Abstract
The aryl hydrocarbon receptor (AHR) recognises xenobiotics as well as natural compounds such as tryptophan metabolites, dietary components and microbiota-derived factors1–4 and is important for maintenance of homeostasis at mucosal surfaces. AHR activation induces cytochrome P4501 (CYP1) enzymes, which oxygenate AHR ligands, leading to their metabolic clearance and detoxification5. Thus, CYP1 enzymes appear to play an important feedback role that curtails the duration of AHR signalling6, but it remains elusive whether they also regulate AHR ligand availability in vivo. Here we show that dysregulated expression of Cyp1a1 depletes the reservoir of natural AHR ligands, generating a quasi AHR-deficient state. Constitutive expression of Cyp1a1 throughout the body or restricted specifically to intestinal epithelial cells (IECs) resulted in loss of AHR-dependent type 3 innate lymphoid cells (ILC3) and T helper 17 (Th17) cells and increased susceptibility to enteric infection. The deleterious effects of excessive AHR ligand degradation on intestinal immune functions could be counter-balanced by increasing the intake of AHR ligands in the diet. Thus, our data indicate that IECs serve as gatekeepers for the supply of AHR ligands to the host and emphasise the importance of feedback control in modulating AHR pathway activation.
It is increasingly understood that AHR signalling needs to be tightly controlled, since prolonged activation either by ligands that resist metabolic clearance or by constitutively active AHR has deleterious effects7–10. Enzymes of the CYP1A and CYP1B sub-families (CYP1) control AHR signalling due to their capacity for metabolising ligands and thereby terminating AHR activation.
We hypothesized that excessive CYP1A1-mediated metabolic clearance of natural AHR ligands would impact the intestinal immune system, where several immune cell types are dependent on AHR signalling for their survival. To investigate this, we generated a mouse model with constitutive Cyp1a1 expression due to its placement under control of the Rosa26 promoter (R26Cyp1a1) (Extended Data Fig.1). As proof of principle we investigated the effect of constitutive Cyp1a1 expression on Th17 cells, which express AHR and produce IL-22 in an AHR-dependent manner11. Th17 cells generated from R26Cyp1a1 mice had increased CYP1A1 enzymatic activity compared with wild-type B6 (WT) or Ahr-deficient Th17 cells (Fig.1a). CD4+ T cells from WT, Ahr-deficient or R26Cyp1a1 mice were cultured under Th17 cell conditions in the presence of the endogenous, tryptophan-derived AHR ligand 6-formylindolo[3,2-b]carbazole (FICZ) and residual cellular FICZ levels following metabolic clearance were determined. FICZ did not decay in cultures of Ahr-deficient Th17 cells due to absence of CYP1A1 induction, but cultures of R26Cyp1a1 Th17 cells showed accelerated clearance of FICZ compared with those from WT Th17 cells (Fig.1b). Furthermore, Th17 cells generated from R26Cyp1a1 mice produced less IL-22 to baseline AHR ligands present in culture medium12 and to low, but not higher FICZ concentrations (Fig.1c and Extended Data Fig.2). Ahr-deficient Th17 cells did not produce IL-22 under any condition (Fig.1c). Thus, constitutive CYP1A1 activity decreased AHR ligands and compromised IL-22 production.
Figure 1. CYP1A1 controls AHR ligand availability.
a, CYP1A1 enzyme activity in Th17 cells (n=3 per group). b, Intracellular levels of FICZ in Th17 cell cultures (n=2 per time point). c, IL-22 protein in supernatants of in vitro differentiated Th17 cells (n=4 per group). d,e, Absolute numbers of ILC subsets in the small intestine or colon (n=3 per group). Results are representative of three independent experiments. Error bars represent mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, as calculated by one-way ANOVA with Tukey post-test a, c, d and e or two-way ANOVA with Dunnett’s post-test in b. ns = not significant.
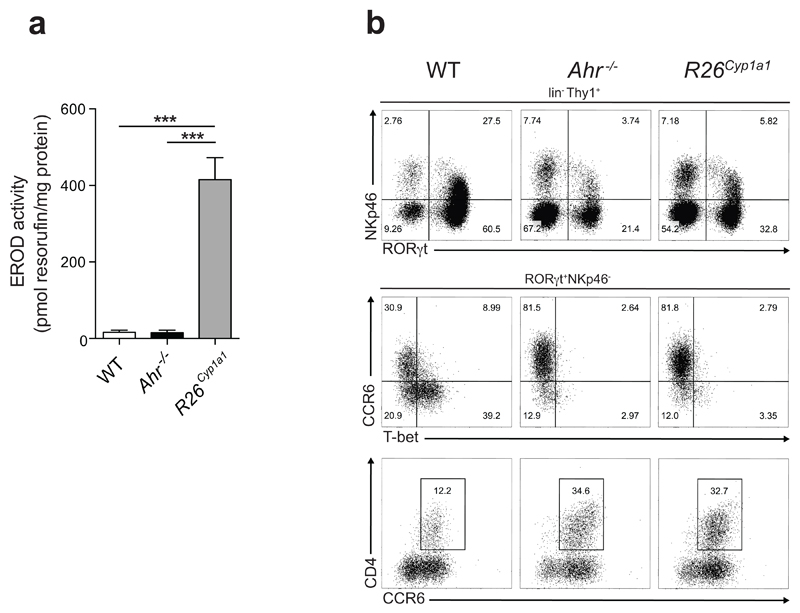
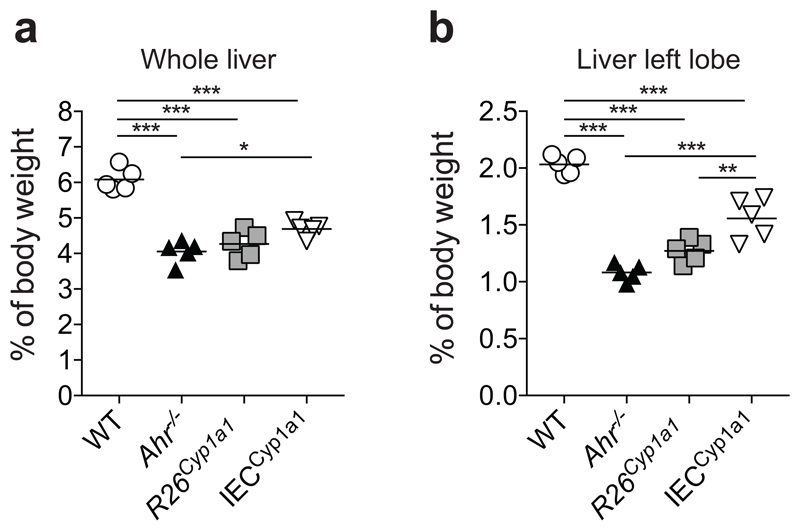
Ahr deficiency in haematopoietic cells causes the disappearance of intraepithelial lymphocytes13 and innate lymphoid cells (ILC3) in the intestinal immune system 14–16. R26Cyp1a1 mice exhibited enhanced intestinal CYP1A1 enzyme activity (Extended Data Fig.3a) and loss of ILC3 in colon and small intestine similar to Ahr-deficient mice (Fig.1d,e and Extended Data Fig.3b), suggesting that rapid metabolic clearance of natural AHR ligands mimics an Ahr-deficient state. In addition, R26Cyp1a1 mice displayed similar developmental abnormalities to Ahr-deficient mice, such as decreased liver weight indicative of patent ductus venosus17 (Extended Data Fig.4).
