Type IV pili (T4P) are filamentous appendages found on many bacteria and archaea. They are helical fibers of pilin proteins assembled by a multi-component macromolecular machine we call the basal body. Based on pilin features, T4P are classified into type IVa pili (T4aP) and type IVb pili (T4bP)1,2. T4aP are more widespread and are involved in cell motility3, DNA transfer4, host predation5 and electron transfer6. T4bP are less prevalent and are mainly found in enteropathogenic bacteria, where they play key roles in host colonization7. Following previous similar work on T4aP machines8,9, here we use electron cryotomography (ECT)10 to reveal the three-dimensional in situ structure of a T4bP machine in its piliated and non-piliated states. The specific machine we analyze is the Vibrio cholerae toxin-coregulated pilus machine (TCPM). While only about half of the components of the TCPM show sequence homology to components of the previously analyzed Myxococcus xanthus T4aP machine (T4aPM), we find that their structures are nevertheless remarkably similar. Based on homologies with components of the M. xanthus T4aPM and additional reconstructions of TCPM mutants in which the non-homologous proteins are individually deleted, we propose locations for all eight TCPM components within the complex. Non-homologous proteins in the T4aPM and TCPM are found to form similar structures, suggesting new hypotheses for their functions and evolutionary histories.
Intact T4aPMs have already been visualized by ECT in M. xanthus and Thermus thermophilus, and were seen to consist of interconnected rings spanning the cell envelope8,9. In M. xanthus, the locations of the various T4aPM components were determined by imaging mutants with individual components either deleted or fused to protein tags8. Fitting atomic models of components11–24 into the reconstructions produced new insights into the overall structure and function of the T4aPM8. While approximately half (5 out of 9) of the structural components of the T4bPM exhibit sequence homology to T4aPM components, and are therefore expected to have similar structures and functions, the other half are novel, and so their positions and roles remain unclear.
Perhaps the best-characterized T4bP is the toxin-coregulated pilus (TCP) of V. cholerae, the causative agent of the diarrheal disease cholera. TCP adhere bacterial cells to gut epithelia and play an essential role in micro-colony formation25, and so are required for host colonization26. Moreover, the TCP is a receptor for the CTXφ bacteriophage which carries the genes encoding cholera toxin; thus, the presence of TCP on V. cholerae is a crucial difference between toxigenic and non-toxigenic strains.
The TCPM is encoded by an operon containing 11 genes27. TcpA is the major pilin subunit and polymerizes to form the TCP fiber28,29. TcpB is predicted to be a minor pilin30. TcpC is a member of the secretin family31 and likely forms an outer membrane (OM) pore similar to that formed by the PilQ of the T4aP system, GspD of the type II secretion system (T2SS), or PscC of the type III secretion system (T3SS). The stability and proper OM localization of TcpC is dependent on the associated periplasmic protein TcpQ31. TcpC, TcpQ, and TcpS are thought to form the OM-associated portion of the TCPM. Little is known about TcpS, except that decreased levels of the cytoplasmic ATPase TcpT are observed in a ΔtcpS deletion strain32. TcpD, TcpE, TcpJ, TcpR, and TcpT comprise the inner membrane (IM)-associated portion of the TCPM. TcpD is an IM protein with unknown function. TcpE is a conserved IM platform protein homologous to PilC of the T4aPM and GspF of the T2SS. TcpJ is the prepilin peptidase that processes pilins into a mature form33. TcpR is another IM protein which localizes TcpT, the assembly ATPase that powers extension of the pilus, to the IM32,34. The IM tethering of TcpT by TcpR is unstable in the absence of other TCP proteins, suggesting that TcpR is likely to be stabilized by the other IM proteins TcpD and/or TcpE32. Finally, TcpF is a soluble colonization factor crucial for V. cholerae virulence whose secretion into the environment requires the TCPM35,36.
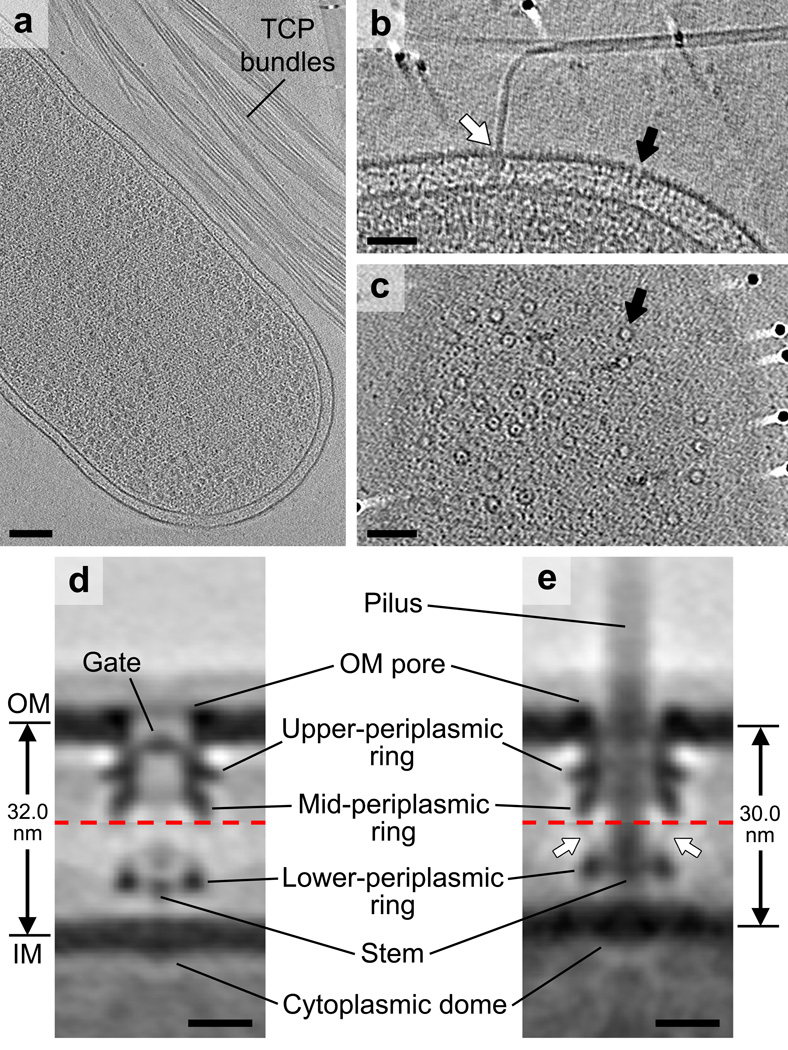
To determine the structure of the TCPM in vivo, we used ECT to image V. cholerae cells expressing abundant TCP37. Bundles of TCP with diameters of ~8 nm were observed in the vicinity of the cells (Fig. 1a). Basal bodies spanning the cell envelope were observed anchoring the pili (Fig. 1b, white arrow). We also observed non-piliated basal bodies with similar structures (Fig. 1b, c, black arrows). Several lines of evidence revealed that all these structures were TCPMs rather than any other cellular complex (see Supplementary Discussion). To reveal structural details, we generated sub-tomogram averages of non-piliated and piliated TCPMs (Fig. 1d, e). We found that both structures comprise two major complexes associated with the OM and IM, respectively (Supplementary Fig. 1), which were not rigidly attached to one another. Due to this flexibility, these two complexes were aligned separately to produce clearer averages (Supplementary Fig. 2). The resulting averages of the OM and IM complexes were then combined to generate composite reconstructions of complete non-piliated and piliated TCPMs with local resolutions of 3–4 nm (Supplementary Fig. 2d, h).
