Abstract
Development of new drug regimens that allow rapid, sterilizing treatment of tuberculosis has been limited by the complexity and time required for genetic manipulations in Mycobacterium tuberculosis. CRISPR interference (CRISPRi) promises to be a robust, easily engineered, and scalable platform for regulated gene silencing. However, in M. tuberculosis, the existing Streptococcus pyogenes Cas9-based CRISPRi system is of limited utility because of relatively poor knockdown efficiency and proteotoxicity. To address these limitations, we screened eleven diverse Cas9 orthologues and identified four that are broadly functional for targeted gene knockdown in mycobacteria. The most efficacious of these proteins, the CRISPR1 Cas9 from Streptococcus thermophilus (dCas9Sth1), typically achieves 20–100 fold knockdown of endogenous gene expression with minimal proteotoxicity. In contrast to other CRISPRi systems, dCas9Sth1-mediated gene knockdown is robust when targeted far from the transcriptional start site, thereby allowing high-resolution dissection of gene function in the context of bacterial operons. We demonstrate the utility of this system by addressing persistent controversies regarding drug synergies in the mycobacterial folate biosynthesis pathway. We anticipate that the dCas9Sth1 CRISPRi system will have broad utility for functional genomics, genetic interaction mapping, and drug-target profiling in M. tuberculosis.
Introduction
Treatment of drug-susceptible M. tuberculosis requires a regimen of four drugs taken for six months. One of the overarching goals of the M. tuberculosis research community is to develop a regimen of novel agents that can treat both drug-susceptible and drug-resistant M. tuberculosis in less than 2 weeks. This will necessitate the development of new genetic tools to identify drug target synergies that accelerate pathogen clearance while avoiding drug target antagonisms. Current genetic approaches for drug target characterization include promoter replacement and inducible protein degradation systems that allow regulation of target protein levels over two orders of magnitude1–3, but can take months to target a single gene. Transposon-sequencing (Tn-seq) allows the simultaneous assessment of hundreds of thousands of loss-of-function mutants, but as currently implemented in M. tuberculosis is not conditional and thus does not allow manipulation of essential genes4. Furthermore, neither method provides a simple mechanism to modulate multiple genes simultaneously and thereby elucidate genetic interactions.
The re-purposing of the bacterial CRISPR-Cas (clustered regularly interspersed short palindromic repeats-CRISPR associated proteins) system as a site-specific transcriptional repressor could fill this methodological gap5–7. CRISPR systems use small RNAs to target and cleave foreign nucleic acid in a sequence-specific manner8–11. The best-characterized system is the type IIA system from Streptococcus pyogenes. Here, a single nuclease Cas9 and two small RNAs, a CRISPR RNA (crRNA) and a partially complementary trans-acting RNA (tracrRNA), are necessary and sufficient for RNA-guided silencing of foreign DNA9,11,12. This dual RNA system can be further simplified by the fusion of the crRNA and tracrRNA to generate a chimeric single guide RNA (sgRNA)11. Targeting specificity is determined both by Watson-Crick base pairing of the sgRNA and target DNA as well as a short DNA motif (protospacer adjacent motif (PAM)) within the target DNA sequence8,13,14. To repurpose Cas9 for transcriptional repression (CRISPR interference or CRISPRi), Cas9 nuclease activity is inactivated by point mutations within its HNH and RuvC nuclease domains5,7. Co-expression of the nuclease-dead S. pyogenes Cas9 protein (dCas9Spy) with an sgRNA designed with 20 nucleotides of complementarity to the target gene juxtaposed to an appropriate PAM yields specific silencing of the gene of interest (Figure 1A)5,7.
Figure 1. In vivo screen for functional Cas9 orthologues in mycobacteria.
(A) Schematic of CRISPRi-mediated transcriptional repression (adapted from reference15). Anhydrotetracycline (ATc)-inducible (PTet) dcas9 is directed to specific DNA targets by ATc-inducible or constitutively (Pcon) expressed sgRNA, which prevents transcription initiation or elongation.
(B) Phylogenetic tree of selected Cas9 orthologues. Listed are the Cas9 name abbreviations, size (amino acids), and sub-type classification.
(C) dCas9 orthologues show variable efficacy in mycobacteria. The Renilla reporter 5’UTR was engineered such that the optimal PAM for each dCas9 protein could be tested with an identical sgRNA targeting sequence (UTR1) for all dCas9 proteins. The x-axis denotes each dCas9 protein tested; the y-axis shows the ratio of the luminescence repression observed with the UTR1 targeting sgRNA normalized to the luminescence repression observed with a non-targeting control sgRNA. The fold inhibition is shown in white type for each column.
(D) sgRNAs targeting NGG PAMs were co-expressed with dCas9Spy (+ATc) and the effects on the Renilla target measured by luciferase assay. Shown above the graph is a schematic depicting the Renilla gene (yellow arrow) and transcriptional start site (black arrow) and the relative targeting position of each sgRNA (P = promoter; UTR = 5’ untranslated region, NT = Renilla non-template strand). The ΔRL promoter sample depicts basal levels of luminescence.
(E) sgRNAs targeting NNAGAAW PAMs were co-expressed with dCas9Sth1 (the cas9 allele derived from the CRISPR1 locus; +ATc) and luminescence repression measured as in Figure 1D. Fewer sgRNAs were tested for dCas9Sth1 because of the less frequent PAM occurrence in the Renilla target.
(F) sgRNAs targeting NGGNG PAMs were co-expressed with dCas9Sth3 (the cas9 allele derived from the CRISPR3 locus; +ATc) and luminescence repression measured as in Figure 1D.
(G) sgRNAs targeting NNGTGA PAMs were co-expressed with dCas9Spa (+ATc) and luminescence repression measured as in Figure 1D. Fewer sgRNAs were tested for dCas9Spa because of the less frequent PAM occurrence in the Renilla target.
Error bars are standard deviations of three technical replicates (C–G).
