Abstract
Proliferating Cell Nuclear Antigen (PCNA) lies at the center of the faithful duplication of eukaryotic genomes. With its distinctive doughnut-shaped molecular structure, PCNA was originally studied for its role in stimulating DNA polymerases. However, we now know that PCNA does much more than promote processive DNA synthesis. Because of the complexity of the events involved, cellular DNA replication poses major threats to genomic integrity. Whatever predicament lies ahead for the replication fork, PCNA is there to orchestrate the events necessary to handle it. Through its many protein interactions and various post-translational modifications, PCNA has far-reaching impacts on a myriad of cellular functions.
General structural and functional characteristics of PCNA
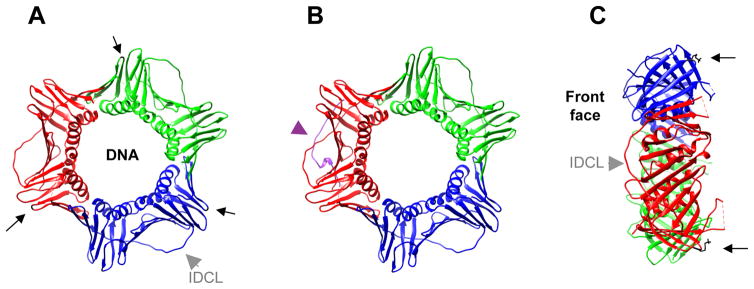
PCNA is a ring-shaped homotrimer that encircles the DNA (Figure 1), hence it is also referred to as a sliding clamp (Boehm et al., 2016a; Dieckman et al., 2012; Moldovan et al., 2007; Warbrick, 2000). PCNA has a highly conserved six-fold symmetry since each of the monomers are composed of two similarly folded globular regions united by a flexible interdomain connecting loop (IDCL). The face of PCNA pointing in the direction of DNA synthesis is known as the front face, and it is the site of interaction for most binding partners. A staggering number of proteins interact directly with PCNA. Thus, they must compete with each other for dancing with the sliding clamp (De Biasio and Blanco, 2013; Maga and Hubscher, 2003; Mailand et al., 2013). Most interacting partners bind PCNA through a conserved sequence termed the PIP (PCNA-interacting peptide)-box, which inserts itself into a hydrophobic pocket on the front face, beneath the IDCLs. The consensus PIP sequence is Q-x-x-ψ-x-x-ϑ- ϑ, in which ψ is a moderately hydrophobic amino acid (L, V, I, or M) and ϑ is an aromatic residue (Y or F). Many partners have degenerate sequences missing some of the core amino acids, and are still able to bind PCNA. A reverse PIP-box is also able to support PCNA interaction (Pedley et al., 2014). In addition, a second PCNA interacting motif, termed APIM (AlkB homologue 2 PCNA-interacting motif) is widespread among DNA repair proteins and is defined as K/R-F/Y/W-L/I/V/AL/I/V/A-K/R (Gilljam et al., 2009). The APIM interaction surface on PCNA partially overlaps with that used by the PIP-box.
Figure 1. PCNA structure.
A. Front view of PCNA. Each monomer is presented in a different color. Arrows indicate the position of K164, the residue targeted by ubiquitination and SUMOylation. Grey arrowhead indicates the interdomain connecting loop (IDCL) on one of the monomers. B. Front view of PCNA showing the interaction of one of the monomers with a p21-derived PIP-box peptide (in purple, marked by arrowhead). C. Side view. Arrows indicate K164 residue on two of the monomers. Grey arrowhead indicates the IDCL on one of the monomers. The Front face is also indicated.
The diversity of sequence variations of PCNA binding motifs (Table 1) allows for differential binding strengths. For example, the specialized PIP box Q-x-x-ψ-T-D-ϑ- ϑ provides a higher affinity for PCNA (Havens and Walter, 2009). This indicates that the PIP-box motifs lacking TD are suboptimal, since they provide lower binding affinity. The TD-containing PIP-box of the CDK inhibitor p21 is one of the strongest PCNA-interacting peptide (Bruning and Shamoo, 2004). This ability to finely tune the interaction affinity through residues other than those conserved in the consensus sequence provides the first level of regulation of the competition for PCNA binding. Indeed, modifying the PIP-box of crucial replication and repair proteins to increase the strength of PCNA interaction results in impaired replication and repair (Fridman et al., 2010), indicating that competition through binding strength is a critical regulatory mechanism.
Table 1. PCNA interacting motifs.
The consensus residues are shown in bold font.
| Interaction motif | Sequence | Example |
|---|---|---|
| PIP-box | Q x x L/V/I/M x x F/Y F/Y | QRSIMSFF (in DNA Ligase I) |
| Degenerate PIP-box | Missing one or two of the core amino acids | KHTLDIFF (in Polκ) QRNHETAF (in MCL1) QTKVEFPE (in PDIP1) |
| Specialized PIP-box for strong interaction | Q x x L/V/I/M T D F/Y F/Y | QTSITDFF (in p21) |
| PIP-degron | Q x x L/V/I/M T D F/Y F/Y x x x K/R | QRRVTDFF ARRR (in CDT1) |
| Inverted PIP-box | F/Y F/Y x x L/V/I/M M x x Q | FFAGIVWQ (in Akt) |
| APIM | K/R F/Y/W L/I/V/A L/I/V/A K/R | NKFLARE (in RAD51B) |
PCNA is an essential co-factor for DNA polymerases during replication (O'Donnell et al., 2013; Siddiqui et al., 2013). PCNA tethers polymerases to DNA and dramatically increases their processivity (the average number of nucleotides added before dissociation from DNA). Similar to other DNA replication factors, PCNA is essential for viability in all organisms. PCNA also participates in non-replicative DNA synthesis events, such as those occurring during DNA repair. Repair DNA synthesis events are common to a variety of DNA repair processes including nucleotide excision repair, homologous recombination, and mismatch repair. But the functions of PCNA extend well beyond DNA synthesis. While devoid of enzymatic activity itself, PCNA exerts a strong influence on the metabolism of DNA and chromatin by recruiting various enzymes to DNA. PCNA not only participates in localizing these factors to their sites of action but, in many instances, it also directly activates their enzymatic activities.
The repertoire of PCNA functions is dramatically amplified by its post-translational modifications (Hoege et al., 2002; Moldovan et al., 2007). Since PCNA lacks enzymatic activity and instead exerts its effects through protein-protein interactions, regulated addition and removal of these moieties to PCNA provides an ideal mechanism for controlling PCNA functions. These modifications generally do not affect PCNA structure. Instead, they provide an additional surface for interaction with specific binding factors –the second level of regulating PCNA-interactions (Mailand et al., 2013). This is the case even for bulky modifications such as ubiquitination or the ubiquitin-like protein SUMO (Tsutakawa et al., 2015). Structures of mono-ubiquitinated PCNA using X-ray crystallography, electron microscopy, and Small angle X-ray scattering (Lau et al., 2015; Tsutakawa et al., 2015; Zhang et al., 2012) revealed several different conformations including one with ubiquitin extending away from PCNA, as well as a “docked” conformation in which ubiquitin interacts extensively with the PCNA surface forming a contiguous binding interface. It is possible that these different conformations allow ubiquitinated PCNA the necessary plasticity to interact with different binding partners. In contrast, SUMO is simply tethered to PCNA without additional interactions, resulting in an extended, flexible tag, which acts as an independent interaction module (Armstrong et al., 2012; Tsutakawa et al., 2015). As detailed below, PCNA post-translational modifications are generally associated with particular functional outcomes (Table 2).
Table 2.
Post-translational modifications of human PCNA
| Modification | Target site | Enzyme | Outcome at the molecular level | Biological function |
|---|---|---|---|---|
| Phosphorylation | Y211 | EGFR, c-Abl (kinases) | Protects against PCNA degradation | Promotes cell proliferation |
| Inhibits MutS binding | Inhibits MMR | |||
| Mono-ubiquitination | K164 (and potentially others) | RAD18, CDT2 (E3 ligases) | Recruits TLS polymerases | Promotes TLS |
| K63-linked multi-ubiquitination | K164 | HLTF, SHPRH (E3 ligases) | Recruits the translocase ZRANB3 | Promotes error-free bypass? |
| ISGylation | K168 | EFP (E3 ligase) | Recruits USP10 to de-ubiquitinate PCNA | Turns off TLS |
| SUMOylation | K164 | UBC9 (E2 conjugating enzyme; No E3 ligase necessary?) | Recruits HR inhibitor PARI | Inhibits HR |
| Acetylation | K13, K14, K20, K77, K80, K284 | CBP, p300 (acetyl- transferases) | Promotes removal of PCNA from DNA after NER | Promotes genomic stability |
| S-Nitrosylation | C81, C162 | (nitrosylases ?) | Blocks interaction with caspase-9 | Promotes apoptosis |
DNA replication
DNA replication is initiated at distinct replication origins, which are marked by binding of the pre-replicative protein complex in G1 phase of the cell cycle (O'Donnell et al., 2013; Siddiqui et al., 2013). Origin firing during S-phase involves the assembly of two back-to-back replication forks that proceed in opposing directions. Each replication fork is headed by the CMG helicase, which unwinds the DNA. This allows synthesis of the primer by the DNA Polymerase α complex, which contains one catalytic subunit responsible for synthesis of the initial ~10nt RNA primer, and a separate DNA polymerase subunit that extends the RNA primer with about ~20nt. PCNA is loaded at the primer-template junction by the multi-subunit Replication Factor C (RFC) complex, which can open the PCNA ring and clamp it on the DNA. While the fork assembly takes place on a DNA region devoid of nucleosomes, PCNA loading seems to require a chromatin context marked by methylation of histone H3 at lysine K56, which is catalyzed by the methyltransferase G9a (Yu et al., 2012). In vitro reconstitution of the replisome complex assembly showed that, in the presence of PCNA-RFC, the primase is switched with the high fidelity replicative DNA Polymerases ε (on the leading strand) and δ (on the lagging strand) (Georgescu et al., 2014; Georgescu et al., 2015). On the leading strand, DNA synthesis catalyzed by Polε proceeds continuously. PCNA interaction with Polε is weak and PCNA may even dissociate and remain behind the fork, to mark the leading strand for post-replicative events such as mismatch repair.