Figure 1. Visualizing the TCPM in intact V. cholerae cells.
(a) Slice through a cryotomogram of a frozen-hydrated V. cholerae cell on an EM grid adjacent to bundles of TCP. (b) A tomographic slice through the cell envelope. White arrow: a piliated TCPM basal body with its pilus fiber joining a TCP bundle; black arrow: a non-piliated TCPM basal body. (c) A tomographic slice through the cell surface showing top views of TCPM OM pores (black arrow). Images in (a–c) are representative of at least 50 examples. (d, e) Composite sub-tomogram averages of wild-type non-piliated (d) and piliated (e) TCPM basal bodies, with upper and lower halves (separated by red dashed lines) aligned based on the OM- or IM-associated parts, respectively. The generation of the composite sub-tomogram averages is demonstrated in Supplementary Figure 2. Arrows in (d): densities connecting the mid- and lower-periplasmic rings. Scale bars (a) 100 nm, (b, c) 50 nm, (d, e) 10 nm.
The structure of the non-piliated TCPM comprises an OM-spanning pore with a gate, a periplasmic vestibule containing two distinct ring layers (upper- and mid-periplasmic rings), a ring in the periplasm close to the IM (lower-periplasmic ring), a short stem in between the IM and the lower-periplasmic ring, a light density above the short stem, and a cytoplasmic dome on the IM (Fig. 1d). The structure of the piliated TCPM similarly exhibits an OM pore connected to the upper- and mid-periplasmic rings, a lower-periplasmic ring close to the IM, and a cytoplasmic dome on the IM (Fig. 1e). In addition, the stem is much longer and passed through the periplasmic structures and the OM pore to the extracellular space. A fainter density connecting the mid- and lower-periplasmic rings is also present (Fig. 1e, arrows). Aligning the structures of non-piliated and piliated TCPMs with reference to the OM revealed clear conformational changes associated with piliation (Supplementary Video 1).
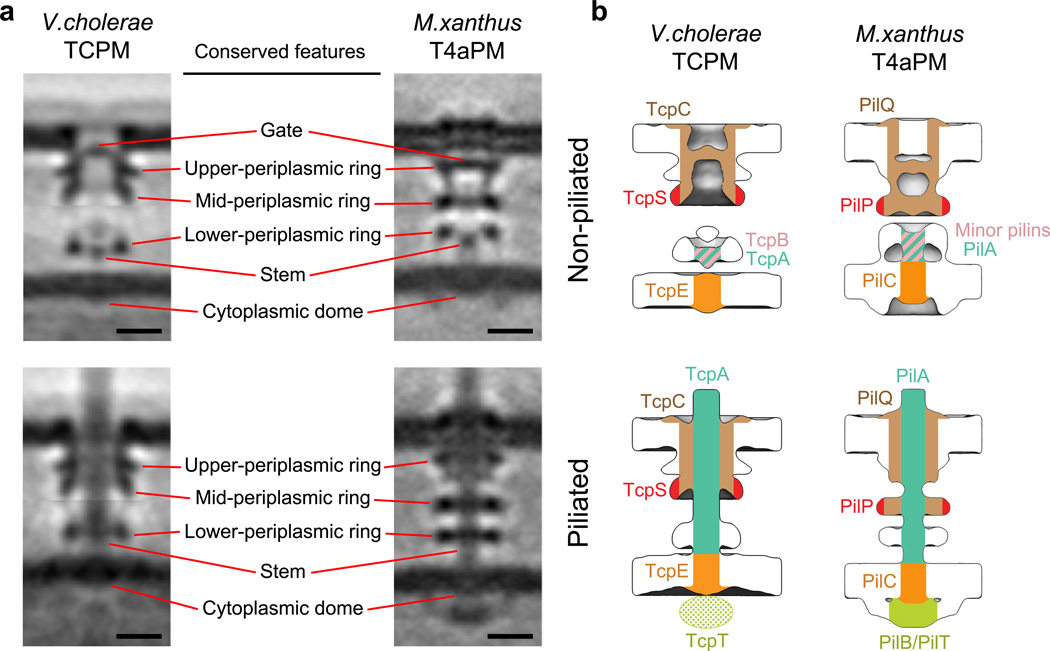
Despite major differences in gene arrangement and limited component homology, the TCPM structures look very similar to those of the M. xanthus T4aPM (Fig. 2a)8. Comparing the non-piliated structures, both exhibit a clear gate density in the OM pore, three periplasmic rings, a short stem surrounded by the lower-periplasmic ring, and a cytoplasmic dome. The main differences are that in the TCPM, the gate density is located closer to the OM, the upper- and mid-periplasmic rings are more tightly associated, there is an additional density above the short stem, there is no visible connection between the IM and the lower-periplasmic ring, and there is no cytoplasmic ring. Comparing the piliated structures, both the TCPM and T4aPM contain a long stem originating at the IM and passing through the periplasmic structures and OM pore, three periplasmic rings surrounding the long stem, and a cytoplasmic dome. The main differences are that in the TCPM, the long stem exhibits a larger diameter (8 nm vs. 6 nm in the T4aPM), the upper- and mid-periplasmic rings remain tightly associated (rather than disengaging from one another as seen in the M. xanthus T4aPM), and the cytoplasmic ring and disc are lacking. Interestingly, TCP assembly in V. cholerae draws the OM and IM 2 nm closer together, but T4aP formation in M. xanthus pushes them apart by an additional 2 nm. We also compared our V. cholerae TCPM and M. xanthus T4aPM structures with the other available ECT in situ T4aPM structure from T. thermophilus9 (Supplementary Fig. 3). Despite the substantially longer length of the T. thermophilus T4aPM due to its additional secretin N domains, the key structural features of the OM-pore, gate and three periplasmic rings were seen to be conserved among all three systems.
Figure 2. Comparison between V. cholerae TCPM and M. xanthus T4aPM structures, and the inferred TCPM component locations based on the T4aPM component map.
(a) Left column: slices through the composite sub-tomogram averages of V. cholerae wild-type non-piliated and piliated TCPM structures. Right column: slices through sub-tomogram averages of M. xanthus ΔpilB non-piliated and wild-type piliated T4aPM structures8. Scale bars, 10 nm. (b) Inferred TCPM component locations of TcpC, TcpS, TcpB, TcpA and TcpE (left column) based on their identified analogy to PilQ, PilP, minor pilins, PilA and PilC, respectively, in the reported T4aPM component map8 (right column).