CRISPRi has been extensively characterized in the model bacteria Escherichia coli and Bacillus subtilis5,7,15. In these organisms, the dCas9Spy-sgRNA complex represses target gene transcription by either occluding RNA polymerase from the target promoter or by causing a steric block to transcription elongation (Figure 1A). When targeting the promoter, sgRNAs specific for either the template or non-template strand can be used, whereas an elongation block is most effective when targeting the non-template strand, producing up to 300-fold repression5,7. However, despite successes in E. coli and B. subtilis, first attempts to introduce a similar dCas9Spy-based CRISPRi system in mycobacteria were less impressive, typically resulting in ~4-fold knockdown of gene expression16,17.
Results
Limited utility of first-generation dCas9Spy CRISPRi in mycobacteria
We sought to develop a CRISPRi system for targeted gene knockdown in mycobacteria. Given the efficacy of dCas9Spy CRISPRi in model bacteria5,7,15, we first attempted to build a similar dCas9Spy–based system for mycobacteria. While some degree of dCas9Spy toxicity has been reported, dCas9Spy is in general well tolerated in E. coli18. In contrast, however, high-level expression of dCas9Spy with a non-genome targeting control sgRNA was lethal in M. smegmatis, and this lethality was not relieved by site-specific mutations that abrogate PAM interaction19 (Supplementary Figure 1A). While lowering dCas9Spy expression levels largely eliminated the growth defect, this first-generation dCas9Spy CRISPRi system resulted in poor target gene knockdown (~3–4 fold repression; Supplementary Figure 1B,C), consistent with published data16,17.
Identification of efficacious Cas9 alleles
While 4-fold gene knockdown is sufficient to generate loss-of-function phenotypes for some mycobacterial genes, many genes appear to require higher-level knockdown to reveal a phenotype3,20–23. Thus, we sought to develop a more efficacious CRISPRi system for mycobacteria. Given the extensive sequence diversity across the Cas9 family24, we hypothesized that orthologous Cas9 proteins might function more robustly than dCas9Spy in mycobacteria. We selected 11 diverse Cas9 proteins from the Type IIA and Type IIC subfamilies based on their relatively small size (range 984–1388 amino acids) and availability of data to predict crRNA, tracrRNA, and PAM sequences (Figure 1B, Supplementary Figure 2)24–26. We built single-plasmid platforms that expressed a mycobacterial codon-optimized nuclease-dead dcas9 allele under the control of an anhydrotetracycline (ATc)-inducible promoter and the cognate sgRNA under the control of a strong, constitutive promoter. To evaluate the performance of each dCas9 protein, we developed a Renilla luciferase-based reporter assay. The Renilla 5’ untranslated region (UTR) was engineered such that the optimal PAM for each dCas9 protein could be tested with an identical sgRNA targeting sequence (UTR1) for all dCas9 proteins. Each dCas9 orthologue was then co-expressed with its cognate sgRNA (UTR1) and gene silencing was measured by luciferase assay.
We observed a wide-range of gene silencing mediated by the dCas9 proteins (Figure 1C). Of the 11 tested dCas9 proteins, only five produced >5-fold gene knockdown. The effective dCas9 proteins included all five Type IIA but none of the Type IIC proteins, which may reflect the recently described limited double-stranded DNA helicase activity of Type IIC Cas9 proteins27. We next tested multiple Renilla-targeting sgRNA sequences to control for variation in sgRNA efficacy and, where relevant, variations in the PAM sequence. While there was variability in the fold knockdown observed across different sgRNA sequences for each dCas9 protein, this more extensive evaluation broadly confirmed the results obtained with the UTR1 sgRNA (Figure 1D–G, Supplementary Figure 3). The dCas9 protein derived from the CRISPR1 locus of S. thermophilus28 (dCas9Sth1) produced by far the most robust and consistent gene silencing in our Renilla reporter system (Figure 1C,E), although interestingly the expression systems used in this second-generation platform improved even dCas9Spy gene silencing relative to our first-generation system (Figure 1D, Supplementary Figure 1B,C).
dCas9Sth1 enables robust gene knockdown in mycobacteria
We next extended our analysis of this second-generation CRISPRi platform by targeting endogenous M. smegmatis genes with dCas9Sth1 and, as a point of comparison, dCas9Spy. We initially targeted two essential genes, dnaE1 and pptT21,29. In both cases, dCas9Sth1 allowed robust target gene knockdown with resulting inhibition of bacterial growth (Figure 2A,B). In the case of dnaE1, our second-generation dCas9Spy platform was capable of achieving high-level knockdown of endogenous gene expression that was roughly comparable to levels seen with dCas9Sth1 (Figure 2A). However, targeting pptT, which requires ~95% knockdown in M. tuberculosis in order to reveal a growth defect21, was successful with 7 out of 8 sgRNAs with dCas9Sth1 but only 1 out of 8 sgRNAs with dCas9Spy (Figure 2B).
Figure 2. dCas9Spy sensitizes M. smegmatis to sub-lethal drug treatment.
(A) dCas9Spy can mediate high-level knockdown of endogenous targets in M. smegmatis. sgRNAs targeting dnaE1 (Ms3178) were co-expressed with dCas9Spy or dCas9Sth1 (+ATc). Gene knockdown was quantified by qRT-PCR; the consequences of dnaE1 knockdown were monitored by spotting dilutions of each culture on the indicated media. Error bars are 95% confidence intervals of three technical replicates.
(B) sgRNAs targeting pptT (Ms2648) in M. smegmatis were co-expressed with the indicated dCas9 protein (+ATc) and plated as in Figure 2A.
(C) The indicated dCas9 proteins were HA-tagged, co-expressed with a non-targeting control sgRNA (+ATc), and monitored by western blot. RpoB protein levels are shown as loading controls. *: background α-HA cross-reactive band.
(D) dCas9Spy sensitizes M. smegmatis to sub-minimum inhibitory concentration (sub-MIC) drug treatment. Non-targeting control sgRNAs were co-expressed with dCas9Spy or dCas9Sth1 (+ATc); the consequences of dCas9 expression in the presence of a sub-MIC concentrations of the indicated drugs were monitored by spotting serial dilutions of each culture as in Figure 2A. Approximately 5,000 cells are deposited in the first spot and each subsequent spot is a 10-fold serial dilution. AMK: amikacin; NAT: nourseothricin; STR: streptomycin; RIF: rifampicin. Data shown in B–D are representative of three independent experiments.