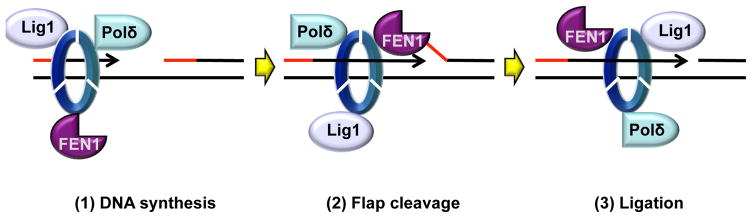
Since DNA synthesis can only occur in the 5’-3’ direction, replication on the lagging strand is discontinuous. On this strand, PCNA interacts with multiple PIP-box containing subunits of Polδ, as well as with the Polδ-associated protein PDIP46 (Wang et al., 2016). Primers are being synthesized every 100–200nt to generate Okazaki fragments. The end of the previous Okazaki fragment functions as a molecular break that slows down Polδ tenfold. The polymerase partly displaces the primer end forming a flap structure that is cleaved off by FEN1, one nucleotide at a time (Stodola and Burgers, 2016). Eventually, DNA ligase I seals the resulting nick. Both enzymes interact with PCNA, which activates their activities (Zheng and Shen, 2011). Formation of a stable, active Okazaki fragment maturation complex in which different monomers of the PCNA trimer interact with Polδ, FEN1, and DNA ligase I respectively (a so-called “PCNA tool belt” - Figure 2) have been proposed many years ago, and recently demonstrated experimentally (Stodola and Burgers, 2016). On the other hand, mutant PCNA trimers with a single binding site are perfectly capable of supporting Okazaki fragment maturation (Dovrat et al., 2014) arguing that tool belts may not be absolutely required. Interestingly, post-translational modifications of FEN1 seem to play an important role in regulating these interactions (Zheng and Shen, 2011).
Figure 2. Example of PCNA tool-belt: the Okazaki fragment maturation complex.
Polδ, FEN1, and DNA Ligase I bind PCNA simultaneously and each interact with a monomer of the PCNA trimer to facilitate enzyme switching during Okazaki fragment maturation.
Because it is loaded on the primer template junction at each Okazaki fragment, PCNA accumulates on the lagging strand. Unloading of PCNA requires the activity of an RFC-like complex in which the catalytic subunit RFC1 is replaced by ATAD5 (Elg1 in yeast) (Kubota et al., 2013; Lee et al., 2013). Underlining the importance of removing PCNA from DNA, loss of Elg1 unloading activity results in DNA repair defects and chromosome instability. Re-expression of Elg1 during G2 and M-phase, but not during S-phase, can restore genomic stability, indicating that PCNA retention on DNA beyond DNA replication and into G2/M is toxic (Johnson et al., 2016). Interestingly, Elg1 is itself inhibited during G1 to allow PCNA loading on DNA at the G1/S transition (Huang et al., 2016).
PCNA as co-factor for regulated protein degradation during replication
One essential activity of PCNA is to promote degradation of a subset of its binding partners. A large number of substrates have been identified for this PCNA-targeted degradation. The CDK inhibitors p21 and Xic1 are degraded to allow normal cell cycle progression (Abbas et al., 2008; Kim et al., 2010). Degradation of the accessory p12 subunit of Polδ alters its enzymatic activity (Zhang et al., 2013). The thymine DNA glycosylase TDG is degraded to prevent unwanted DNA demethylation (Shibata et al., 2014; Slenn et al., 2014). Degradation of the anti-recombinogenic FBH1 helicase regulates DNA repair (Bacquin et al., 2013). The replication licensing factors CDC6 and CDT1 are degraded to prevent re-initiation of DNA replication (re-replication) (Arias and Walter, 2006; Clijsters and Wolthuis, 2014). Degradation of the histone methyltransferase Set8 prevents untimely chromatin compaction (Centore et al., 2010; Oda et al., 2010).
Degradation of these substrates occurs only when they are bound to PCNA on chromatin during S-phase or after DNA damage exposure, and requires ubiquitination of the substrate by the CRL4-CDT2 ubiquitin ligase. The PCNA interactors degraded through this mechanism possess a special type of PIP-box, termed PIP-degron: Q-x-x-ψ-T-D-ϑ- ϑ-x-x-x-B, where B is a basic residue (K or R) (Havens and Walter, 2009). Importantly, this basic residue recruits the CRL4-CDT2 ubiquitin ligase, but only when the substrate is bound to chromatin-loaded DNA. The mechanistic basis for this is still unclear. Once ubiquitinated by CDT2, the substrates are removed from chromatin by the ubiquitin-targeted chaperone p97, a hexameric AAA-ATPase complex known to remodel ubiquitin complexes, and delivered to the proteasome for degradation (Raman et al., 2011). In order to allow re-expression of the substrates in G2 phase, CDK1 phosphorylates CDT2 in late S-phase preventing its recruitment to chromatin-bound PCNA (Rizzardi et al., 2015).
PCNA and chromatin assembly
Nascent DNA is rapidly coated with nucleosomes in a co-replicational process that requires binding of the histone chaperone CAF-1 to PCNA at replication forks (Zhang et al., 2000). Interestingly, chromatin assembly can regulate the speed of replication fork progression: reducing histone pools results in decreased replication speed. Under these conditions, PCNA is retained on chromatin to allow CAF-1 recruitment once histones become available (Mejlvang et al., 2014). Not only do nucleosomes need to be assembled on nascent DNA, but their histone marks also need to be preserved so that the parental epigenetic status is maintained in the daughter cells. Parental histones ahead of replication forks are relocated to nascent DNA behind the replication fork, providing a possible mechanism for transferring epigenetic modifications. However, there is evidence that most histones loaded on the nascent DNA lack modifications present on the nucleosomes ahead of the fork, such as H3K4me3 or H3K27me3 modifications. Instead, epigenetic inheritance relies on the presence of the enzymes responsible for such modifications (histone methylation, acetylation, ubiquitination) in close proximity to PCNA during replication (Petruk et al., 2013; Petruk et al., 2012). It is likely that these enzymes interact with PCNA. Moreover, DNMT1, an enzyme responsible for DNA methylation at CpG islands, binds directly to PCNA (Mortusewicz et al., 2005).
Regulation of cell proliferation and apoptosis
Because of its essential role in DNA replication, regulation of PCNA steady state levels represents an important mechanism to control proliferation in response to extracellular conditions. Growth factor signaling (EGF through EGFR kinase, and HGFL through c-Abl kinase) results in phosphorylation of PCNA at Tyr211, which increases its chromatin association and protects it from degradation (Wang et al., 2006; Zhao et al., 2014). Several other signaling molecules have been shown to regulate proliferation through controlling PCNA levels, including ERK8 (Groehler and Lannigan, 2010) and 14-3-3ζ (Gao et al., 2015). In addition, p21, one of the most potent negative regulators of proliferation, inhibits not only CDKs but also PCNA, through a tight PIP-box mediated association that out-competes DNA polymerases and other interactors (Gulbis et al., 1996; Moldovan et al., 2007).
PCNA is an important modulator of apoptosis, through interactions with both pro-apoptotic (ING1b) and anti-apoptotic (p53, Gadd45, MyD118, CR6) regulators (Moldovan et al., 2007). More recently, an unexpected role of cytoplasmic PCNA in regulating apoptosis has emerged where PCNA binds to procaspases −3, −8, −9, and −10, blocking their activation and inhibiting apoptosis (Witko-Sarsat et al., 2010). This anti-apoptotic activity is regulated by S-Nitrosylation of cytoplasmic PCNA, which blocks interaction with caspase-9, thus allowing activation of this caspase (Yin et al., 2015). Interestingly, cytosolic PCNA also participates in the activation of the Akt pathway, possibly through its interactions with APIM motif-containing components of this pathway. Indeed, inhibition of these interactions reduces Akt activation and cell growth (Olaisen et al., 2015).
Translesion Synthesis
One of the major activities of PCNA is to promote tolerance of DNA damage during DNA replication. DNA lesions block the progression of high fidelity replicative DNA polymerases δ and ε. To bypass them, cells employ specialized low fidelity polymerases (such as Polη, Polκ, and Rev1)–a process termed translesion synthesis (TLS) (Cipolla et al., 2016; Jansen et al., 2015). TLS is a double-edged sword: while bypass of DNA lesions allows cells to continue their proliferation program without replication arrest, low-fidelity polymerases are mutagenic on both normal and damaged DNA templates, and thus must be kept in check. The regulation of TLS relies on post-translational modification of PCNA (Bienko et al., 2005; Guo et al., 2006; Hoege et al., 2002; Kannouche et al., 2004; Plosky et al., 2006; Watanabe et al., 2004). Upon fork stalling at DNA lesions, PCNA mono-ubiquitination at Lys 164 by the ubiquitin ligase RAD18, recruits TLS polymerases to bypass the lesion (Figure 3). The molecular basis for this recruitment is the presence of separate PCNA-interacting motifs and ubiquitin-interacting domains in most TLS polymerases, providing higher binding affinity for the ubiquitinated form of PCNA. In contrast, replicative DNA polymerases only possess PCNA-binding motifs and lack ubiquitin-interacting domains. Indeed, it was recently shown that PCNA ubiquitination does not affect the assembly or processivity of the Polδ complex (Hedglin et al., 2016).
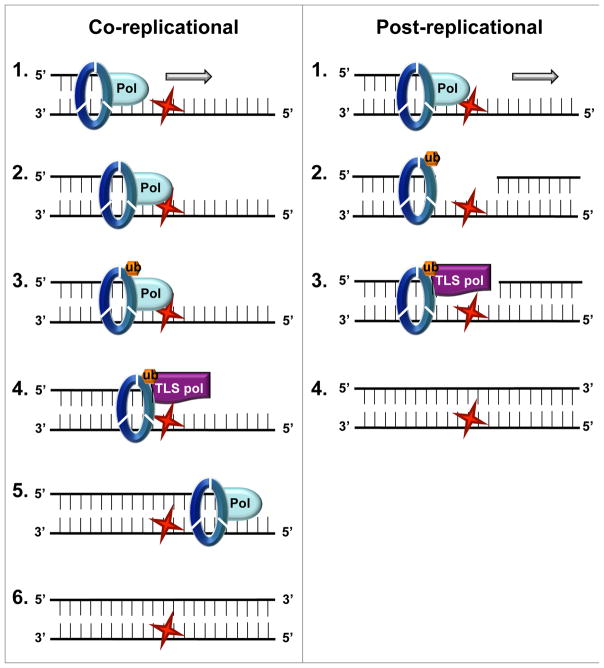
Figure 3. Two non-exclusive models for the timing of TLS.
