Abstract
AZD6738 is an orally active ATR inhibitor (ATRi) currently in phase I clinical trials. We found in vitro growth inhibitory activity of this ATRi in a panel of human cancer cell lines. We demonstrated radiosensitization by AZD6738 to single radiation fractions in multiple cancer cell lines independent of both p53 and BRCA2 status by the clonogenic assay. Radiosensitization by AZD6738 to clinically relevant doses of fractionated radiation was demonstrated in vitro using a 3D tumor spheroid model and, in vivo, AZD6738 radiosensitized by abrogating the radiation-induced G2 cell-cycle checkpoint and inhibiting homologous recombination. Mitosis with damaged DNA resulted in mitotic catastrophe as measured by micronucleus formation by live-cell fluorescent-ubiquitination cell-cycle imaging of cell-cycle progression and nuclear morphology. Induction of micronuclei was significantly more prominent for AZD6738 compared with inhibition of the downstream kinase CHK1 alone at isoeffective doses. Micronuclei were characterized as acentric chromosomal fragments, which displayed characteristics of increased DNA damage and cell-cycle dyssynchrony when compared with the primary nucleus.
Introduction
There is a need for tumor-selective radiosensitizers to increase tumor control without increasing normal tissue toxicities. Numerous novel agents have demonstrated promising preclinical activity in this context but few have progressed beyond the earliest stages of clinical development (1).
Most tumor cells have a defective G1 checkpoint (2), leaving the G2 checkpoint critical for cell-cycle arrest and DNA repair following damage. The G2 checkpoint, through its principal effectors ATR and Chk1, is an attractive target for selective sensitization of cancer cells (3). Normal cells have an intact G1 checkpoint, rendering them less sensitive to G2 checkpoint abrogation.
Ionizing radiation causes a variety of DNA lesions: direct double-strand breaks (DSB), one-ended secondary DSB as a result of collapsed replication forks, single-strand breaks (SSB), base and sugar damage, and intra-strand crosslinks. ATR is one of the principal kinases of the DNA damage response (DDR). It is activated through lesions that result in expanses of single-stranded DNA (ssDNA) that become coated with replication protein A, leading to binding and activation of ATR through its partner ATRIP. The most well-described ATR target is Chk1, which effects cell-cycle arrest at the intra-S and G2 checkpoints. Chk1 inhibitors have been shown to radiosensitize in preclinical studies (4). ATR also targets replication fork factors under normal and perturbed DNA replication (5), and DNA repair pathways including homologous recombination (HR), cross-link repair, and nucleotide excision repair (6). ATR inhibition has been shown to potentiate DNA-damaging chemotherapy (platinum and gemcitabine; ref. 7), and it has been proposed that ATR may be important for the radioresistance of hypoxic cells (8–10).
ATR inhibitors (ATRi; ref. 7) are currently in early-phase clinical trials as monotherapy, in combination with DNA-damaging chemotherapy and radiotherapy and in combination with other novel agents (NCT02157792, NCT01955668, NCT02223923, and NCT02264678). Here, we demonstrate in vitro and in vivo radiosensitization by the orally active, specific ATR inhibitor AZD6738 (11), which is in phase I clinical development.
Materials and Methods
Cell culture
CAL27, FaDu, A549 (obtained 2013), PJ34, PJ41 (obtained 2014), T24, A2780, RT4 (obtained 2012), NCI-H1838 (2013), NCI-H1373 (2015), DU-4475 (year unknown, STR profiled 2016) cell lines were purchased from ATCC. SCC090, and SCC131 were purchased from DSMZ (2014). SK-OV-3 (2011) was obtained from HPA culture collection (Porton Down). HCT116 p53 Xman isogenic cell lines were purchased from Horizon Discovery (2014), DLD1 BRCA2 wild-type and deficient cells were provided to the ICR as part of a collaborative agreement. HT-29-luc2 was purchased from Caliper (2015). LON-LICR-HN4 and LON-LICR-HN5 cell lines from Professor Sue Eccles (ICR, London, 2012). STR profiling was carried out by Bio-Synthesis Inc. Mycoplasma testing used the e-Myco PCR kit (Intron Biotechnology). Cell lines received from noncommercial sources were STR profiled prior to freezing, all cell lines were mycoplasma tested, and experiments were carried out within 3 months of resuscitation.
Drugs and irradiation
AZD6738 (12) was obtained from AstraZeneca. The ATM inhibitor KU-55933 (13) was obtained from Stratech Scientific Ltd. CCT244747 (14) was obtained from Professor Ian Collins (The Institute of Cancer Research, UK). Irradiation was carried out using an AGO 250 kV X-ray machine (AGO).
MTT, clonogenic, and spheroid assay
Cells were seeded in 96-well plates, drugged the following day, and viability was determined at 72 hours by the MTT assay. Absorbance at 550 nm was measured and normalized to DMSO-treated cells. For clonogenic assays, cells were seeded at appropriate densities and left overnight. Drug was added 1 hour before and removed 48 hours after irradiation. Colonies were fixed in 5% glutaraldehyde-0.5% crystal violet. Counting was performed both manually or automated using CellProfiler or GelCount (Oxford Optronix). Colony counts were expressed as surviving fraction relative to plating efficiency of DMSO-only treated control. Curves were corrected for drug only toxicity and the enhancement ratio estimated by linear quadratic curve fitting using Y = e−(αX + βX2) and interpolating the radiation dose required to achieve an SF of 0.37 using GraphPad Prism 6 (GraphPad Software). For spheroid assays, cells were plated in ultra-low attachment plates (Corning). After 1 week, 20 Gy in ten daily fractions of 2 Gy was administered. Drug was added 1 hour prior to irradiation and replaced thrice weekly during irradiation. Spheroids were imaged using a Celigo cytometer (Nexcelom) and cross-sectional area assessed using CellProfiler (15).