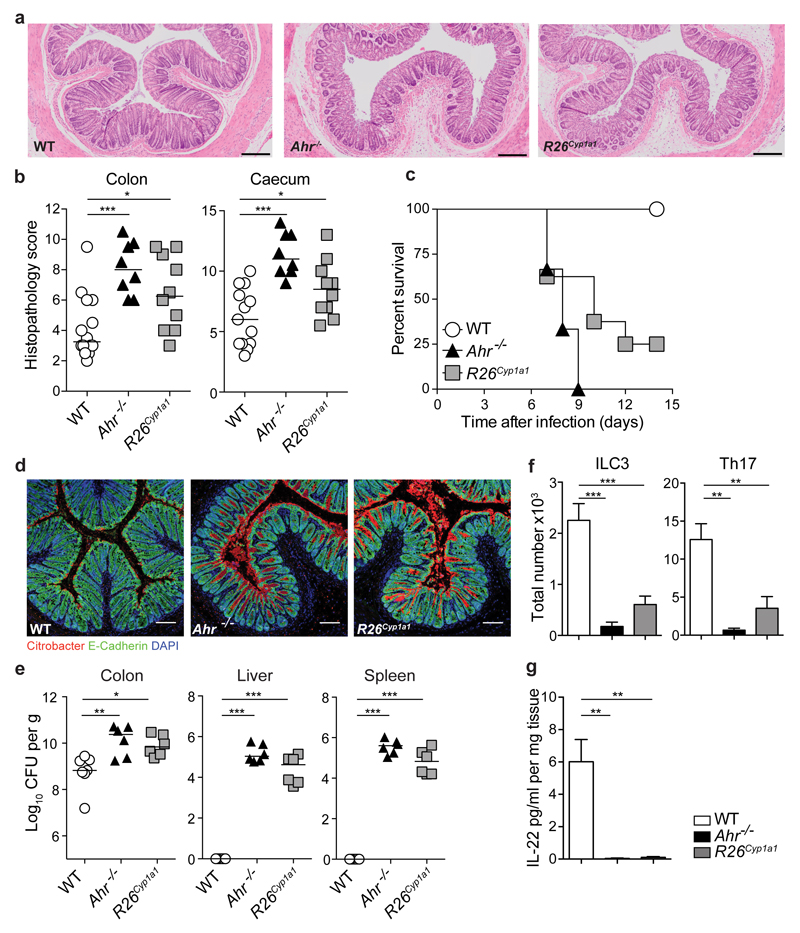
We next investigated the response of R26Cyp1a1 mice to infection with the intestinal pathogen Citrobacter rodentium, which causes severe pathology in Ahr-deficient14–16, compared with WT mice. R26Cyp1a1 mice also had increased pathology, albeit not as extensive as that seen in Ahr-deficient mice (Fig.2a,b). While all WT mice survived the infection, Ahr-deficient mice reached endpoint by day 9 after infection and R26Cyp1a1 mice survived only a few days longer (Fig.2c). C. rodentium penetrated deeply into the base of the crypts in Ahr-deficient and R26Cyp1a1 mice, but mainly attached to the luminal surface of the epithelium in WT mice (Fig.2d). The bacterial burden in the colon of Ahr-deficient and R26Cyp1a1 mice was higher and failure to clear the infection resulted in dissemination of bacteria to liver and spleen (Fig.2e). IL-22 is essential in the defence against C. rodentium18 and the extreme susceptibility of Ahr-deficient mice can be attributed to their lack of ILC3 as well as failure to mount a T cell IL-22 response. This deficiency in IL-22 as well as Th17 cells and ILC3 was mirrored in R26Cyp1a1 mice (Fig.2f, g). Administration of recombinant IL-22–Fc fusion protein significantly reduced bacterial burden and crypt invasion and prolonged survival of R26Cyp1a1 mice (Extended Data Fig.5).
Figure 2. Depletion of natural AHR ligands leads to impaired immunity to C. rodentium.
a, Representative photomicrographs of H&E stained colon sections of C. rodentium infected mice (day7). Scale bars represent 200μm. b, Pathology scores. Bars show the mean and each symbol represents an individual mouse. c, Survival plot (WT: n=8, R26Cyp1a1: n=8, Ahr-/-: n=6). d, Colon sections stained for E-Cadherin (green), C. rodentium (red) and DAPI (blue). Scale bars represent 100μm. e, C. rodentium burdens in colon, liver and spleen. Bars show the median and each symbol represents an individual mouse. f, Absolute numbers of colonic RORγt+ ILC3 (WT: n=7, R26Cyp1a1: n=7, Ahr-/-: n=3) and IL-17A-producing TCRβ+CD4+ T cells (WT: n=5, R26Cyp1a1: n=6, Ahr-/-: n=4), mean ± s.e.m. g, IL-22 protein colon explant cultures (WT: n=10, R26Cyp1a1: n=6, Ahr-/-: n=5), mean ± s.e.m. Data represent pooled results of at least two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, as calculated by one-way ANOVA with Tukey post-test.
Thus, constitutive Cyp1a1 expression severely impairs the intestinal immune response against an enteric pathogen.