In the co-replication model, also termed “on-the-fly”, the polymerase switch occurs instantaneously upon stalling, following PCNA ubiquitination. In the post-replicational model, the fork is reassembled downstream of the lesion leaving behind a gap which is filled later in S phase or G2 by TLS polymerases. Whether PCNA is left behind at the gap or is loaded de novo is not clear.
Fine-tuning TLS
The various TLS polymerases differ in their abilities to bypass different lesions; some of them can even ensure accurate bypass of specific lesions, such as in the case of Polη-mediated bypass of UV-induced thymidine dimers. Mechanisms for specific recruitment of a polymerase to a lesion are not yet known. Recent models suggest that this may be a stochastic process: since PCNA is a trimer, tool-belts can be formed in which each monomer binds a different polymerase, albeit with low affinity. This ensures a high local concentration of various TLS polymerases that can individually sample the lesion and eventually engage it (Boehm et al., 2016b). Such tool-belts have been observed even in the absence of ubiquitination. Indeed, under certain conditions polymerases can perform TLS in the absence of PCNA ubiquitination (Acharya et al., 2010; Despras et al., 2012; Hendel et al., 2011; Krijger et al., 2011; Wit et al., 2015). However, PCNA ubiquitination not only increases efficiency, but also alters the mutation spectra of TLS (Hendel et al., 2011). It is likely that the addition of ubiquitin provides an additional surface that the polymerases can attach to with varying affinities.
Turning off TLS is an important aspect of the pathway, since unrestrained use of low fidelity TLS polymerases would introduce mismatches throughout the genome. PCNA deubiquitination by USP1 was one of the first mechanisms described as a negative regulator of eukaroytic TLS (Huang et al., 2006), and since then, several other mechanisms have been described. The protein SPARTAN binds ubiquitinated PCNA at stalled forks and is important for maintaining ubiquitin-PCNA levels through a yet unclear mechanism (Centore et al., 2012; Ghosal et al., 2012; Juhasz et al., 2012). At the same time, SPARTAN plays a crucial role in switching off TLS, by recruiting the ubiquitin-selective chaperone p97 to remove Polη from DNA (Davis et al., 2012; Mosbech et al., 2012). The small PCNA interacting protein PAF15 also participates in turning off TLS by tightly associating with PCNA following lesion bypass, out-competing TLS polymerases (De Biasio et al., 2015; Povlsen et al., 2012).
TLS is also regulated by additional PCNA modification. After DNA damage, PCNA can be modified with the ubiquitin-like molecule ISG15 (ISGylation) through the recruitment of the ISG15 E3 ligase EFP to ubiquitinated PCNA. This modification then recruits the deubiquitinating enzyme USP10 to remove ubiquitin from PCNA and restore high fidelity DNA synthesis (Park et al., 2014).
Additional layers of complexity have been added in recent years. The CRL4-CDT2 ubiquitin ligase can also ubiquitinate PCNA at K164 to promote TLS, but unlike RAD18, it seems to be associated with endogenous replication stress rather than DNA damage exposure (Terai et al., 2010). PCNA ubiquitination can also be increased by a variety of factors, including: the repair proteins FANCD2 (Chen et al., 2016) and PTIP (Gohler et al., 2008), the ADP-ribosyltransferase PARP10 (Nicolae et al., 2014), the signaling molecules SIVA1 (Han et al., 2014), and MAGE-A4 (Gao et al., 2016), the ubiquitin ligases BRCA1 (Tian et al., 2013) and RNF8 (Zhang et al., 2008), the checkpoint proteins CHK1 and claspin (Yang et al., 2008), and others. While it is unclear how each of these factors fit in the bigger picture, they likely provide fine-tuning mechanisms ensuring that the proper balance of TLS is achieved, depending on cell type, growth conditions and proliferation signals, cell cycle stage, chromatin status, amount of damage etc.
Timing of PCNA ubiquitination and TLS
PCNA ubiquitination is normally detected during S-phase in both yeast and human cells (Hoege et al., 2002; Huang et al., 2006; Terai et al., 2010). Mechanistic studies have shown that uncoupling between the helicase machinery and the stalled polymerase results in accumulation of single stranded DNA that recruits RAD18 to ubiquitinate PCNA (Chang and Cimprich, 2009; Davies et al., 2008). Chromatin remodeling by the ZBTB1-KAP1 complex promotes RAD18 recruitment and PCNA ubiquitination, perhaps by increasing RAD18 accessibility to DNA (Kim et al., 2014). Switching the replicative polymerase with a TLS polymerase seems to be a straightforward and time-saving mechanism for restarting a stalled replication fork and continuation of DNA replication. Thus, PCNA ubiquitination and TLS were thought to occur co-replicationally, taking place instantaneously following fork stalling –a process sometimes referred to as “on the fly” (Hedglin and Benkovic, 2015; Yang et al., 2013a). While there is evidence for PCNA ubiquitination-mediated polymerase switching in vitro (Masuda et al., 2010), the process can occur independent of PCNA ubiquitination in vivo. (Edmunds et al., 2008). Consistent with this, genetic experiments in yeast showed that PCNA ubiquitination can also occur in G2 phase, without impacting damage tolerance (Daigaku et al., 2010; Karras and Jentsch, 2010). These results instead point towards a post-replicational model for TLS, where the fork is re-established downstream of the lesion leaving behind a short unreplicated region with the lesion, to be repaired or bypassed at a later time in S-phase or even G2 (Figure 3). Mechanistic details of such a process are missing and many important questions remain unanswered: Is the integrity of the stalled fork maintained? Is PCNA ubiquitinated upon stalling or rather later, right before bypass? Why resort to TLS bypass when behind the fork, the lesion can be bypassed in an error-free manner for example through recombination? Nevertheless, such a post-replicative mechanism would provide the cell with additional flexibility to deal with a larger number of lesions, and would perhaps provide the opportunity to sample the local environment of each individual lesion before deciding on error-free versus error-prone bypass. The two possibilities may not be mutually exclusive (Waters et al., 2009). Interestingly, p53 was recently shown to bind PCNA at stalled forks and suppress the extension from these forks (Hampp et al., 2016), perhaps providing a mechanism for marking those particular stalled forks which will be resolved post-replicationally.
Non-canonical roles of PCNA ubiquitination
PCNA ubiquitination can be detected outside of S-phase in human cells, under certain conditions. For example PCNA ubiquitination can be induced by UV in quiescent (G0-arrested) cells (Ogi et al., 2010; Yang et al., 2013b). This event seems to promote recruitment of the TLS Polκ to UV lesions for repair synthesis (Ogi et al., 2010). Oxidative damage also induces PCNA ubiquitination outside S-phase (Zlatanou et al., 2011). While still dependent on RAD18, this ubiquitination event involves a non-canonical activity of the mismatch repair complex MSH2-MSH6, which is required to initiate removal of the damaged strand by the nuclease EXO1. In case DNA lesions are present on the remaining strand, which may frequently be the case, PCNA ubiquitination promotes a TLS-like lesion bypass event using low fidelity polymerases. Interestingly, even though this process still uses the canonical RAD18 ubiquitin ligase, an alternative de-ubiquitinting enzyme, namely USP7, is employed to remove ubiquitin from PCNA under these conditions (Kashiwaba et al., 2015).
PCNA ubiquitination was recently shown to participate in activation of a novel replication stress checkpoint, which is mediated by ATM (unlike the classic replication stress checkpoint activated by ATR). Ubiquitinated PCNA recruits WRNIP protein, which in turn binds to the ATM activating cofactor ATMIN. This pathway is important for coordinating replication completion with cell cycle progression and, in its absence, cells enter mitosis with under-replicated DNA (Kanu et al., 2016). Finally, recent genetic experiments in yeast suggest a role for PCNA ubiquitination and TLS in Okazaki fragment maturation, at least in the context of FEN1 inactivation (Becker et al., 2015).
PCNA poly-ubiquitination
The PCNA ubiquitination events described above to regulate TLS are mono-ubiquitination events. However, PCNA can also be ubiquitinated by K63-linked multi-ubiquitin chains at Lys 164. This modification is significant in yeast, but present at much lower levels in human cells (Hoege et al., 2002). PCNA poly-ubiquitination is believed to initiate an alternative, error-free lesion bypass mechanism involving the use of the newly replicated sister chromatid as template (known as Template Switching; TS). While many details of TS are still missing, recent electron microscopy studies identified structures resembling double Holliday junctions as critical intermediates of TS –suggesting a mechanism analogous to recombination (Giannattasio et al., 2014). Since DNA replication is more faithful in early S-phase, it has been proposed that TS may be preferentially used over TLS early in replication (Branzei and Psakhye, 2016; Stamatoyannopoulos et al., 2009).
How PCNA poly-ubiquitination promotes template switching is not yet understood. The ZRANB3 translocase is recruited to poly-ubiquitinated PCNA and remodels the DNA structure at the stalled fork (Ciccia et al., 2012; Weston et al., 2012; Yuan et al., 2012). Mgs1 (the yeast homolog of WRNIP1) also binds poly-ubiquitinated PCNA, and appears to interfere with Polδ functions (Saugar et al., 2012). It is still unclear how these activities promote error-free lesion bypass. Further complicating the picture, the E3 ubiquitin ligases SHPRH and HLTF that are involved in PCNA poly-ubiquitination in human cells (Motegi et al., 2008) also participate in TLS (Lin et al., 2011).
Homologous Recombination
Homologous Recombination (HR) repairs DNA double strand breaks. Following DNA end resection at the break, the resulting single stranded DNA overhang is coated by multiple molecules of the recombinase RAD51 (Prakash et al., 2015). This nucleoprotein filament then invades the sister chromatid. When homology is found, the target strand is unwound and a displacement loop is formed. This structure is extended by DNA polymerases past the break site, after which the elongated strand is re-annealed to its original partner. The resulting gap is filled by regular repair DNA synthesis. Both replicative and TLS polymerases can perform DNA synthesis in the D-loop extension step. Processivity of Polδ and Polκ is enhanced by PCNA, while it has no effect on the extension by Polη. This suggests that PCNA can affect the outcome of HR by regulating the polymerase choice for D-loop extension (Sebesta et al., 2013). PCNA also promotes HR at the end resection step. PCNA loads onto DNA strands at double strand breaks and serves as a processivity factor for EXO1 during its end resection activity (Chen et al., 2013).
