Abstract
The loop-mediated isothermal amplification (LAMP) assay with its advantages of cost effectiveness, rapidity, and simplicity, has evolved as a sensitive and specific method for the detection of African trypanosomes. Highly sensitive LAMP reactions specific for Trypanosoma brucei rhodesiense or that recognize but do not discriminate between Trypanosoma brucei brucei, T. b. rhodesiense, Trypanosoma brucei gambiense, and Trypanosoma evansi have been developed. A sensitive LAMP assay targeting the T. b. gambiense 5.8S ribosomal RNA internal transcribed spacer 2 (5.8S-ITS2) gene is also available but this assay does not target binding sites that span the CCCA (C3A) (557–560 bps) insertion site that further differentiates T. b. gambiense from T. b. brucei. Here we describe 5.8S-ITS2-targeted LAMP assay that fit these criteria. The LAMP primer sets containing the T. b. gambiense-specific C3A tetranucleotide at the start of the outer forward primer sequences showed high specificity and sensitivity down to at least 0.1 fg T. b. gambiense genomic DNA.
Trypanosoma brucei rhodesiense and Trypanosoma brucei gambiense are tsetse fly-transmitted protozoan parasites and human pathogens that cause human African trypanosomiasis (HAT; sleeping sickness). Causing substantial morbidity throughout sub-Saharan Africa, death by HAT is likely if a patient is untreated.1–3 The trypanosomes replicate at the tsetse fly bite site before disseminating from the skin through the hemolymphatic system to the spleen, liver, lymph nodes, skin, eyes, endocrine system, and the heart (stage 1).
The development of clinical neurological signs and symptoms that characterize stage 2 HAT can take weeks to months with T. b. rhodesiense (east African) and months to years with T. b. gambiense (west African) infections. Once central nervous system disease is established, the parasites are shielded from most trypanocidal drugs, the majority of which do not penetrate the blood–brain barrier (BBB). For treatment of stage 2 neurologic HAT, toxic BBB-permeable trypanocidal drugs are used.1–3 Melarsoprol, effective for stage 2 T. b. gambiense and T. b. rhodesiense HAT, can cause a lethal severe posttreatment reactive encephalopathy brain inflammation. Less toxic eflornithine is active only against gambiense HAT and resistance to both drugs is increasing.
The early identification and treatment of the largely asymptomatic human reservoir is absolutely critical if any program to control gambiense HAT is to be successful. Although parasite detection in the blood by wet blood film examination is frequently successful in T. b. rhodesiense infections because of the high levels of parasitemia, this method is insensitive in T. b. gambiense infections that constitute 90–95% of all HAT cases.1–3 Because of the often-silent neurological involvement, stage determination still relies on lumbar puncture to examine cerebrospinal fluid (CSF) for trypanosomes and/or changes suggestive of chronic meningoencephalitis. Single or double centrifugation of CSF is required to concentrate parasites for microscopic detection. In the absence of visible parasites, the conventional criterion for stage 2 classification requires CSF lymphocyte cell counts that are scored according to arbitrary cutoffs ranging from > 5, > 10, or even 20 cells/μL.4,5
Loop-mediated isothermal amplification (LAMP) is a proven cost-effective, simple, and rapid DNA amplification method that uses four or six primers for the detection of DNA with high sensitivity and specificity.6,7 The basic four LAMP primers consist of the outer forward (F3), outer backward (B3), forward inner (FIP), and backward inner (BIP) primers, and when present, the loop forward (LF) and loop backward (LB) primers. The major advantages of LAMP include 1) the reaction proceeds under isothermal conditions and detection can be conducted within 60 minutes, 2) it requires simple heating devices such as a water bath, laboratory heat block, or portable tube-LAMP scanners8, and 3) target amplification is indicated by i) turbidimetric measurements,9 or ii) by the addition of fluorescent dyes,10–12 or iii) after addition of hydroxynaphthol blue to yield a reaction product color change from purple (no DNA) to blue (DNA positive).12
Based on protocols for polymerase chain reaction (PCR) diagnosis of HAT, and recognizing the potential of the use for the diagnosis of HAT and other infectious diseases, we described LAMP for the detection of African trypanosomes.13 Highly sensitive, T. brucei subspecies LAMP reactions that recognize Trypanosoma brucei brucei, T. b. rhodesiense, T. b. gambiense, and Trypanosoma evansi,13,14 or are specific for T. b. rhodesiense,15 have been developed. Although specific for T. b. gambiense, the detection limit of LAMP targeting the T. b. gambiense-specific glycoprotein (TgsGP) is ∼100 fg (equivalent to one parasite).16 Trypanosoma brucei gambiense LAMP (TBG1) targeting the high copy number T. b. gambiense 5.8S ribosomal RNA internal transcribed spacer 2 (5.8S-ITS2) gene (GenBank accession no. AF306777) has high analytical sensitivity to as little as 1 fg DNA (equivalent to 0.01 parasite)17; however, none of the TBG1 primer binding sites span the CCCA (C3A) (557–560 bps) insertion site described by Agbo and others.18 Sequence analysis revealed two regions in the 5.8S-ITS2 gene containing C3A sequences (Figure 1A ). The first C3A fragment (461–464 bp) on the 5.8S-ITS2 gene is conserved among T. brucei species infective to animals and humans. However, the second C3A fragment (557–560) is conserved in T. b. gambiense.18 Using PrimerExplorer software version 4 software (http://primerexplorer.jp/e/), we identified a 5.8S-ITS2-targeted LAMP primer set that targets the parasite-specific C3A tetranucleotide at the 5′ end of primer F3 in TBG4 (Figure 1B). Comparisons between the TBG4 LAMP primers to the published TBG1 LAMP sequences that target the same 5.8S-ITS2 gene show that only TBG4 targeted the specific second T. b. gambiense C3A fragment (Figure 1C).