If CYP1A1-mediated clearance of natural ligands produces a quasi Ahr-deficient state, the absence of metabolic clearance should increase ligand availability. To test this, we investigated mice with deletion of the three AHR controlled CYP enzymes CYP1A1, CYP1A2 and CYP1B1, termed Cyp1 knockout19. Th17 cells from Cyp1 knockout mice failed to metabolise FICZ and showed increased Cyp1a1 expression as well as enhanced IL-22 production in the absence of added AHR ligands with reduced histopathology scores and bacterial burdens upon infection (Extended Data Fig.6).
Increased ligand availability in Cyp1-deficient mice was visualized in a Cyp1a1 fate-reporter mouse that reports AHR activity via induction of an eYFP reporter activated via targeted insertion of Cre recombinase in the murine Cyp1a1 locus20. Although Cyp1-deficient mice failed to produce this enzyme, they still reported AHR stimulation with induction of Cre mediated eYFP expression. In the intestine of WT Cyp1a1 reporter mice eYFP expression was scarce, reflecting the transient nature of AHR signalling that limits Cre induction (Fig.3a left). However, Cyp1-deficient mice showed enhanced eYFP expression primarily in EpCAM+ IECs (Fig.3a left). Dietary supplementation with indole-3-carbinol (I3C), a tryptophan-derived phytochemical that is converted to high affinity AHR ligands by exposure to stomach acid21, further enhanced the eYFP signal in EpCAM+ IECs of Cyp1-deficient mice (Fig.3a,b).
Figure 3. IECs serve as gatekeeper for the supply of AHR ligands to the host.
a, Cyp1a1 expression gated on live colonic cells of mice fed control or I3C containing diet for two weeks (left panel) and colon sections stained for E-Cadherin (red), Cyp1a1 (green) and DAPI (blue). Scale bars represent 100μm (right panel). b, Cyp1a1 expression in the colon of WT Cyp1a1 reporter (WT) and Cyp1-/- Cyp1a1 reporter mice treated with control or I3C diet. c, Absolute numbers of RORγt+ ILC3 in mice of indicated genotypes, mean ± s.e.m. (n=3 per group). d, Survival plot of C. rodentium infected mice (n=4 per group). e, Colon sections stained for E-Cadherin (green), C. rodentium (red) and DAPI (blue) of infected mice (day 7). Scale bars represent 100μm. f, Representative photomicrographs of (H&E) stained colon sections of C. rodentium infected mice (day 7). Scale bars represent 100μm. g, C. rodentium burdens. Bars show the median, symbols represent individual mice. h, Absolute numbers of colonic RORγt+ ILC3 and Th17 cells (WT: n=6, IECCyp1a1: n=3) and IL-22 protein in colon explant cultures (n=6 per group), mean ± s.e.m.. i, Pathology scores (day 6) of distal colon and caecum in bone marrow chimeras of wildtype (WT→WT) or Cyp1 deficient (WT→Cyp1-/-) recipients. Bars show the mean, symbols represent individual mice. j, C. rodentium burdens in the colon. Bars are the median and symbols represent individual mice. k, Absolute numbers of WT donor-derived colonic RORγt+ ILC3 and Th17 cells (n=7 per group) and IL-22 protein in distal colon organ explant cultures (WT: n=7, Cyp1-/-: n=6), mean ± s.e.m.. Results are representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 as calculated by Student’s t-test or one-way ANOVA with Tukey post-test. ns = not significant.
It has been shown previously that IEC specific deletion of the AHR partner ARNT causes systemic upregulation of Cyp1a122. Thus, we next tested Villin-cre-R26LSL-Cyp1a1 mice (termed IECCyp1a1) in which Cyp1a1 expression was restricted to IECs (Extended Data Fig.1b). Unexpectedly, IECCyp1a1 mice had drastically reduced numbers of ILC3 similar to Ahr-deficient mice and mice with constitutive Cyp1a1 expression throughout the body (Fig. 3c).
In contrast, mice with constitutive Cyp1a1 expression restricted to adaptive immune cells via Rag1-Cre (Rag1Cyp1a1), had normal numbers of ILC3 under steady state conditions (Fig.3c and Extended Data Fig.1b).
This suggests that constitutive CYP1A1 activity in IECs, but not in adaptive immune cells, restricts the availability of AHR ligands to cells in the intestinal lamina propria, resulting in loss of AHR ligand-dependent ILC3.
Upon infection with C. rodentium IECCyp1a1 mice rapidly succumbed (Fig. 3d) with increased bacterial crypt invasion (Fig. 3e) and intestinal pathology (Fig. 3f).
C. rodentium disseminated into liver and spleen of IECCyp1a1 mice (Fig.3g) and the reduction of ILC3 and Th17 cells collectively affected the amount of IL-22 that was detectable in colon tissue (Fig.3h). In contrast, C. rodentium infected Rag1Cyp1a1 survived similarly to WT mice over 14 days after infection and had no significant differences in bacterial load, pathology score or dissemination of bacteria (Extended Data Fig.7a-c). While ILC3 were not affected, Th17 cells and their IL-22 production were strongly reduced (Extended Data Fig.7d). Thus, enhanced AHR ligand degradation in T cells compromises T cell derived IL-22 responses, but spares ILC3. In contrast, enhanced AHR ligand degradation in IECs deprives ILC3 as well as T cells of AHR stimulation, thereby causing a more severe deficiency in immune defence against C. rodentium.
Next we investigated whether lack of AHR ligand metabolism in non-haematopoietic cells affects the immune response to C. rodentium infection. We generated bone marrow chimeras by transferring WT donor bone marrow into irradiated WT (WT→WT) or Cyp1-deficient (WT→Cyp1-/-) recipients, followed by infection with C. rodentium. WT→Cyp1-/- chimeras showed significantly reduced pathology (Fig.3i) as well as bacterial loads (Fig.3j) compared with WT recipients (WT→WT). Furthermore, Cyp1-/- recipients had increased numbers of WT donor-derived ILC3 and Th17 cells and markedly higher IL-22 levels (Fig.3k). Thus, absence of CYP1-mediated AHR ligand metabolism in IECs results in increased ligand availability to intestinal immune cells and promotes resistance to enteric infection.
These data are consistent with a crucial role for IECs as gatekeepers for the availability of AHR ligands in the intestinal immune system.
To further substantiate the role of CYP1A1 in controlling ligand availability, we investigated whether increasing AHR ligands by exogenous application of I3C in the diet might mitigate the loss of AHR signalling in R26Cyp1a1 mice.