PCNA can also inhibit HR through its SUMO modification, which is a predominant modification in yeast. While it has been much harder to detect in human cells, it is clear that PCNA SUMOylation has a conserved role in inhibiting homologous recombination. (Gali et al., 2012; Hoege et al., 2002; Moldovan et al., 2012; Moldovan et al., 2007). This activity may serve as a means to regulate how stalled replication forks restart. HR-based mechanisms can restart stalled replication forks, but this process has the potential to initiate chromosome rearrangements (Carr and Lambert, 2013) and likely requires prolonged replication arrest. In contrast, TLS, while locally mutagenic, would provide a faster and more straightforward way to restart the forks. Since both ubiquitination and SUMOylation occur at the same lysine residue, K164, they would be mutually exclusive on a PCNA monomer (though not necessarily on a trimeric molecule). In any case, the two modifications functionally cooperate to promote TLS over HR. Since PCNA SUMOylation is found constitutively during S-phase (and not only in response to DNA damage treatment as is the case for ubiquitination), SUMOylation-mediated HR suppression may also be involved in suppressing unwanted recombination events between newly synthesized sister chromatids during normal fork progression.
In yeast, PCNA SUMOylaton recruits the helicase Srs2, a recombination inhibitor, which contains separate modules for interacting with PCNA (PIP-box) and SUMO (SIM motif) (Armstrong et al., 2012; Hoege et al., 2002; Papouli et al., 2005; Pfander et al., 2005). Srs2 inhibits recombination by removing RAD51 from single stranded DNA, thereby disassembling an essential recombination intermediate structure. In human cells, SUMOylated PCNA recruits the recombination inhibitor PARI, an Srs2 homolog that lacks helicase activity. Similar to Srs2, PARI binds to RAD51 and has an inhibitory effect toward RAD51 nucleofilaments (Moldovan et al., 2012). PARI also inhibits HR at a later stage, during the DNA synthesis step (Burkovics et al., 2016). Independent of its SUMOylation, PCNA also interacts with a number of other recombination inhibitors. This includes RTEL1, a telomeric recombination inhibitor that disrupts RAD51 post-invasion filaments (D-loops) (Vannier et al., 2013), and FBH1, which promotes degradation of RAD51 (Chu et al., 2015)
Other genome stability mechanisms
Nucleotide excision repair (NER)
Nucleotide excision repair removes helix-distorting DNA lesions and bulky adducts (Marteijn et al., 2014). This is achieved through the concerted efforts of about 30 proteins that act sequentially to detect the lesion (XPC-RAD23B complex), open up the helix (TFIIH, XPB, and XPD), dually incise the damaged strand ~30nt apart (endonucleases ERCC1-XPF on the 5’ side, and XPG on the 3’ side), and remove the lesion-containing oligonucleotide. PCNA-dependent repair DNA synthesis fills the gap to complete the reaction. PCNA also interacts, through an APIM motif, with XPA, a scaffold protein involved in organizing events downstream of helix opening (Gilljam et al., 2012). Moreover, PCNA interacts with XPG and targets it for CDT1-mediated degradation, to allow repair DNA synthesis which bulky XPG molecules may sterically hinder (Han et al., 2015). Finally, PCNA activates the enzymatic activity of XPF (Hutton et al., 2010).
Following completion of NER by DNA synthesis, PCNA is acetylated by CBP and p300 at lysine residues that lie in the DNA-interacting inner hole of PCNA. Acetylated PCNA is removed from chromatin and subjected to proteasomal degradation (Cazzalini et al., 2014). This prevents retention of PCNA on chromatin after NER events, which may be damaging, as noted above.
Base excision repair (BER)
BER repairs small lesions and damaged bases (Krokan and Bjoras, 2013). BER is initiated by the recognition and excision of damaged bases by damage-specific DNA glycosylases, followed by cleavage 5’ of the abasic site by the AP-endonuclease 1 (APE1). PCNA interacts with, and stimulates the activities of several glycosylases (including UNG2, NTH1, and MPG) as well as APE1 (Moldovan et al., 2007). Repair of the gap is achieved by two distinct mechanisms: Short-patch repair involves a one-nucleotide re-synthesis step by DNA polymerase β and nick sealing by XRCC1-DNA ligase III. On the other hand, long-patch repair proceeds by synthesis of a stretch of 2-20nt by Polδ in concert with PCNA and RFC, creating a flap structure that is engaged by the FEN-1-DNA ligase I-PCNA complex.
Mismatch repair (MMR)
MMR repairs replication-generated errors such as mismatches and insertion/deletions loops. Crucial for this process is the ability to discriminate between the template and the nascent strands, so that the nascent strand can be corrected (Ganai and Johansson, 2016). The recognition of the nascent strand is based on the presence of nicks generated during replication, or during removal of erroneously incorporated ribonucleotides, which have not yet been sealed. Indeed, mismatch correction occurs faster on the lagging strand, where more nicks are present because of discontinuous DNA synthesis. Replication errors are recognized by MutSα (MSH2-MSH6; identifies base mismatches and small insertion/deletion loops) and MutSβ (MSH2-MSH3; recognizes larger insertion/deletion loops) heterodimers. Once the mismatch is recognized, the MutS heterodimers form clamps that dissociate from DNA and slide away. Eventually, a complex with MutLα (MLH1-PMS2) and PCNA is formed. PCNA activates the endonuclease activity of MutLα on the nascent strand; the resulting nick is further processed by the EXO1 exonuclease, or through repeat nickase activity by MutLα (Ganai and Johansson, 2016; Goellner et al., 2014). Repair DNA synthesis completes the process. PCNA also binds MutSα, which seems to promote PCNA retention on chromatin after replication to facilitate MMR (Kawasoe et al., 2016). Intriguingly, PCNA phosphorylation at Y211 by EGFR was shown to inhibit binding of PCNA to MutS complexes, resulting in reduced MMR efficiency and increased mutagenesis (Ortega et al., 2015). Since, as described above, this phosphorylation is a proliferation signal, these findings provide another example of proliferation taking place at the expense of genome fidelity.
The replication stress response
Prolonged replication fork stalling generates aberrant fork structures containing long stretches of single stranded DNA. Such structures activate a stress response, mediated by checkpoint kinases, which helps stabilize and restart the forks thus preventing generation of DNA damage and genomic instability (Zeman and Cimprich, 2014). In addition to the ubiquitinated PCNA dependent ATM checkpoint described above, PCNA interacts with two ubiquitin ligases, TRAIP and HUWE1, and these interactions are required for efficient fork progression under replication stress conditions (Choe et al., 2016; Hoffmann et al., 2016). In both cases, PCNA was shown to not be a substrate for these ligases. Instead, HUWE1 ubiquitinates the histone H2AX, a variant of H2A that is essential for DNA damage detection and signaling. Indeed, ubiquitination of H2AX by this novel PCNA-HUWE1 pathway promotes its phosphorylation by ATR. Phosphorylated H2AX, also known as γH2AX, in turn recruits repair proteins to deal with broken forks (Choe et al., 2016). A relevant substrate for TRAIP has not been identified so far, although its inactivation also reduces γH2AX formation (Harley et al., 2016). It was proposed that TRAIP may participate in removing PCNA from stalled forks under certain conditions (Feng et al., 2016). Indeed, It has been previously shown that upon treatment with replication inhibitors such as hydroxyurea, ATR-dependent fork remodeling results in active unloading of PCNA (Dungrawala et al., 2015; Yu et al., 2014). The exact role of this unloading is unclear, but it may facilitate formation of novel DNA structures important for fork restart under these conditions. It is not known how common these events are under endogenous replication stress conditions.
PCNA and human disease
Reflecting its role in promoting genomic stability, several close companions of PCNA have been found to be associated with cancer predisposition syndromes, including ATAD5 for ovarian cancer (Kuchenbaecker et al., 2015), and SPARTAN for hepatocellular carcinoma (Lessel et al., 2014). High PARI expression in tumors is associated with increased sensitivity to genotoxic cancer therapy, presumably through its HR-inhibiting activity (Pitroda et al., 2014). Mice expressing the K164R mutant as the only source of PCNA show altered somatic hypermutation at the IgG loci (Roa et al., 2008), suggesting that human B-cell deficiency syndromes may also be caused by defective PCNA ubiquitination. However, until very recently, no PCNA mutation has been reported in human disease, reflecting perhaps the fact that most mutations are expected to destroy the complex fold of PCNA and thus result in inactivation of this essential protein.
A recent report presented the first human syndrome caused by PCNA mutation (Baple et al., 2014). The patients, from a single extended family, share a homozygous missense PCNA mutation, Ser228Ile. Ser228 lies close to the IDCL and the mutation results in a large conformational change that affects interactions with PIP-box proteins including Fen1, DNA ligase 1, and XPG (Duffy et al., 2016). For reasons still not completely clear, the mutation affects DNA repair rather than DNA replication functions of PCNA (Green et al., 2014). This helps explain why the patients with this mutation survive, but suffer from immunodeficiency, photosensitivity, and neurodegeneration symptoms (including: hearing loss, ataxia, cognitive decline, and other brain defects). Indeed, many DNA repair-related syndromes, such as ataxia telangiectasia (ATM mutations), exhibit clinical characteristics that include neurodegeneration (Rass et al., 2007). More recently, homozygous missense mutations in the PCNA binding partners TRAIP and PARP10 have been associated with microcephaly (Harley et al., 2016; Shahrour et al., 2016). Similar to the PCNA mutations, cells from these patients show a strong DNA repair defect but a relatively minor proliferation defect.
Targeting PCNA in cancer therapy
Because of its essential role in promoting proliferation, PCNA is an obvious target for cancer therapy (Wang, 2014) However, since PCNA is not an enzyme and instead acts through its protein interactions, it has been very difficult to design drugs that inhibit PCNA. In recent years, a number of studies identified small molecules that block PCNA-Polδ interaction or PCNA trimer formation, with proliferation inhibitory effects in vitro (Punchihewa et al., 2012; Tan et al., 2012; Zhao et al., 2011) (Table 3). A cell permeable peptide containing the APIM motif, which blocks binding of partners with the APIM motif, showed promising results in treating myeloma and bladder cancer, in mouse xenograft studies (Müller et al., 2013). Similarly, a peptide spanning the IDCL, which presumably inhibits PCNA interaction with PIP-box containing partners, could inhibit cancer growth in neuroblastoma and breast cancer mouse xenograft models (Smith et al., 2015). These studies suggest that PCNA targeting in cancer may not be far away (Wang, 2014).