Through sequence analyses we confirmed that TcpA, TcpT, TcpE, and TcpC are homologs of PilA, PilB, PilC, and PilQ, respectively. In further consistency with previous reports38,39, TcpB was also identified as a homolog of the enterotoxigenic Escherichia coli (ETEC) T4bP minor pilin CofB. Using more advanced techniques to identify remote relationships (see Methods), two previously unrecognized homologies were also identified. First, TcpS was found to be a homolog of PilP. Interestingly, the conserved region is the domain of PilP that binds the base of the secretin PilQ in the mid-periplasmic ring of the T4aPM (Supplementary Fig. 5), suggesting that TcpS does the same in the TCPM. Second, TcpQ was found to be a homolog of Xanthomonas citri VirB7, part of the OM complex of the type IV secretion system (T4SS). Interestingly, the region of X. citri VirB7 that is homologous to TcpQ is an unusual C-terminal domain not found in other canonical T4SS VirB7s, and has the same fold as secretin N0 domains40. Thus, TcpQ likely associates with the OM pore like other secretin N0 domains. No relationships to T4aPM components were found for TcpB, TcpQ, TcpR and TcpD.
To begin assigning locations of TCPM components, we compared the TCPM OM complex to single particle cryo-EM structures of purified secretin channels. There are multiple structures from T4aPMs and the closely related T2SSs41–43, but only one from a T4bPM, the BfpB channel from the enteropathogenic E. coli bundle-forming pilus (EPEC BFP) system (Supplementary Fig. 4b)44. Since T4bP are in general thought to be thicker than T4aP (~8 vs. ~6 nm), their secretin pores must have different diameters to accommodate the pilus fiber. The EPEC BfpB structure is therefore the most relevant. Overlaying the outline of BfpB onto the V. cholerae TCPM OM complex and aligning their gates (Supplementary Fig. 4) identified the region occupied by the secretin TcpC, but also revealed the presence of two additional densities forming the peripheries of the upper- and mid-periplasmic rings, respectively (Supplementary Fig. 4c, arrows). These additional densities are probably not extensions of the secretin TcpC because (i) BfpB and TcpC are sequence homologs and therefore likely have the same fold (Supplementary Fig. 4d), and (ii) BfpB has a higher molecular weight than TcpC (Supplementary Fig. 4d), but has a smaller outline than the TCPM OM complex. In the EPEC BFP system, the homologs of TcpQ and TcpS were found to bind directly to the secretin45,46, so TcpQ and TcpS are the most likely candidates for these ring densities. A notable difference between the BfpB and TCPM OM complex structures is that the transmembrane region of the purified BfpB channel is shorter, potentially due to disruption by detergent solubilization, as was seen for the purified T4aPM, T2SS and T3SS secretins8.
Using the recognized homologies between the TCPM and T4aPM systems and the known locations of components of the M. xanthus T4aPM (Fig. 2b, right), we assigned the major pilin TcpA to the long stem in the piliated structure and part of the short stem in the non-piliated structure (Fig. 2b, left), the minor pilin TcpB to the short stem of the non-piliated structure, the secretin TcpC to the OM pore and the central core of the upper- and mid-periplasmic rings (Supplementary Fig. 4c), the IM protein TcpE to the cytoplasmic dome, and the periplasmic protein TcpS to the periphery of the mid-periplasmic ring (Supplementary Fig. 5). We did not observe clear densities in the cytoplasm of the wild-type structure to which to assign the ATPase TcpT. Based on images of the ΔtcpR mutant described below, however, we propose that just like their homologs PilB and PilC, TcpT binds the TcpE cytoplasmic dome at the bottom of the TCPM, but it was not observed in the wild-type structure due to low occupancy.
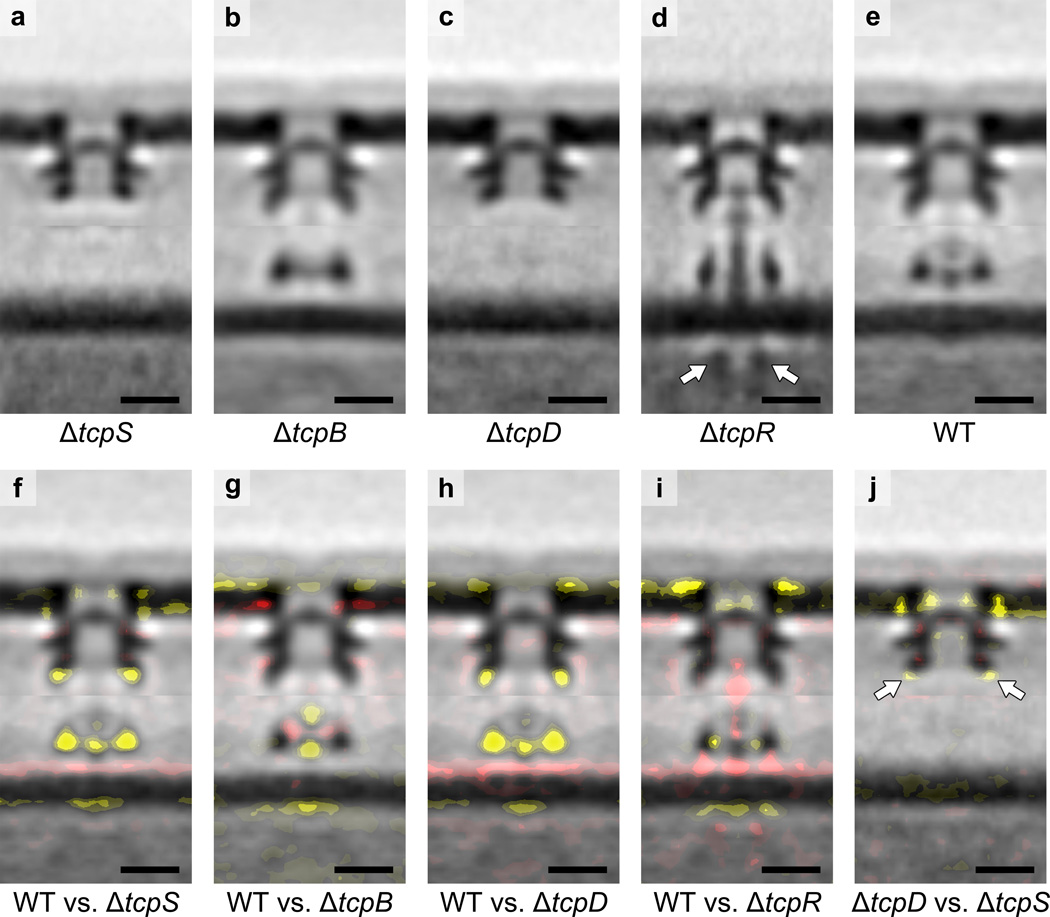
To complete the assignment of TCPM components in the structures, we imaged mutants lacking TcpQ, TcpB, TcpD, TcpR or TcpS. In these mutants, no pili were observed, but non-piliated TCPM basal bodies were detected on the cell surface in every case except ΔtcpQ (in which none were found, as expected, providing clear evidence that all the structures we were analyzing were in fact TCPMs, see Supplementary Discussion). From the resulting sub-tomogram averages and difference maps (Fig. 3) and other knowns about each protein, we propose that TcpS resides in the mid-periplasmic ring, TcpQ occupies the upper-periplasmic ring, TcpB contributes to both the short stem and the density above, and TcpD forms the lower-periplasmic ring and extends to the mid-periplasmic ring (see details in Methods). Moreover, the ΔtcpR structure revealed an interesting “piliation-stalled” conformation in which the short stem extends to the periplasmic vestibule entrance and an assembly ATPase density is visible in the cytoplasm. This confirmed the location of TcpT, and also suggested that TcpR is required for TCP extension through the OM channel. The resulting complete TCPM component maps in both non-piliated and piliated states are shown in Figure 4. Finally, we placed all available atomic models into the maps to show that they satisfy known protein sizes, connectivities and cellular locations (Supplementary Fig. 6b). Higher resolution imaging will be required to determine component orientations and interactions.