R26Cyp1a1 mice infected with C. rodentium that were fed on control diet all succumbed to infection by day 12 and failed to clear the infection, whereas all mice on the I3C supplemented diet survived and had cleared the infection by day 14 (Fig.4a,b). Penetration of C.rodentium to the crypts was abrogated (Fig.4c) and mice on I3C diet had drastically reduced pathology (Fig.4d,e). IL-22 producers in R26Cyp1a1 mice on I3C diet had recovered (Fig.4f,g), whereas blockade of IL-22 during I3C diet exposure abrogated the protective effect (Fig.4h). The beneficial effect of I3C is due to the combination of strong inhibition of CYP1A1 activity by its acid-condensation products DIM and ICZ (Fig. 4i) and the agonist activity of DIM and ICZ, which are known AHR ligands23. ICZ like FICZ, but unlike DIM is also efficiently metabolised by CYP1A1 (Extended Data Fig.8).
Figure 4. Dietary AHR ligands restore immunity to C. rodentium.
a, Survival plot C. rodentium infected R26Cyp1a1 mice fed control or I3C diet (n=6 per group). b, C. rodentium burdens in faeces (Ctrl: n=5, I3C: n=6). c, Colon sections of C. rodentium infected mice stained for E-Cadherin (green), C. rodentium (red) and DAPI (blue). Scale bars represent 100μm. d, Representative photomicrographs of H&E stained colon sections. Scale bars represent 100μm. e, Pathology scores. Bars show the mean and symbols represent individual mice. f, Absolute numbers of cytokine-producing TCRβ+CD4+ T cells in the colon of C. rodentium infected mice (n=4 per group), mean ± s.e.m.. g, IL-17A and IL-22 expression in TCRβ+CD4+ T cells from the colon of C. rodentium infected mice. h, Survival plot of R26Cyp1a1 mice fed I3C diet treated with anti-IL-22 blocking antibody (n=6) or isotype control (n=5). i, Concentration-dependent inhibition of human recombinant CYP1A1 enzyme activity by I3C, DIM and FICZ.
Results are representative of three independent experiments. Error bars, mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, as calculated by Student’s t-test.
Thus, the excessive clearance of natural ligands, which produces a quasi Ahr-deficient state in R26Cyp1a1 mice can be ameliorated by the supply of dietary AHR ligands, allowing sufficient AHR signalling to restore IL-22 production by Th17 cells and successful eradication of C. rodentium.
The AHR pathway is of crucial importance in the intestinal immune system and genome-wide association studies have identified AHR as a susceptibility locus in inflammatory bowel disease24. While the main focus has been on AHR itself or on its ligands2,4,25, we show here that feedback regulation of the AHR pathway by CYP1 enzymes controls ligand availability and thereby activation of the AHR pathway, with a critical role for IECs in controlling availability of ligands to the intestinal immune system. Thus, constitutive CYP1A1 activity in IECs reduced the availability of AHR ligands in the body, causing the decay of intestinal immune cells such as ILC3 that depend on AHR signals for survival and impairment of the adaptive IL-22 response upon infection with intestinal pathogens. Conversely, lack of CYP1A1-mediated metabolism confined to IECs had the opposite effect, increasing protection against intestinal infection.
As previously shown in an in vitro setting inhibition of CYP1A1 results in a boost to AHR pathway activation due to increased availability of AHR ligand FICZ26. Here we demonstrate that genetic deletion of CYP1 enzymes delays ligand metabolism, resulting in increased AHR signals in vivo. Sources of AHR ligands are abundant and include dietary compounds21, microbial virulence factors4 as well as AHR ligands derived via microbiota or host cell-mediated tryptophan metabolism2. Given that environmental factors such as oxidative stress, chemical pollutants and dietary factors modulate CYP1 enzyme activity26, interference with CYP1-mediated AHR ligand degradation has potential consequences for AHR pathway activation and intestinal homeostasis.
Our finding that the intestinal pathology resulting from constitutive CYP1A1 could be counter-balanced by increasing the intake of AHR ligands in the diet suggests that dysregulated CYP1A1 activity is potentially amenable to therapeutic manipulation via dietary supplementation.
Online Material and Methods
Mice
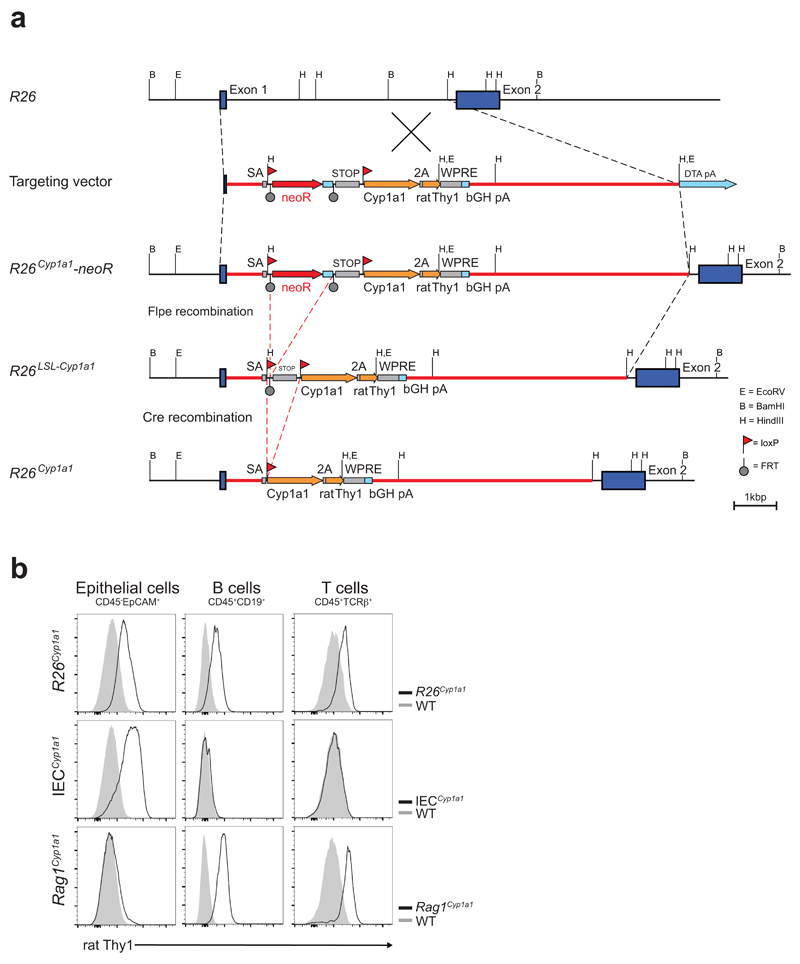
