Abstract
Campylobacter is a major cause of foodborne illnesses worldwide. Campylobacter infections, commonly caused by ingestion of undercooked poultry and meat products, can lead to gastroenteritis and chronic reactive arthritis in humans. Whole genome sequencing (WGS) is a powerful technology that provides comprehensive genetic information about bacteria and is increasingly being applied to study foodborne pathogens: e.g., evolution, epidemiology/outbreak investigation, and detection. Herein we report the complete genome sequence of Campylobacter coli strain YH502 isolated from retail chicken in the United States. WGS, de novo assembly, and annotation of the genome revealed a chromosome of 1,718,974 bp and a mega-plasmid (pCOS502) of 125,964 bp. GC content of the genome was 31.2% with 1931 coding sequences and 53 non-coding RNAs. Multiple virulence factors including a plasmid-borne type VI secretion system and antimicrobial resistance genes (beta-lactams, fluoroquinolones, and aminoglycoside) were found. The presence of T6SS in a mobile genetic element (plasmid) suggests plausible horizontal transfer of these virulence genes to other organisms. The C. coli YH502 genome also harbors CRISPR sequences and associated proteins. Phylogenetic analysis based on average nucleotide identity and single nucleotide polymorphisms identified closely related C. coli genomes available in the NCBI database. Taken together, the analyzed genomic data of this potentially virulent strain of C. coli will facilitate further understanding of this important foodborne pathogen most likely leading to better control strategies. The chromosome and plasmid sequences of C. coli YH502 have been deposited in GenBank under the accession numbers CP018900.1 and CP018901.1, respectively.
Keywords: Campylobacter coli (C. coli), Whole genomes sequencing (WGS), Type VI secretion system (T6SS), Plasmid
| Specifications | |
|---|---|
| Organism/cell line/tissue | Campylobacter coli |
| Strain | YH502 |
| Sequencer or array type | PacBio RS II, Illumina Miseq |
| Data format | Analyzed |
| Experimental factors | Campylobacter coli isolated from retail chicken |
| Experimental features | Genome sequencing, de novo assembly, annotation, and analysis |
| Consent | Not applicable |
| Sample source location | Wyndmoor, Pennsylvania, USA and Latitude & Longitude 40.08 N 75.19 W |
1. Direct link to deposited data
2. Experimental design, materials and methods
Among all of the species of Campylobacter, C. jejuni and C. coli are responsible for a large majority of cases of campylobacteriosis [1]. Considering the importance of Campylobacters and the advantages of whole genome sequencing (WGS) for studying foodborne bacteria, many researchers have reported genome sequences of these organisms in public databases. However, high quality complete genomes are relatively rare: e.g., there are only 17 complete genomes of C. coli against a total submission of 797 genomes (https://www.ncbi.nlm.nih.gov/genome/genomes/1145 accessed on Dec. 12th, 2016). In the present study, we report the complete and analyzed genome of C. coli strain YH502, recently isolated from retail chicken, using PacBio single molecule real-time and Illumina MiSeq sequencing technologies.
The strain C. coli YH502 was isolated from chicken collected from a local supermarket using a passive filtration method [2]. Briefly, a 450-g chicken carcass was rinsed with 200 mL of 0.1% buffered peptone water. The chicken rinse was concentrated and enriched in Bolton broth containing horse blood and antibiotic supplement (20 mg/L cefoperazone, 20 mg/L trimethoprim, 20 mg/L vancomycin, and 50 mg/L cyclohexamide) for 24 h. Passive filtration was performed for the isolation of Campylobacter cells. The strain was routinely grown in Brucella broth or on Brucella agar plates under microaerophilic conditions (5% O2, 10% CO2 and 85% N2) at 42 °C.
Genomic DNA was extracted from the isolate using the Qiagen Genomic-tip 100/G kit (Qiagen, Valencia, CA) as per manufacturer's instruction and quantified with a Qubit 3.0 fluorometer (Thermo Fisher Scientific, Waltham, MA). The identity of the strain was initially established by 16S rRNA sequencing [3] and was further confirmed by a multiplex real-time PCR assay targeting the hipO and cdtA genes [4].
