Abstract
Background
Lung squamous cell carcinoma (lung SCC) is a common type of malignancy. Its pathogenesis mechanism of tumor development is unclear. The aim of this study was to identify key genes for diagnosis biomarkers in lung SCC metastasis.
Methods
We searched and downloaded mRNA expression data and clinical data from The Cancer Genome Atlas (TCGA) database to identify differences in mRNA expression of primary tumor tissues from lung SCC with and without metastasis. Gene co-expression network analysis, protein-protein interaction (PPI) network, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and quantitative real-time polymerase chain reactions (qRT-PCR) were used to explore the biological functions of the identified dysregulated genes.
Results
Four hundred and eighty-two differentially expressed genes (DEGs) were identified between lung SCC with and without metastasis. Nineteen modules were identified in lung SCC through weighted gene co-expression network analysis (WGCNA). Twenty-three DEGs and 26 DEGs were significantly enriched in the respective pink and black module. KEGG pathway analysis displayed that 26 DEGs in the black module were significantly enriched in bile secretion pathway. Forty-nine DEGs in the two gene co-expression module were used to construct PPI network. CFTR in the black module was the hub protein, had the connectivity with 182 genes. The results of qRT-PCR displayed that FIGF, SFTPD, DYNLRB2 were significantly down-regulated in the tumor samples of lung SCC with metastasis and CFTR, SCGB3A2, SSTR1, SCTR, ROPN1L had the down-regulation tendency in lung SCC with metastasis compared to lung SCC without metastasis.
Conclusions
The dysregulated genes including CFTR, SCTR and FIGF might be involved in the pathology of lung SCC metastasis and could be used as potential diagnosis biomarkers or therapeutic targets for lung SCC.
Keywords: Lung squamous cell carcinoma (lung SCC), genes expression profiling, gene regulatory network, neoplasm metastasis, biomarkers
Introduction
Lung squamous cell carcinoma (lung SCC) is a histological subtype of non-small cell lung cancer (NSCLC), which is the second most frequent type of NSCLC after lung adenocarcinoma (1). Lung SCC is a multi-step progressive disease with high rates of morbidity and mortality worldwide.
The initiation of lung SCC is divided into the following five successive stages: normal bronchial epithelium, squamous metaplasia, mild-moderate dysplasia, carcinoma in situ, and invasive carcinoma (2). The prognosis of lung cancer is unfavorable, despite significant therapeutic improvements have been made in recent years. In current, there is no specific molecular targets for therapy have been identified (3), therefore, cisplatin plus gemcitabine is still the first-line treatment for lung SCC (4).
Currently, the tumorigenesis mechanism of lung SCC remains unclear. Numerous published articles demonstrate that dysregulated genes are essential for initiation, progression and development of lung cancer. SMC4 (structural maintenance of chromosome 4) is over-expressed in lung adenocarcinoma tissues and its inhibition significantly suppresses the proliferation and invasion of A549 cells (5). Knocking down the expression of WW45 (salvador family WW domain containing protein 1) promotes cell growth and migration of lung cancer and over-expression of WW5 improves the survival of mice model of lung cancer (6). INO80 (INO80 complex subunit), the SWI/SNF ATPase in the complex, is highly expressed in NSCLC cells compared with normal lung epithelia cells and its expression level is negatively correlated with the disease prognosis of patients with lung cancer (7).
Weighted gene co-expression network analysis (WGCNA) offers an effective approach to quantitatively assess the interconnectedness of genes, investigate the expression patterns of gene co-activity and evaluate the importance of genes within the network. It benefit to provide potential malignancy diagnostic molecular and connecting them together for disease (8,9). Shi et al. identifies four co-expression modules significantly correlated with clinic trait, the hub gene of each module including RPS15A, PTGDS, CD53 and MSI2, which might play a vital role in progress of uveal melanoma (10).
To our knowledge, this is the first report of WGCNA of lung SCC expression profiles. In this study, bioinformatics methods were applied to integrate mRNA expression data of lung SCC in The Cancer Genome Atlas (TCGA) database and construct gene co-expression module for pathogenesis mechanism elucidation and identification of the diagnostic biomarkers and therapeutic targets of lung SCC metastasis.
Methods
TCGA gene expression profiles
The level 3 mRNA expression data of lung SCC and the corresponding clinical records in TCGA database (Oct 26, 2015) were obtained from the data portal (https://tcga-data.nci.nih.gov/tcga/). Total of 504 lung SCC patients were available in TCGA. The inclusion criteria of expression profiling in our study were shown as below: (I) the dataset was from primary solid tumor of patients with lung SCC; (II) patients had no other malignancy history; (III) patients received no treatment before collection of tumor samples. The mRNA expression datasets of 163 lung SCC patients with lymph node metastasis or distant metastasis, and mRNA expression datasets of 222 patients without metastasis were available in TCGA database. The datasets contained 20,531 genes and the sequencing of expression profiles was based on the platform of IlluminaHiSeq_RNASeqV2.