Figure 1.
LAMP primer sequences, sequence alignment comparisons, and LAMP primer sequence overlaps. (A) The Trypanosoma brucei gambiense (Tbg) and Trypanosoma brucei brucei (Tbb) isolates with each ribosomal RNA-internal transcribed spacer 2 (5.8S-ITS2) sequence used for their alignment include Tbg DAL 972 (GenBank accession no. AF306774), Tbg Suzena (AF406775), Tbg NW2 (AF306776), Tbg TH2 (AF306777), Tbb KP2 (AF306773), Tbb B8/18 (AF30677), Tbb STIB215 (AF306771), and Tbb H3 (AF306770). Using the online CLUSTAL-W program (www.genome.jp/tools/clustalw/), the first C3A fragment (461–464) on the 5.8S-ITS2 gene is conserved among T. brucei species isolated from humans and animals. The second fragment of C3A (557–560) is only conserved among Trypanosoma brucei gambiense subspecies. (B) The basic TBG4 LAMP primers consisting F3, B3, FIP, and BIP primers and the LF and LB loop primers are shown. (C) The 186-bp target area (gray shading) on the 5.8S-ITS2 gene where TBG4 (shown in blue) and TBG1 primers overlap. The C3A sequences (shown in red) and the dashed and solid lines show the TBG4 LAMP and TBG1 LAMP primer sets, respectively. The primers from each set greatly overlap except for TBG4-F3. B3 = outer backward primer; BIP = backward inner primer; FIP = forward inner primer; F3 = outer forward primer; LAMP = loop-mediated isothermal amplification; LB = loop backward; LF = loop forward.
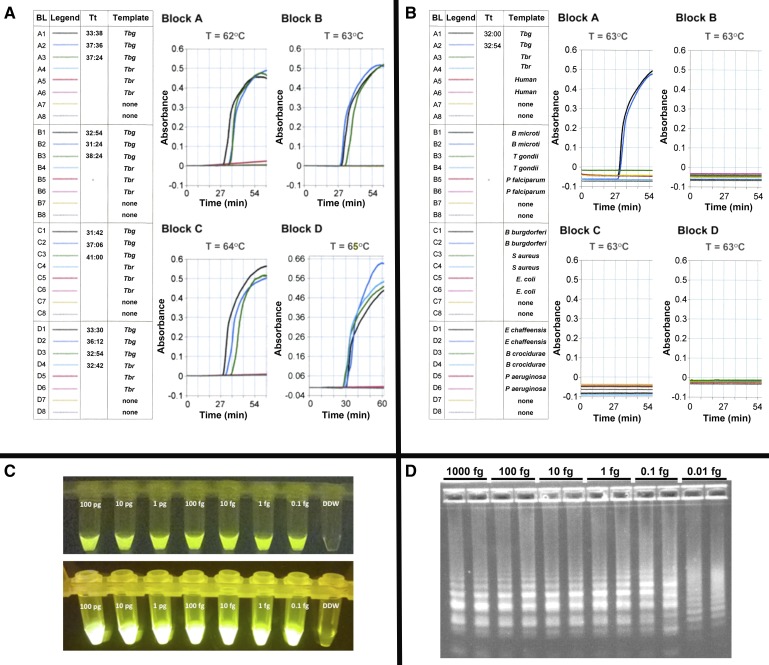
LAMP reactions using a commercially available kit (Eiken Chemical Co., Tochigi, Japan) were optimized for reagent concentration, reaction time, and temperature in real time in a Loopamp real-time turbidimeter LA320C (Teramecs, Tokyo, Japan) as previously described.19 Briefly, the reaction contained 12.5 μL of 2 × LAMP reaction buffer (40 mM Tris-HCl [pH 8.8], 20 mM KCl, 16 mM MgSO4, 20 mM [NH4]2SO4, 0.2% Tween 20, 1.6 M Betaine, 2.8 mM of each deoxyribonucleotide triphosphates [dNTPs]), 1.0 μL TBG1,17 or TBG4 (Figure 1) primer mix (5 pmol each of F3 and B3, 40 pmol each of FIP and BIP, and for TBG4 20 pmol each of LF and LB), 1 μL (8 units) Bst DNA polymerase (New England Biolabs, Tokyo, Japan), and template DNA. Final volumes were adjusted to 25 μL with distilled water and the reaction temperature was kept at 63°C, optimal for TBG1 LAMP and TBG4 LAMP (Figure 2 ). We considered precipitation occurring after a reaction time of 60 minutes to be nonspecific artifacts. It is important to note that TBG4 LAMP specificity appears to diminish with assay temperatures ≥65°C monitored in real time by turbidity and after addition of hydroxynaphthol blue (not shown). Furthermore, the reactions were assayed in four blocks with each block containing eight samples with two no template controls for every six samples assayed. A positive response from any no template control negated the entire 32-reaction run and the samples were reassayed.