Figure 3. Structures of TCPM in ΔtcpS, ΔtcpB, ΔtcpD and ΔtcpR cells.
(a, b, c, d, e) Central slices of sub-tomogram averages of TCPM basal bodies from ΔtcpS, ΔtcpB, ΔtcpD, ΔtcpR and wild-type cells, respectively. Arrows in (d) indicate the cytoplasmic disc-like density. (f, g, h, i) Differences in the ΔtcpS, ΔtcpB, ΔtcpD and ΔtcpR structures compared to the wild-type, respectively (addition and omission of densities are shown by red or yellow color, respectively, with weak to strong intensities corresponding to density differences from one to three standard deviations, overlaid on the wild-type sub-tomogram average). (j) Differences in the ΔtcpS structure compared to the ΔtcpD structure (difference maps overlaid on the ΔtcpD sub-tomogram average). Arrows indicate a larger decrease of the mid-periplasmic ring density caused by ΔtcpS than by ΔtcpD. Scale bars, 10 nm.
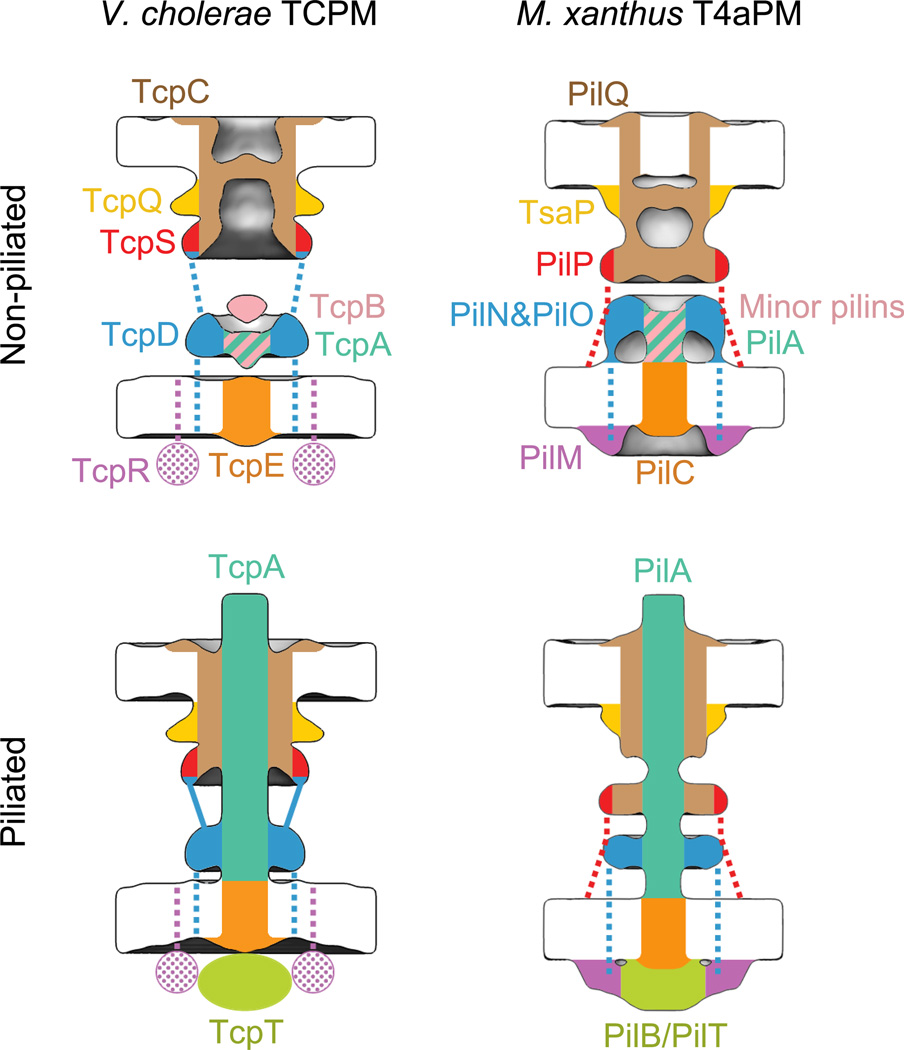
Figure 4. Locations of TCPM components and the comparison with M. xanthus T4aPM.
Left column: schematics showing the component locations assigned for the piliated and non-piliated V. cholerae TCPM (this study). Right column: schematics showing the component locations identified in the piliated and non-piliated M. xanthus T4aPM8. Dashed lines indicate known connections without visible electron density in sub-tomogram averages. Dotted colors indicate proposed protein locations.
While T4aPM exhibits at least two clear states, assembly and retraction, and utilizes two different ATPases to drive those activities, there is no evidence that T4bP retract, and a dedicated pilus disassembly ATPase is not found in the TCP system. In our sub-tomogram averages, the assembly ATPase TcpT was observed in the “piliation-stalled” ΔtcpR structure (Fig. 3d), but not the wild-type TCPM (Fig. 3e). Our interpretation is that when a TCP fiber contacts a surface/bundle, the assembly ATPase is released, and the pilus stops growing. This scenario agrees with the observation that TCP gather cells together to form micro-colonies, rather than push cells apart, as would occur if bundled pili kept growing indefinitely. Because we routinely cultured cells to high density before preparing ECT samples, the vast majority of the pili imaged were likely in contact with surfaces or bundled together, and therefore would have released their assembly ATPase. Accordingly, we observed numerous large bundles of pili in our samples (Fig. 1a).
The structures suggest new ideas about how the TCPM might sense pili bundling and then release the assembly ATPase. In the T4aPM, the PilN/PilO heterodimer forms the lower-periplasmic ring surrounding the pilus. We proposed that the ring might sense conformational changes in the pilus and select the appropriate ATPase accordingly by controlling the conformation of the cytoplasmic PilM ring8. This idea has since gained support from the observation that PilN does indeed modulate PilM’s quaternary structure47. Even though the TCPM TcpD shares no sequence homology with PilN/PilO, here we find that it forms a remarkably similar lower-periplasmic ring structure. Interestingly, the structure is disrupted in the ΔtcpB mutant, in which the short stem is missing, suggesting that like the PilN/PilO ring, the TcpD ring is also sensitive to the state of the pilus passing through it. On the cytoplasmic side of the IM, the TCPM TcpR shares no sequence homology with the T4aPM PilM, but both recruit the cytoplasmic ATPases in their system, and here we point out that TcpR’s association with both the IM and TcpT place it in the same location with respect to the machine as PilM32,47,48. We therefore speculate that conformational changes in the TCP fiber that may occur upon binding a host cell or bundling with other pili could also be sensed by the lower-periplasmic ring and communicated to a cytoplasmic TcpR structure responsible for releasing the assembly ATPase. Since there is no sequence homology among PilN/PilO/PilM and TcpD/TcpR, if true this would be an interesting example of convergent evolution. A testable prediction of this model is that the transmembrane segments of TcpD and TcpR interact within the IM.