Identification of differentially expressed genes (DEGs)
The raw expression data of lung SCC patients in our study were downloaded. The significantly DEGs were identified between lung SCC patients with metastasis and lung SCC patients without metastasis through DESeq2 repackage in R language (11). The false discovery rate (FDR) was performed for multiple testing corrections of raw P value through the Benjamin and Hochberg method (12,13). The threshold of DEGs was set as FDR <0.05.
Construct gene co-expression network
To explore the interactions between the genes, a system biology approach, WGCNA, which converts co-expression measure into connections weight or topology overlap measure, was applied for gene co-expression network construction (14). Co-expression methodology is typically used for explore correlation between gene expression level. Genes involved in the same pathway or same functional compound tend to demonstrate a similar expression pattern (15). Therefore, the construction of a gene co-expression network facilitates the identification of genes with similar biological functions (16). In our work, all of genes of lung SCC patients with metastasis and lung SCC patients without metastasis were inputted to construct weighted co-expression modules using the WGCNA package in R language (17). The threshold of co-expression module was set as P<0.05.
Functional annotation
To obtain the biological function and signaling pathways of DEGs, the online software, GeneCodis3 was used for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment and Gene Ontology (GO) annotation of DEGs (18). The threshold of GO function and KEGG pathway of DEGs was set as FDR <0.0001 and FDR <0.05, respectively.
Protein-protein interaction (PPI)
In order to obtain insights into the interaction between DEGs of the black module and the pink module, PPI network was constructed by BioGRID, a database of known and predicted protein interactions (19). Then, PPI was visualized by Cytoscape software (http://cytoscape.org/) (20).
The expression level of DEGs in lung SCC tumor samples were validated by quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA of fresh tumor samples from lung SCC patients with metastasis and lung SCC patients without metastasis were extracted by using TRIzol reagent (Invitrogen, CA, USA) according to the manual instructions. The SuperScript III Reverse Transcription Kit (Invitrogen, CA, USA) was used to synthesize the cDNA. qRT-PCR reactions were performed using Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) on the Applied Biosystems 7,500 (Applied Biosystems, Foster City, CA). 18s rRNA was used as internal control for mRNA detection. The relative expression of genes was calculated using the 2−ΔΔCT equation (21). The PCR primers used were shown as Table S1. The GraphPad Prism version 6.0 software packages (GraphPad Software, San Diego, CA, USA) was used to output figures.
Ethics statement
The study was approved by the ethics committee board of Linyi People’s Hospital (No. lyll2015N67). Written informed consent was obtained from the patient for publication of this manuscript.
Results
DEGs between lung SCC patients with metastasis and lung SCC patients without metastasis
The raw expression profiles of lung SCC patients and corresponding clinical records were downloaded from the data portal of TCGA database. All of patients were divided into lung SCC with metastasis group or lung SCC without metastasis according to AJCC pathologic tumor stage. DEGs analysis was performed to the two groups. Total of 482 DEGs were identified as the threshold of FDR <0.05 (supplementary Table S2), consisting of 312 up-regulated DEGs and 170 down-regulated DEGs. As Table 1 shown, VEG, VSIG10L and MFAP5 were the most significantly up-regulated DEGs; ZNF208, C8orf46 and TNF were the most significantly down-regulated DEGs.
Table 1. The top ten DEGs in lung SCC with metastasis.
| Gene ID | Gene symbol | Log2FC | FDR |
|---|---|---|---|
| Up-regulated DEGs | |||
| 7425 | VGF | 6.302272 | 1.90E-06 |
| 147645 | VSIG10L | 5.870895 | 1.28E-05 |
| 8076 | MFAP5 | 5.335057 | 0.000187 |
| 6927 | HNF1A | 5.184579 | 0.000382 |
| 57521 | RPTOR | 5.125631 | 0.000449 |
| 2147 | F2 | 5.062773 | 0.000535 |
| 22809 | ATF5 | 4.959337 | 0.000714 |
| 51083 | GAL | 4.944154 | 0.000714 |
| 3706 | ITPKA | 4.839489 | 0.001009 |
| 84057 | MND1 | 4.835881 | 0.001009 |
| Down-regulated DEGs | |||
| 7757 | ZNF208 | −6.99816 | 5.04E-08 |
| 254778 | C8orf46 | −6.37244 | 1.81E-06 |
| 7124 | TNF | −5.93751 | 1.28E-05 |
| 2890 | GRIA1 | −5.90534 | 1.28E-05 |
| 339398 | LINGO4 | −5.8607 | 1.28E-05 |
| 338324 | S100A7A | −5.42097 | 0.000144 |
| 4693 | NDP | −5.33403 | 0.000187 |
| 116379 | IL22RA2 | −5.12306 | 0.000449 |
| 25833 | POU2F3 | −5.09145 | 0.000493 |
| 8190 | MIA | −5.04813 | 0.000542 |
DEG, differentially expressed genes; lung SCC, lung squamous cell carcinoma; FC, fold change; FDR, false discovery rate.