For the generation of a constitutively active as well as an inducible Cyp1a1 allele the coding sequence (CDS) of Cyp1a1 was linked to the CDS of rat Thy1 using a 2A sequence. In order to stabilize the transcriptional activity of the inserted genes, a woodchuck hepatitis virus derived regulatory element in conjunction with a bovine growth hormone polyadenylation site was inserted 3’ of the CDSs. This module was inserted downstream of a cassette containing an FRT-flanked neomycin-resistance gene and loxP-flanked transcriptional stop elements. The targeting vector did not contain any artificial promoter for the expression of both Cyp1a1 and rat Thy1 to avoid toxicity caused by Cyp1a1 overexpression (details of the gene targeting and Rosa26-targeting strategy, Extended Data Figure 1). The targeting vector was generated by Biocytogen and used to establish R26Cyp1a1-neoR mice by homologous recombination in BCGEN B61-6 (C57BL/6) embryonic stem cells. R26Cyp1a1-neoR mice were generated after successful germ line transmission and backcrossed to C57BL/6J. This strain was further bred with mice expressing the Cre recombinase under the control of the mouse protamine (Prm1) promoter to obtain constitutive Cyp1a1 expression in all cell-types (referred to as R26Cyp1a1 mice) or with ACTB:FLPe mice to obtain mice with a conditional floxed Cyp1a1 allele (referred to as R26LSL-Cyp1a1). These were crossed with Villin-cre mice to generate the IECCyp1a1 strain (constitutive Cyp1a1 expression specific to intestinal epithelial cells) and Rag1-cre mice to generate the Rag1Cyp1a1 strain (constitutive Cyp1a1 expression specific to T and B cells).
To generate the Cyp1a1 reporter strain Cyp1a1Cre mice20 (with targeted insertion of Cre recombinase in the murine Cyp1a1 locus) were crossed with R26ReYFP mice27,expressing eYFP from the ubiquitous Rosa26 promoter downstream of a cassette containing a loxP-flanked transcriptional stop element. This strain was further bred with Cyp1-/- mice19 to obtain Cyp1-/- Cyp1a1 reporter mice. All mice used in this study were either generated in C57BL/6 ES cells or backcrossed to C57BL/6 mice for at least 10 generations. All mice were bred in the Francis Crick Institute animal facility under specified pathogen–free conditions. All animal procedures were conducted under a Project Licence granted by the UK Home Office. Mice were age and sex-matched and more than 6 weeks old when first used. Both female and male mice were used in experiments. Generally, each mouse of the different experimental groups is reported. Exclusion criteria such as inadequate staining or low cell yield due to technical problems were pre-determined. Animals were assigned randomly to experimental groups.
In vitro T cell differentiation
CD4+ T cells were isolated using EasySep mouse CD4+ T cell isolation kit (Stemcell Technologies) with the addition of biotinylated anti-CD25 antibody (BioLegend). Cells were cultured in Iscove’s modified Dulbecco medium (IMDM, Sigma) supplemented with 2mM L-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, 0.05mM β-mercaptoethanol and 5% fetal calf serum (biosera). CD4+ T cells were differentiated in 48 well plates coated with 1 µg/ml anti-CD3 (clone 145-2C11, eBioscience) and 10 µg/ml soluble anti-CD28 (clone 37.51, BioLegend) in the presence of 2ng/ml TGF-β1, 20ng/ml IL-6, 10ng/ml IL-1β (all R&D Systems) and 10μg anti-IFN-γ (BioXCell). In some cultures 6-formylindolo[3,2-b]carbazole (FICZ, EnzoLifeSciences) was added from the start of culture. IL-22 cytokine levels in culture supernatants were determined by ELISA (eBioscience).
Isolation of lamina propria cells and flow cytometry
Colon and small intestine were cut open longitudinally and incubated in wash buffer (IMDM 1%FCS, 5mM EDTA, 10mM HEPES, penicillin/streptomycin, and 2mM DTT) for 20min at 37°C with 200rpm shaking. Colon tissue was collected, cut into small pieces and incubated in digestion buffer (IMDM supplemented with 1% FCS, 10mM HEPES, penicillin/streptomycin, 50μg/ml DNAse I (Roche)) containing 0.4mg/ml Liberase TL (Roche) and incubated for 30min at 37°C with 200rpm shaking. Small intestinal tissue was collected, cut into small pieces and incubated in digestion buffer (IMDM supplemented with 1% FCS, 10mM HEPES, penicillin/streptomycin, 50μg/ml DNAse I (Roche)) containing 1mg/ml Collagenase VIII (Sigma) and incubated for 10min at 37°C with 200rpm shaking. Single cell suspensions from colon and small intestine were further subjected to 40% Percoll (Amersham) density gradient centrifugation to remove debris. For surface staining, cell suspensions the lamina propria were incubated with anti-CD16/CD32 (eBioscience) and fixable live/dead cell dye (ThermoFisher) followed by staining with antibodies against CD11b, CD3, TCRγδ, Gr1, CD11b, CD19, NKp46, CCR6, TCR-β, Thy1.1 (all BioLegend) and Thy1.2, CD45, CD4 (all BD Biosciences). For intracellular staining, single cell suspension were re-stimulated for 2hrs and 15min in the presence of 1ng/ml phorbol-12-myristate-13-acetate (PMA), 1 μg/ml ionomycin and 10 μg/ml Brefeldin A (all Sigma), washed and stained for surface markers as described above. Cell were then fixed in eBioscience Fix/Perm buffer or 4% formaldehyde (for preservation of eYFP fluorescence) for 30min on ice followed by permeabilization in eBioscience permeabilization buffer for 45min in the presence of antibodies against IL-17A, IL-22, T-bet (all ebioscience) and RORγt (BD Biosciences). Cells were acquired with a BD Fortessa X20 and analysis was performed with FlowJo v10 (Tree Star) software.