Immunoblotting
Cells were scraped in PBS with 2 mmol/L Na3VO4 and cell pellets lysed in 50 mmol/L Tris–HCl pH 7.5, 150 mmol/L NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 2 mmol/L Na3VO4, and protease inhibitors. In vivo samples were processed by homogenization using a Precellys24 homogenizer (Bertin Technologies). Protein supernatants, quantified by the BCA assay (Pierce), were separated by SDS-PAGE, transferred to PVDF membrane (Thermo Scientific) and blocked in TBS with 5% non-fat dry milk and 0.1% Tween-20. Membranes were probed using the following antibodies: phospho-ATM Ser1981, #5883; ATM, #2873; p21 #2946; phospho-p53 Ser15, #2528; GAPDH, #2118; Chk1, #2360; phospho-Chk1 Ser345, #2341; Chk2, #2662; phospho-Chk2 Thr68, #2197; γH2AX, #9718; cleaved caspase 3, #9661 and cleaved PARP #5625, were all obtained from Cell Signaling Technology. p53 DO-7 mAb #M7001 was obtained from Dako and PARP-1 from Santa Cruz Biotechnology. For confocal microscopy on fixed cells, γH2AX JBW-301 (Merck-Millipore) and Rad51 H-92 (Santa Cruz 8349) were used.
Flow cytometry
Cell-cycle analysis: cells were fixed in ice-cold ethanol for at least 8 hours, then rehydrated in PBS before staining in PBS with 1% BSA, 0.1% Tween-20, 0.5 mg/mL RNAse A ,and 10 μg/mL PI (Sigma). Mitotic index: cells were fixed in 10% formalin for 20 minutes, rinsed in PBS, blocked in 1% BSA in PBS for 15 minutes and stained with phospho-S10 histone-H3 Alexa-647 (Cell Signaling Technology) for 15 to 30 minutes. DNA content and phospho-histone H3-positive cells were measured using an LSR II flow cytometer (BD Biosciences).
Confocal microscopy
FFPE tissue antigen retrieval was performed at pH 6 in a pressure cooker. Samples were treated with DNAse I (Roche) 1,000 units/mL in PBS for 1 hour at 37 degrees before washing in PBS, blocking in PBS with 1% BSA, 2% FBS. Slides were stained for anti-Rad51 (GeneTex 70230) and anti-γH2AX (Cell Signaling Technology) overnight at 4 degrees and incubated with fluorescent secondary antibodies, counterstained with DAPI, mounted in mounting medium (Dako S3023), coverslipped and sealed before imaging with Zeiss LSM710 inverted confocal microscope (Carl Zeiss). Images were acquired as z-stacks, processed as a maximum-intensity projection and quantified with a minimum of three fields of view with an average of 150 nuclei per field. In vitro cells were left to attach overnight on 35-mm glass bottom dishes (Mattek). At the indicated time points, cells were fixed in 10% formalin and permeabilized with 0.2% Triton X-100 with blocking and staining as above. Images were quantified using ImageJ or CellProfiler.
In vivo xenografts
All experiments were approved by the Institutional Review Board and complied with National Cancer Research Institute guidelines (16). Six-week old female NOD scid gamma (NSG) mice were obtained from Charles River, 2 million HCT116 p53−/− were implanted subcutaneously. When tumors reached approximately 5 mm diameter, animals were divided into 4 equal groups. AZD6738 was administered at a dose of 75 mg/kg once daily by oral gavage, in 10% DMSO, 40% propylene glycol, 50% water. Three orthogonal tumor diameters were measured by vernier calipers and volume calculated as V = π/6 d1·d2·d3. The experimental endpoint was a tumor diameter of 15 mm. For hypoxia studies, pimonidazole (Hypoxyprobe, HPI) 60 mg/kg was injected intraperitoneally 1 hour prior to collection.
High content microscopy
Cal27 cells were transfected with Fluorescent Ubiquitination-based Cell-Cycle Indicator (FUCCI) cell-cycle vectors pRetroX-G1-Red and pRetroX-SG2M according to the manufacturer's instructions using MMLV retrovirus and GP2-293 packaging cell line (Clontech). G1 red expresses mCherry hCdt1(30-120aa); S–G2–M cyan expresses AmCyan hGeminin(1-110aa). Cells were seeded in 96-well optical plastic bottomed plates and imaged using an Operetta microscope (PerkinElmer). Drug treatments were added 1 hour prior to irradiation and imaging started immediately after irradiation. Data were collected every 15 minutes for 60 hours and analyzed using Columbus software (PerkinElmer).
Comet assay
Trypsinized cells were combined 1:8 with 1% low melting point agarose, placed onto a 1% normal melting point agarose precoated slide, coverslipped on ice for 5 minutes before lysis (NaCl 2.5 mol/L, EDTA disodium salt 100 mmol/L and Tris base 10 mmol/L in distilled water, adjusted to pH10.5 with NaOH, 1% DMSO, and 1% Triton X-100) for 1 hour at 4 degrees in the dark. Slides were placed in an electrophoresis tank with 300 mmol/L NaOH, 1 mmol/L EDTA, and 1% DMSO for 30 minutes before electrophoresis at 25 V, 300 mA, neutralized with 500 mmol/L Tris–HCl, pH 8.0; DNA was stained using SYBR-safe (Thermo-Fisher) and imaged using a Zeiss LSM710 confocal microscope. A minimum of 115 cells were analyzed from 2 independent replicates and percent tail DNA was calculated using OpenComet 1.3 plugin for ImageJ (17).
Statistical analyses
Statistical analysis was performed using Prism 6. Means were compared using Student unpaired t test. Individual cell data were compared using one-way ANOVA with Tukey multiple comparisons test. An asterisk indicates P < 0.05.