This work also rationalizes the previous finding that in the absence of TcpS, less TcpT is found in the cytoplasm32. Here we report that TcpS is a homolog of PilP and likely resides in the mid-periplasmic ring. When TcpS was deleted, not only its own density in the mid-periplasmic ring but also the whole IM-associated part of TCPM were missing, indicating that TcpS is crucial for stabilizing the IM-associated TCPM components, including the cytoplasmic ATPase TcpT. More broadly, our imaging demonstrates that like the T4aPM8,49, the TCPM assembles in an “outside-in” manner: the secretin channel is stabilized by an upper-periplasmic ring (TcpQ), and this complex forms independent of other components. As just mentioned, the mid-periplasmic ring TcpS is necessary for all lower components. The lower-periplasmic TcpD ring is not necessary for TcpS to localize, but it is required for all lower components. The minor pilin TcpB short stem is not required for the localization of TcpD (though it effects its structure), but it is required to recruit the IM platform/cytoplasmic dome TcpE. Finally, the ATPase TcpT can localize, perhaps transiently, in the absence of TcpR, but TcpR stabilizes TcpT and allows the pilus to penetrate the OM channel.
Finally, all the TCPM components with sequence homology to M. xanthus T4aPM components showed higher homology to the M. xanthus T2SS, suggesting a closer evolutionary relationship between the TCPM and the T2SS than the T4aPM. The secretion of the colonization factor TcpF through the TCPM also suggests a functional relationship between the TCPM and T2SS. We found that another TCPM component (TcpQ) is related to the T4SS and two others (TcpD and TcpR) seem to be special innovations of the V. cholerae species, since they show no detectable homology to components of other known secretin-containing machines. One interpretation for these results is that within the superfamily of secretin-containing machines, after the T4aPM branch split from the T2SS, the TCPM evolved from the T2SS, and then acquired new components that converged on similar functions that were either already present or developed independently in T4aPM families.
METHODS
Bacterial strains and growth conditions
V. cholerae O395 N1 strains (wild-type or TCPM component knockout mutants) harboring the pMT5-ToxT plasmid were grown in 5 mL LB media pH 7 with 100 µg/ml ampicillin and 1 mM IPTG. ToxT is the master virulence regulator protein that transcriptionally activates production of the TCPM, therefore facilitating our ECT imaging analysis. Cultures were grown at 30 °C with 250 rpm shaking in glass tubes to avoid adhesion of the TCP to the sides.
Sample preparation
After 24 hours of growth, the tube of V. cholerae culture was placed on a bench at room temperature for ~1 min to sediment visible microcolonies. After the pellet was formed the supernatant was gently removed by pipetting and the pellet was resuspended in fresh LB. The cells were incubated for 15 minutes prior to mixing with 10-nm colloidal gold (Sigma-Aldrich, St. Louis, MO) pretreated with bovine serum albumin and subsequently gently applied to freshly glow-discharged Quantifoil copper R2/2 200 EM grid (Quantifoil Micro Tools GmbH, Jena, Germany). The grids were plunge-frozen in a liquid ethane propane mixture50 using an FEI Vitrobot Mark IV (FEI Company, Hillsboro, OR). Grids were stored in liquid nitrogen prior to imaging.
ECT
The frozen grids were imaged in an FEI Polara 300 keV FEG transmission electron microscope (FEI Company, Hillsboro, OR) equipped with a Gatan energy filter (Gatan, Pleasanton, CA) and a Gatan K2 Summit direct detector (Gatan, Pleasanton, CA). Energy-filtered tilt-series of images of cells were collected automatically from −60° to +60° at 1° intervals using the UCSF Tomography data collection software51 with a total dosage of 160 e−/Å2, a defocus of −6 µm and a pixel size of 3.9 Å. Data collection parameters are summarized in Supplementary Table 1. The images were then binned by 2, aligned and contrast transfer function corrected using the IMOD software package52. SIRT reconstructions were then produced using the TOMO3D program53. TCPM structures on cell envelopes were located by visual inspection. Sub-tomogram averages with 2-fold symmetrization along the particle Y-axis were produced using the PEET program54. Due to the flexibility observed between the OM- and IM-associated parts, binary masks were applied to cover one or the other for local alignment in PEET. The number of cryotomograms and particles used in the process are summarized in Supplementary Table 2.
Identifying corresponding components in the V. cholerae TCPM and M. xanthus T4aPM and related machines
V. cholerae strain O395 N1 protein sequences were selected from the MiST database55. To identify corresponding components between V. cholerae TCPM and M. xanthus T4aPM, we first performed a BLAST search using the package BLAST 2.2.2956 for each component of the V. cholerae TCPM against the M. xanthus proteome. Surprisingly, no TCPM component returned a best BLAST hit (BBH) to a component of the M. xanthus T4aPM. Instead, TcpT and TcpE returned BBHs to the T2SS proteins GspE and GspF, respectively, and second-best BLAST hits to the T4aPM proteins PilB and PilC. To find more remote relationships, we used CDvist57 (a search tool based on HHSearch58) to identify known protein domains within the TCPM components which are also present in T4aPM components. Consistent with the previous report31, TcpC was found to be a homolog of PilQ. A previously unrecognized homology was also identified: TcpS was found to be a homolog of PilP. Interestingly, the homologous region is the domain of PilP that binds to the base of the secretin PilQ in the mid-periplasmic ring of the T4aPM (Supplementary Fig. 5)8,15, suggesting that TcpS does the same in the TCPM. Unlike PilP, however, TcpS was not predicted to be a lipoprotein by the LipoP software59. To find available atomic models of TCPM components, CDvist was used to search against the Protein Data Bank database. The results further revealed that TcpB is a homolog of the ETEC T4bP minor pilin CofB, and TcpQ is a homolog of the X. citri VirB7 N0 domain. Identified component relationships are summarized in Supplementary Table 3.
Mapping components by imaging knockout mutants
-TcpS
To locate TcpS within the TCPM structures, we produced a sub-tomogram average of TCPMs from cells lacking TcpS (Fig. 3a). The entire IM complex and the periphery of the mid-periplasmic ring were missing (Fig. 3f). As described above, because (i) the homolog of TcpS binds directly to the secretin in the EPEC BFP system45, and (ii) TcpS has homology to PilP of T4aPM, which binds the secretin at the periphery of the mid-periplasmic ring and is critical for assembly of the IM complex8, we propose that like PilP, TcpS forms the periphery of the mid-periplasmic ring and is critical for assembly of the IM complex in TCPM.
-TcpQ
In an attempt to locate TcpQ, we imaged cells lacking TcpQ, but could not find any TCPM-like particles, consistent with the previous finding that TcpQ is crucial for TcpC channel formation31. As described in Results, the overlay of the purified EPEC BfpB on the OM complex of the TCPM suggested that the peripheries of the upper- and mid-periplasmic rings were likely formed by TcpQ and TcpS. Having proposed that TcpS is in the periphery of the mid-periplasmic ring, and noting our finding here that TcpQ shows homology to the N0 domain of X. citri T4SS VirB7, which was previously proposed to produce an extra layer on the perimeter of the T4SS OM pore40, we propose that TcpQ forms the periphery of the upper-periplasmic ring.
