30% (v/v) glycerol as cryoprotectant before being flash-frozen in liquid nitrogen for data collection at 100K. For phasing, a gold derivative was obtained by soaking crystals in mother liquor with 2 mM KAu(CN)2 for 5 hours and transferring to cryoprotectant for 2 min. The crystals belong to space group P3221 with one separase-securin complex in the asymmetric unit.
Data collection and structure determination
X-ray diffraction data sets up to 3.0 Å resolution were collected at beamline 24-ID-E of the Advanced Photon Source (APS). The data were processed with the HKL package 32. The structure was solved using the single isomorphous replacement with anomalous scattering method with the gold derivative with AutoSharp 33, which located 10 gold sites. The atomic model was built manually with Coot 34 and refined with PHENIX 35. The crystallographic information is summarized in Extended Data Table 1.
From the same crystallization condition, crystals belonging to space group C2 were also obtained, with unit cell parameters of a=238.4 Å, b=89.5 Å, c=119.0 Å and β=105.2°. There is one separase-securin complex in the asymmetric unit. The best diffraction data set that we could collect on this crystal form extended to 3.7 Å resolution. The structure could be readily solved with the molecular replacement method using the structure in P3221 as the model, and no significant structural differences were observed.
X-ray diffraction data sets for the crystal containing residues 71–1630 of separase were collected at beamline 24-ID-C of the Advanced Photon Source (APS). The structure was refined at 2.6 Å resolution, and the crystallographic information is summarized in Extended Data Table 1.
For the 2.6 Å structure, 94.8% of the residues are in the favored region of the Ramachandran plot. For the 3.0 Å structure, 96.9% of the residues are in the favored region of the Ramachandran plot. No residues are in the disallowed region in either structure.
Mutagenesis studies
Site-specific and deletion mutations were introduced with the QuikChange kit (Agilent) and sequenced for confirmation. The mutants were expressed in insect cells following the same protocol as that for the wild-type protein.
Data availability
The atomic coordinates and the diffraction data sets have been deposited at the Protein Data Bank, with accession codes 5U1S and 5U1T.
Extended Data
Extended Data Fig. 1.
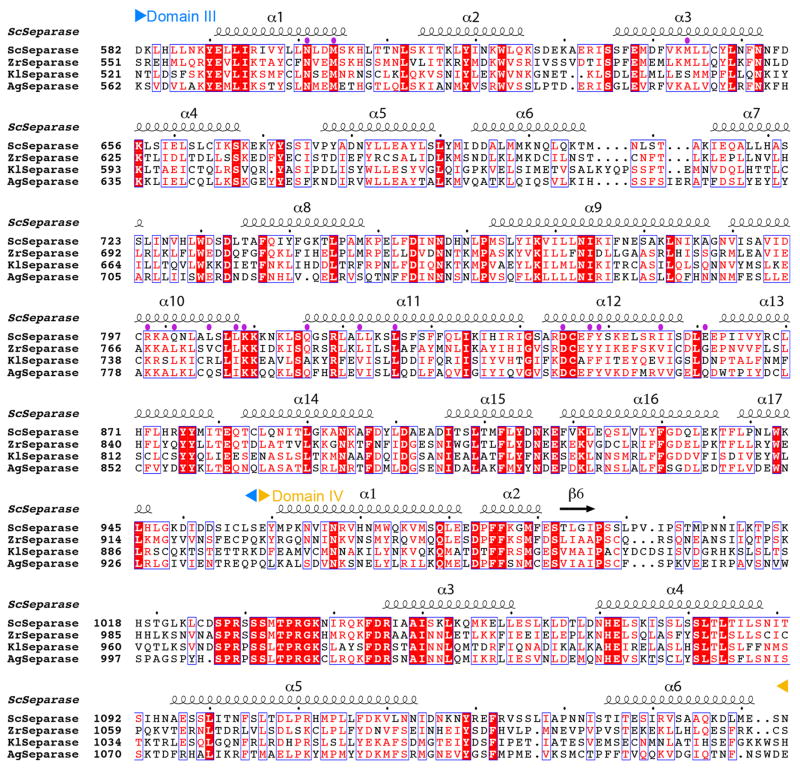
Sequence alignment of domains I and II of separase. The secondary structure elements in the S. cerevisiae separase structure are shown and labeled. The boundaries of the domains are indicated. Residues in contact with securin are indicated with the purple dots. Sc: Saccharomyces cerevisiae, Zr: Zygosaccharomyces rouxii, Kl: Kluyveromyces lactis; Ag: Ashbya gossypii. Modified from an output from ESPript 36.