Functional annotation of DEGs between lung SCC with and without metastasis
To explore the functional significance of the identified DEGs in lung SCC metastasis, 482 DEGs were performed to unbiased GO term and KEGG pathway enrichment analyses. For DEGs related to lung SCC metastasis, transmembrane transport (FDR =8.76E-06), calcium ion binding (FDR =5.18E-06) and extracellular region (FDR =1.20E-17) were the most significant enrichment in biological process, molecular function and cellular component, respectively (Table 2); PPAR signaling pathway (KEGG ID: hsa03320, FDR =1.64E-05), cytokine-cytokine receptor interaction (KEGG ID: hsa04060, FDR =1.05E-03) and neuroactive ligand-receptor interaction (KEGG ID: hsa04080, FDR =3.95E-03) were the most significantly enriched pathways (Table 3).
Table 2. GO term enrichment analyses of DEGs.
| GO ID | GO term | Count | FDR |
|---|---|---|---|
| Biological process | |||
| GO:0055085 | Transmembrane transport | 24 | 8.76E-06 |
| GO:0006810 | Transport | 27 | 9.31E-06 |
| GO:0050995 | Negative regulation of lipid catabolic process | 4 | 2.29E-05 |
| GO:0007165 | Signal transduction | 4 | 4.67E-05 |
| GO:0051091 | Positive regulation of sequence-specific DNA binding transcription factor activity | 4 | 4.67E-05 |
| GO:0070374 | Positive regulation of ERK1 and ERK2 cascade | 4 | 4.67E-05 |
| GO:0042493 | Response to drug | 4 | 4.67E-05 |
| GO:0001666 | Response to hypoxia | 4 | 4.67E-05 |
| Molecular function | |||
| GO:0005509 | Calcium ion binding | 29 | 5.18E-06 |
| GO:0005216 | Ion channel activity | 9 | 7.99E-05 |
| Cellular component | |||
| GO:0005576 | Extracellular region | 81 | 1.20E-17 |
| GO:0005615 | Extracellular space | 43 | 3.18E-12 |
| GO:0005576 | Extracellular region | 39 | 1.52E-11 |
| GO:0005737 | Cytoplasm | 119 | 5.21E-07 |
| GO:0016021 | Integral to membrane | 101 | 3.11E-06 |
| GO:0005886 | Plasma membrane | 87 | 3.69E-06 |
| GO:0016020 | Membrane | 70 | 7.56E-05 |
GO, Gene Ontology; FDR, false discovery rate; DEG, differentially expressed genes.
Table 3. KEGG pathway enrichment analyses of DEGs.
| KEGG ID | KEGG term | Count | FDR | Genes |
|---|---|---|---|---|
| hsa03320 | PPAR signaling pathway | 9 | 1.64E-05 | ACSBG1, FABP7, UCP1, APOA2, RXRG, ACSL4, APOC3, MMP1, OLR1 |
| hsa04060 | Cytokine-cytokine receptor interaction | 13 | 1.05E-03 | IL1B, KIT, CRLF2, TNFRSF19, CX3CR1, IL22RA2, CCL17, PDGFA, FIGF, LEP, PF4, IL20, CCR6 |
| hsa04080 | Neuroactive ligand-receptor interaction | 12 | 3.95E-03 | DRD2, DRD5, NTSR1, SCTR, SSTR1, AGTR2, GRIA1, GRIK4, LEP, PRSS1, F2, GRIK1 |
| hsa04920 | Adipocytokine signaling pathway | 6 | 3.01E-03 | ACSBG1, G6PC2, AGRP, LEP, RXRG, ACSL4 |
| hsa04972 | Pancreatic secretion | 7 | 3.05E-03 | CFTR, SCTR, BST1, CPB2, PRSS1, PLA2G2F, ATP2B3 |
| hsa04976 | Bile secretion | 5 | 1.67E-02 | CFTR, SCTR, AQP1, ABCC2, BAAT |
| hsa05160 | Hepatitis C | 3 | 1.50E-02 | CLDN2, CLDN19, CLDN23 |
| hsa04514 | Cell adhesion molecules (CAMs) | 3 | 1.53E-02 | CLDN2, CLDN19, CLDN23 |
| hsa04670 | Leukocyte transendothelial migration | 3 | 1.50E-02 | CLDN2, CLDN19, CLDN23 |
| hsa04530 | Tight junction | 3 | 1.50E-02 | CLDN2, CLDN19, CLDN23 |
| hsa04964 | Proximal tubule bicarbonate reclamation | 3 | 1.53E-02 | SLC25A10, AQP1, CA4 |
| hsa04724 | Glutamatergic synapse | 3 | 1.78E-02 | GRIA1, GRIK4, GRIK1 |
| hsa04150 | mTOR signaling pathway | 4 | 2.10E-02 | RICTOR, RPS6KB1, RPTOR, FIGF |
| hsa05200 | Pathways in cancer | 3 | 2.18E-02 | KIT, PDGFA, FIGF |
KEGG, Kyoto Encyclopedia of Genes and Genomes; FDR, false discovery rate; DEGs, differentially expressed genes.