These results were surprising in light of two published reports in E. coli and human cells where Cas9Spy is more efficacious than Cas9Sth126,30. Thus, we next sought to explore the basis for the relative underperformance of dCas9Spy relative to dCas9Sth1 in mycobacteria. To determine if this difference in efficacy could be the result of differences in dCas9 expression, we performed western blots to monitor the expression of both dCas9 proteins. Despite induction from identical promoters and 5’UTRs, we consistently saw higher levels of full-length dCas9Sth1 than dCas9Spy (Figure 2C), although both proteins showed some degree of degradation. However, this relatively poor expression of dCas9Spy was not phenotypically silent, since expression of dCas9Spy sensitized cells to sub-minimum inhibitory concentrations (sub-MIC) of aminoglycosides and rifampicin, which cause proteotoxic stress by inducing mis-translation and inhibiting transcription, respectively (Figure 2D). Thus, it appears that under ideal conditions (e.g. defined medium at 37°C) the second-generation dCas9Spy platform can produce high-level gene knockdown, but it also sensitizes mycobacteria to stress, which is likely to affect both bacterial viability and gene knockdown levels under desired screening conditions. Such target-independent dCas9Spy phenotypes could confound the identification of target-dependent phenotypes.
Identification of functional PAM variants for dCas9Sth1
Given its robust performance, we chose to move forward with a dCas9Sth1-based CRISPRi system. The published Cas9Sth1 PAM consensus sequence (NNAGAAW) allows limited targeting opportunities in GC-rich mycobacterial genomes13,14,24. However, recent work has highlighted the ability of Cas9Sth1 to tolerate deviations from this consensus sequence26,31–33. Thus, we defined the repertoire of PAMs available to dCas9Sth1 in vivo. Leveraging our Renilla reporter system, we created single-base mutations in the Renilla 5’UTR within the context of the consensus NNAGAAT PAM while maintaining the same UTR1 sgRNA sequence. We found that dCas9Sth1 functions with “non-canonical” PAM sequences (Figure 3), tolerating a G at position 3, G or C at position 4, and any base at position 7. Given that each possible permissive PAM position variant may not be functional with all other variants, we next tested all 24 possible PAM combinations (Table 1). These PAM variants produced a range of target knockdown, spanning 2.7–216.7 fold. The more divergent the PAM sequence was from the consensus, the less well the PAM functioned in gene silencing (Table 1). By defining a cutoff score of >25-fold knockdown, this analysis increased the number of possible dCas9Sth1 PAMs from 2 to 15, thereby increasing the number of possible target sites in the M. tuberculosis genome from 4,624 to 83,091, or approximately one targetable site every 53 base pairs.
Figure 3. In vivo identification of permissive PAM position variants for dCas9Sth1.
The Renilla 5’UTR from Figure 1C was modified to contain the indicated PAM sequences listed below the x-axis. Each PAM contains a single-base mutation within the context of the consensus NNAGAAT dCas9Sth1 PAM. dCas9Sth1 was co-expressed with the UTR1 sgRNA and luminescence repression measured as in Figure 1C.
Table 1. Permissive PAM sequences for dCas9Sth1.
Parameters for silencing endogenous genes
We next sought to assess our dCas9Sth1 system by silencing a larger panel of endogenous genes. To confirm the functionality of the alternative dCas9Sth1 PAMs, we chose to benchmark our system by targeting the same genes investigated by Choudhary et al. which produced, on average, 3–4 fold knockdown with a first generation dCas9Spy CRISPRi system16. In our first attempts to target these essential genes with dCas9Sth1, we found that for some sgRNAs, leaky expression of dCas9Sth1 from the ATc-regulated promoter resulted in growth inhibition even in the absence of CRISPRi induction. We therefore built and optimized a new set of ATc-regulated promoters that minimize leaky expression without compromising high-level induction and used these to control expression of dCas9Sth1 and the sgRNA (Supplementary Figure 4).
Consistent with our Renilla assay results, the optimized dCas9Sth1 system produced robust knockdown (11.1–226 fold) of all five M. smegmatis genes tested (Figure 4A–E). This high-level knockdown is quantitatively similar to that produced with CRISPR-based gene knockdown platforms in both E. coli and B. subtilis5–7,15. The robust knockdown achieved with our system was sufficient to silence genes that had previously been difficult to target. For example, we found that yidC is indeed essential in M. smegmatis as it is in M. tuberculosis, in contrast to published results achieved with a dCas9Spy system (Figure 4B)16. In addition, target mRNA knockdown kinetics were rapid, with near complete knockdown occurring in 1–2 hours or less than one cell cycle after ATc addition (Supplementary Figure 5A,B).
Figure 4. dCas9Sth1 CRISPRi is highly active against endogenous genes in mycobacteria.
(A–E) dCas9Sth1 achieves high-level knockdown of endogenous targets in M. smegmatis. sgRNAs targeting the indicated genes were co-expressed with dCas9Sth1 (+ATc). Gene knockdown was quantified and visualized as in Figure 2A.
(F) dCas9Sth1-mediated target knockdown is specific. Three sgRNAs targeting M. smegmatis dnaE1 (Ms3178) were co-expressed with dCas9Sth1 (+ATc) in wild-type M. smegmatis strains or strains complemented with M. tuberculosis dnaE1 (Rv1547). All three M. smegmatis dnaE1 targeting sgRNAs have perfect PAM matches to M. tuberculosis dnaE1 and either one, two, or four mismatches between the sgRNA targeting sequence and M. tuberculosis dnaE1, as indicated to the right of each strain. Positions of the sgRNA-M. tuberculosis dnaE1 mismatches (position 1 is closest to the target PAM): sgRNA1 (5), sgRNA2 (6,21), sgRNA3 (7,9,15,19). Strains were spotted as in Figure 2D.