Extended Data Fig. 2.
Sequence alignment of domains III and IV of separase. The secondary structure elements in the S. cerevisiae separase structure are shown.
Extended Data Fig. 3.
Sequence alignment of the SD and CD of separase. The catalytic Cys1531 and His1505 residues are indicated with the red dots. Hs: Homo sapiens; Sp: Schizosaccharomyces pombe; Ct: Chaetomium thermophilum.
Extended Data Fig. 4.
Sequence alignment of securin. The separase interaction segment (SIS) is indicated. Residues in contact with separase are indicated with the blue dots. Residue 263 is equivalent to the P1 residue of separase substrates, and is indicated with the red asterisk. Residues 317–360 of S. cerevisiae securin are disordered in the current structures and are poorly conserved in sequence.
Extended Data Fig. 5.
Overlay of the structures of the separase-securin complexes. (a). The complex formed by residues 71–1630 of separase and 258–373 of securin is shown in color, and that by residues 51–1630 of separase and 258–373 of securin in gray. Residues 73–80 from another molecule of separase in the crystal is shown in orange, forming a β-sheet with the N-terminal segment of separase. (b). panel a viewed after a 50° rotation around the vertical axis.
Extended Data Fig. 6.
Additional structural information on the separase-securin complex. (a). 2Fo–Fc electron density for helices 4 and 5 of domain I at 2.6 Å resolution, contoured at 1σ. Helices 6 and 7 are also shown for reference. The directions of the helices are indicated with the red arrows. (b). The β4A-β4B segment of CD (cyan) is immediately after the catalytic Cys1531 residue, and has interactions with domain III (light blue). The equivalent segment in the C. thermophilum SD-CD free enzyme structure is a loop (L4), shown in dark gray. (c). Interactions between residues 290–296 of securin SIS (magenta) with domains III (light blue) and II (light brown) of separase. (d). Interactions between the C-terminal segment of securin SIS (magenta) and domain I of separase (green). Deletion of the first 155 residues of separase 12 would remove helix α3 in this binding site.
Extended Data Fig. 7.
Structural homologs of domains III and SD of separase. (a). Overlay of the structures of domain III of separase (color ramp from N- (blue) to C-terminus (red)) and the TPR domain of LGN (gray, PDB entry 4WNG; 11% sequence identity, Z score of 14.4) 23–25. The Frmpd4 ligand (black) of LGN is bound to a different region of the structure compared to securin. (b). Overlay of the structures of domain III of separase and the subunit 7 of the APC/C (PDB entry 5G04; 5% identity, 14.3 Z score) 37. (c). Overlay of the structures of the SD of separase (green) and a part of the PIWI domain of Argonaute (gray, PDB entry 4N76; 10% sequence identity, 5.5 Z score. As a comparison, matching this β-sheet to that in C. thermophilum separase produced a Z score of 5.7.) 27,38. Residues in the helical insert between β3 and β4 of separase are removed for clarity. (d). Overlay of the structures of the SD of separase and the YqgF domain of Tex (gray, PDB entry 3BZK; 4% sequence identity, 5.5 Z score) 39.
Extended Data Fig. 8.
Possible binding groove for the P′ residues of the substrate. (a). Alignment of the separase cleavage sites in Scc1 substrates. The two cleavage sites in each protein are named a and b. The equivalent residues in securin are also shown. The asterisks indicate securin mutants (mutations in green) that become substrates of separase. The P and P′ residues are labeled at the top, and the cleavage site is indicated with the vertical line. (b). The overall binding mode of residues 258–271 of securin in separase. The β4A-β4B segment of CD (cyan) is a loop (L4, dark gray) in the C. thermophilum SD-CD structure. (c). A groove in the active site of separase (red arrows) can accommodate the P′ residues. The blue arrow indicates another groove in this region, but the binding mode of securin suggests that the groove indicated by the red arrows is more likely.
Extended Data Fig. 9.