-TcpB
We inferred that TcpB forms at least part of the short stem in the non-piliated TCPM basal body based on analogy to the minor pilins in T4aPM. Interestingly, in the TCPM there is an additional density above the short stem not seen in the M. xanthus T4aPM structure. To test whether this density was also formed by TcpB, we imaged a tcpB knockout mutant. The sub-tomogram average of ΔtcpB non-piliated basal bodies clearly lacked both the short stem and the additional density above (Fig. 3b, g). Moreover, as was observed in the T4aPM, in the absence of the minor pilin the structure of the lower-periplasmic ring was perturbed and the cytoplasmic dome was missing (Fig. 3g), suggesting the same structural/functional links among these components in the two systems8. TcpB is homologous to the CofB minor pilin in the ETEC T4bP system. CofB was shown to be necessary to initiate pilus assembly and was detected in purified pilus fibers38. Two crystal structures of CofB with the N-terminal transmembrane region truncated have been reported recently38,60. As the truncated soluble CofB forms a homo-trimer in crystals38,60 as well as in solution60, and the hydrophobic core of the helical pilus is stabilized by four pilin subunits11,28, it was proposed that a homo-trimeric full-length CofB initiates piliation by forming a binding pocket that accommodates an incoming major pilin60. The model provided in that study correlated nicely with not only the short stem but also the additional density seen above it in our TCPM structure (Supplementary Fig. 6b, left). Since our previous study of the M. xanthus T4aPM also showed that the short stem of the non-piliated basal body comprises both major and minor pilins, we propose that the short pilus-like stem and the density above it both represent a priming complex that involves TcpB and likely at least one TcpA subunit, and that this priming complex is necessary to stabilize the lower-periplasmic ring and recruit the cytoplasmic dome.
-TcpD
TcpD is predicted to be a bitopic IM protein with nearly all its mass located in the periplasm32. To identify its location in the TCPM structures, we imaged a tcpD knockout mutant and generated a sub-tomogram average of the non-piliated basal bodies (Fig. 3c). The ΔtcpD structure looked like the ΔtcpS structure in that the entire IM complex was missing, but in the case of ΔtcpD a smaller portion of the mid-periplasmic ring was missing (Fig. 3h, j). Since in the IM complex the short stem and the density above it are accounted for by TcpA and TcpB, and TcpD is the only other unassigned protein with a large periplasmic domain, we propose that TcpD occupies the lower-periplasmic ring that is absent in this mutant. Interestingly, the molecular weight of the TcpD periplasmic domain is similar to that of M. xanthus PilN and PilO globular domains, which together form the similarly-sized lower-periplasmic ring in the T4aPM8. Unlike the known connection between PilP in the mid-periplasmic ring and PilN/PilO in the lower-periplasmic ring in the M. xanthus T4aPM, no connections between TcpS and TcpD have yet been proposed. Densities were observed, however, in the sub-tomogram average connecting the mid- and lower-periplasmic rings of the piliated TCPM structure (Fig. 1e, arrows). Furthermore, in the absence of TcpD the very tip of the mid-periplasmic ring was missing. While it is possible that TcpS forms the entire mid-periplasmic ring, and loss of TcpD somehow destabilized the tip, the combination of the connections between the rings and the missing tip lead us to propose that part of TcpD extends from the lower-periplasmic ring to the tip of the mid-periplasmic ring where it interacts with TcpS.
-TcpR
TcpR is predicted to be a bitopic IM protein with nearly all its mass in the cytoplasm (with just a single periplasmic residue at the C-terminus)32. Even though we did not observe any potential cytoplasmic TcpR density in the wild-type TCPM structures, we nevertheless imaged a tcpR knockout mutant for completeness and to evaluate other effects of its absence. Surprisingly, the sub-tomogram average of the ΔtcpR non-piliated basal body showed a much longer stem, which extended all the way to the entrance of the periplasmic vestibule (Fig. 3d, i). A new cytoplasmic disc-like density (Fig. 3d, arrows) also appeared below the cytoplasmic dome. As the TCPM components containing cytoplasmic domains are TcpE, TcpR and TcpT, and since TcpE forms the cytoplasmic dome and TcpR was knocked out, our interpretation is that the cytoplasmic disc is TcpT. The shape and size of the density matches that of a hexameric assembly ATPase, and the location is the same as was found for the TcpT homolog PilB in the T4aPM8. By carefully analyzing the individual TCPM particles used to generate the sub-tomogram average, we found that many of them contain the “extended” short stem into the entrance of the periplasmic vestibule (Supplementary Fig. 7a, arrows), and many others do not (Supplementary Fig. 7b). These two populations resemble the piliated and non-piliated TCPMs in wild-type cells. We therefore propose that in the absence of TcpR, the assembly ATPase TcpT can associate with the cytoplasmic dome transiently and initiate pilus growth, but the growing pilus fails to penetrate the OM channel, resulting in the “piliation-stalled” state captured in Supplementary Figure 7a. This could be because TcpR is required to either stabilize TcpT or open the OM gate. These observations agree nicely with the previous findings that TcpR interacts with TcpT through interactions between their N-terminal domains, and TcpR helps recruit TcpT to the IM, but TcpR cannot tether TcpT on the IM stably without additional components32. We have therefore depicted TcpR touching both the IM (it is an IM protein) and TcpT in Figure 4.
Placing available atomic models into the TCPM component map
In our previous work on the M. xanthus T4aPM structure we examined the plausibility of our component maps by placing available atomic models of the components into the map, satisfying known protein connectivities and cellular locations. While the resolution was not sufficient to unambiguously orient the atomic models within the densities, the exercise nevertheless demonstrated that the components were appropriately sized, and more importantly, dramatically advanced our hypotheses about how the machine worked. Following that work, here we searched for atomic homology models for TCPM components using the CDvist program against the Protein Data Bank database (Supplementary Fig. 6a). Structural models for five out of nine structural components were found: TcpA, TcpB, TcpE, TcpT and TcpQ. (i) V. cholerae TcpA has a crystal structure available with its N-terminal transmembrane helix truncated, and the structure has been fit into a cryo-EM helical reconstruction of purified TCP fibers to generate a pseudo-atomic model of the pilus28. We placed this model directly into the long stem region of the piliated TCPM structure. The diameters matched well (Supplementary Fig. 6b, right). (ii) As described above, TcpB is homologous to the CofB minor pilin in the ETEC T4bP system. The model of a CofB homo-trimer complexed with a major pilin60 was placed in the short stem of the non-piliated TCPM (Supplementary Fig. 6b, left). (iii) TcpE is a highly conserved IM protein among T4aPM, T4bPM and T2SS, and atomic models are available for its two soluble domains17,18,24. In our previous work on the M. xanthus T4aPM structure we generated a hypothetical pseudo-atomic model of a full-length PilC dimer8. Here we adopted the same model and placed it at the bottom of the TCP fiber model to occupy the cytoplasmic dome density (Supplementary Fig. 6b). (iv) TcpT is a homolog of the ATPase PilB of the M. xanthus T4aPM. We therefore placed the same hexameric model8 below the cytoplasmic dome where we observed the cytoplasmic disk-like density in the ΔtcpR piliation-stalled structure and where the ATPase PilB was seen in the M. xanthus T4aPM (Supplementary Fig. 6b, right). (v) TcpQ is a homolog of the X. citri T4SS VirB7 N0 domain. We placed its atomic model40 in the upper-periplasmic ring of both the piliated and non-piliated TCPM structures (Supplementary Fig. 6b) and saw that its size matched the density.