High performance liquid chromatography (HPLC) analysis of FICZ clearance and CYP1A1-mediated metabolism of DIM, ICZ and FICZ
Cellular clearance: CD4+ T cells were isolated and cultured under Th17-cell-inducing conditions as described above and 5nM FICZ (EnzoLifeSciences) added at the start of culture. At various time-points cells were collected, washed in PBS and re-suspended in 200µl distilled water and stored at -20°C. Prior to HPLC analysis the cells were homogenized by sonication on ice (4 x 5 seconds) using a MSE Soniprep 150 equipped with an exponential probe (Measuring and Scientific Equipment). Extraction and chemical analysis of the homogenates, using an in-line solid-phase extraction column coupled to a reverse-phase C18 analytical column, was performed as previously described26. FICZ quantity was determined according to a standard curve of FICZ and the results were normalized to total protein contents determined by Pierce Coomassie protein assay kit according to the manufacturer´s instructions.
CYP1A1-mediated metabolism: Time-dependent metabolism of DIM, ICZ and FICZ was studied in the presence of human recombinant CYP1A1 (3.5nM) and the co-factor NADPH (1.0mM). Each compound (0.1µM) was incubated with CYP1A1 and NADPH in Tris-HCl (0.1M, pH 7.4) with EDTA (1mM) at 37°C. At various time-points samples were collected and acetonitrile was added to each sample at a final concentration of 20%, followed by vortexing (30 seconds) and centrifugation (10 minutes at 12x106 rpm and 4°C). Chemical analysis of the supernatants was performed using the same HPLC equipment as for the cellular clearance measurements. Separation of respective compound was achieved using a reverse-phase C18-AR column (ACE, 4.6 × 150mm, 5μm particle size) with a mobile phase consisting of water (A) and acetonitrile (B), both containing 1.5mM formic acid. Initially, the solvent contained 30% B, with a linear increase to 100% B during a period of 20 min, at a flow rate of 0.8 mL/min. DIM, ICZ and FICZ was detected using excitation/emission wavelengths of 230/460 (DIM and ICZ) or 390/525nm (FICZ). All compounds were quantified according to separate standard curves. To determine non-enzymatic degradation of each compound parallel incubations were performed where phosphate buffer was substituted for nadph.
Ethoxyresorufin-O-Deethylase (EROD) Assay
Intestinal tissue homogenates or CD4+ T cells cultured for 48 hours under Th17-cell-inducing conditions were washed in PBS followed by incubation with 2 μM 7-ethoxyresorufin in sodium phosphate buffer (50 mM, pH 8.0) at 37 °C for 30 minutes. The reaction was terminated by adding fluorescamine (Sigma) dissolved in acetonitrile. Formation of resorufin (excitation/emission of 535/590nm) and fluorescamine (excitation/emission of 390/485nm) was quantified using a plate reader.
Inhibition of CYP1A1 activity by I3C, DIM and ICZ
Effects on the ethoxyresorufin deethylase (EROD) activity of human recombinant CYP1A1 (2.5nM) was assayed by first pre-incubating the enzyme with the compound to be tested for 5 min in Tris-HCl (0.1M, pH 7.4) with EDTA (1mM) at 37°C followed by addition of ethoxyresorufin (0.1 μM) and NADPH (0.4 mM). Formation of resorufin was quantified using a multiwell plate reader with the excitation/emission wavelengths of 535/590 nm) and activity of the enzyme was determined by the rate of resorufin formation. ICZ and FICZ were purchased from Syntastic AB (Stockholm, Sweden). DIM, I3C, 7-ethoxyresorufin, β-Nicotinamide adenine dinucleotide 2′-phosphate (NADPH, N7505) and human recombinant cytochrome P4501A1 with P450 reductase (C3735) was purchased from Sigma-Aldrich.
Immunofluorescence Microscopy
Tissues were fixed in 4% phosphate-buffered formaldehyde solution (Fisher Scientific) for 24 hours. Fixed tissue sections were de-paraffinised and antigen retrieval performed in 0.01M sodium citrate buffer. Slides were blocked with goat serum, stained with mouse anti-E-cadherin (BD, 610181) and rabbit anti-C.rodentium antiserum followed by staining with secondary antibodies (AF555-conjugated goat-anti-rabbit and AF488-conjugated goat-anti-mouse from ThermoFisher). Slides were further stained with DAPI (Sigma) and mounted in Fluoromount-G (SouthernBiotech) and visualized using a Leica Confocal SP5-Invert microscope. For staining of eYFP, tissue sections were fixed in 4% paraformaldehyde at 4°C for 16 hours followed by incubation in 30% sucrose for 24 hours. Tissues were embedded in O.C.T. compound (VWR) followed by cryosectioning. Slides were blocked with rabbit serum and stained with AF488-conjugated rabbit-anti-GFP (ThermoFisher, A21311) and AF647-conjugated mouse-anti-E-cadherin (BD, 560062) and visualized using a Leica Confocal SP5-Invert microscope. Image analysis was performed in ImageJ.
Infection with Citrobacter rodentium
For C. rodentium infection a single colony of strain DBS100 (ATCC 51459; American Type Culture Collection) was transferred to Luria–Bertani (LB) broth and grown to log phase followed by centrifugation and resuspension in PBS. Mice were orally gavaged with 200μl of PBS containing 2x109 C. rodentium. To determine bacterial load, intestinal tissue pieces or faecal pellets were weighed and homogenized in sterile PBS and serial dilutions were plated onto Brilliance E. coli/coliform Selective Agar (Fisher Scientific) or LB agar plates (liver and spleen) for measurement of colony-forming units (CFU). For neutralization of IL-22, mice were injected intraperitoneally (i.p.) three times per week with 150μg/mouse per dose monoclonal anti-IL-22 (clone 8E11, Genentech) or mouse IgG1 isotype control (BioXCell/2BScientific). Where indicated, mice were injected i.p. three times per week with 125μg/ml per dose IL-22–Fc (PRO312045, Genentech) or mouse IgG2a isotype control (BioXCell/2BScientific).
Diet studies
For diet studies mice were fed purified diet AIN-93M (TestDiet-IPS) or AIN-93M supplemented with 200ppm indole-3-carbinol (Sigma). Mice were put on purified diets shortly after weaning for at least 4 weeks and maintained on the purified diets throughout the duration experiments.
Generation of bone marrow chimeric mice
Bone marrow was injected intravenously (2.5x106 cells per mouse) into recipient mice irradiated with two doses of 5Gy using a 137Cs-source. Donor bone marrow and recipient mice were distinguished on the basis of congenic markers. Chimeric mice were used in experiments 6-8 weeks following reconstitution.