Biochemical characterizations of the interactions between separase and securin. Z. rouxii separase was co-expressed with various segments of Z. rouxii securin, with truncations at the N- and/or C-terminus. The insoluble fraction was run on SDS PAGE. The position of separase (with an N-terminal His tag) is indicated with the black arrowhead. WT: full-length Z. rouxii separase; WT-CS: full-length Z. rouxii separase with C1497S mutation; WT′: Z. rouxii separase with an internal deletion of residues 952–1010, corresponding to a poorly conserved, disordered loop in domain IV of the S. cerevisiae separase structure. For gel source data, see Supplementary Figure 1.
Extended Data Table 1.
Data collection and refinement statistics
| Separase (51–1630)-securin (258–373) complex crystal form I | Separase (71–1630)-securin (258–373) complex crystal form I | Separase (51–1630)-securin (258–373) complex crystal form II | |
|---|---|---|---|
| Data collection | |||
| Space group | P3221 | P3221 | C2 |
| Cell dimensions | |||
| a, b, c (Å) | 126.3, 126.3, 273.9 | 125.9, 125.9, 271.9 | 238.4, 89.5, 119.0 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 105.2, 90 |
| Resolution (Å) | 50–3.0 (3.11–3.0)* | 50–2.6 (2.69–2.6)* | 50–3.7 (3.93–3.7) |
| Rmerge (%) | 14.4 (>100) | 14.5 (>100) | 16.6 (232) |
| CC1/2 | (0.480) | (0.476) | (0.271) |
| I/σI | 15.0 (1.5) | 14.4 (1.2) | 4.43 (0.49) |
| Completeness (%) | 99.8 (99.6) | 100 (99.9) | 96.2 (90.9) |
| Redundancy | 10.4 (7.6) | 10.0 (8.2) | 2.7 (2.5) |
| Refinement | |||
| Resolution (Å) | 50–3.0 (3.11–3.0) | 50–2.6 (2.69–2.6) | |
| No. reflections | 51,130 (4,818) | 77,428 (7,588) | |
| Rwork/Rfree (%) | 19.5 (31.4)/25.2 (36.3) | 22.1 (34.6)/26.4 (37.0) | |
| No. atoms | |||
| Protein | 12,609 | 12,633 | |
| Ligand/ion | 0 | 0 | |
| Water | 0 | 16 | |
| B-factors | |||
| Protein | 97.1 | 72.0 | |
| Ligand/ion | - | - | |
| Water | - | 63.2 | |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.003 | 0.011 | |
| Bond angles (°) | 0.65 | 1.2 | |
Two crystals were used for data collection for crystal form I, and one crystal for crystal form II.
Highest resolution shell is shown in parenthesis.
Supplementary Material
Acknowledgments
We thank Hailong Zhang, Song Xiang and Luis Cunha for carrying out initial studies on this project; S. Banerjee, K. Perry, R. Rajashankar, J. Schuermann, N. Sukumar for access to NE-CAT 24-C and 24-E beamlines at the Advanced Photon Source. This research is supported by grants R35GM118093 and S10OD012018 from the NIH to LT. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, funded by the NIH (P41 GM103403). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Footnotes
Author Contributions. SL carried out cloning, protein expression, purification, crystallization, data collection, structure determination and refinement, and site-directed mutagenesis. LT initiated the project, supervised the entire research, and analyzed the results. SL and LT wrote the paper.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 2.Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- 3.Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- 4.Peters JM. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell. 2002;9:931–943. doi: 10.1016/s1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- 5.Uhlmann F. Separase regulation during mitosis. Biochem Soc Trans. 2003;70:243–251. doi: 10.1042/bss0700243. [DOI] [PubMed] [Google Scholar]
- 6.Yanagida M. Cell cycle mechanisms of sister chromatid separation; role of Cut1/separin and Cut2/securin. Genes to Cells. 2000;5:1–8. doi: 10.1046/j.1365-2443.2000.00306.x. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan M, Hornig NCD, Porstmann T, Uhlmann F. Studies on substrate recognition by the budding yeast separase. J Biol Chem. 2004;279:1191–1196. doi: 10.1074/jbc.M309761200. [DOI] [PubMed] [Google Scholar]
- 8.Zhang N, et al. Identification and characterization of separase inhibitors (Sepins) for cancer therapy. J Biomol Screen. 2014;19:878–889. doi: 10.1177/1087057114520972. [DOI] [PubMed] [Google Scholar]
- 9.Ciosk R, et al. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- 10.Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- 11.Waizenegger IC, Gimenez-Abian JF, Wernic D, Peters JM. Regulation of human separase by securin binding and autocleavage. Curr Biol. 2002;12:1368–1378. doi: 10.1016/s0960-9822(02)01073-4. [DOI] [PubMed] [Google Scholar]