Data availability
The sub-tomogram averages of TCPMs that support the findings of this study have been deposited in the Electron Microscopy Data Bank with accession codes EMD-8492 (wild-type, non-piliated, aligned on the OM-part); EMD-8493 (wild-type, non-piliated, aligned on the IM-part); EMD-8494 (wild-type, piliated, aligned on the OM-part); EMD-8495 (wild-type, piliated, aligned on the IM-part); EMD-8496 (ΔtcpS, non-piliated, aligned on the OM-part); EMD-8497 (ΔtcpS, non-piliated, aligned on the IM-part); EMD-8498 (ΔtcpB, non-piliated, aligned on the OM-part); EMD-8499 (ΔtcpB, non-piliated, aligned on the IM-part); EMD-8500 (ΔtcpD, non-piliated, aligned on the OM-part); EMD-8501 (ΔtcpD, non-piliated, aligned on the IM-part); EMD-8502 (ΔtcpR, non-piliated, aligned on the OM-part); and EMD-8503 (ΔtcpR, non-piliated, aligned on the IM-part).
Supplementary Material
Acknowledgments
We thank Dr. Catherine Oikonomou and Dr. Carrie Shaffer for discussions and editorial assistance. This work was supported by NIH grant R01 GM094800B to G.J.J., the Howard Hughes Medical Institute, and the John Templeton Foundation.
Footnotes
AUTHOR CONTRIBUTIONS
Y.-W.C. and A.K. collected, processed and analyzed the ECT data. D.R.O. performed the bioinformatics analyses. L.A.R. assisted with ECT data processing. G.K., J.A.S. and R.K.T. provided the V. cholerae strains. Y.-W.C., A.K., D.R.O. and G.J.J. wrote the paper.
REFERENCES
- 1.Strom MS, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 2.Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 3.Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 4.Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat Rev Microbiol. 2004;2:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 5.Evans KJ, Lambert C, Sockett RE. Predation by Bdellovibrio bacteriovorus HD100 requires type IV pili. J Bacteriol. 2007;189:4850–4859. doi: 10.1128/JB.01942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reguera G, et al. Extracellular electron transfer via microbial nanowires. Nature. 2005;435:1098–1101. doi: 10.1038/nature03661. [DOI] [PubMed] [Google Scholar]
- 7.Roux N, Spagnolo J, de Bentzmann S. Neglected but amazingly diverse type IVb pili. Res Microbiol. 2012;163:659–673. doi: 10.1016/j.resmic.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 8.Chang YW, et al. Architecture of the type IVa pilus machine. Science. 2016;351:aad2001. doi: 10.1126/science.aad2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gold VA, Salzer R, Averhoff B, Kuhlbrandt W. Structure of a type IV pilus machinery in the open and closed state. Elife. 2015;4 doi: 10.7554/eLife.07380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oikonomou CM, Jensen GJ. A new view into prokaryotic cell biology from electron cryotomography. Nat Rev Microbiol. 2016;14:205–220. doi: 10.1038/nrmicro.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craig L, et al. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Molecular cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Tammam S, et al. Characterization of the PilN, PilO and PilP type IVa pilus subcomplex. Molecular microbiology. 2011;82:1496–1514. doi: 10.1111/j.1365-2958.2011.07903.x. [DOI] [PubMed] [Google Scholar]
- 13.Sampaleanu LM, et al. Periplasmic domains of Pseudomonas aeruginosa PilN and PilO form a stable heterodimeric complex. Journal of molecular biology. 2009;394:143–159. doi: 10.1016/j.jmb.2009.09.037. [DOI] [PubMed] [Google Scholar]
- 14.Karuppiah V, Derrick JP. Structure of the PilM-PilN inner membrane type IV pilus biogenesis complex from Thermus thermophilus. J Biol Chem. 2011;286:24434–24442. doi: 10.1074/jbc.M111.243535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korotkov KV, et al. Structural and functional studies on the interaction of GspC and GspD in the type II secretion system. PLoS Pathog. 2011;7:e1002228. doi: 10.1371/journal.ppat.1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karuppiah V, Collins RF, Thistlethwaite A, Gao Y, Derrick JP. Structure and assembly of an inner membrane platform for initiation of type IV pilus biogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E4638–E4647. doi: 10.1073/pnas.1312313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karuppiah V, Hassan D, Saleem M, Derrick JP. Structure and oligomerization of the PilC type IV pilus biogenesis protein from Thermus thermophilus. Proteins. 2010;78:2049–2057. doi: 10.1002/prot.22720. [DOI] [PubMed] [Google Scholar]
- 18.Abendroth J, et al. The three-dimensional structure of the cytoplasmic domains of EpsF from the type 2 secretion system of Vibrio cholerae. J Struct Biol. 2009;166:303–315. doi: 10.1016/j.jsb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abendroth J, Murphy P, Sandkvist M, Bagdasarian M, Hol WG. The X-ray structure of the type II secretion system complex formed by the N-terminal domain of EpsE and the cytoplasmic domain of EpsL of Vibrio cholerae. Journal of molecular biology. 2005;348:845–855. doi: 10.1016/j.jmb.2005.02.061. [DOI] [PubMed] [Google Scholar]
- 20.Yamagata A, Tainer JA. Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. EMBO J. 2007;26:878–890. doi: 10.1038/sj.emboj.7601544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Misic AM, Satyshur KA, Forest K. T P. aeruginosa PilT structures with and without nucleotide reveal a dynamic type IV pilus retraction motor. Journal of molecular biology. 2010;400:1011–1021. doi: 10.1016/j.jmb.2010.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satyshur KA, et al. Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility. Structure. 2007;15:363–376. doi: 10.1016/j.str.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu C, et al. Hexamers of the type II secretion ATPase GspE from Vibrio cholerae with increased ATPase activity. Structure. 2013;21:1707–1717. doi: 10.1016/j.str.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolappan S, Craig L. Structure of the cytoplasmic domain of TcpE, the inner membrane core protein required for assembly of the Vibrio cholerae toxin-coregulated pilus. Acta crystallographica. Section D, Biological crystallography. 2013;69:513–519. doi: 10.1107/S0907444912050330. [DOI] [PubMed] [Google Scholar]
- 25.Krebs SJ, Taylor RK. Protection and attachment of Vibrio cholerae mediated by the toxin-coregulated pilus in the infant mouse model. J Bacteriol. 2011;193:5260–5270. doi: 10.1128/JB.00378-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. Identification of a pilus colonization factor that is coordinately regulated with cholera toxin. Ann Sclavo Collana Monogr. 1986;3:51–61. [PubMed] [Google Scholar]