Table 3.
PCNA inhibitors that are being tested for cancer therapy.
| Inhibitor | Properties | Mechanism of action | Effects |
|---|---|---|---|
|
T2AA
(Punchihewa et al 2012) |
T3 thyroid hormone derivative | - Binds the PCNA cavity that interacts with PIP-box motifs; - Interferes with PCNA-PIP-box interactions; ; - Does not affect PCNA-DNA binding. |
Inhibits PCNA interaction with p21 (in vitro) and Polδ (in cells). Treatment results in inhibition of cell growth and proliferation, increased replication stress, and reduced TLS. Combination with cisplatin increases cellular DNA damage and reduces cell growth in U2OS cells. |
|
PCNA-I1
(Tan et al 2012) |
Small molecule inhibitor | - Stabilizes the PCNA trimer by binding Arg146 through an O-N hydrogen bond on one monomer of PCNA, and to Asp86 through an N-O hydrogen bond on its adjacent monomer. | Treatment reduces PCNA binding to chromatin, possibly through trimer stabilization. Inhibits growth in various human and mouse cancer cell lines with a potency ~9-fold higher than in untransformed cells. Leads to cell cycle arrest. |
|
ATX-101
(Müller et al 2013) |
APIM- containing peptide | Interferes with PCNA-APIM motif interactions. | Induces apoptosis in multiple myeloma and primary cancer cells in a caspase-dependent manner. Potentiates the cytotoxicity of Melphalan in multiple myeloma cell lines and in a xenograft multiple myeloma mouse model. |
|
R9-caPeptide
(Smith et al 2015) |
Cell permeable peptide | Interferes with PCNA-PIP motif interactions. | Preferentially targets PCNA-protein interactions in cancer cells over normal cells. Disrupts PCNA association to chromatin and Polδ, inhibiting DNA replication in vitro and cell growth. Enhances sensitivity to cisplatin in cancer cells. Inhibits tumor growth in mice. |
|
Y211F Peptide
(Zhao et al 2011) |
Peptide covering the Y211 region of PCNA | Inhibits PCNA phosphorylation at Y211. | Induces S-phase arrest, inhibition of DNA synthesis, and enhanced cell death in human prostate cancer cell lines. Decreases tumor growth in PC3-derived xenograft tumors in nude mice |
Final remarks
Studies over the past decade have unveiled many hidden aspects of PCNA. Identification of new binding partners and new functions including non-canonical roles outside the nucleus greatly expanded the range of PCNA activities. Moreover, it has become clear that within the same functional pathway, PCNA can play both activating and inhibitory roles, as detailed above for HR and MMR. The switch from activation to inhibition is achieved, at least in these two cases, through posttranslational modifications (SUMOylation and phosphorylation, respectively).
How the numerous PCNA interactions are regulated continues to puzzle. It is likely that dynamic exchanges of the functional complexes loaded on PCNA are required throughout replication. The composition of PCNA complexes will vary from fork to fork depending on chromatin structure, DNA sequence, and replication timing. For example, common fragile sites in the genome act as natural fork barriers and their replication, which generally takes place in late S-phase, requires TLS rather than replicative polymerases (Bergoglio et al., 2013). Other polymerase switching events may be required during the replication of heavily chromatinized DNA sequences and of other regions replicating late in S-phase, when mutation rates are higher (Stamatoyannopoulos et al., 2009). Moreover, replication of various types of chromatin may require a different set of chromatin factors bound to PCNA. What regulates the dynamics of PCNA complexes is not clear, but it is likely that post-translational modifications play some role. Quantitative proteomic studies may be able to reveal the composition of the various PCNA networks during replication and repair. Next generation sequencing approaches may be used to investigate the impact of these complexes on replication fidelity. Finally, molecular mechanistic studies are needed to shed light on the more enigmatic PCNA –dependent processes including template switching, and strand discrimination during MMR. Without a doubt, secret facets of PCNA are waiting to be discovered.
Acknowledgments
We would like to thank Michael O’Connor, Mark Hedglin, Tanay Thakar, Kristen Clements, and Claudia Nicolae for comments on the manuscript. GLM is supported by: NIH 1R01ES026184, Department of Defense CA140303, and the St. Baldrick’s Foundation. This paper is dedicated to the memory of Prof. Stefan Jentsch, irreplaceable maestro of cellular biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N, Yoon JH, Hurwitz J, Prakash L, Prakash S. DNA polymerase eta lacking the ubiquitin-binding domain promotes replicative lesion bypass in humans cells. Proc Natl Acad Sci U S A. 2010;107:10401–10405. doi: 10.1073/pnas.1005492107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias EE, Walter JC. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol. 2006;8:84–90. doi: 10.1038/ncb1346. [DOI] [PubMed] [Google Scholar]
- Armstrong AA, Mohideen F, Lima CD. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature. 2012;483:59–63. doi: 10.1038/nature10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacquin A, Pouvelle C, Siaud N, Perderiset M, Salome-Desnoulez S, Tellier-Lebegue C, Lopez B, Charbonnier JB, Kannouche PL. The helicase FBH1 is tightly regulated by PCNA via CRL4(Cdt2)-mediated proteolysis in human cells. Nucleic Acids Res. 2013;41:6501–6513. doi: 10.1093/nar/gkt397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baple EL, Chambers H, Cross HE, Fawcett H, Nakazawa Y, Chioza BA, Harlalka GV, Mansour S, Sreekantan-Nair A, Patton MA, et al. Hypomorphic PCNA mutation underlies a human DNA repair disorder. J Clin Invest. 2014;124:3137–3146. doi: 10.1172/JCI74593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JR, Pons C, Nguyen HD, Costanzo M, Boone C, Myers CL, Bielinsky AK. Genetic Interactions Implicating Postreplicative Repair in Okazaki Fragment Processing. PLoS Genet. 2015;11:e1005659. doi: 10.1371/journal.pgen.1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergoglio V, Boyer AS, Walsh E, Naim V, Legube G, Lee MY, Rey L, Rosselli F, Cazaux C, Eckert KA, et al. DNA synthesis by Pol eta promotes fragile site stability by preventing under-replicated DNA in mitosis. J Cell Biol. 2013;201:395–408. doi: 10.1083/jcb.201207066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- Boehm EM, Gildenberg MS, Washington MT. The Many Roles of PCNA in Eukaryotic DNA Replication. Enzymes. 2016a;39:231–254. doi: 10.1016/bs.enz.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm EM, Spies M, Washington MT. PCNA tool belts and polymerase bridges form during translesion synthesis. Nucleic Acids Res. 2016b doi: 10.1093/nar/gkw563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Psakhye I. DNA damage tolerance. Curr Opin Cell Biol. 2016;40:137–144. doi: 10.1016/j.ceb.2016.03.015. [DOI] [PubMed] [Google Scholar]
- Bruning JB, Shamoo Y. Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-delta p66 subunit and flap endonuclease-1. Structure. 2004;12:2209–2219. doi: 10.1016/j.str.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Burkovics P, Dome L, Juhasz S, Altmannova V, Sebesta M, Pacesa M, Fugger K, Sorensen CS, Lee MY, Haracska L, et al. The PCNA-associated protein PARI negatively regulates homologous recombination via the inhibition of DNA repair synthesis. Nucleic Acids Res. 2016;44:3176–3189. doi: 10.1093/nar/gkw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr AM, Lambert S. Replication stress-induced genome instability: the dark side of replication maintenance by homologous recombination. J Mol Biol. 2013;425:4733–4744. doi: 10.1016/j.jmb.2013.04.023. [DOI] [PubMed] [Google Scholar]
- Cazzalini O, Sommatis S, Tillhon M, Dutto I, Bachi A, Rapp A, Nardo T, Scovassi AI, Necchi D, Cardoso MC, et al. CBP and p300 acetylate PCNA to link its degradation with nucleotide excision repair synthesis. Nucleic Acids Res. 2014;42:8433–8448. doi: 10.1093/nar/gku533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centore RC, Havens CG, Manning AL, Li JM, Flynn RL, Tse A, Jin J, Dyson NJ, Walter JC, Zou L. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol Cell. 2010;40:22–33. doi: 10.1016/j.molcel.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centore RC, Yazinski SA, Tse A, Zou L. Spartan/C1orf124, a reader of PCNA ubiquitylation and a regulator of UV-induced DNA damage response. Mol Cell. 2012;46:625–635. doi: 10.1016/j.molcel.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DJ, Cimprich KA. DNA damage tolerance: when it's OK to make mistakes. Nat Chem Biol. 2009;5:82–90. doi: 10.1038/nchembio.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Bosques L, Sung P, Kupfer GM. A novel role for non-ubiquitinated FANCD2 in response to hydroxyurea-induced DNA damage. Oncogene. 2016;35:22–34. doi: 10.1038/onc.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Paudyal SC, Chin RI, You Z. PCNA promotes processive DNA end resection by Exo1. Nucleic Acids Res. 2013;41:9325–9338. doi: 10.1093/nar/gkt672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KN, Nicolae CM, Constantin D, Imamura Kawasawa Y, Delgado-Diaz MR, De S, Freire R, Smits VA, Moldovan GL. HUWE1 interacts with PCNA to alleviate replication stress. EMBO Rep. 2016;17:874–886. doi: 10.15252/embr.201541685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WK, Payne MJ, Beli P, Hanada K, Choudhary C, Hickson ID. FBH1 influences DNA replication fork stability and homologous recombination through ubiquitylation of RAD51. Nat Commun. 2015;6:5931. doi: 10.1038/ncomms6931. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Nimonkar AV, Hu Y, Hajdu I, Achar YJ, Izhar L, Petit SA, Adamson B, Yoon JC, Kowalczykowski SC, et al. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol Cell. 2012;47:396–409. doi: 10.1016/j.molcel.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolla L, Maffia A, Bertoletti F, Sabbioneda S. The Regulation of DNA Damage Tolerance by Ubiquitin and Ubiquitin-Like Modifiers. Front Genet. 2016;7:105. doi: 10.3389/fgene.2016.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clijsters L, Wolthuis R. PIP-box-mediated degradation prohibits re-accumulation of Cdc6 during S phase. J Cell Sci. 2014;127:1336–1345. doi: 10.1242/jcs.145862. [DOI] [PubMed] [Google Scholar]