Campylobacter has a wide genetic diversity due to frequent inter- and intra-species DNA recombination [5], [6]. To determine if the C. coli pan-genome was open (As per Heap's Law a pan-genome is the set of non-redundant genes obtained from already sequenced genomes and the pan-genome is considered open if sequencing of new genomes are expected to add new genes to the existing pangenome; conversely a pan-genome is considered closed if sequencing of new genomes does not add to the growth of the existing pan-genome.), all of the 17 complete genomes of C. coli were downloaded from GenBank in NCBI. These genomes were analyzed for their gene content using the Micropan R-package which estimated the openness of the population following Heap's law (https://www.r-project.org/) [7], [8]. Results of the analysis indicated an open pan-genome for C. coli (decay parameter, α = 0.579; if ‘α < 1.0’ pan-genome is open, if ‘α > 1.0’ the pan-genome is closed), which indicated the possibility of finding new gene(s) in newly sequenced C. coli genomes. Therefore, a recently isolated C. coli strain YH502 was processed for WGS analysis.
To generate high quality sequence data from the strain, two parallel next-generation sequencing technologies were employed—Single Molecule Real-Time (SMRT) sequencing (Pacific Bioscience, Menlo Park, CA) and the MiSeq System (Illumina, San Diego, CA). SMRT library preparation and sequencing were provided by the University of Delaware Sequencing & Genotyping Center performed using the SMRTbell template prep kit and PacBio RS II. For MiSeq sequencing, a genomic DNA library was prepared using the Nextera XT sample preparation kit and subsequent reactions were run as per manufacturer's guidelines (Illumina, San Diego, CA). PacBio reads were de novo assembled using software Canu v 1.3 [9] and Illumina data were assembled with Spades v 3.7.1 [10]. The contigs from both assemblies were joined and corrected using Pilon to produce a draft genome (https://sepsis-omics.github.io/tutorials/modules/cmdline_assembly/). The genome was manually edited, ordered, oriented, and then confirmed by mapping reads back using CLC Genomics Workbench 9.5 (Qiagen Bioinformatics, Redwood, CA). The depth coverages of SMRT sequencing and MiSeq reads were 506 × and 48 ×, respectively.
3. Data description
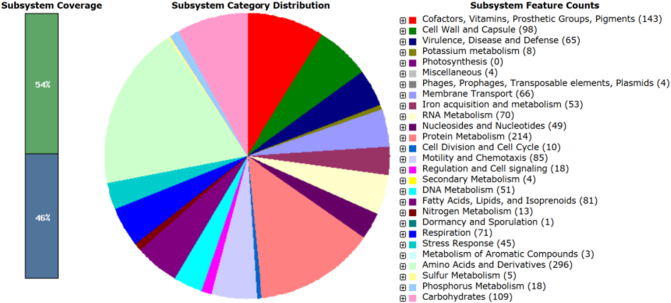
The complete genome of C. coli YH502 contains a 1,718,974 bp chromosome and 125,964 bp plasmid with a GC content of 31.2%, which corresponded well with that of other C. coli genomes available in the NCBI database. Annotation of the genome via Rapid Annotation using Subsystems Technology (RAST, http://rast.nmpdr.org/) [11] revealed 318 subsystems, 1931 coding sequences and 53 RNAs. Among the subsystems identified by RAST there were 67 genes associated with virulence, disease causation and defense of the organism (Fig. 1). Four phage related genes were also present. In view of the large number of virulence associated genes in C. coli YH502, the genome was assessed for its pathogenic potential by the PathogenFinder tool [12]. The analysis showed the probability of being a human pathogen was 80.5%, indicating a high potential for this organism to cause human disease.
Fig. 1.
Subsystem categories of the C. coli YH502 genome annotated by RAST. Bar diagram on the left shows subsystem coverage. The pie diagram shows the distribution of subsystem features.
Among the virulence factors identified by RAST, there were a number of genes associated with antimicrobial resistance. These included beta-lactamase (1), CmeABC multidrug efflux pump (4), various families of multidrug resistance efflux pumps (11; RND, MATE, MFS), and fluoroquinolone resistance (2). Further investigation of the genome with the ResFinder tool [13] identified the signatures of beta-lactam resistance mediated by blaOXA-61 (encoding beta-lactamase) and aminoglycoside resistance mediated by aph(3′)-VIIa (encoding aminoglycoside kinase).