Figure 2.
Optimal reaction temperature, analytical specificity, and analytical sensitivity of TBG4 LAMP. (A) Temperature optimization. Real time TBG4 LAMP was conducted at four different temperatures (62–65°C) and 0.01 pg Trypanosoma brucei gambiense DAL 972 (Tbg) and Trypanosoma brucei rhodesiense LouTat 1A (Tbr) genomic DNA as template. LAMP in the absence of DNA templates served as controls. The data are plotted as reaction time vs. absorbance. The measurement result Tt, represents the time any sample shown in the measurement block (BL) equals or exceeds the threshold (i.e., 0.1 absorbance unit). Our data show that 0.01 pg Tbg DNA can be amplified within a time range 31–41 minutes when reactions are conducted at 62–65°C. However, a faster reaction time (31′24″) was observed at 63°C, hence its selection for further experiments. The slight time differences for the LAMP positive wells are not significant. Note that specificity for Tbg DNA diminished with assay temperatures ≥ 65°C. (B) Analytical Specificity. Real time LAMP was conducted at 63°C (60 minutes) using 0.01 pg genomic DNA from T. b. gambiense DAL 972 (Tbg), T. b. rhodesiense LouTat 1A (Tbr), human, Babesia microti, Toxoplasma gondii, Plasmodium falciparum, Borrelia burgdorferi, Staphylococcus aureus, Ehrlichia Chaffeensis, and Pseudomonas aeruginosa as template. LAMP in the absence of DNA templates served as controls. Analytical sensitivity. TBG4 LAMP was run at 63°C (60 minutes) using 1:10 serial dilutions of T. b. gambiense DAL 972 and 348BT DNA. (C) Shows the detection of LAMP amplified T. b. gambiense 348BT DNA in reaction tubes containing calcein (Eiken Chemical Co., Tochigi, Japan) as seen on (upper panel) a black background with normal lighting and (lower panel) under a BLOOK LED transilluminator (Gene Direx, Taqkey Science Co., Ltd., Miaoli County, Taiwan) are shown. In both cases, TBG4 LAMP was able to detect 0.1 fg of parasite DNA. (D) The detection of LAMP-amplified T. b. gambiense DAL 972 DNA by agarose gel electrophoresis. Generally, there is no specific band pattern observed in LAMP gel electrophoresis, however, in this study we observed that the density of amplified fragments is greater (∼6- to 7-fold) at higher concentrations (1,000–1 fg) and are reduced (∼4–5) at lower concentrations (0.1–0.01 fg). Although 0.01 fg DNA could be detected, the altered electrophoretic banding pattern using 0.01 fg DNA prompts us to conservatively set the sensitivity limit to be at least 0.1 fg DNA. LAMP = loop-mediated isothermal amplification.