- 12.Hornig NCD, Knowles PP, McDonald NQ, Uhlmann F. The dual mechanism of separase regulation by securin. Curr Biol. 2002;12:973–982. doi: 10.1016/s0960-9822(02)00847-3. [DOI] [PubMed] [Google Scholar]
- 13.Jallepalli PV, et al. Securin is required for chromosomal stability in human cells. Cell. 2001;105:445–457. doi: 10.1016/s0092-8674(01)00340-3. [DOI] [PubMed] [Google Scholar]
- 14.Nagao K, Yanagida M. Securin can have a separase cleavage site by substitution mutations in the domain required for stabilization and inhibition of separase. Genes to Cells. 2006;11:247–260. doi: 10.1111/j.1365-2443.2006.00941.x. [DOI] [PubMed] [Google Scholar]
- 15.Wirth KG, et al. Separase: a universal trigger for sister chromatid disjunction but not chromosome cycle progression. J Cell Biol. 2006;172:847–860. doi: 10.1083/jcb.200506119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Csizmok V, Felli IC, Tompa P, Banci L, Bertini I. Structural and dynamic characterization of intrinsically disordered human securin by NMR spectroscopy. J Amer Chem Soc. 2008;130:16873–16879. doi: 10.1021/ja805510b. [DOI] [PubMed] [Google Scholar]
- 17.Lin Z, Luo X, Yu H. Structural basis of cohesin cleavage by separase. Nature. 2016;532:131–134. doi: 10.1038/nature17402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viadiu H, Stemmann O, Kirschner MW, Walz T. Domain structure of separase and its binding to securin as determined by EM. Nat Struct Mol Biol. 2005;12:552–553. doi: 10.1038/nsmb935. [DOI] [PubMed] [Google Scholar]
- 19.Bachmann G, et al. A closed conformation of the Caenorhabditis elegans separase-securin complex. Open Biol. 2016;6:160032. doi: 10.1098/rsob.160032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Y, et al. Separase is recruited to mitotic chromosomes to dissolve sister chromatid cohesion in a DNA-dependent manner. Cell. 2009;137:123–132. doi: 10.1016/j.cell.2009.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takayanagi H, Yuzawa S, Sugimoto H. Structural basis for the recognition of the scaffold protein Frmpd4/Preso1 by the TPR domain of the adaptor protein LGN. Acta Cryst. 2015;F71:175–183. doi: 10.1107/S2053230X14028143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carminati M, et al. Concomitant binding of Afadin to LGN and F-actin directs planar spindle orientation. Nat Struct Mol Biol. 2016;23:155–163. doi: 10.1038/nsmb.3152. [DOI] [PubMed] [Google Scholar]
- 25.Pan Z, et al. Structural and biochemical characterization of the interaction between LGN and Frmpd1. J Mol Biol. 2013;425:1039–1049. doi: 10.1016/j.jmb.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Winter A, Schmid R, Bayliss R. Structural insights into separase architecture and substrate recognition through computational modelling of caspase-like and death domains. PLoS Comp Biol. 2015;11:e1004548. doi: 10.1371/journal.pcbi.1004548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 28.Rao H, Uhlmann F, Nasmyth K, Varshavsky A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature. 2001;410:955–959. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- 29.Holt LJ, et al. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agarwal R, Cohen-Fix O. Phosphorylation of the mitotic regulator Pds1/securin by Cdc28 is required for efficient nuclear localization of Esp1/separase. Genes Dev. 2002;16:1371–1382. doi: 10.1101/gad.971402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fitzgerald DJ, et al. Protein complex expression by using multigene baculovirus vectors. Nat Methods. 2006;3:1021–1032. doi: 10.1038/nmeth983. [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan KD. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Adams PD, et al. PHENIX: building a new software for automated crystallographic structure determination. Acta Cryst. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 36.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 37.Zhang S, et al. Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature. 2016;533:260–264. doi: 10.1038/nature17973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson SJ, et al. Crystal structure and RNA binding of the Tex protein from Pseudomonas aeruginosa. J Mol Biol. 2008;377:1460–1473. doi: 10.1016/j.jmb.2008.01.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and the diffraction data sets have been deposited at the Protein Data Bank, with accession codes 5U1S and 5U1T.