Interestingly, of the 66 genes identified as part of a membrane transport subsystem, 15 were involved in the Type VI Secretion System (T6SS), and 4 were associated with the Type IV Secretion System (T4SS). This strain harbored hallmark T6SS genes namely, vgrG (encoding valine-glycine repeats) and hcp (encoding hemolysin correlated protein). Recently, T6SS has been reported in C. jejuni strains [14], [15], and to our knowledge this is the first report of a plasmid-borne putative T6SS in C. coli. The presence of T6SS in a mobile genetic element (plasmid) facilitates horizontal transfer of these virulence genes to other organisms. Other important virulence factors identified by RAST included genes encoding cytolethal distending toxins (cdtA, cdtB, and cdtC), which were confirmed by real-time PCR experiments during identity establishment of the strain.
In addition to the virulence factors, C. coli YH502 also harbored a distinct CRISPR (clustered regularly interspaced palindromic repeats) system between 1,469,763 and 1,476,435 bp, including 14 CRISPR repeats (GTTTTAGTCCCTTTTTAAATTTCTTTATGGTAAAAT) and genes encoding Cas1, Cas2 and Csn family proteins.
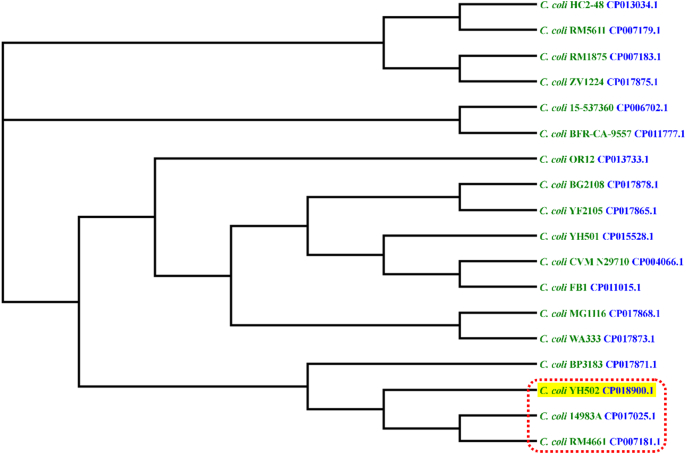
RAST identified C. coli RM2228 as the closest neighbor of the YH502 strain. However, analysis of Average Nucleotide Identity [16], [17] of YH502 against other complete C. coli genomes in the NCBI database revealed the highest nucleotide identity with C. coli strain 14983A isolated from a turkey farm housefly in North Carolina (Table 1) [18]. Therefore, we performed a phylogenetic analysis based on single nucleotide polymorphisms (SNPs) among these complete genomes of C. coli including our strain YH502. Results indicated that the closest neighbor of the YH502 strain was C. coli strain 14983A (Fig. 2) as was revealed by ANI calculation. Moreover, multilocus sequence typing (http://pubmlst.org/) analysis of the complete genome showed that C. coli YH502 and 14983A belonged to the same clonal complex (ST-828), confirming the close relatedness of these two strains [19].
Table 1.
Average nucleotide identity (ANI) of C. coli YH502 compared to other complete C. coli genomes at NCBI genome database.
| C. coli strain | Two way ANI%a |
|---|---|
| ZV1224 | 98.46 |
| 14983A | 99.1 |
| 15537360 | 98.94 |
| BFRCA9557 | 98.95 |
| BG2108 | 98.99 |
| BP3183 | 98.97 |
| CVMN29710 | 98.96 |
| FB1 | 98.97 |
| HC248 | 98.87 |
| MG1116 | 99.07 |
| OR12 | 98.98 |
| RM1875 | 98.52 |
| RM4661 | 96.82 |
| RM5611 | 98.93 |
| WA333 | 99.08 |
| YF2105 | 99.05 |
| YH501 | 98.95 |
ANI was calculated using ANI calculator (http://enve-omics.ce.gatech.edu/ani/).
Fig. 2.
Phylogenetic tree of C. coli strains based on whole genome single nucleotide polymorphism analysis. The tree was drawn from single nucleotide polymorphism data generated by the comparison of complete C. coli genome sequences using CSI Phylogeny 1.4 (https://cge.cbs.dtu.dk/services/CSIPhylogeny/).
Taken together, we believe that the genomic data of this potentially virulent strain of C. coli YH502 with T6SS on a plasmid and multidrug resistant genes will facilitate further understanding of this important foodborne pathogen likely leading to better control strategies.
4. Accession number of nucleotide sequence
The complete genome and plasmid sequences of C. coli YH502 have been deposited to NCBI under the accession CP018900.1 and CP018901.1, respectively.
Conflict of interest
The authors declare no conflict of interest about the work published in this paper.