(G) dCas9Sth1 achieves high-level knockdown of endogenous targets in M. tuberculosis. sgRNAs targeting the indicated genes were co-expressed with dCas9Sth1 (+ATc). Gene knockdown was quantified as in Figure 2A (fold knockdown +/− 95% confidence interval is shown beneath the +ATc plates). The consequences of gene knockdown were monitored by plating dilutions of each culture on the indicated media.
(H) dCas9Sth1 achieves robust target knockdown far from the transcriptional start site (TSS) in M. smegmatis. sgRNAs targeting the pptT operon were co-expressed with dCas9Sth1. Gene knockdown of pptT was quantified as in Figure 2A. The first two genes of the pptT gene operon are depicted at scale below the graph; the black bar beneath the pptT gene marks the site of the qPCR amplicon.
(I) dCas9Sth1 achieves robust target knockdown far from the TSS in M. tuberculosis. sgRNAs targeting the groES-groEL1 operon were co-expressed with dCas9Sth1. Gene knockdown of groEL1 was quantified as in Figure 2A. The groES-groEL1 operon is depicted at scale below the graph; the black bar beneath the groEL1 gene marks the site of the qPCR amplicon.
Data shown in A–G are representative of three independent experiments.
Given that the dCas9Sth1 system effectively silenced endogenous targets, we next sought to test its specificity. To do this, we tested the ability of the homologous M. tuberculosis dnaE1 gene29 to complement the dCas9Sth1-mediated silencing of the endogenous M. smegmatis dnaE1 gene (Figure 4F). All three M. smegmatis dnaE1 targeting sgRNAs (sgRNA1–3) have perfect PAM matches to M. tuberculosis dnaE1 and either one (sgRNA1), two (sgRNA2), or four (sgRNA3) mismatches between the sgRNA targeting sequence and M. tuberculosis dnaE1. Our results indicate that as few two mismatches between the sgRNA and the target are sufficient to abrogate phenotypic silencing (Figure 4F)5. Thus, off-target silencing with the dCas9Sth1 system would require near-perfect sgRNA sequence complementarity, an appropriately spaced PAM, as well as requiring targeting of the promoter or non-template strand (Supplementary Figure 5C), the combination of which is likely to be exceedingly rare in the context of compact bacterial genomes.
Gene silencing with CRISPRi in M. tuberculosis
Having developed this system in the model mycobacterium M. smegmatis, we then confirmed that our dCas9Sth1 system was effective in M. tuberculosis. As in M. smegmatis, the use of optimized ATc-regulated promoters was essential to prevent leaky expression of CRISPRi from inhibiting bacterial growth (Supplementary Figure 4). We designed sgRNAs to target eight essential genes in H37Rv4, including genes known or predicted to require high-level gene knockdown in order to reveal a growth defect3,21. Consistent with our results in M. smegmatis, dCas9Sth1-mediated silencing prevented growth and produced robust gene knockdown for all genes and PAM variants tested (Figure 4G, Supplementary Figure 5D). Moreover, our dCas9Sth1 system again reached maximal mRNA knockdown within one-to-two cell divisions after addition of ATc (Supplementary Figure 5E).
Interestingly, in both M. smegmatis and M. tuberculosis, dCas9Sth1 achieved high-level gene knockdown when targeted far from the predicted transcriptional start site (TSS); for example, yidC is predicted to be the fourth gene in an operon in M. tuberculosis and shows a syntenic operon structure in M. smegmatis34. This high-level silencing was somewhat unexpected given published observations describing an inverse correlation between repression efficacy and sgRNA targeting site distance from the TSS5,35. To explore this relationship, we quantified knockdown for 26 sgRNAs tiled along the pptT gene operon in M. smegmatis and 60 sgRNAs tiled along the groES-groEL1 operon or dnaE1 gene in M. tuberculosis34. First, these results further confirmed that the permissive PAM variants identified in our Renilla assay (Table 1) were also functional in the context of endogenous gene targeting in both M. smegmatis and M. tuberculosis (Figure 4H–I, Supplementary Figure 5F, Supplementary Table 2). Second, in contrast to published work with dCas9Spy, we observed no correlation between repression efficacy and the sgRNA targeting site distance from the TSS (Figure 4H–I, Supplementary Figure 5F)5. However, consistent with published work, CRISPRi is polar: any operonic gene downstream of the dCas9 binding site will be silenced in addition to the targeted gene (Figure 4H,I)5. In addition to a “downstream” polar effect, an “upstream” polar effect of CRISPRi was also recently reported in B. subtilis, in which the targeting of a downstream gene in an operon represses the transcription of the upstream genes15. To query for this effect, we quantified the expression level of the upstream gene in the pptT and groES-groEL1 operon targeting sgRNAs (Figure 4H,I). Knockdown of the upstream gene occurred only when the sgRNAs were targeted within or very close to the 3’ end of the upstream gene coding sequence (Supplementary Figure 5G,H).
Identification of synergistic interactions in the folate biosynthesis pathway
Finally, we tested the ability of our optimized CRISPRi system to interrogate genetic and chemical interactions in an essential metabolic pathway (Figure 5A). In both prokaryotes and eukaryotes, reduced folate species serve as essential co-factors in the transfer of one-carbon groups in pathways for the synthesis of DNA, RNA, and protein. While mammals lack the de novo folate synthesis pathway, many microbes are unable to acquire folates from their environment and rely entirely on de novo folate biosynthesis36. The dichotomy in essentiality of this pathway makes it an ideal target for the development of antibiotics. Two such antifolate antibiotics, trimethoprim (TMP) and sulfamethoxazole (SMX), have been used clinically for 40 years. TMP and SMX inhibit successive steps of the folate synthesis pathway (Figure 5A). SMX inhibits dihydropteroate synthase FolP1, which catalyzes the addition of dihydropterin diphosphate to p-aminobenzoic acid. The product of FolP1, 7,8-dihydropteroate, reacts with glutamate in a FolC-catalyzed reaction to form dihydrofolate. Dihydrofolate is then reduced to tetrahydrofolate by the dihydrofolate reductase FolA, the target of TMP. The combination of SMX and TMP has been demonstrated to be synergistic in many bacteria37. However, the efficacy of TMP, SMX, and whether or not these drugs act synergistically is controversial in mycobacteria. Initially, M. tuberculosis was reported to be resistant to the combination of TMP+SMX37. Consistent with this, TMP is a relatively weak in vitro inhibitor of M. tuberculosis FolA38. However, subsequent studies demonstrated that the combination of TMP and SMX is effective against M. tuberculosis39. One study evaluated the synergy between these two drugs and found that sub-MIC concentrations of SMX sensitized cells to TMP40; in contrast, a parallel study found that SMX alone was responsible for the observed M. tuberculosis inhibition, with no improvement by the addition of TMP41.