Colon explant cultures
Intestinal tissue pieces (0.5-1cm length) were cultured for 24 hours in complete IMDM medium. IL-22 cytokine levels in the supernatants were determined by ELISA (eBioscience) and concentrations were normalized to the weight of the explants.
Histological Assessment
Tissues from distal colon and caecum were fixed in 4% phosphate-buffered formaldehyde solution (Fisher Scientific) for 24 hours, cut and stained with hematoxylin and eosin. Slides were blinded and scored (0-15) for parameters of inflammation and tissue damage as described in28.
Statistical Analysis
For comparisons between two groups unpaired, two-tailed Student’s t-test was used or when appropriate a two-way ANOVA with Dunnett’s post-test. For the comparison of three groups a one-way ANOVA followed by Tukey multiple comparison test was performed. All statistical analysis was calculated in Prism (GraphPad 6). No statistical methods were used to predetermine sample size.
Data availability statement
The authors declare that the source data supporting the findings of this study are available within the paper and its extended data files.
Extended Data
Extended Data Figure 1. Generation of R26Cyp1a1 allele.
a, The endogenous Rosa26 locus, the gene targeting vector, the targeted Rosa26 allele including the Neo resistance gene cassette (R26Cyp1a1-neoR), the targeted allele (R26LSL-Cyp1a1) after FLPe-mediated recombination and the ubiquitously expressed R26Cyp1a1 are schematically depicted to scale. A minigene composed of the coding sequences of mouse Cyp1a1 and rat Thy1 connected by a 2A sequence followed by the woodchuck hepatitis virus derived regulatory element (WPRE) and a bovine growth hormone polyadenylation site (bGH pA). b, Expression of rat THY1 in indicated cell types in the colon of R26Cyp1a1 , IECCyp1a1 (Villin-Cre), Rag1Cyp1a1 (Rag1-Cre) strains.
Extended Data Figure 2. Altered AHR ligand availability affects IL-22 production.
Flow cytometry analysis of IL-17A and IL-22 expression in in vitro differentiated Th17 cells from indicated genotypes (day 4) exposed to DMSO, 0.01nM FICZ or 1nM FICZ from the start of culture.
Extended Data Figure 3. Altered AHR ligand affects intestinal AHR-dependent ILC populations.
a, CYP1A1 enzyme activity, measured by EROD assay, in intestinal tissue homogenates of steady state mice. b, Flow cytometry analysis of NKp46 and RORγt expression in CD45+ lineage negative (TCRβ-CD3-TCRγδ-CD19-CD11b-Gr1-) Thy1+ live cells in the small intestine (upper panel) and phenotypic analysis of RORγt+NKp46- innate lymphoid cells (lower panel).
Results are representative of three independent experiments (n=3).
Error bars, mean ± s.e.m. ***P < 0.001, as calculated by one-way ANOVA with Tukey post-test.
Extended Data Figure 4. Effects of Cyp1a1 overexpression on liver development.
a, Liver weight represented as percentage of body weight. b, Liver (left lobe) weight represented as percentage of body weight. Results are representative of two independent experiments. Bars are the mean and each symbol represents an individual mouse.
*P < 0.05, **P < 0.01, ***P < 0.001, as calculated by one-way ANOVA with Tukey post-test.
Extended Data Figure 5. IL-22-Fc improves resistance to C. rodentium infection in R26Cyp1a1 mice.
Mice of indicated genotypes were infected orally with ~2x109 C. rodentium and killed 7 days after infection or monitored for survival. a, C. rodentium burdens in the colon and caecum. Bars are the median and each symbol represents an individual mouse. b, Colon sections stained for E-Cadherin (green), C. rodentium (red) and DAPI (blue). Scale bars represent 100μm. c, Survival plot (WT: n=4, R26Cyp1a1 +Ctrl Ig: n=6, R26Cyp1a1 +IL-22Fc: n=5).
Results are representative of three independent experiments.
NS = not significant, **P < 0.01, ***P < 0.001, as calculated by one-way ANOVA with Tukey post-test.
Extended Data Figure 6. Cyp1-deficiency enhances AHR pathway activation.
a, CD4+ T cells from indicated genotypes were cultured under Th17-cell-inducing conditions and exposed to FICZ from the start of culture. Intracellular levels of FICZ were determined by HPLC and normalized to total protein content at the indicated time points (n=3 per time point), mean ± s.e.m.. b, Frequencies of IL-22-producing cells after 4 days of culture under Th17-cell-inducing conditions in presence of indicated concentrations of FICZ. Results are representative of three independent experiments, mean ± s.e.m.. **P < 0.01, ***P < 0.001 as calculated by two-way ANOVA with Dunnett’s post-test a, or one-way ANOVA with Tukey post-test b. ns = not significant. c, Flow cytometry analysis of Cyp1a1 (eYFP) expression by Th17 cells differentiated from indicated genotypes. Plots are gated on IL-17A+ cells and numbers indicate frequencies. d, Representative flow cytometry plots of IL-17A and IL-22 expression in in vitro differentiated Th17 cells from indicated genotypes (day 4) exposed to DMSO, 0.01nM FICZ or 1nM FICZ from the start of culture. e, Mice of indicated genotypes were infected orally with ~2x109 C. rodentium and bacterial burdens measured in the faeces at various time points. f, Pathology scores of distal colon. Bars are the mean and each symbol represents an individual mouse.
Results are representative of at least two independent experiments. NS = not significant, **P < 0.01, ***P < 0.001, as calculated by Student’s t-test.
Extended Data Figure 7. Effects of Rag1-Cre mediated Cyp1a1 expression on immunity to C. rodentium.
Mice of indicated genotypes were infected orally with ~2x109 C. rodentium and killed 14 days after infection or monitored for survival. a, Survival plot. b, C. rodentium burdens in the colon, cecum, liver and spleen. Bars are the median and each symbol represents an individual mouse. c, Pathology scores of distal colon and cecum. Bars are the mean and each symbol represents an individual mouse. d, Absolute numbers of cytokine-producing TCRβ+CD4+ T cells in the colon of C. rodentium infected mice.
Results are representative of two independent experiments (n=5 per group).
NS = not significant, **P < 0.01, as calculated by Student’s t-test.