- Daigaku Y, Davies AA, Ulrich HD. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 2010;465:951–955. doi: 10.1038/nature09097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol Cell. 2008;29:625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EJ, Lachaud C, Appleton P, Macartney TJ, Nathke I, Rouse J. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat Struct Mol Biol. 2012;19:1093–1100. doi: 10.1038/nsmb.2394. [DOI] [PubMed] [Google Scholar]
- De Biasio A, Blanco FJ. Proliferating cell nuclear antigen structure and interactions: too many partners for one dancer? Adv Protein Chem Struct Biol. 2013;91:1–36. doi: 10.1016/B978-0-12-411637-5.00001-9. [DOI] [PubMed] [Google Scholar]
- De Biasio A, de Opakua AI, Mortuza GB, Molina R, Cordeiro TN, Castillo F, Villate M, Merino N, Delgado S, Gil-Carton D, et al. Structure of p15(PAF)-PCNA complex and implications for clamp sliding during DNA replication and repair. Nat Commun. 2015;6:6439. doi: 10.1038/ncomms7439. [DOI] [PubMed] [Google Scholar]
- Despras E, Delrieu N, Garandeau C, Ahmed-Seghir S, Kannouche PL. Regulation of the specialized DNA polymerase eta: revisiting the biological relevance of its PCNA- and ubiquitin-binding motifs. Environ Mol Mutagen. 2012;53:752–765. doi: 10.1002/em.21741. [DOI] [PubMed] [Google Scholar]
- Dieckman LM, Freudenthal BD, Washington MT. PCNA structure and function: insights from structures of PCNA complexes and post-translationally modified PCNA. Subcell Biochem. 2012;62:281–299. doi: 10.1007/978-94-007-4572-8_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovrat D, Stodola JL, Burgers PM, Aharoni A. Sequential switching of binding partners on PCNA during in vitro Okazaki fragment maturation. Proc Natl Acad Sci U S A. 2014;111:14118–14123. doi: 10.1073/pnas.1321349111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy CM, Hilbert BJ, Kelch BA. A Disease-Causing Variant in PCNA Disrupts a Promiscuous Protein Binding Site. J Mol Biol. 2016;428:1023–1040. doi: 10.1016/j.jmb.2015.11.029. [DOI] [PubMed] [Google Scholar]
- Dungrawala H, Rose KL, Bhat KP, Mohni KN, Glick GG, Couch FB, Cortez D. The Replication Checkpoint Prevents Two Types of Fork Collapse without Regulating Replisome Stability. Mol Cell. 2015;59:998–1010. doi: 10.1016/j.molcel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds CE, Simpson LJ, Sale JE. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol Cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- Feng W, Guo Y, Huang J, Deng Y, Zang J, Huen MS. TRAIP regulates replication fork recovery and progression via PCNA. Cell Discov. 2016;2:16016. doi: 10.1038/celldisc.2016.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman Y, Palgi N, Dovrat D, Ben-Aroya S, Hieter P, Aharoni A. Subtle alterations in PCNA–partner interactions severely impair DNA replication and repair. PLoS Biol. 2010;8:e1000507. doi: 10.1371/journal.pbio.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gali H, Juhasz S, Morocz M, Hajdu I, Fatyol K, Szukacsov V, Burkovics P, Haracska L. Role of SUMO modification of human PCNA at stalled replication fork. Nucleic Acids Res. 2012;40:6049–6059. doi: 10.1093/nar/gks256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganai RA, Johansson E. DNA Replication-A Matter of Fidelity. Mol Cell. 2016;62:745–755. doi: 10.1016/j.molcel.2016.05.003. [DOI] [PubMed] [Google Scholar]
- Gao X, Dan S, Xie Y, Qin H, Tang D, Liu X, He QY, Liu L. 14-3-3zeta reduces DNA damage by interacting with and stabilizing proliferating cell nuclear antigen. J Cell Biochem. 2015;116:158–169. doi: 10.1002/jcb.24955. [DOI] [PubMed] [Google Scholar]
- Gao Y, Mutter-Rottmayer E, Greenwalt AM, Goldfarb D, Yan F, Yang Y, Martinez-Chacin RC, Pearce KH, Tateishi S, Major MB, et al. A neomorphic cancer cell-specific role of MAGE-A4 in trans-lesion synthesis. Nat Commun. 2016;7:12105. doi: 10.1038/ncomms12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu RE, Langston L, Yao NY, Yurieva O, Zhang D, Finkelstein J, Agarwal T, O'Donnell ME. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat Struct Mol Biol. 2014;21:664–670. doi: 10.1038/nsmb.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu RE, Schauer GD, Yao NY, Langston LD, Yurieva O, Zhang D, Finkelstein J, O'Donnell ME. Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation. Elife. 2015;4:e04988. doi: 10.7554/eLife.04988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal G, Leung JW, Nair BC, Fong KW, Chen J. Proliferating cell nuclear antigen (PCNA)-binding protein C1orf124 is a regulator of translesion synthesis. J Biol Chem. 2012;287:34225–34233. doi: 10.1074/jbc.M112.400135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Zwicky K, Follonier C, Foiani M, Lopes M, Branzei D. Visualization of recombination-mediated damage bypass by template switching. Nat Struct Mol Biol. 2014;21:884–892. doi: 10.1038/nsmb.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilljam KM, Feyzi E, Aas PA, Sousa MM, Muller R, Vagbo CB, Catterall TC, Liabakk NB, Slupphaug G, Drablos F, et al. Identification of a novel, widespread, and functionally important PCNA-binding motif. J Cell Biol. 2009;186:645–654. doi: 10.1083/jcb.200903138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilljam KM, Muller R, Liabakk NB, Otterlei M. Nucleotide excision repair is associated with the replisome and its efficiency depends on a direct interaction between XPA and PCNA. PLoS One. 2012;7:e49199. doi: 10.1371/journal.pone.0049199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goellner EM, Smith CE, Campbell CS, Hombauer H, Desai A, Putnam CD, Kolodner RD. PCNA and Msh2-Msh6 activate an Mlh1-Pms1 endonuclease pathway required for Exo1-independent mismatch repair. Mol Cell. 2014;55:291–304. doi: 10.1016/j.molcel.2014.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohler T, Munoz IM, Rouse J, Blow JJ. PTIP/Swift is required for efficient PCNA ubiquitination in response to DNA damage. DNA Repair (Amst) 2008;7:775–787. doi: 10.1016/j.dnarep.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Green CM, Baple EL, Crosby AH. PCNA mutation affects DNA repair not replication. Cell Cycle. 2014;13:3157–3158. doi: 10.4161/15384101.2014.969994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groehler AL, Lannigan DA. A chromatin-bound kinase, ERK8, protects genomic integrity by inhibiting HDM2-mediated degradation of the DNA clamp PCNA. J Cell Biol. 2010;190:575–586. doi: 10.1083/jcb.201002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol. 2006;26:8892–8900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampp S, Kiessling T, Buechle K, Mansilla SF, Thomale J, Rall M, Ahn J, Pospiech H, Gottifredi V, Wiesmuller L. DNA damage tolerance pathway involving DNA polymerase iota and the tumor suppressor p53 regulates DNA replication fork progression. Proc Natl Acad Sci U S A. 2016;113:E4311–4319. doi: 10.1073/pnas.1605828113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Wani G, Zhao R, Qian J, Sharma N, He J, Zhu Q, Wang QE, Wani AA. Cdt2-mediated XPG degradation promotes gap-filling DNA synthesis in nucleotide excision repair. Cell Cycle. 2015;14:1103–1115. doi: 10.4161/15384101.2014.973740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Liu T, Huen MS, Hu L, Chen Z, Huang J. SIVA1 directs the E3 ubiquitin ligase RAD18 for PCNA monoubiquitination. J Cell Biol. 2014;205:811–827. doi: 10.1083/jcb.201311007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley ME, Murina O, Leitch A, Higgs MR, Bicknell LS, Yigit G, Blackford AN, Zlatanou A, Mackenzie KJ, Reddy K, et al. TRAIP promotes DNA damage response during genome replication and is mutated in primordial dwarfism. Nat Genet. 2016;48:36–43. doi: 10.1038/ng.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens CG, Walter JC. Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol Cell. 2009;35:93–104. doi: 10.1016/j.molcel.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedglin M, Benkovic SJ. Regulation of Rad6/Rad18 Activity During DNA Damage Tolerance. Annu Rev Biophys. 2015;44:207–228. doi: 10.1146/annurev-biophys-060414-033841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedglin M, Pandey B, Benkovic SJ. Stability of the human polymerase delta holoenzyme and its implications in lagging strand DNA synthesis. Proc Natl Acad Sci U S A. 2016;113:E1777–1786. doi: 10.1073/pnas.1523653113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A, Krijger PH, Diamant N, Goren Z, Langerak P, Kim J, Reissner T, Lee KY, Geacintov NE, Carell T, et al. PCNA ubiquitination is important, but not essential for translesion DNA synthesis in mammalian cells. PLoS Genet. 2011;7:e1002262. doi: 10.1371/journal.pgen.1002262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Hoffmann S, Smedegaard S, Nakamura K, Mortuza GB, Raschle M, Ibanez de Opakua A, Oka Y, Feng Y, Blanco FJ, Mann M, et al. TRAIP is a PCNA-binding ubiquitin ligase that protects genome stability after replication stress. J Cell Biol. 2016;212:63–75. doi: 10.1083/jcb.201506071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Saraf A, Florens L, Kusch T, Swanson SK, Szerszen LT, Li G, Dutta A, Washburn MP, Abmayr SM, et al. The Enok acetyltransferase complex interacts with Elg1 and negatively regulates PCNA unloading to promote the G1/S transition. Genes Dev. 2016;30:1198–1210. doi: 10.1101/gad.271429.