- 27.Manning PA. The tcp gene cluster of Vibrio cholerae. Gene. 1997;192:63–70. doi: 10.1016/s0378-1119(97)00036-x. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Egelman EH, Craig L. Structure of the Vibrio cholerae Type IVb Pilus and stability comparison with the Neisseria gonorrhoeae type IVa pilus. Journal of molecular biology. 2012;418:47–64. doi: 10.1016/j.jmb.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y, Hauke CA, Marles JM, Taylor RK. Effects of tcpB mutations on biogenesis and function of the toxin-coregulated pilus, the type IVb pilus of Vibrio cholerae. J Bacteriol. 2016;198:2818–2828. doi: 10.1128/JB.00309-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bose N, Taylor RK. Identification of a TcpC-TcpQ outer membrane complex involved in the biogenesis of the toxin-coregulated pilus of Vibrio cholerae. J Bacteriol. 2005;187:2225–2232. doi: 10.1128/JB.187.7.2225-2232.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tripathi SA, Taylor RK. Membrane association and multimerization of TcpT, the cognate ATPase ortholog of the Vibrio cholerae toxin-coregulated-pilus biogenesis apparatus. J Bacteriol. 2007;189:4401–4409. doi: 10.1128/JB.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strom MS, Nunn DN, Lory S. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:2404–2408. doi: 10.1073/pnas.90.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iredell JR, Manning PA. Translocation failure in a type-4 pilin operon: rfb and tcpT mutants in Vibrio cholerae. Gene. 1997;192:71–77. doi: 10.1016/s0378-1119(97)00040-1. [DOI] [PubMed] [Google Scholar]
- 35.Megli CJ, Taylor RK. Secretion of TcpF by the Vibrio cholerae toxin-coregulated pilus biogenesis apparatus requires an N-terminal determinant. J Bacteriol. 2013;195:2718–2727. doi: 10.1128/JB.01122-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirn TJ, Bose N, Taylor RK. Secretion of a soluble colonization factor by the TCP type 4 pilus biogenesis pathway in Vibrio cholerae. Molecular microbiology. 2003;49:81–92. doi: 10.1046/j.1365-2958.2003.03546.x. [DOI] [PubMed] [Google Scholar]
- 37.Hall RH, Vial PA, Kaper JB, Mekalanos JJ, Levine MM. Morphological studies on fimbriae expressed by Vibrio cholerae 01. Microbial pathogenesis. 1988;4:257–265. doi: 10.1016/0882-4010(88)90086-1. [DOI] [PubMed] [Google Scholar]
- 38.Kolappan S, Ng D, Yang G, Harn T, Craig L. Crystal Structure of the Minor Pilin CofB, the Initiator of CFA/III Pilus Assembly in Enterotoxigenic Escherichia coli. J Biol Chem. 2015;290:25805–25818. doi: 10.1074/jbc.M115.676106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taniguchi T, et al. Gene cluster for assembly of pilus colonization factor antigen III of enterotoxigenic Escherichia coli. Infection and immunity. 2001;69:5864–5873. doi: 10.1128/IAI.69.9.5864-5873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Souza DP, et al. A component of the Xanthomonadaceae type IV secretion system combines a VirB7 motif with a N0 domain found in outer membrane transport proteins. PLoS Pathog. 2011;7:e1002031. doi: 10.1371/journal.ppat.1002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berry JL, et al. Structure and assembly of a trans-periplasmic channel for type IV pili in Neisseria meningitidis. PLoS Pathog. 2012;8:e1002923. doi: 10.1371/journal.ppat.1002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reichow SL, Korotkov KV, Hol WG, Gonen T. Structure of the cholera toxin secretion channel in its closed state. Nature structural & molecular biology. 2010;17:1226–1232. doi: 10.1038/nsmb.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tosi T, et al. Structural similarity of secretins from type II and type III secretion systems. Structure. 2014;22:1348–1355. doi: 10.1016/j.str.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 44.Lieberman JA, et al. Outer membrane targeting, ultrastructure, and single molecule localization of the enteropathogenic Escherichia coli type IV pilus secretin BfpB. J Bacteriol. 2012;194:1646–1658. doi: 10.1128/JB.06330-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daniel A, et al. Interaction and localization studies of enteropathogenic Escherichia coli type IV bundle-forming pilus outer membrane components. Microbiology. 2006;152:2405–2420. doi: 10.1099/mic.0.28860-0. [DOI] [PubMed] [Google Scholar]
- 46.Gomez-Duarte OG, et al. Genetic diversity of the gene cluster encoding longus, a type IV pilus of enterotoxigenic Escherichia coli. J Bacteriol. 2007;189:9145–9149. doi: 10.1128/JB.00722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCallum M, et al. PilN binding modulates the structure and binding partners of the Pseudomonas aeruginosa Type IVa Pilus protein PilM. J Biol Chem. 2016 doi: 10.1074/jbc.M116.718353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bischof LF, Friedrich C, Harms A, Sogaard-Andersen L, van der Does C. The type IV pilus assembly ATPase PilB of Myxococcus xanthus interacts with the inner membrane platform protein PilC and the nucleotide-binding protein PilM. J Biol Chem. 2016;291:6946–6957. doi: 10.1074/jbc.M115.701284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Friedrich C, Bulyha I, Sogaard-Andersen L. Outside-in assembly pathway of the type IV pilus system in Myxococcus xanthus. J Bacteriol. 2014;196:378–390. doi: 10.1128/JB.01094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tivol WF, Briegel A, Jensen GJ. An improved cryogen for plunge freezing. Microscopy and microanalysis : the official journal of Microscopy Society of America, Microbeam Analysis Society, Microscopical Society of Canada. 2008;14:375–379. doi: 10.1017/S1431927608080781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng SQ, et al. UCSF tomography: an integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. J Struct Biol. 2007;157:138–147. doi: 10.1016/j.jsb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 52.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 53.Agulleiro JI, Fernandez JJ. Fast tomographic reconstruction on multicore computers. Bioinformatics. 2011;27:582–583. doi: 10.1093/bioinformatics/btq692. [DOI] [PubMed] [Google Scholar]
- 54.Nicastro D, et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 55.Ulrich LE, Zhulin IB. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res. 2010;38:D401–D407. doi: 10.1093/nar/gkp940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adebali O, Ortega DR, Zhulin IB. CDvist: a webserver for identification and visualization of conserved domains in protein sequences. Bioinformatics. 2015;31:1475–1477. doi: 10.1093/bioinformatics/btu836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 59.Juncker AS, et al. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kawahara K, et al. Homo-trimeric structure of the type IVb minor pilin CofB suggests mechanism of CFA/III pilus assembly in human enterotoxigenic Escherichia coli. Journal of molecular biology. 2016;428:1209–1226. doi: 10.1016/j.jmb.2016.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sub-tomogram averages of TCPMs that support the findings of this study have been deposited in the Electron Microscopy Data Bank with accession codes EMD-8492 (wild-type, non-piliated, aligned on the OM-part); EMD-8493 (wild-type, non-piliated, aligned on the IM-part); EMD-8494 (wild-type, piliated, aligned on the OM-part); EMD-8495 (wild-type, piliated, aligned on the IM-part); EMD-8496 (ΔtcpS, non-piliated, aligned on the OM-part); EMD-8497 (ΔtcpS, non-piliated, aligned on the IM-part); EMD-8498 (ΔtcpB, non-piliated, aligned on the OM-part); EMD-8499 (ΔtcpB, non-piliated, aligned on the IM-part); EMD-8500 (ΔtcpD, non-piliated, aligned on the OM-part); EMD-8501 (ΔtcpD, non-piliated, aligned on the IM-part); EMD-8502 (ΔtcpR, non-piliated, aligned on the OM-part); and EMD-8503 (ΔtcpR, non-piliated, aligned on the IM-part).