Our initial analytical specificity studies showed that TBG4 LAMP recognized genomic DNA from T. b. gambiense (DAL 069, DAL 972, IL 3258, 348BT) but not genomic DNA from T. b. rhodesiense (Type 1 T. b. rhodesiense strains LouTat 1A, GYBO, IL1501) or T. b. brucei (427, 927). TBG4 LAMP also did not recognize DNA from the eukaryotic protozoan parasites (Babesia microti, Plasmodium falciparum, and Toxoplasma gondii) and prokaryotic bacteria (Borrelia burgdorferi, Borrelia crocidurae, Ehrlichia chaffeensis, Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus), DNA from normal human blood and blood spiked with DNA of pathogens whose clinical presentation might mimic HAT (E. coli) or DNA from clinical blood samples of bacteremic patients from The Johns Hopkins Hospital Microbiology laboratory (with approval of the Johns Hopkins Medicine Institutional Review Board) (Figure 2B). TBG1 LAMP has high analytical sensitivity and is capable of detecting 1 fg DNA (equivalent to 0.01 parasite).17 We tested TBG1 and TBG4 LAMP using the same assay conditions. We found the detection limit of TBG1 LAMP for T. b. gambiense genomic DNA was ≥1 fg (not shown). TBG4 LAMP was 10-fold more sensitive than TBG1 LAMP; that is, the detection limit for T. b. gambiense genomic DNA was at least 0.1 fg of T. b. gambiense DNA, equivalent to 0.001 parasite (Figure 2C and D).
Early and accurate HAT diagnosis presents the highest likelihood that effective anti-trypanosome treatment can be administered before the onset of neurological signs. While based on TBG1 LAMP, TBG4 LAMP was designed to target the T. b. gambiense-specific C3A tetranucleotide sequence present in the 5.8S-ITS2 gene. With sequence information for the 5.8S-ITS2 gene region in T. b. rhodesiense being unavailable, it is remotely possible that some rhodesiense isolates may contain the T. b. gambiense-specific C3A tetranucleotide. Just how well TBG4 LAMP compares to other LAMP17,20 or PCR-based assays under actual clinical and field situations with regard to sensitivity and specific identification of T. b. gambiense remains to be determined. Based on previous work,19 we predict that the detection of intact free-swimming T. b. gambiense in blood or CSF by TBG4 LAMP will be dramatically enhanced after detergent lysis to facilitate release of their DNA prior to assay. This is especially important when sample size is limited. Circulating cell-free DNA (cfDNA) released into the bloodstream as a result of cell death, necrosis, or by release by viable cells has been found in many conditions including a variety of inflammatory and autoimmune diseases, cancer,20 and during trypanosome infections.16 The detection of low levels of T. b. gambiense-derived cfDNA by TBG4 released from damaged and dying African trypanosomes into the circulation may help provide for very early HAT detection.
In conclusion, further development and controlled laboratory and field validation of LAMP-based tests will facilitate HAT diagnosis and monitoring during the early stages of disease prior to serological reactivity in tests such as card agglutination test for trypanosomiasis, as well as in the early detection of neurological HAT. We believe the studies outlined here define the next steps in achieving prompt, accurate diagnosis of HAT. We hope that improved diagnostic testing could ultimately inform appropriate treatment of HAT and thereby reduce mortality from both disease and treatment.
ACKNOWLEDGMENTS
We thank John Mansfield (University of Wisconsin at Madison, WI) for the gift of Trypanosoma brucei rhodesiense LouTat 1A.
Footnotes
Financial support: This research was supported in part by grants from the National Institutes of Health (5R01AI082695 and 1R21AI079282) to Dennis J. Grab.
Authors' addresses: Olga V. Nikolskaia and Dennis J. Grab, Department of Pathology, The Johns Hopkins University School of Medicine, Baltimore, MD, E-mails: onikols1@jhmi.edu and dgrab1@jhmi.edu. Oriel M. M. Thekisoe, Unit for Environmental Sciences and Management, North-West University, Potchefstroom, South Africa, E-mail: oriel.Thekisoe@nwu.ac.za. J. Stephen Dumler, Department of Pathology, University of Maryland School of Medicine, Baltimore, MD, E-mail: sdumler@som.umaryland.edu.