Results
AZD6738 single-agent activity is p53-independent
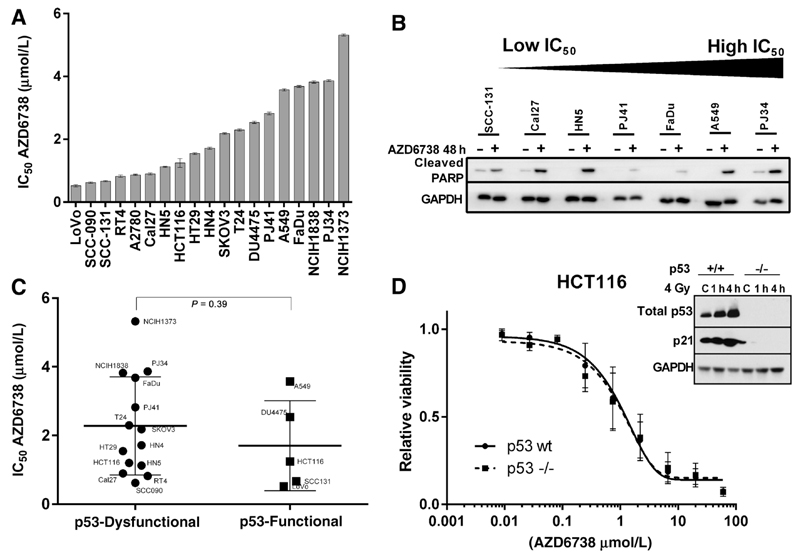
IC50 values for AZD6738 were calculated from 72-hour MTT dose–response curves (Fig. 1A). Sensitivity ranged from 0.52 μmol/L for LoVo, a colorectal cancer cell line, to 5.32 μmol/L for NCI-H1373, a lung cancer cell line. Evidence of apoptotic cell death by PARP cleavage was observed for cell lines throughout the sensitivity range (Fig. 1B).
Figure 1.
A, Cell line IC50 values assessed for growth inhibition in a 72-hour MTT assay. B, Evidence of PARP cleavage (detected using antibody specific to the cleaved fragment) with ATRi monotherapy for 48 hours at 0.5 µmol/L AZD6738. C, Cell line IC50 sorted by p53 functional status. Differences between groups were compared using the Mann–Whitney test. D, Comparison of dose–response curves to AZD6738 treatment of HCT116 p53 wild-type and HCT116 p53-null cells. Dose–response curves were plotted based on 72 hours MTT. Inset, Western blot represents assessment of p53 function by exposing to radiation.
Dysfunctional p53 has previously been reported as a putative marker of sensitivity to ATR inhibition. We tested the panel for p53 functional status by probing for Ser15 phospho-p53, total-p53, and the downstream transcriptional target p21 after 4-Gy irradiation (Supplementary Fig. S1). No significant difference in sensitivity was observed between p53 functional and dysfunctional cell lines (Fig. 1C). To further investigate the effect of p53 status, the sensitivity of HCT116 cells with and without functional p53 was compared. Sensitivity to AZD6738 was similar across a range of concentrations (Fig. 1D).
Radiosensitization by AZD6738 in vitro
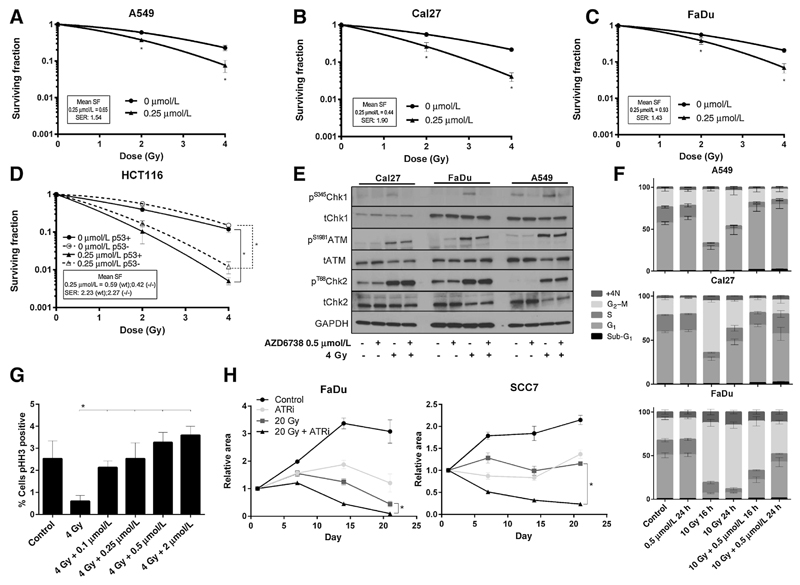
Clonogenic assays showed radiosensitization by AZD6738 in both p53 wild-type A549 (Fig. 2A) and p53 mutant Cal27 and FaDu cell lines (Fig. 2B and C). HCT116 cells were equally radiosensitized by AZD6738 irrespective of p53 status (Fig. 2D). Western analysis showed that radiation activates the ATR-Chk1 (as shown by phosphorylation of Chk1 on the ATR-specific S345 site) and ATM-Chk2 pathways. AZD6738 inhibited radiation-induced Chk1 phosphorylation but had no impact on ATM-Chk2 signaling (Fig. 2E). ATRi promoted mitotic entry following irradiation, as measured by the proportion of cells positive for phospho-histone H3 (Fig. 2F). Cell-cycle analysis indicated abrogation of the radiation-induced G2 checkpoint by AZD6738 (Fig. 2G). To investigate the effect on fractionated treatment, tumor spheroids of FaDu and SCC7 (a murine squamous carcinoma cell line) were treated with 20 Gy in 10 fractions in the presence or absence of AZD6738. Significant difference was seen between the radiation and radiation–ATRi curves (Fig. 2H; Supplementary Fig. S2). Bliss analysis for synergy between radiation and ATRi was calculated using the equation Eexp = Ex + Ey − (ExEy). Eexp is the expected effect if two treatments are additive with Ex and Ey corresponding to the effect of each treatment individually. ΔE = Eobserved − Eexp. Evidence of synergy was observed in both cell lines with ΔE ± 95% CI values of 0.22 ± 0.025 for FaDu and 0.234 ± 0.01 for SCC7, both at 21 days.
Figure 2.