Extended Data Figure 8. CYP1A1-mediated metabolism of DIM, ICZ and FICZ.
a-c, CYP1A1-mediated metabolism of a, DIM b, ICZ and c, FICZ was studied over time in the presence of human recombinant CYP1A1 (3.5nM) and the co-factor NADPH (1.0mM). At indicated timepoints, samples from respective incubations were extracted and analysed by means of HPLC. All chemicals were quantified according to separate standard curves. Left panels show relative amount of compound remaining at each given timepoint compared to parallel incubations without co-factor present. Right panels show HPLC chromatograms at 40 minutes enzyme-incubation, with and without co-factor present. All three compounds were detected based on their fluorescence properties (fluorescence units, FLU).
Results are representative of two independent experiments with two biological replicates at each experiment.
Acknowledgements
This work was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK, The UK Medical Research Council and the Wellcome Trust.
We would like to acknowledge the Biological Research Facility at the Francis Crick Institute for expert breeding and maintenance of our mouse strains, the Histopathology Facility for help with sections and staining and the Flow Cytometry Facility for cell sorting. We thank Prof. Gad Frankel (Imperial College) for the gift of antiserum to Citrobacter rodentium and Genentech, South San Francisco, California, USA for kindly providing anti-IL-22 and IL-22-Fc.
The study was supported by a Wellcome Advanced Investigator Grant (B.S.) and a Sir Henry Wellcome Fellowship and the Fondation Acteria (C.S.). C.H. and C.R.W. are funded by Cancer Research UK Programme Grant C4639/A10822. E.W. is funded by the Swedish Research Council FORMAS and D.N. by National Institute of Environmental Health Sciences, NIH grant R01 ES014403.
Footnotes
Author information:
The authors declare no competing financial interests.
Contributions:
C.S. designed, performed and analysed most of the experiments with input from A.M., A.I. Y.L.S.O. E.W. performed the metabolic studies, A.P. assisted in designing the construct for Rosa26 Cyp1a1 mice, C.J.H. and C.R.W. provided Cyp1a1 reporter mice and D.W.N. provided Triple Cyp KO mice. B.S. conceived the project and wrote the manuscript together with C.S.
References
- 1.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annual review of pharmacology and toxicology. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- 2.Zelante T, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 3.McMillan BJ, Bradfield CA. The aryl hydrocarbon receptor sans xenobiotics: endogenous function in genetic model systems. Mol Pharmacol. 2007;72:487–498. doi: 10.1124/mol.107.037259. [DOI] [PubMed] [Google Scholar]
- 4.Moura-Alves P, et al. AhR sensing of bacterial pigments regulates antibacterial defence. Nature. 2014;512:387–392. doi: 10.1038/nature13684. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt JV, Bradfield CA. Ah receptor signaling pathways. Annual review of cell and developmental biology. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- 6.Chiaro CR, Patel RD, Marcus CB, Perdew GH. Evidence for an aryl hydrocarbon receptor-mediated cytochrome p450 autoregulatory pathway. Mol Pharmacol. 2007;72:1369–1379. doi: 10.1124/mol.107.038968. [DOI] [PubMed] [Google Scholar]
- 7.Bock KW, Kohle C. Ah receptor: dioxin-mediated toxic responses as hints to deregulated physiologic functions. Biochemical pharmacology. 2006;72:393–404. doi: 10.1016/j.bcp.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Kerkvliet NI. Recent advances in understanding the mechanisms of TCDD immunotoxicity. Int Immunopharmacol. 2002;2:277–291. doi: 10.1016/s1567-5769(01)00179-5. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell KA, Elferink CJ. Timing is everything: consequences of transient and sustained AhR activity. Biochemical pharmacology. 2009;77:947–956. doi: 10.1016/j.bcp.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson P, et al. A constitutively active dioxin/aryl hydrocarbon receptor induces stomach tumors. Proc Natl Acad Sci U S A. 2002;99:9990–9995. doi: 10.1073/pnas.152706299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veldhoen M, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 12.Veldhoen M, Hirota K, Christensen J, O'Garra A, Stockinger B. Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J Exp Med. 2009;206:43–49. doi: 10.1084/jem.20081438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 14.Kiss EA, et al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- 15.Lee JS, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2012;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu J, et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lahvis GP, et al. The aryl hydrocarbon receptor is required for developmental closure of the ductus venosus in the neonatal mouse. Mol Pharmacol. 2005;67:714–720. doi: 10.1124/mol.104.008888. [DOI] [PubMed] [Google Scholar]
- 18.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 19.Dragin N, et al. Phenotype of the Cyp1a1/1a2/1b1-/- triple-knockout mouse. Mol Pharmacol. 2008;73:1844–1856. doi: 10.1124/mol.108.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson CJ, et al. Application of a novel regulatable Cre recombinase system to define the role of liver and gut metabolism in drug oral bioavailability. The Biochemical journal. 2015;465:479–488. doi: 10.1042/BJ20140582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bjeldanes LF, Kim JY, Grose KR, Bartholomew JC, Bradfield CA. Aromatic hydrocarbon responsiveness-receptor agonists generated from indole-3-carbinol in vitro and in vivo: comparisons with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Proc Natl Acad Sci U S A. 1991;88:9543–9547. doi: 10.1073/pnas.88.21.9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito S, Chen C, Satoh J, Yim S, Gonzalez FJ. Dietary phytochemicals regulate whole-body CYP1A1 expression through an arylhydrocarbon receptor nuclear translocator-dependent system in gut. J Clin Invest. 2007;117:1940–1950. doi: 10.1172/JCI31647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen LP, Bradfield CA. The search for endogenous activators of the aryl hydrocarbon receptor. Chem Res Toxicol. 2008;21:102–116. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soshilov AA, Denison MS. Ligand promiscuity of aryl hydrocarbon receptor agonists and antagonists revealed by site-directed mutagenesis. Molecular and cellular biology. 2014;34:1707–1719. doi: 10.1128/MCB.01183-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wincent E, et al. Inhibition of cytochrome P4501-dependent clearance of the endogenous agonist FICZ as a mechanism for activation of the aryl hydrocarbon receptor. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:4479–4484. doi: 10.1073/pnas.1118467109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song-Zhao GX, et al. Nlrp3 activation in the intestinal epithelium protects against a mucosal pathogen. Mucosal Immunol. 2014;7:763–774. doi: 10.1038/mi.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors declare that the source data supporting the findings of this study are available within the paper and its extended data files.