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D'Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- Hutton RD, Craggs TD, White MF, Penedo JC. PCNA and XPF cooperate to distort DNA substrates. Nucleic Acids Res. 2010;38:1664–1675. doi: 10.1093/nar/gkp1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JG, Tsaalbi-Shtylik A, de Wind N. Roles of mutagenic translesion synthesis in mammalian genome stability, health and disease. DNA Repair (Amst) 2015;29:56–64. doi: 10.1016/j.dnarep.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Johnson C, Gali VK, Takahashi TS, Kubota T. PCNA Retention on DNA into G2/M Phase Causes Genome Instability in Cells Lacking Elg1. Cell Rep. 2016;16:684–695. doi: 10.1016/j.celrep.2016.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz S, Balogh D, Hajdu I, Burkovics P, Villamil MA, Zhuang Z, Haracska L. Characterization of human Spartan/C1orf124, an ubiquitin-PCNA interacting regulator of DNA damage tolerance. Nucleic Acids Res. 2012;40:10795–10808. doi: 10.1093/nar/gks850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- Kanu N, Zhang T, Burrell RA, Chakraborty A, Cronshaw J, DaCosta C, Gronroos E, Pemberton HN, Anderton E, Gonzalez L, et al. RAD18, WRNIP1 and ATMIN promote ATM signalling in response to replication stress. Oncogene. 2016;35:4009–4019. doi: 10.1038/onc.2015.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- Kashiwaba S, Kanao R, Masuda Y, Kusumoto-Matsuo R, Hanaoka F, Masutani C. USP7 Is a Suppressor of PCNA Ubiquitination and Oxidative-Stress-Induced Mutagenesis in Human Cells. Cell Rep. 2015;13:2072–2080. doi: 10.1016/j.celrep.2015.11.014. [DOI] [PubMed] [Google Scholar]
- Kawasoe Y, Tsurimoto T, Nakagawa T, Masukata H, Takahashi TS. MutSalpha maintains the mismatch repair capability by inhibiting PCNA unloading. Elife. 2016;5 doi: 10.7554/eLife.15155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Budhavarapu VN, Herrera CR, Nam HW, Kim YS, Yew PR. The CRL4Cdt2 ubiquitin ligase mediates the proteolysis of cyclin-dependent kinase inhibitor Xic1 through a direct association with PCNA. Mol Cell Biol. 2010;30:4120–4133. doi: 10.1128/MCB.01135-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Dejsuphong D, Adelmant G, Ceccaldi R, Yang K, Marto JA, D'Andrea AD. Transcriptional repressor ZBTB1 promotes chromatin remodeling and translesion DNA synthesis. Mol Cell. 2014;54:107–118. doi: 10.1016/j.molcel.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijger PH, van den Berk PC, Wit N, Langerak P, Jansen JG, Reynaud CA, de Wind N, Jacobs H. PCNA ubiquitination-independent activation of polymerase eta during somatic hypermutation and DNA damage tolerance. DNA Repair (Amst) 2011;10:1051–1059. doi: 10.1016/j.dnarep.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Krokan HE, Bjoras M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T, Nishimura K, Kanemaki MT, Donaldson AD. The Elg1 replication factor C-like complex functions in PCNA unloading during DNA replication. Mol Cell. 2013;50:273–280. doi: 10.1016/j.molcel.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Kuchenbaecker KB, Ramus SJ, Tyrer J, Lee A, Shen HC, Beesley J, Lawrenson K, McGuffog L, Healey S, Lee JM, et al. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat Genet. 2015;47:164–171. doi: 10.1038/ng.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau WC, Li Y, Zhang Q, Huen MS. Molecular architecture of the Ub-PCNA/Pol eta complex bound to DNA. Sci Rep. 2015;5:15759. doi: 10.1038/srep15759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Fu H, Aladjem MI, Myung K. ATAD5 regulates the lifespan of DNA replication factories by modulating PCNA level on the chromatin. J Cell Biol. 2013;200:31–44. doi: 10.1083/jcb.201206084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessel D, Vaz B, Halder S, Lockhart PJ, Marinovic-Terzic I, Lopez-Mosqueda J, Philipp M, Sim JC, Smith KR, Oehler J, et al. Mutations in SPRTN cause early onset hepatocellular carcinoma, genomic instability and progeroid features. Nat Genet. 2014;46:1239–1244. doi: 10.1038/ng.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JR, Zeman MK, Chen JY, Yee MC, Cimprich KA. SHPRH and HLTF act in a damage-specific manner to coordinate different forms of postreplication repair and prevent mutagenesis. Mol Cell. 2011;42:237–249. doi: 10.1016/j.molcel.2011.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J Cell Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- Mailand N, Gibbs-Seymour I, Bekker-Jensen S. Regulation of PCNA-protein interactions for genome stability. Nat Rev Mol Cell Biol. 2013;14:269–282. doi: 10.1038/nrm3562. [DOI] [PubMed] [Google Scholar]
- Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- Masuda Y, Piao J, Kamiya K. DNA replication-coupled PCNA mono-ubiquitination and polymerase switching in a human in vitro system. J Mol Biol. 2010;396:487–500. doi: 10.1016/j.jmb.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Mejlvang J, Feng Y, Alabert C, Neelsen KJ, Jasencakova Z, Zhao X, Lees M, Sandelin A, Pasero P, Lopes M, et al. New histone supply regulates replication fork speed and PCNA unloading. J Cell Biol. 2014;204:29–43. doi: 10.1083/jcb.201305017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Dejsuphong D, Petalcorin MI, Hofmann K, Takeda S, Boulton SJ, D'Andrea AD. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol Cell. 2012;45:75–86. doi: 10.1016/j.molcel.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc Natl Acad Sci U S A. 2005;102:8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosbech A, Gibbs-Seymour I, Kagias K, Thorslund T, Beli P, Povlsen L, Nielsen SV, Smedegaard S, Sedgwick G, Lukas C, et al. DVC1 (C1orf124) is a DNA damage-targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nat Struct Mol Biol. 2012;19:1084–1092. doi: 10.1038/nsmb.2395. [DOI] [PubMed] [Google Scholar]
- Motegi A, Liaw HJ, Lee KY, Roest HP, Maas A, Wu X, Moinova H, Markowitz SD, Ding H, Hoeijmakers JH, et al. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc Natl Acad Sci U S A. 2008;105:12411–12416. doi: 10.1073/pnas.0805685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R, Misund K, Holien T, Bachke S, Gilljam KM, Våtsveen TK, Rø TB, Bellacchio E, Sundan A, Otterlei M. Targeting proliferating cell nuclear antigen and its protein interactions induces apoptosis in multiple myeloma cells. PLoS One. 2013;8:e70430. doi: 10.1371/journal.pone.0070430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae CM, Aho ER, Vlahos AH, Choe KN, De S, Karras GI, Moldovan GL. The ADP-ribosyltransferase PARP10/ARTD10 interacts with proliferating cell nuclear antigen (PCNA) and is required for DNA damage tolerance. J Biol Chem. 2014;289:13627–13637. doi: 10.1074/jbc.M114.556340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell M, Langston L, Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell. 2010;40:364–376. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, et al. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell. 2010;37:714–727. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Olaisen C, Muller R, Nedal A, Otterlei M. PCNA-interacting peptides reduce Akt phosphorylation and TLR-mediated cytokine secretion suggesting a role of PCNA in cellular signaling. Cell Signal. 2015;27:1478–1487. doi: 10.1016/j.cellsig.2015.03.009. [DOI] [PubMed] [Google Scholar]
- Ortega J, Li JY, Lee S, Tong D, Gu L, Li GM. Phosphorylation of PCNA by EGFR inhibits mismatch repair and promotes misincorporation during DNA synthesis. Proc Natl Acad Sci U S A. 2015;112:5667–5672. doi: 10.1073/pnas.1417711112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Park JM, Yang SW, Yu KR, Ka SH, Lee SW, Seol JH, Jeon YJ, Chung CH. Modification of PCNA by ISG15 plays a crucial role in termination of error-prone translesion DNA synthesis. Mol Cell. 2014;54:626–638. doi: 10.1016/j.molcel.2014.03.031. [DOI] [PubMed] [Google Scholar]
- Pedley AM, Lill MA, Davisson VJ. Flexibility of PCNA-protein interface accommodates differential binding partners. PLoS One. 2014;9:e102481. doi: 10.1371/journal.pone.0102481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Black KL, Kovermann SK, Brock HW, Mazo A. Stepwise histone modifications are mediated by multiple enzymes that rapidly associate with nascent DNA during replication. Nat Commun. 2013;4:2841. doi: 10.1038/ncomms3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Johnston DM, Hodgson JW, Black KL, Kovermann SK, Beck S, Canaani E, Brock HW, Mazo A. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell. 2012;150:922–933. doi: 10.1016/j.cell.2012.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- Pitroda SP, Pashtan IM, Logan HL, Budke B, Darga TE, Weichselbaum RR, Connell PP. DNA repair pathway gene expression score correlates with repair proficiency and tumor sensitivity to chemotherapy. Sci Transl Med. 2014;6:229ra242. doi: 10.1126/scitranslmed.3008291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plosky BS, Vidal AE, Fernandez de Henestrosa AR, McLenigan MP, McDonald JP, Mead S, Woodgate R. Controlling the subcellular localization of DNA polymerases iota and eta via interactions with ubiquitin. EMBO J. 2006;25:2847–2855. doi: 10.1038/sj.emboj.7601178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povlsen LK, Beli P, Wagner SA, Poulsen SL, Sylvestersen KB, Poulsen JW, Nielsen ML, Bekker-Jensen S, Mailand N, Choudhary C. Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nat Cell Biol. 2012;14:1089–1098. doi: 10.1038/ncb2579. [DOI] [PubMed] [Google Scholar]
- Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol. 2015;7:a016600. doi: 10.1101/cshperspect.a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punchihewa C, Inoue A, Hishiki A, Fujikawa Y, Connelly M, Evison B, Shao Y, Heath R, Kuraoka I, Rodrigues P, et al. Identification of small molecule proliferating cell nuclear antigen (PCNA) inhibitor that disrupts interactions with PIP-box proteins and inhibits DNA replication. J Biol Chem. 2012;287:14289–14300. doi: 10.1074/jbc.M112.353201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman M, Havens CG, Walter JC, Harper JW. A genome-wide screen identifies p97 as an essential regulator of DNA damage-dependent CDT1 destruction. Mol Cell. 2011;44:72–84. doi: 10.1016/j.molcel.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell. 2007;130:991–1004. doi: 10.1016/j.cell.2007.08.043. [DOI] [PubMed] [Google Scholar]