Acknowledgement
This research was supported by the U.S. Department of Agriculture, Agricultural Research Service (USDA-ARS), Current Research Information System number 8072-42000-084. The first author (SG) is thankful to Department of Biotechnology, New Delhi, India for Overseas Associateship Grant for NER (2015-16) and to ICAR for necessary support. We thank Dr. Pina Fratamico at the USDA-ARS-ERRC for her constructive comments on the manuscript.
References
- 1.van Vliet A.H., Ketley J.M. 2001. Pathogenesis of enteric Campylobacter infection; pp. 45S–56S. (Symp Ser Soc Appl Microbiol). [DOI] [PubMed] [Google Scholar]
- 2.He Y., Reed S., Bhunia A.K., Gehring A., Nguyen L.H., Irwin P.L. Rapid identification and classification of Campylobacter spp. using laser optical scattering technology. Food Microbiol. 2015;47:28–35. doi: 10.1016/j.fm.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Irwin P., Nguyen T.L., Chen C.Y. Binding of nontarget microorganisms from food washes to anti-Salmonella and anti-E. coli O157 immunomagnetic beads: minimizing the errors of random sampling in extreme dilute systems. Anal. Bioanal. Chem. 2008;391:515–524. doi: 10.1007/s00216-008-1961-8. [DOI] [PubMed] [Google Scholar]
- 4.He Y., Yao X., Gunther N.W., IV, Xie Y., Tu S.-I., Shi X. Simultaneous detection and differentiation of Campylobacter jejuni, C. coli, and C. lari in chickens using a multiplex real-time PCR assay. Food Anal. Methods. 2010;3:321–329. [Google Scholar]
- 5.Vidal A.B., Colles F.M., Rodgers J.D., McCarthy N.D., Davies R.H., Maiden M.C., Clifton-Hadley F.A. Genetic diversity of Campylobacter jejuni and Campylobacter coli isolates from conventional broiler flocks and the impacts of sampling strategy and laboratory method. Appl. Environ. Microbiol. 2016;82:2347–2355. doi: 10.1128/AEM.03693-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suerbaum S., Lohrengel M., Sonnevend A., Ruberg F., Kist M. Allelic diversity and recombination in Campylobacter jejuni. J. Bacteriol. 2001;183:2553–2559. doi: 10.1128/JB.183.8.2553-2559.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snipen L., Liland K.H. micropan: an R-package for microbial pan-genomics. BMC Bioinforma. 2015;16:79. doi: 10.1186/s12859-015-0517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tettelin H., Masignani V., Cieslewicz M.J., Donati C., Medini D., Ward N.L., Angiuoli S.V., Crabtree J., Jones A.L., Durkin A.S. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M. bioRxiv; 2017. Canu: Scalable and Accurate Long-read Assembly Via Adaptive k-mer Weighting and Repeat Separation; p. 071282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overbeek R., Olson R., Pusch G.D., Olsen G.J., Davis J.J., Disz T., Edwards R.A., Gerdes S., Parrello B., Shukla M. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosentino S., Voldby Larsen M., Moller Aarestrup F., Lund O. PathogenFinder—distinguishing friend from foe using bacterial whole genome sequence data. PLoS One. 2013;8 doi: 10.1371/journal.pone.0077302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bleumink-Pluym N.M., van Alphen L.B., Bouwman L.I., Wosten M.M., van Putten J.P. Identification of a functional type VI secretion system in Campylobacter jejuni conferring capsule polysaccharide sensitive cytotoxicity. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lertpiriyapong K., Gamazon E.R., Feng Y., Park D.S., Pang J., Botka G., Graffam M.E., Ge Z., Fox J.G. Campylobacter jejuni type VI secretion system: roles in adaptation to deoxycholic acid, host cell adherence, invasion, and in vivo colonization. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richter M., Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goris J., Konstantinidis K.T., Klappenbach J.A., Coenye T., Vandamme P., Tiedje J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 18.Miller W.G., Huynh S., Parker C.T., Niedermeyer J.A., Kathariou S. Complete genome sequences of multidrug-resistant Campylobacter jejuni strain 14980A (turkey feces) and Campylobacter coli strain 14983A (housefly from a turkey farm), harboring a novel gentamicin resistance mobile element. Genome Announc. 2016;4 doi: 10.1128/genomeA.01175-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dingle K.E., Colles F.M., Ure R., Wagenaar J.A., Duim B., Bolton F.J., Fox A.J., Wareing D.R., Maiden M.C. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 2002;8:949–955. doi: 10.3201/eid0809.02-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]