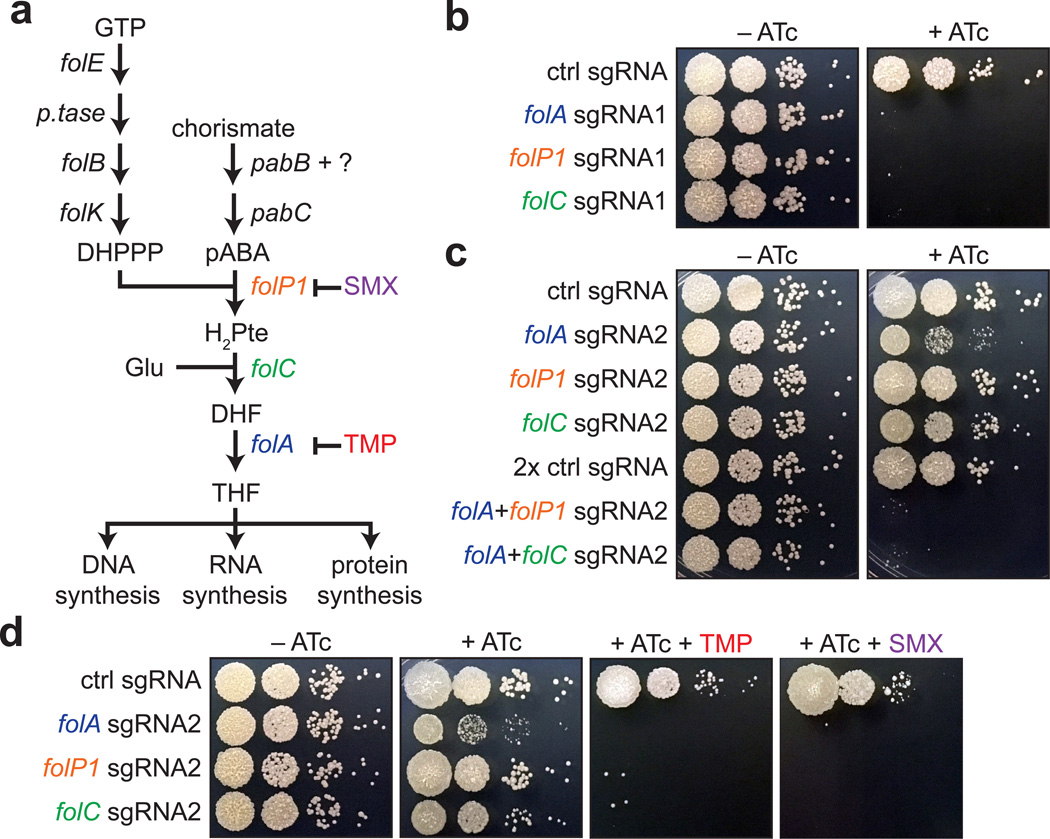
Figure 5. Functional profiling of the mycobacterial folate synthesis pathway.
(A) Schematic of mycobacterial folate metabolism. The targeted genes (folP1 Ms6103, folC-incorrectly annotated as pseudogene in M. smegmatis, and folA Ms2671) and the inhibitors of those genes (SMX = sulfamethoxazole, TMP = trimethoprim) are shown colored.
(B) dCas9Sth1-induced growth inhibition due to knockdown of folate pathway targets. sgRNAs targeting the indicated genes were co-expressed with dCas9Sth1 (+ATc) and spotted as in Figure 2D.
(C) Multiplexed targeting reveals synthetic lethal interactions in the folate pathway. The consequences of partial knockdown of folate pathway targets, individually or in combination, were monitored by co-expressing “hypomorphic” sgRNAs targeting the indicated genes and dCas9Sth1 (+ATc). Strains were spotted as in Figure 2D.
(D) Partial knockdown of folate pathway targets sensitizes cells to TMP and SMX. sgRNAs from Figure 5C were co-expressed with dCas9Sth1 (+ATc) and spotted on plates containing sub-MIC concentrations of TMP or SMX.
Data shown in B–D are representative of three independent experiments.
Elucidation of the mycobacterial folate synthesis pathway has been hampered by the inability to generate conditional loss-of-function alleles of folate biosynthesis genes. Previous attempts to target FolA with an inducible protein degradation system achieved ~97% knockdown but no significant reduction in growth rate3. We therefore first sought to determine whether our optimized CRISPRi system would be robust enough to block growth when targeting folate biosynthesis genes. We designed a panel of sgRNAs against folP1, folC, and folA. Due to the operonic nature of these genes, the targeting of folP1 is also expected to knockdown the expression of folB and folK (Supplementary Figure 6). Strikingly, we identified sgRNAs for all three genes that produced knockdown of sufficient magnitude to inhibit growth, consistent with these genes being essential in M. smegmatis (Figure 5B).
To investigate genetic synergy within this pathway, we tuned gene knockdown by using “weaker” dCas9Sth1 PAM variants to generate “hypomorphic” sgRNAs, which could then be combined to assess pairwise knockdown phenotypes. These hypomorphic sgRNAs cause partial knockdown of their targeted genes but not to levels high enough to completely inhibit growth (Figure 5C). Whereas folA, folP1, and folC hypomorphic sgRNAs induced moderate growth defects individually, they were synthetic lethal when co-expressed (Figure 5C). These genetic data suggest that knockdown of these targets should sensitize cells to TMP and SMX. Consistent with this hypothesis, partial knockdown of all three targets sensitized M. smegmatis to TMP and SMX (Figure 5D). Thus, CRISPRi-mediated silencing demonstrates that drugs targeting successive steps within the mycobacterial folate biosynthesis pathway (folP1, folC, and folA) can interact to produce a synergistic antimicrobial effect.