A, Radiosensitization of A549 cells by AZD6738 demonstrated by a clonogenic assay. Number of colonies expressed as a fraction of the number on untreated plates. Surviving fractions normalized to the cell kill by AZD6738 alone, expressed in the box on each graph. B, Cal27cells, as per (A); C, FaDu cells, as per (A). D, clonogenic assay demonstrating radiosensitization of HCT116 wild-type and p53-null cells by AZD6738. Clonogenic data were analyzed by ANOVA with multiple comparisons; *, P < 0.05. A minimum of 3 replicates; error bars represent SEM. SER, sensitizer enhancement ratio obtained by fitting a linear quadratic curve and obtaining the ratio for sensitized and unsensitized conditions to achieve survival of 0.37. E, Western blot for ATM, Chk1, Chk2 function 1 hour after 4 Gy in the presence or absence of AZD6738. F, Cell-cycle distribution effect of AZD6738 and radiation, propidium iodide flow cytometry. Cells exposed to 0.5 µmol/L AZD6738 or DMSO control, 1 hour prior to 4 Gy or mock irradiation, harvested 16 or 24 hours later. Percentage cell-cycle distributions are displayed. G, Cal27 cells were stained for mitotic fraction using phospho-histone-H3. AZD6738 was added 1 hour prior to irradiation and cells harvested 6 hours after. Percentage of cells staining positive for phospho-histone-H3 are displayed. Error bars, SEM. Minimum of 3 replicates. H, Analysis of tumor spheroid growth for FaDu and SCC7 cells treated with AZD6738 (1 µmol/L), fractionated radiation (20 Gy in 10 fractions over 2 weeks), or the combination. Drug was added 1 hour prior to radiation and removed after the final fraction of radiation. Spheroids were imaged weekly. Error bars: SEM, 3 replicates; *, P < 0.05 between RT-only and RT-ATRi curves.
Radiosensitization by AZD6738 in an in vivo model
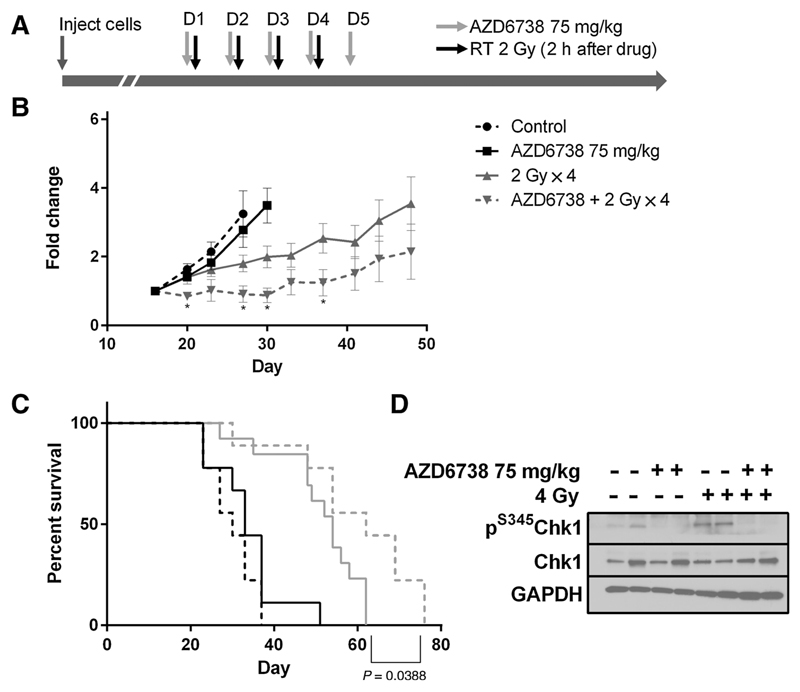
Mice-bearing HCT116 p53-null xenograft tumors received 4 fractions of 2 Gy over 4 days, with vehicle or AZD6738 administered 2 hours before irradiation and for one day following irradiation (Fig. 3A). Radiation delayed tumor growth and combination of AZD6738 and radiation significantly delayed tumor growth compared with radiation alone (Fig. 3B) and significantly prolonged survival over vehicle control, AZD6738 alone and radiation alone (Fig. 3C). There were no deaths due to treatment-related toxicity. Western analysis on tumor samples harvested 2 hours after the first dose of radiation confirmed drug-on-target effect in vivo, with reduction of radiation-induced S345Chk1 phosphorylation (Fig. 3D).
Figure 3.
A, Experimental schedule: NSG mice bearing HCT116 p53−/− tumor xenografts. N = 9 control, 9 AZD6738, 13 radiation only, 9 radiation + AZD6738. B, Tumor growth curves, normalized to the first tumor measurement after treatment began. Error bars, SEM. C, Survival probabilities, P value: difference between radiation and radiation + AZD6738. D, Western analysis of xenograft samples for p(S345)Chk1 after irradiation or mock irradiation in the presence or absence of 75 mg/kg AZD6738 administered 2 hours prior to radiation treatment, samples collected 2 hours after radiation.
AZD6738 enhances radiation-induced DNA damage by reducing homologous recombination
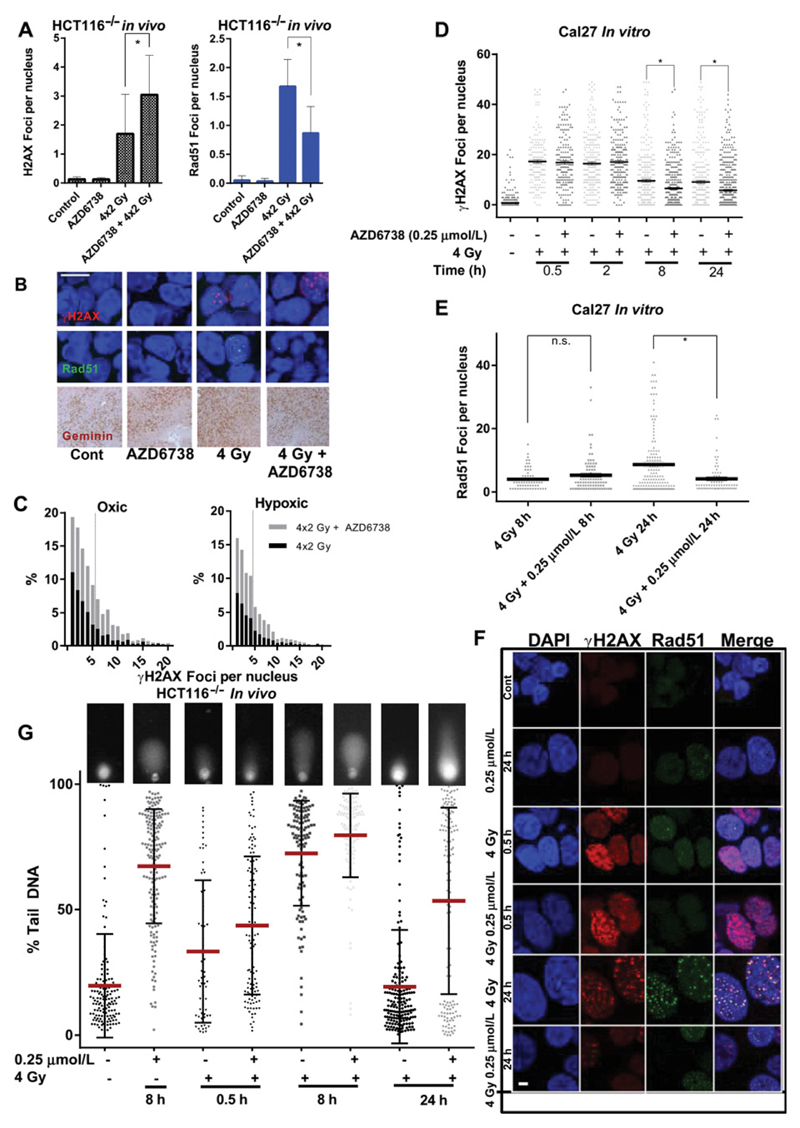
To investigate the effect of AZD6738 on radiation-induced DNA damage and repair, FFPE xenograft specimens were taken 4 hours after the final of 4 fractions of radiation with vehicle or AZD6738. Nuclei were probed for γH2AX and Rad51 foci by confocal microscopy. More γH2AX foci were observed when radiation was combined with AZD6738 with a corresponding decrease in RAD51 foci (Fig. 4A and B). Variations in geminin staining indicate that the substantial reduction in Rad51 foci was partially due to cell-cycle distribution (Fig. 4B). Rad51 and γH2AX foci were also quantified in oxic and hypoxic conditions using pimonidazole to identify these regions. As expected, fewer γH2AX foci were observed in hypoxic areas after radiation. AZD6738 increased γH2AX foci in both oxic and hypoxic conditions (Fig. 4C).