References
- 1.Blum JA, Neumayr AL, Hatz CF. Human African trypanosomiasis in endemic populations and travellers. Eur J Clin Microbiol Infect Dis. 2012;31:905–913. doi: 10.1007/s10096-011-1403-y. [DOI] [PubMed] [Google Scholar]
- 2.Kennedy PG. Human African trypanosomiasis of the CNS: current issues and challenges. J Clin Invest. 2004;113:496–504. doi: 10.1172/JCI21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molyneux DH, Pentreath V, Doua F. African Trypanosomiasis in Man. London, United Kingdom: WB Saunders; 1996. [Google Scholar]
- 4.World Health Organization . Control and Surveillance of African Trypanosomiasis. Geneva, Switzerland: World Health Organization; 1998. p. 114. Report of a WHO Expert Committee. Technical Report Series No. 881. [Google Scholar]
- 5.Bisser S, Lejon V, Preux PM, Bouteille B, Stanghellini A, Jauberteau MO, Buscher P, Dumas M. Blood-cerebrospinal fluid barrier and intrathecal immunoglobulins compared to field diagnosis of central nervous system involvement in sleeping sickness. J Neurol Sci. 2002;193:127–135. doi: 10.1016/s0022-510x(01)00655-4. [DOI] [PubMed] [Google Scholar]
- 6.Mori Y, Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J Infect Chemother. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 8.Surabattula R, Vejandla MP, Mallepaddi PC, Faulstich K, Polavarapu R. Simple, rapid, inexpensive platform for the diagnosis of malaria by loop mediated isothermal amplification (LAMP) Exp Parasitol. 2013;134:333–340. doi: 10.1016/j.exppara.2013.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori Y, Nagamine K, Tomita N, Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 10.Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 11.Qiao YM, Guo YC, Zhang XE, Zhou YF, Zhang ZP, Wei HP, Yang RF, Wang DB. Loop-mediated isothermal amplification for rapid detection of Bacillus anthracis spores. Biotechnol Lett. 2007;29:1939–1946. doi: 10.1007/s10529-007-9472-9. [DOI] [PubMed] [Google Scholar]
- 12.Wastling SL, Picozzi K, Kakembo AS, Welburn SC. LAMP for human African trypanosomiasis: a comparative study of detection formats. PLoS Negl Trop Dis. 2010;4:e865. doi: 10.1371/journal.pntd.0000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuboki N, Inoue N, Sakurai T, Di Cello F, Grab DJ, Suzuki H, Sugimoto C, Igarashi I. Loop-mediated isothermal amplification for detection of African trypanosomes. J Clin Microbiol. 2003;41:5517–5524. doi: 10.1128/JCM.41.12.5517-5524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Njiru ZK, Mikosza AS, Matovu E, Enyaru JC, Ouma JO, Kibona SN, Thompson RC, Ndung'u JM. African trypanosomiasis: sensitive and rapid detection of the sub-genus Trypanozoon by loop-mediated isothermal amplification (LAMP) of parasite DNA. Int J Parasitol. 2008;38:589–599. doi: 10.1016/j.ijpara.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Njiru ZK, Mikosza AS, Armstrong T, Enyaru JC, Ndung'u JM, Thompson AR. Loop-mediated isothermal amplification (LAMP) method for rapid detection of Trypanosoma brucei rhodesiense. PLoS Negl Trop Dis. 2008;2:e147. doi: 10.1371/journal.pntd.0000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mansfield JM, Paulnock DM. Regulation of innate and acquired immunity in African trypanosomiasis. Parasite Immunol. 2005;27:361–371. doi: 10.1111/j.1365-3024.2005.00791.x. [DOI] [PubMed] [Google Scholar]
- 17.Thekisoe OM, Kuboki N, Nambota A, Fujisaki K, Sugimoto C, Igarashi I, Yasuda J, Inoue N. Species-specific loop-mediated isothermal amplification (LAMP) for diagnosis of trypanosomosis. Acta Trop. 2007;102:182–189. doi: 10.1016/j.actatropica.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Agbo EC, Majiwa PA, Claassen EJ, Roos MH. Measure of molecular diversity within the Trypanosoma brucei subspecies Trypanosoma brucei brucei and Trypanosoma brucei gambiense as revealed by genotypic characterization. Exp Parasitol. 2001;99:123–131. doi: 10.1006/expr.2001.4666. [DOI] [PubMed] [Google Scholar]
- 19.Grab DJ, Nikolskaia OV, Inoue N, Thekisoe MM, Morrison L, Gibson W, Dumler JS. Using detergent to enhance detection sensitivity of African trypanosomes in human CSF and blood by loop-mediated isothermal amplification (LAMP) PLoS Negl Trop Dis. 2011;5:e1249. doi: 10.1371/journal.pntd.0001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aarthy R, Mani S, Velusami S, Sundarsingh S, Rajkumar T. Role of circulating cell-free DNA in cancers. Mol Diagn Ther. 2015;6:339–350. doi: 10.1007/s40291-015-0167-y. [DOI] [PubMed] [Google Scholar]