Discussion
We report here an optimized CRISPRi platform for mycobacteria and believe that this system presents a number of advantages relative to more conventional methods of gene perturbation. First, our system represents the simplest and fastest method for programmable gene regulation in mycobacteria. With our single-plasmid platform, all that is required for gene knockdown is the cloning of a unique ~20-bp targeting region into the sgRNA scaffold. Second, CRISPRi is scalable. With advances in array-based oligo synthesis, generating large pools of oligos that contain unique ~20-bp targeting regions is fast and inexpensive. Third, unlike complementary methods like Tn-seq, CRISPRi is inducible, thereby allowing the facile manipulation of essential genes. Fourth, the magnitude of CRISPRi knockdown is tunable, either by varying targeted PAM “strength” in the case of dCas9Sth1 (Table 1, Figure 5C,D), varying the concentration of the small molecule CRISPRi inducer15, modulating the degree of sgRNA-target complementarity5, or truncating the sgRNA targeting region5. Finally, CRISPRi can be multiplexed, which is of particular importance in slow-growing pathogens like M. tuberculosis in which serial genetic manipulation is prohibitively time consuming.
The primary disadvantage of CRISPRi is the polar effect- any operonic gene downstream of the dCas9 binding site will be silenced in addition to the targeted gene (Figure 4H,I)5. The fact that genes in an operon typically function in the same biological pathway partially mitigates this problem. Moreover, the fact that dCas9Sth1 can mediate robust gene knockdown irrespective of distance from the TSS (Figure 4H,I, Supplementary Figure 5F) with a minimal upstream polar effect (Supplementary Figure 5G,H) means that dCas9Sth1 CRISPRi can be used to target deep into bacterial operons and thereby increase the resolution of phenotyping.
In addition to facilitating study of Mtb biology, our dCas9Sth1 CRISPRi system will have broad utility in Mtb drug discovery. High-level gene knockdown can be used to define essential drug targets, and here CRISPRi will be a complementary and confirmatory approach to Tn-seq. The ability to tune the level of gene knockdown can then be used to define drug target vulnerability (i.e. the degree of knockdown required to inhibit growth) as well as serve as a platform for target-based whole cell screening to identify novel small molecules active against the desired drug target. Furthermore, the ability to generate hypomorphic alleles of essential genes will enable chemical and genetic interaction studies to define the interactions amongst essential drug targets, including the identification of drug target synergies. Using large-scale chip synthesis of unique sgRNA targeting regions, it will be possible to perform these studies at genome-scale. The identification of such drug target synergies will be a significant step towards the goal of developing novel drug regimens to shorten the duration of tuberculosis chemotherapy.
Methods
Bacterial strains
All M. tuberculosis strains are derivatives of H37Rv; all M. smegmatis strains are derivatives of mc2155.
Media
M. tuberculosis and M. smegmatis were grown at 37°C in Middlebrook 7H9 broth or 7H10 plates supplemented with 0.5% glycerol, 0.05% Tween80, 1× ADC (M. smegmatis) or OADC (M. tuberculosis), and the appropriate antibiotics. Where indicated, antibiotics or small molecules were used at the following concentrations: ATc (100 ng/ml anhydrotetracycline); AMK (25 ng/ml amikacin); NAT (800 ng/ml nourseothricin); STR (100 ng/ml streptomycin); RIF (1.25 µg/ml rifampicin); TMP (5 µg/ml trimethoprim); SMX (625 ng/ml sulfamethoxazole).
Plasmid construction and cloning
Cas9 sequences were identified by Blast42 or obtained from published studies24–26. Cas9 nuclease activity was inactivated by making the homologous D10A and H840A (S. pyogenes numbering) mutations in the RuvC and HNH nuclease domains for each Cas9 protein. dcas9 alleles were then codon optimized for mycobacterial expression with Jcat43 and synthesized by Genscript. dCas9 protein sequences used in this study are listed in Supplementary Table 1. Single-plasmid platforms were generated for all dcas9 alleles that contained: 1) the dcas9 allele under the control of a Tet repressor (TetR)-regulated UV15-Tet promoter1 or an optimized, synthetic TetR-regulated promoter (Supplementary Figure 4, Supplementary Table 1); 2) the cognate sgRNA under the control of a minimal synthetic constitutive promoter or an optimized, synthetic TetR-regulated promoter (Supplementary Figure 4, Supplementary Table 1); 3) a Tet repressor; 4) a single-copy L5-integrating backbone44; 5) a pBR322-derived E. coli replication origin; and 6) a kanamycin-selectable marker. sgRNA scaffold sequences were obtained from published studies25,26 or engineered by fusing the crRNA direct repeat to the tracrRNA in an analogous manner to the S. pyogenes sgRNA11. We attempted to enrich for highly active dCas9Spy sgRNAs by selecting top-scoring sgRNAs based on the algorithm described in Doench et al.45. All sgRNA scaffold sequences used in this study are listed in Supplementary Table 1.
To clone sgRNA targeting sequences, the sgRNA scaffolds were designed with two unique BsmBI restriction sites immediately 5’ to the sgRNA scaffold sequence. Complementary sgRNA targeting oligos (N20–25) were then annealed and ligated (T4 DNA ligase, NEB) into the BsmBI-digested CRISPRi vector backbone. To clone multiple sgRNAs into the same vector, the CRISPRi vector backbones were designed with a SapI-based Golden Gate cloning site 3’ to the first sgRNA scaffold. sgRNA targeting sequences are listed in Supplementary Table 2.
To construct the Renilla luciferase reporter plasmid, the Renilla gene was cloned downstream of the G13 promoter from Mycobacterium marinum46 with a synthetic 5’ untranslated region (UTR) in a single-copy Giles integrating vector47. The Renilla 5’UTR was engineered such that the optimal PAM for each dCas9 protein could be tested with an identical sgRNA targeting sequence (UTR1) for all dCas9 proteins. The Renilla reporter sequences used for each dCas9 protein are shown in Supplementary Table 1.