Figure 4.
A, Quantification of γH2AX (A) and Rad51 (B) foci in formalin-fixed, paraffin-embedded HCT116 (p53 null) xenograft tumors at the indicated time points after a fractionated course of 2 × 4 Gy (daily fractions) given with vehicle or 75 mg/kg AZD6738 daily by oral gavage, 2 hours prior to irradiation. B, Representative images showing cells quantified in A and B. Scale bar, 10 µm; bottom row, immunohistochemical staining with anti-geminin antibody of xenografts shown in A. C, Histograms of γH2AX foci distribution in oxic and hypoxic conditions (determined by pimonidazole staining) for radiation alone (4 × 2 Gy) and radiation + AZD6738, for formalin-fixed paraffin-embedded specimens from the in vivo fractionated regimen used in A and B. D, Quantification of γH2AX foci at the indicated time points after a single fraction of 4 Gy in the presence or absence of 0.25 µmol/L AZD6738 in Cal27 cells. A minimum of 3 high-power fields were analyzed from each of 2 independent experiments, an average of 356 (range, 174–653) cells per condition were analyzed. E, Quantification of Rad51 foci per nucleus at 8 and 24 hours after 4 Gy in the presence or absence of 0.25 µmol/L AZD6738. Only Rad51-positive cells were quantified (average 89.5 cells per condition, range 63–144). D, E, Data per cell presented, analyzed by one-way ANOVA with multiple comparisons. Error bars, SEM. At least 2 independent experiments. F, Representative images showing nuclear foci presented in D and E; scale bar, 1 µm. G, Alkaline comet assay analysis for the indicated conditions. Cal27 cells were treated with drug or DMSO 1 hour prior to irradiation and lysed at the indicated time points before alkaline comet assay was performed. Single-cell data for percent tail DNA are presented; inset images are representative comets for each condition; line represents mean; error bars, SD. Data from 2 independent experiments.
To assess DNA damage and repair kinetics, we analyzed in vitro response to single radiation fractions. Fewer γH2AX and RAD51 foci were observed 8 hours after radiation in combination with AZD6738 (Fig. 4D–F). Single-cell analysis of Rad51 foci numbers at 8 and 24 hours found that there were significantly fewer Rad51 foci in the combination group at 24 hours after irradiation (Fig. 4E), as well as a greater proportion of cells without foci (Supplementary Fig. S3).
Due to the discrepancy seen between the γH2AX foci in in vivo fractionated and in vitro single-fraction experiments, we hypothesized that reduced γH2AX at early time points after a single fraction could be due to reduction in ATR-mediated phosphorylation of H2AX (18) at these times, rather than a reflection of decreased DNA damage per se, with more γH2AX foci after fractionated radiation reflective of persistent DNA damage. We used the alkaline comet assay as a sensitive direct method to assess levels of unrepaired DNA damage. We found that a reduction in γH2AX due to AZD6738 at 8 hours after irradiation did not indicate a reduction in DNA damage, but that DNA damage present was marginally higher than radiation alone (Fig. 4G). Significantly more residual DNA damage was observed at 24 hours in the cells treated with AZD6738 and radiation compared with radiation alone. Together, these results indicate that the combination treatment leads to increased DNA damage, but decreased repair signaling through γH2AX and HR.
Abrogation of radiation-induced G2 arrest by AZD6738 results in acentric chromosome fragments
To determine the role of HR inhibition, AZD6738-mediated radiosensitization was assessed by clonogenic assay in a DLD1 BRCA2−/− isogenic cell line model. HR deficiency was confirmed by confocal microscopy (Supplementary Fig. S4). Although the BRCA2 null cell line was extremely sensitive to radiation, it was still profoundly radiosensitized by the addition of AZD6738 (Fig. 5A). This strongly indicated that while loss of HR repair sensitizes to radiation, AZD6738 sensitization may be mediated by both inhibition of DNA repair and abrogation of the G2 checkpoint.
Figure 5.