- Rizzardi LF, Coleman KE, Varma D, Matson JP, Oh S, Cook JG. CDK1-dependent inhibition of the E3 ubiquitin ligase CRL4CDT2 ensures robust transition from S Phase to Mitosis. J Biol Chem. 2015;290:556–567. doi: 10.1074/jbc.M114.614701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roa S, Avdievich E, Peled JU, Maccarthy T, Werling U, Kuang FL, Kan R, Zhao C, Bergman A, Cohen PE, et al. Ubiquitylated PCNA plays a role in somatic hypermutation and class-switch recombination and is required for meiotic progression. Proc Natl Acad Sci U S A. 2008;105:16248–16253. doi: 10.1073/pnas.0808182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saugar I, Parker JL, Zhao S, Ulrich HD. The genome maintenance factor Mgs1 is targeted to sites of replication stress by ubiquitylated PCNA. Nucleic Acids Res. 2012;40:245–257. doi: 10.1093/nar/gkr738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebesta M, Burkovics P, Juhasz S, Zhang S, Szabo JE, Lee MY, Haracska L, Krejci L. Role of PCNA and TLS polymerases in D-loop extension during homologous recombination in humans. DNA Repair (Amst) 2013;12:691–698. doi: 10.1016/j.dnarep.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahrour MA, Nicolae CM, Edvardson S, Ashhab M, Galvan AM, Constantin D, Abu-Libdeh B, Moldovan G-L, Elpeleg O. PARP10 deficiency manifests by severe developmental delay and DNA repair defect. neurogenetics. 2016:1–6. doi: 10.1007/s10048-016-0493-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata E, Dar A, Dutta A. CRL4Cdt2 E3 ubiquitin ligase and proliferating cell nuclear antigen (PCNA) cooperate to degrade thymine DNA glycosylase in S phase. J Biol Chem. 2014;289:23056–23064. doi: 10.1074/jbc.M114.574210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui K, On KF, Diffley JF. Regulating DNA replication in eukarya. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slenn TJ, Morris B, Havens CG, Freeman RM, Jr, Takahashi TS, Walter JC. Thymine DNA glycosylase is a CRL4Cdt2 substrate. J Biol Chem. 2014;289:23043–23055. doi: 10.1074/jbc.M114.574194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SJ, Gu L, Phipps EA, Dobrolecki LE, Mabrey KS, Gulley P, Dillehay KL, Dong Z, Fields GB, Chen YR, et al. A Peptide mimicking a region in proliferating cell nuclear antigen specific to key protein interactions is cytotoxic to breast cancer. Mol Pharmacol. 2015;87:263–276. doi: 10.1124/mol.114.093211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Adzhubei I, Thurman RE, Kryukov GV, Mirkin SM, Sunyaev SR. Human mutation rate associated with DNA replication timing. Nat Genet. 2009;41:393–395. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stodola JL, Burgers PM. Resolving individual steps of Okazaki-fragment maturation at a millisecond timescale. Nat Struct Mol Biol. 2016;23:402–408. doi: 10.1038/nsmb.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Wortman M, Dillehay KL, Seibel WL, Evelyn CR, Smith SJ, Malkas LH, Zheng Y, Lu S, Dong Z. Small-molecule targeting of proliferating cell nuclear antigen chromatin association inhibits tumor cell growth. Mol Pharmacol. 2012;81:811–819. doi: 10.1124/mol.112.077735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai K, Abbas T, Jazaeri AA, Dutta A. CRL4(Cdt2) E3 ubiquitin ligase monoubiquitinates PCNA to promote translesion DNA synthesis. Mol Cell. 2010;37:143–149. doi: 10.1016/j.molcel.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Sharma S, Zou J, Lin SY, Wang B, Rezvani K, Wang H, Parvin JD, Ludwig T, Canman CE, et al. BRCA1 promotes the ubiquitination of PCNA and recruitment of translesion polymerases in response to replication blockade. Proc Natl Acad Sci U S A. 2013;110:13558–13563. doi: 10.1073/pnas.1306534110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Yan C, Xu X, Weinacht CP, Freudenthal BD, Yang K, Zhuang Z, Washington MT, Tainer JA, Ivanov I. Structurally distinct ubiquitin- and sumo-modified PCNA: implications for their distinct roles in the DNA damage response. Structure. 2015;23:724–733. doi: 10.1016/j.str.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier JB, Sandhu S, Petalcorin MI, Wu X, Nabi Z, Ding H, Boulton SJ. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science. 2013;342:239–242. doi: 10.1126/science.1241779. [DOI] [PubMed] [Google Scholar]
- Wang SC. PCNA: a silent housekeeper or a potential therapeutic target? Trends Pharmacol Sci. 2014;35:178–186. doi: 10.1016/j.tips.2014.02.004. [DOI] [PubMed] [Google Scholar]
- Wang SC, Nakajima Y, Yu YL, Xia W, Chen CT, Yang CC, McIntush EW, Li LY, Hawke DH, Kobayashi R, et al. Tyrosine phosphorylation controls PCNA function through protein stability. Nat Cell Biol. 2006;8:1359–1368. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang S, Zheng R, Yue F, Lin SH, Rahmeh AA, Lee EY, Zhang Z, Lee MY. PDIP46 (DNA polymerase delta interacting protein 46) is an activating factor for human DNA polymerase delta. Oncotarget. 2016;7:6294–6313. doi: 10.18632/oncotarget.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warbrick E. The puzzle of PCNA's many partners. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston R, Peeters H, Ahel D. ZRANB3 is a structure-specific ATP-dependent endonuclease involved in replication stress response. Genes Dev. 2012;26:1558–1572. doi: 10.1101/gad.193516.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wit N, Buoninfante OA, van den Berk PC, Jansen JG, Hogenbirk MA, de Wind N, Jacobs H. Roles of PCNA ubiquitination and TLS polymerases kappa and eta in the bypass of methyl methanesulfonate-induced DNA damage. Nucleic Acids Res. 2015;43:282–294. doi: 10.1093/nar/gku1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witko-Sarsat V, Mocek J, Bouayad D, Tamassia N, Ribeil JA, Candalh C, Davezac N, Reuter N, Mouthon L, Hermine O, et al. Proliferating cell nuclear antigen acts as a cytoplasmic platform controlling human neutrophil survival. J Exp Med. 2010;207:2631–2645. doi: 10.1084/jem.20092241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Weinacht CP, Zhuang Z. Regulatory Role of Ubiquitin in Eukaryotic DNA Translesion Synthesis. Biochemistry. 2013a doi: 10.1021/bi400194r. [DOI] [PubMed] [Google Scholar]
- Yang XH, Shiotani B, Classon M, Zou L. Chk1 and Claspin potentiate PCNA ubiquitination. Genes Dev. 2008;22:1147–1152. doi: 10.1101/gad.1632808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Durando M, Smith-Roe SL, Sproul C, Greenwalt AM, Kaufmann W, Oh S, Hendrickson EA, Vaziri C. Cell cycle stage-specific roles of Rad18 in tolerance and repair of oxidative DNA damage. Nucleic Acids Res. 2013b;41:2296–2312. doi: 10.1093/nar/gks1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Xie Y, Yin S, Lv X, Zhang J, Gu Z, Sun H, Liu S. The S-nitrosylation status of PCNA localized in cytosol impacts the apoptotic pathway in a Parkinson's disease paradigm. PLoS One. 2015;10:e0117546. doi: 10.1371/journal.pone.0117546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Gan H, Han J, Zhou ZX, Jia S, Chabes A, Farrugia G, Ordog T, Zhang Z. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol Cell. 2014;56:551–563. doi: 10.1016/j.molcel.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Song C, Zhang Q, DiMaggio PA, Garcia BA, York A, Carey MF, Grunstein M. Histone H3 lysine 56 methylation regulates DNA replication through its interaction with PCNA. Mol Cell. 2012;46:7–17. doi: 10.1016/j.molcel.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Ghosal G, Chen J. The HARP-like domain-containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol Cell. 2012;47:410–421. doi: 10.1016/j.molcel.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Chea J, Meng X, Zhou Y, Lee EY, Lee MY. PCNA is ubiquitinated by RNF8. Cell Cycle. 2008;7:3399–3404. doi: 10.4161/cc.7.21.6949. [DOI] [PubMed] [Google Scholar]
- Zhang S, Zhao H, Darzynkiewicz Z, Zhou P, Zhang Z, Lee EY, Lee MY. A novel function of CRL4(Cdt2): regulation of the subunit structure of DNA polymerase delta in response to DNA damage and during the S phase. J Biol Chem. 2013;288:29550–29561. doi: 10.1074/jbc.M113.490466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Shibahara K, Stillman B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 2000;408:221–225. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhang S, Lin SH, Wang X, Wu L, Lee EY, Lee MY. Structure of monoubiquitinated PCNA: implications for DNA polymerase switching and Okazaki fragment maturation. Cell Cycle. 2012;11:2128–2136. doi: 10.4161/cc.20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Chen MS, Lo YH, Waltz SE, Wang J, Ho PC, Vasiliauskas J, Plattner R, Wang YL, Wang SC. The Ron receptor tyrosine kinase activates c-Abl to promote cell proliferation through tyrosine phosphorylation of PCNA in breast cancer. Oncogene. 2014;33:1429–1437. doi: 10.1038/onc.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Lo YH, Ma L, Waltz SE, Gray JK, Hung MC, Wang SC. Targeting tyrosine phosphorylation of PCNA inhibits prostate cancer growth. Mol Cancer Ther. 2011;10:29–36. doi: 10.1158/1535-7163.MCT-10-0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Shen B. Okazaki fragment maturation: nucleases take centre stage. J Mol Cell Biol. 2011;3:23–30. doi: 10.1093/jmcb/mjq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanou A, Despras E, Braz-Petta T, Boubakour-Azzouz I, Pouvelle C, Stewart GS, Nakajima S, Yasui A, Ishchenko AA, Kannouche PL. The hMsh2-hMsh6 complex acts in concert with monoubiquitinated PCNA and Pol eta in response to oxidative DNA damage in human cells. Mol Cell. 2011;43:649–662. doi: 10.1016/j.molcel.2011.06.023. [DOI] [PubMed] [Google Scholar]