Phenotying
To visually monitor the effects of CRISPRi-mediated gene silencing, cultures were grown to log-phase and plated on solid media with or without ATc. Plates were incubated at 37°C for 3–4 days (M. smegmatis) or 21 days (M. tuberculosis). For the complementation experiment shown in Figure 4F, M. smegmatis was complemented with a single-copy Giles integrating vector expressing M. tuberculosis dnaE1 (Rv1547) under the control of the endogenous Rv1547 promoter.
Western blots
HA-dCas9Sth1 and HA-dCas9Spy were detected using an anti-HA antibody (Cell Signaling, C29F4) at a 1:1,000 dilution; RpoB was detected using an anti-RpoB antibody (Neoclone, WP023) at a 1:1,000 dilution. IRDye-800 anti-mouse and IRDye-680 anti-rabbit were used at 1:15,000 dilution (Licor). Immunoblots were imaged on a Licor Odyssey scanner. The full scans for the western blot shown in Figure 2C are provided as Supplementary Figure 7.
Cas9 phylogeny
Representative Cas9 sequences were pooled from published sources48,49 and aligned with FastTree50 with the default parameters. Tree visualization was done with FigTree.
Luciferase assays
For each sample, cultures were grown to log phase and then diluted back with or without 100 ng/ml ATc to maintain the cultures in log phase and induce CRISPRi. Renilla knockdown was allowed to proceed for 24 hours. One OD600 equivalent of cells was harvested by centrifugation and processed for Renilla luciferase assay as per manufacturers instructions (Promega Renilla Luciferase Assay system). Luciferase activity was quantified in 96-well white plates (Costar; 3362) using a VarioSkan Flash plate reader (Thermo Scientific). Error bars represent standard deviations of three technical replicates.
mRNA quantification
For each sample, cultures were grown to log phase and then diluted back in the presence of 100 ng/ml ATc to maintain the cultures in log phase and induce CRISPRi. Target knockdown was allowed to proceed for 14 hours (M. smegmatis) or 4 days (M. tuberculosis). One-to-two OD600 equivalents of cells from each culture were harvested by centrifugation, resuspended in TRIzol (Thermo Fisher), and disrupted by bead beating (Lysing Matrix B, MP Biomedicals). Total RNA was isolated by RNA miniprep (Zymo Research), residual contaminating genomic DNA digested with TURBO DNase (Ambion), and samples cleaned with RNA cleanup columns (Zymo Research). cDNA was prepared with random hexamers as per manufacturer instructions (Life Technologies Superscript IV). RNA was then removed by alkaline hydrolysis and cDNA was purified by spin column (Qiagen). cDNA levels were then quantified by quantitative real-time PCR (qRT-PCR) on a Viia7 light cycler (Applied Biosystems) using iTaq Universal SYBR Green Supermix (BioRad). All qPCR primer pairs were verified to be >95% efficient and cDNA masses tested were experimentally validated to be within the linear dynamic range of the assay. Signals were normalized to the housekeeping sigA transcript for both M. tuberculosis (Rv2703) and M. smegmatis (Ms2758) and quantified by the ΔΔCt method. Error bars are 95% confidence intervals of three technical replicates.
M. smegmatis operon predictions
The homologs of M. tuberculosis folate biosynthesis genes were identified in M. smegmatis by Blast42; note that folC is incorrectly annotated as a pseudogene in mc2155. Operon predictions from the TB Database51 or primary data52 were used to predict syntenic operons in M. smegmatis. Gene essentiality predictions are based on essentially predictions of M. tuberculosis homologs4.
Supplementary Material
Acknowledgments
The authors thank Luke Gilbert for advice. This work was supported by a Helen Hay Whitney fellowship to J.M.R., an NIH Director’s New Innovator Award 1DP20D001378, subcontracts from NIAID U19 AI107774, and a Doris Duke Charitable Foundation Grant 2010054 to S.M.F. A.C. was funded by the National Cancer Institute grant 5T32CA009216-34. G.M.C. acknowledges support from the US National Institutes of Health National Human Genome Research Institute grant P50 HG005550, and the Wyss Institute for Biologically Inspired Engineering.
Footnotes
Author contributions
S.M.F. supervised the project; J.M.R. and J.R.P. conceived the study; J.M.R designed and executed the study; F.F.H. and M.D. cloned CRISPR constructs and assisted with luciferase assays; A.C., M.R.C., E.G., G.M.C., E.R., C.S., D.S. contributed reagents and expertise; J.M.R. and S.M.F. wrote the manuscript with input from all co-authors.
The authors declare no competing financial interests.
Data availability
All data that support the findings of this study are available from the corresponding author upon request.