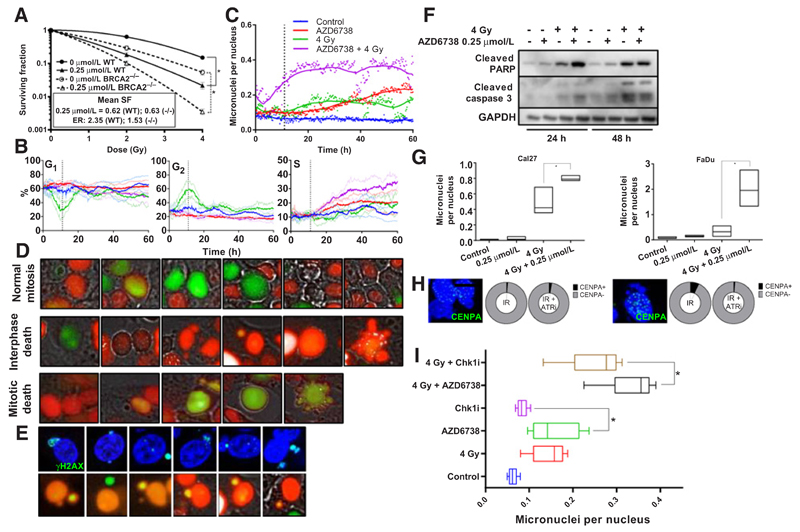
A, Clonogenic survival assay comparing DLD1 cells with wild-type or absent BRCA2. Surviving fractions normalized to the cell kill by AZD6738 alone, expressed in the inset box. Solid lines: wild-type, interrupted lines: BRCA2 null. B, Analysis of cell-cycle distributions over a 60-hour time period following 4 Gy or mock irradiation in the presence of AZD6738 (0.9 µmol/L, the IC50 value derived from 72-hour MTT assay) or DMSO control. Nuclei were automatically quantified and cell-cycle phase assigned to each nucleus depending upon fluorescence. C, Quantification of micronuclei (expressed as number of micronuclei counted divided by number of nuclei counted) over the time course. Points represent mean of 3 measurements and lines represent the smoothed mean. D, Representative images from time lapse microscopy. Top, a cell going through normal mitosis; middle, the first mitosis yielding micronuclei, dyssynchrony of cell-cycle between the micronucleus and nucleus and subsequent apoptosis at the second mitosis; bottom, a cell undergoing apoptosis at the first mitosis. E, Representative images showing micronuclei with γH2AX staining (top) and representative images showing cell-cycle phase dyssynchrony between the main nuclei and micronuclei. F, Western analysis showing levels of cleaved caspase 3 and cleaved PARP, reflective of apoptotic cell death, 24 hours after mock irradiation or 4 Gy in the presence of 0.25 μmol/L AZD6738 or DMSO control in Cal27. G, Quantification of fixed Cal27 and FaDu cells, 24 hours after mock irradiation or 4 Gy in the presence of 0.25 mmol/L AZD6738 or DMSO control; box plots represent range, with line at median number of micronuclei per nucleus. H, Proportion of micronuclei positive or negative for the kinetochore marker CENPA, treated with 4 Gy or 4 Gy in the presence of 0.25 µmol/L AZD6738 in Cal27 (left) and FaDu (right). Images show representative nuclei and micronuclei showing absence of CENPA in micronuclei. Scale bar, 10 µm. I, Quantification of the average number of micronuclei over the 60-hour time course, in cells treated with AZD6738, the Chk1 inhibitor CCT244747 at isoeffective doses for single-agent growth inhibition (0.9 µmol/L AZD6738, 1.3 µmol/L CCT244747).
To study the consequences of mitotic transition with unrepaired DNA damage due to AZD6738 we generated Cal27 FUCCI cells. These express mCherry hCdt1(30–120 aa) in G1–S and AmCyan hGeminin (1–110 aa) in S–G2–M enabling discrimination of cell-cycle progression and nuclear morphology (19). Automated high content microscopy and image analysis software were used to quantify numbers of cells in each phase of the cell-cycle over a 60-hour time course. This showed that G2 arrest peaked 11 hours after radiation and that this was abrogated in the presence of AZD6738. There was a gradual accumulation of cells in the S phase after treatment with radiation and AZD6738, which may reflect higher numbers of stalled replication forks after irradiation in the presence of AZD6738 (Fig. 5B).
Automated quantification of nuclear morphology revealed a sharp increase in micronuclei due to the combination of AZD6738 and radiation (Fig. 5C). This coincided with abrogation of the radiation-induced G2 arrest (Fig. 5B). Prolonged exposure to AZD6738 also gave rise to a less pronounced increase in micronuclei. Timelapse microscopy showed cell death in mitosis or abnormal mitosis resulting in micronuclei and subsequent cell death in interphase (Fig. 5D). We also noted some micronuclei had increased levels of γH2AX compared with the main nucleus or dyssynchrony of cell-cycle phase (Fig. 5E). Death was apoptotic based on a morphology of cell blebbing and apoptotic bodies which was confirmed by increased cleaved PARP and cleaved caspase-3 (Fig. 5F).
To further investigate the mechanism of micronucleus formation, we fixed cells at 24 hours and stained for the presence or absence of the centromere marker CENPA. We found that in both Cal27 and FaDu the majority of micronuclei were acentric chromosomal fragments lacking a centromere (Fig. 5G and H). These data confirmed significantly more micronuclei were present after combination treatment in both cell lines (Fig. 5G and H) cells. Greater numbers of micronuclei were observed when isoeffective (equal effects on monotherapy growth inhibition) doses of ATRi were used compared with Chk1 inhibitors, indicating a potentially different mechanism of radiosensitization between these two types of compound (Fig. 5I).
Discussion
Through detailed mechanistic analysis both in vitro and using in vivo xenograft specimens, we have demonstrated radiosensitization by AZD6738, in multiple cell lines, through reduced HR and abrogation of the radiation-induced G2 cell checkpoint, promoting an aberrant mitosis which results in acentric chromosome fragment-containing dysfunctional micronuclei, indicative of mitotic catastrophe. We also found S-phase accumulation after AZD6738 and RT, suggestive of accumulating stalled replication forks. Our studies have used clinically meaningful doses of 2 Gy and 4 Gy, commonly used in radical and palliative radiotherapy, respectively. The radiosensitizing effect at these doses will be amplified over a course of radiotherapy which uses multiple 2-4 Gy fractions: we have shown significant enhancement of radiation tumor control with the addition of AZD6738 at 20 Gy in 10 fractions (currently being assessed in a phase I study) in an in vitro model, and delayed tumor growth in vivo. The sensitizer enhancement ratios we have obtained would translate into a significant reduction in radiation dose required for tumor control (see Supplementary Table S1).
