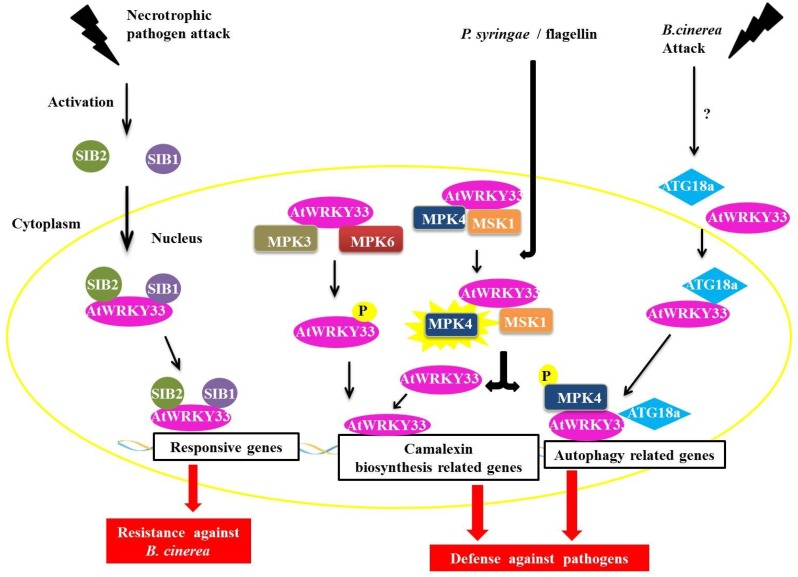
Figure 6.
Overview of AtWRKY33 interactions during biotic stress responses. During an attack by a necrotrophic pathogen, AtWRKY33 interacts with the proteins SIGMA FACTOR-INTERACTING PROTEIN 1 (SIB1) and SIGMA FACTOR-INTERACTING PROTEIN 2 (SIB2) in the nucleus. These interaction leads to transcription of genes responsive to the pathogen, causing an increased resistance in the plant (in this case against B. cinerea, a necrotrophic fungus). In a second interaction, AtWRKY33 can be phosphorylated by two MITOGEN-ACTIVATED PROTEIN (MAP) kinases, MITOGEN-ACTIVATED PROTEIN KINASE 3 (MPK3) and MPK6. This interaction leads to an increase in the transcription of related genes of camalexin biosynthesis, which is an important pathway utilized by the plant defense against pathogens. Another interaction leads to increased transcription of camalexin related genes. After induction by Pseudomonas syringae or flagellin, the protein MPK4 is activated and phosphorylates its substrate, the MAP KINASE SUBSTRATE1 (MSK1) protein. Phosphorylation of MSK1 releases AtWRKY33 of protein complex allowing the protein to exert its role as a transcriptional activator of plant defense genes. Finally, during attack of fungus B. cinerea, AtWRKY33 interacts with ATG18a in the nucleus. ATG18a is an important protein of the autophagy pathway in Arabidopsis, and its interaction with AtWRKY33 along with the activation of the autophagy pathway is important for signaling the response of plant defense against necrotrophic pathogens.