References
- 1.Ehrt S, et al. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Research. 2005;33:e21. doi: 10.1093/nar/gni013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim JH, et al. Protein inactivation in mycobacteria by controlled proteolysis and its application to deplete the beta subunit of RNA polymerase. Nucleic Acids Research. 2011;39:2210–2220. doi: 10.1093/nar/gkq1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei J-R, et al. Depletion of antibiotic targets has widely varying effects on growth. Proc Natl Acad Sci USA. 2011;108:4176–4181. doi: 10.1073/pnas.1018301108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang YJ, et al. Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS Pathog. 2012;8:e1002946. doi: 10.1371/journal.ppat.1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi LS, et al. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rath D, Amlinger L, Hoekzema M, Devulapally PR, Lundgren M. Efficient programmable gene silencing by Cascade. Nucleic Acids Research. 2015;43:237–246. doi: 10.1093/nar/gku1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bikard D, et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Research. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright AV, Nuñez JK, Doudna JA. Biology and Applications of CRISPR Systems: Harnessing Nature's Toolbox for Genome Engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 9.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garneau JE, et al. The CRISPR/Cas bacterial immunesystem cleaves bacteriophage andplasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 11.Jinek M, et al. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horvath P, et al. Diversity, Activity, and Evolution of CRISPR Loci in Streptococcus thermophilus. Journal of Bacteriology. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deveau H, et al. Phage Response to CRISPR-Encoded Resistance in Streptococcus thermophilus. Journal of Bacteriology. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peters JM, et al. A Comprehensive, CRISPR-based Functional Analysis of Essential Genes in Bacteria. Cell. 2016;165:1493–1506. doi: 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choudhary E, Thakur P, Pareek M, Agarwal N. Gene silencing by CRISPR interference in mycobacteria. Nature Communications. 2015;6:6267–6267. doi: 10.1038/ncomms7267. [DOI] [PubMed] [Google Scholar]
- 17.Singh AK, et al. Investigating essential gene function in Mycobacterium tuberculosis using an efficient CRISPR interference system. Nucleic Acids Research. 2016;44:e143. doi: 10.1093/nar/gkw625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen AA, Voigt CA. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Molecular Systems Biology. 2014;10:763–763. doi: 10.15252/msb.20145735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park SW, et al. Evaluating the sensitivity of Mycobacterium tuberculosis to biotin deprivation using regulated gene expression. PLoS Pathog. 2011;7:e1002264–e1002264. doi: 10.1371/journal.ppat.1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leblanc C, et al. 4'-Phosphopantetheinyl transferase PptT, a new drug target required for Mycobacterium tuberculosis growth and persistence in vivo. PLoS Pathog. 2012;8:e1003097–e1003097. doi: 10.1371/journal.ppat.1003097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin K, et al. Mycobacterium tuberculosis Thioredoxin Reductase Is Essential for Thiol Redox Homeostasis but Plays a Minor Role in Antioxidant Defense. PLoS Pathog. 2016;12:e1005675–e1005720. doi: 10.1371/journal.ppat.1005675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy BKK, et al. Assessment of Mycobacterium tuberculosis Pantothenate Kinase Vulnerability through Target Knockdown and Mechanistically Diverse Inhibitors. Antimicrobial Agents and Chemotherapy. 2014;58:3312–3326. doi: 10.1128/AAC.00140-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fonfara I, et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Research. 2014;42:2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015 doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Meth. 2013 doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma E, Harrington LB, O’Connell MR, Zhou K, Doudna JA. Single-stranded DNA cleavage by divergent CRISPR-Cas9 enzymes. Molecular Cell. 2015 doi: 10.1016/j.molcel.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 29.Rock JM, et al. DNA replication fidelity in. Nat Genet. 2015;47:677–681. doi: 10.1038/ng.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chari R, Mali P, Moosburner M, Church GM. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Meth. 2015 doi: 10.1038/nmeth.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015 doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leenay RT, et al. Identifying and Visualizing Functional PAM Diversity across CRISPR-Cas Systems. Molecular Cell. 2016:1–25. doi: 10.1016/j.molcel.2016.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karvelis T, et al. Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol. 2015;16:1–13. doi: 10.1186/s13059-015-0818-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caspi R, et al. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Research. 2007;36:D623–D631. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larson MH, et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minato Y, et al. Mycobacterium tuberculosis Folate Metabolism and the Mechanistic Basis for para-Aminosalicylic Acid Susceptibility and Resistance. Antimicrobial Agents and Chemotherapy. 2015;59:5097–5106. doi: 10.1128/AAC.00647-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bushby S. Trimethoprim-sulfamethoxazole: in vitro microbiological aspects. Journal of Infectious Diseases. 1973 doi: 10.1093/infdis/128.supplement_3.s442. [DOI] [PubMed] [Google Scholar]
- 38.White EL, Ross LJ, Cunningham A. Cloning, expression, and characterization of Mycobacterium tuberculosis dihydrofolate reductase. FEMS microbiology. 2004 doi: 10.1016/S0378-1097(04)00038-2. [DOI] [PubMed] [Google Scholar]
- 39.Forgacs P, Wengenack NL, Hall L. Tuberculosis and trimethoprim-sulfamethoxazole. Antimicrobial agents. 2009 doi: 10.1128/AAC.01658-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vilcheze C, Jacobs WR. The Combination of Sulfamethoxazole, Trimethoprim, and Isoniazid or Rifampin Is Bactericidal and Prevents the Emergence of Drug Resistance in Mycobacterium tuberculosis. Antimicrobial Agents and Chemotherapy. 2012;56:5142–5148. doi: 10.1128/AAC.00832-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ong W, et al. Mycobacterium tuberculosis and Sulfamethoxazole Susceptibility. Antimicrobial Agents and Chemotherapy. 2010;54:2748–2749. doi: 10.1128/AAC.00029-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 43.Grote A, et al. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Research. 2005;33:W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee MH, Pascopella L, Jacobs WR, Hatfull GF. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guérin. Proc Natl Acad Sci USA. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doench JG, et al. rational design of highly active sgrNAs for CrIsPr-Cas9–mediated gene inactivation. Nat. Biotechnol. 2014:1–8. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barker LP, Porcella SF, Wyatt RG, Small PL. The Mycobacterium marinum G13 promoter is a strong sigma 70-like promoter that is expressed in Escherichia coli and mycobacteria species. FEMS Microbiology Letters. 1999;175:79–85. doi: 10.1111/j.1574-6968.1999.tb13604.x. [DOI] [PubMed] [Google Scholar]
- 47.Morris P, Marinelli LJ, Jacobs-Sera D, Hendrix RW, Hatfull GF. Genomic characterization of mycobacteriophage Giles: evidence for phage acquisition of host DNA by illegitimate recombination. Journal of Bacteriology. 2008;190:2172–2182. doi: 10.1128/JB.01657-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Makarova KS, et al. An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Micro. 2015:1–15. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chylinski K, Makarova KS, Charpentier E, Koonin EV. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Research. 2014;42:6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reddy TBK, et al. TB database: an integrated platform for tuberculosis research. Nucleic Acids Research. 2009;37:D499–D508. doi: 10.1093/nar/gkn652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fivian-Hughes AS, Houghton J, Davis EO. Mycobacterium tuberculosis thymidylate synthase gene thyX is essential and potentially bifunctional, while thyA deletion confers resistance to p-aminosalicylic acid. Microbiology (Reading, Engl.) 2012;158:308–318. doi: 10.1099/mic.0.053983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.