ATRi monotherapy has been shown to be more effective when used in cell lines with defects in the DDR (20–24) or activated oncogenes (25, 26), presumably due to the increased replication stress that these generate. In our panel of cell lines, a narrow sensitivity range was observed for AZD6738 alone, with evidence of death by apoptosis. Radiosensitization was uniformly observed in p53 wild-type, null and mutant as well as BRCA2-null cell lines. ATRi have been shown to sensitize to DNA-damaging chemotherapies, an effect that seems to be enhanced by p53 loss (27). The ability of ATRi to radiosensitize irrespective of p53 status has been reported previously (9), and we confirm these findings in multiple cell lines and an isogenic paired model and also report that this lack of selectivity also applies to monotherapy ATRi—both these conclusions will be relevant for patient selection strategies in clinical trials.
AZD6738 reduced HR after RT both in vitro and with fractioned RT in vivo. It has been suggested that there may be a contribution to radiation-induced DNA damage from replication fork stalling after single-strand DNA damage (28). Persistent DNA damage after irradiation due to reduced HR has been observed when ATR activity is reduced by knockdown (29). The observed reduction in γH2AX signaling at later time points after irradiation in vitro suggests a role for ATR in maintenance of DSB signaling, perhaps becoming activated by secondary DSB and replication stress as cells continue to replicate DNA after irradiation. However, after fractionated radiation in vivo, we observed increased numbers of γH2AX foci—this may reflect the difference between DSB-repair kinetics in exponentially growing cells in vitro compared with an in vivo tumor subjected to fractionated radiation and variations in proliferation and oxygenation.
Previous studies have shown that ATRi caused increased DNA damage under hypoxic conditions (9), possibly as a result of replication fork collapse (10). We did not see a significant increase in γH2AX foci in hypoxic regions in the presence of AZD6738 alone; however, in combination with fractionated radiation, we observed persistent γH2AX foci in both oxygenated and hypoxic regions, suggesting that AZD6738 may enhance radiation damage in hypoxic areas. More γH2AX foci were apparent in oxic regions than in hypoxic regions for both fractionated radiation and in combination with AZD6738. This is in keeping with previous observations on DNA damage and hypoxia.
Use of a DLD1 BRCA2-null isogenic model revealed AZD6738 could still potently radiosensitize an already HR-deficient, highly radiosensitive cell line. This suggests G2-checkpoint inhibitory activity is critical for radiosensitization and is likely to act in a combinatorial fashion with DNA repair inhibition by AZD6738. Live cell-cycle imaging suggests that the downstream consequence of DNA-damage repair inhibition and G2 checkpoint abrogation is a profound increase in chromosomal segregation errors.
Analysis revealed that AZD6738 combined with radiation produced daughter cells with micronuclei almost entirely comprising acentric chromosomal fragments. These fragments are likely to be highly heterogeneous and, consequently, the presence of micronuclei alone is not enough to predict cell death or survival. However, they represent increased genomic instability and have been described as a hallmark of mitotic catastrophe. Hence, assessing their accumulation after drug–radiation combinations may represent a method of assessing radiosensitizing potential. We have observed that, combined with radiation, ATRi generates more micronuclei than Chk1i. Micronuclei may represent a loss or a gain of chromosomal material, depending upon which daughter cell the micronucleus is transferred to. Whole chromosome or partial arm aneuploidy occurs at a high frequency in cancer (30). Micronuclei can persist for a number of mitoses or reintegrate into the primary nucleus resulting in increased genomic instability (31).
Micronuclei have also been shown to be dysfunctional relative to the primary nucleus. We observed dyssynchrony between the main nucleus and micronuclei and γH2AX-positive micronuclei following combination therapy with AZD6738 and radiation. Recent evidence has suggested micronuclei lack normal nuclear import function and are susceptible to rupturing of the micronuclei nuclear membrane. Micronuclei can be subject to reduced DNA replication and increased DNA damage during S-phase leading to "pulverization" of the micronuclei (32). This matches our observation of cell-cycle dyssynchrony. Further studying the lethality of these radiosensitizer-induced chromosomal missegregation errors may allow maximization of the therapeutic gains from this approach.
Conclusion
This study demonstrates radiosensitization of a variety of cancer cell lines with different intrinsic radiosensitivities and shows efficacy in vivo. Given the synergy already demonstrated between ATR inhibition and platinum chemotherapy (27), this strongly supports the further development of this approach to investigate ATR inhibition in combination with radiation and chemoradiation in clinical studies.
Supplementary Material
Acknowledgments
We thank Horizon discovery for the provision of cell lines. We would like to thank Alan Lau, Simon Smith and Glen Clack (AstraZeneca) for helpful discussions, and Selva Anbalagan for technical assistance.
Grant Support
This study was supported by Cancer Research United Kingdom (program grant C7224/ A13407), RM/ICR NIHR Biomedical Research Centre, Rosetrees Trust (M.T. Dillon, K.J. Harrington, grant numbers M48 and M444) and Anthony Long Charitable Trust (K.J. Harrington). M.T. Dillon is a Cancer Research United Kingdom Clinical Research Fellow. We thank Horizon discovery for the provision of cell lines.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Disclosure of Potential Conflict of Interest
K.J. Harrington reports receiving a commercial research grant from AstraZeneca. No potential conflicts of interest were disclosed by the other authors.
Authors' Contributions
Conception and design: M.T. Dillon, C.M. Nutting, M. McLaughlin, K.J. Harrington
Development of methodology: M.T. Dillon, H. Hafsi, M. McLaughlin
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): M.T. Dillon, H.E. Barker, K.L. Newbold, M. McLaughlin
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): M.T. Dillon, H.E. Barker, C.M. Nutting, M. McLaughlin, K.J. Harrington
Writing, review, and/or revision of the manuscript: M.T. Dillon, H.E. Barker, S.A. Bhide, K.L. Newbold, C.M. Nutting, M. McLaughlin, K.J. Harrington
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): M.T. Dillon
Study supervision: C.M. Nutting, M. McLaughlin, K.J. Harrington
Other (conducted experiments, acquired data): M. Pedersen
References
- 1.Dillon MT, Harrington KJ. Human papillomavirus-negative pharyngeal cancer. J Clin Oncol. 2015;33:3251–61. doi: 10.1200/JCO.2015.60.7804. [DOI] [PubMed] [Google Scholar]
- 2.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 3.Dillon MT, Good JS, Harrington KJ. Selective targeting of the G2/M cell cycle checkpoint to improve the therapeutic index of radiotherapy. Clin Oncol (R Coll Radiol) 2014;26:257–65. doi: 10.1016/j.clon.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Chen T, Stephens PA, Middleton FK, Curtin NJ. Targeting the S and G2 checkpoint to treat cancer. Drug Discov Today. 2012;17:194–202. doi: 10.1016/j.drudis.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Errico A, Costanzo V. Mechanisms of replication fork protection: a safe-guard for genome stability. Crit Rev Biochem Mol Biol. 2012;47:222–35. doi: 10.3109/10409238.2012.655374. [DOI] [PubMed] [Google Scholar]
- 6.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–27. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fokas E, Prevo R, Hammond EM, Brunner TB, McKenna WG, Muschel RJ. Targeting ATR in DNA damage response and cancer therapeutics. Cancer Treat Rev. 2013 doi: 10.1016/j.ctrv.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Prevo R, Fokas E, Reaper PM, Charlton PA, Pollard JR, McKenna WG, et al. The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol Ther. 2012;13:1072–81. doi: 10.4161/cbt.21093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pires IM, Olcina MM, Anbalagan S, Pollard JR, Reaper PM, Charlton PA, et al. Targeting radiation-resistant hypoxic tumour cells through ATR inhibition. Br J Cancer. 2012;107:291–9. doi: 10.1038/bjc.2012.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammond EM, Dorie MJ, Giaccia AJ. Inhibition of ATR leads to increased sensitivity to hypoxia/reoxygenation. Cancer Res. 2004;64:6556–62. doi: 10.1158/0008-5472.CAN-04-1520. [DOI] [PubMed] [Google Scholar]
- 11.Foote KM, Lau A, MN JW. Drugging ATR: progress in the development of specific inhibitors for the treatment of cancer. Future Med Chem. 2015;7:873–91. doi: 10.4155/fmc.15.33. [DOI] [PubMed] [Google Scholar]
- 12.Guichard SM, Brown E, Odedra R, Hughes A, Heathcote D, Barnes J, et al. Abstract 3343: The pre-clinical in vitro and in vivo activity of AZD6738: A potent and selective inhibitor of ATR kinase. Cancer Res. 2013;73:3343. [Google Scholar]
- 13.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NMB, Orr AI, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 14.Walton MI, Eve PD, Hayes A, Valenti MR, De Haven Brandon AK, Box G, et al. CCT244747 is a novel potent and selective CHK1 inhibitor with oral efficacy alone and in combination with genotoxic anticancer drugs. Clin Cancer Res. 2012;18:5650–61. doi: 10.1158/1078-0432.CCR-12-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:1–11. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Workman P, Aboagye EO, Balkwill F, Balmain A, Bruder G, Chaplin DJ, et al. Guidelines for the welfare and use of animals in cancer research. Br J Cancer. 2010;102:1555–77. doi: 10.1038/sj.bjc.6605642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gyori BM, Venkatachalam G, Thiagarajan PS, Hsu DClement M-V. Open-Comet: an automated tool for comet assay image analysis. Redox Biol. 2014;2:457–65. doi: 10.1016/j.redox.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem. 2001;276:47759–62. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- 19.Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–98. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 20.Peasland A, Wang LZ, Rowling E, Kyle S, Chen T, Hopkins A, et al. Identification and evaluation of a potent novel ATR inhibitor, NU6027, in breast and ovarian cancer cell lines. Br J Cancer. 2011;105:372–81. doi: 10.1038/bjc.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sultana R, Abdel-Fatah T, Perry C, Moseley P, Albarakti N, Mohan V, et al. Ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase inhibition is synthetically lethal in XRCC1 deficient ovarian cancer cells. PloS ONE. 2013;8:e57098. doi: 10.1371/journal.pone.0057098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krajewska M, Fehrmann RS, Schoonen PM, Labib S, de Vries EG, Franke L, et al. ATR inhibition preferentially targets homologous recombination-deficient tumor cells. Oncogene. 2015;34:3474–81. doi: 10.1038/onc.2014.276. [DOI] [PubMed] [Google Scholar]
- 23.Mohni KN, Kavanaugh GM, Cortez D. ATR pathway inhibition is synthetically lethal in cancer cells with ERCC1 deficiency. Cancer Res. 2014;74:2835–45. doi: 10.1158/0008-5472.CAN-13-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohni KN, Thompson PS, Luzwick JW, Glick GG, Pendleton CS, Lehmann BD, et al. A synthetic lethal screen identifies DNA repair pathways that sensitize cancer cells to combined ATR inhibition and cisplatin treatments. PloS One. 2015;10:e0125482. doi: 10.1371/journal.pone.0125482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilad O, Nabet BY, Ragland RL, Schoppy DW, Smith KD, Durham AC, et al. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res. 2010;70:9693–702. doi: 10.1158/0008-5472.CAN-10-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montana MF, et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol. 2011;18:1331–5. doi: 10.1038/nsmb.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall AB, Newsome D, Wang Y, Boucher DM, Eustace B, Gu Y, et al. Potentiation of tumor responses to DNA damaging therapy by the selective ATR inhibitor VX-970. Oncotarget. 2014;5:5674–85. doi: 10.18632/oncotarget.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harper JV, Anderson JA, O'Neill P. Radiation induced DNA DSBs: Contribution from stalled replication forks? DNA Repair. 2010;9:907–13. doi: 10.1016/j.dnarep.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Wang H, Powell SN, Iliakis G, Wang Y. ATR affecting cell radiosensitivity is dependent on homologous recombination repair but independent of nonhomologous end joining. Cancer Res. 2004;64:7139–43. doi: 10.1158/0008-5472.CAN-04-1289. [DOI] [PubMed] [Google Scholar]
- 30.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 31.Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer. 2012;12:663–70. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]
- 32.Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, et al. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–84. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.