Construction of weighted gene co-expression modules
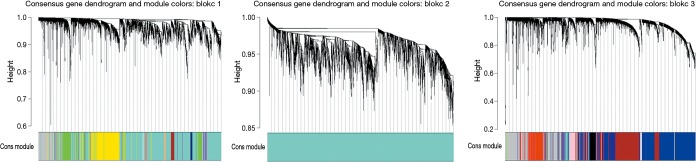
To explore the functional modules in lung SCC patients, the co-expression analysis of the 20,531 genes were performed in WGCNA. Modules for lung SCC were generated using the Scale-free Topology Criterion with a power cutoff of 12 and a minimum module size cutoff of 30. As Figure 1 and Table 4 shown, a total of 19 modules were identified. 23 DEGs and 26 DEGs between lung SCC with metastasis and lung SCC without metastasis were enriched in the respective pink and black modules through Chi-square test at the threshold of P<0.05, besides that, the expression pattern of pink modules, as well as black module, were significantly different between lung SCC with metastasis and lung SCC without metastasis through t-test at the threshold of P<0.05.
Figure 1.
Network analysis of gene expression in lung SCC identifies 19 distinct modules of co-expression genes. The dendrogram produced by average linkage hierarchical clustering of 20,531 genes based on WGCNA package in R. Total of 19 modules were identified. The three panels were the whole dendrogram. lung SCC, lung squamous cell carcinoma; WGCNA, weighted gene co-expression network analysis.
Table 4. The characteristics of gene co-expression modules.
| Module colors consensus | Number of genes | Number of DEGs | T.pval | Chi2.pval |
|---|---|---|---|---|
| Black | 167 | 26 | 3.95E-03 | 5.48E-23 |
| Blue | 1,822 | 61 | 2.06E-01 | 1.23E-02 |
| Brown | 1,272 | 44 | 3.45E-01 | 1.92E-02 |
| Cyan | 56 | 2 | 8.23E-02 | 8.83E-01 |
| Green | 493 | 6 | 6.68E-01 | 1.43E-01 |
| Green yellow | 89 | 0 | 5.25E-01 | 2.76E-01 |
| Grey | 1,652 | 52 | 5.38E-01 | 5.71E-02 |
| Grey60 | 35 | 0 | 6.57E-02 | 7.33E-01 |
| Light cyan | 51 | 0 | 9.71E-01 | 5.32E-01 |
| Light green | 31 | 1 | 3.05E-01 | 1.00E+00 |
| Magenta | 129 | 1 | 3.95E-01 | 3.87E-01 |
| Midnight blue | 54 | 2 | 6.33E-02 | 8.49E-01 |
| Pink | 160 | 23 | 3.42E-02 | 1.04E-18 |
| Purple | 93 | 0 | 3.45E-01 | 2.59E-01 |
| Red | 441 | 5 | 7.91E-01 | 1.38E-01 |
| Salmon | 56 | 3 | 3.54E-01 | 3.21E-01 |
| Tan | 75 | 4 | 1.26E-01 | 2.07E-01 |
| Turquoise | 8,538 | 101 | 5.16E-01 | 3.12E-10 |
| Yellow | 968 | 13 | 9.01E-01 | 5.89E-02 |
DEGs, differentially expressed genes; T.pval, P value of t-test; Chi2.pval, P value of Chi-square.
The functional annotation of DEGs persevered in the black module and pink module
DEGs and biological function preserved in each module were displayed in Tables 5 and 6, respectively. For the black module, Digestion (FDR =4.66E-05) and Extracellular region (FDR =3.96E-06) were the most significant enrichment in biological process and cellular component, respectively; bile secretion (hsa04976, FDR =1.91E-05) were the one significantly enriched pathways. For the pink module, Cilium axoneme (FDR =2.38E-06) was the most significant enrichment in biological process. There was no KEGG pathway was enriched from DEGs of the pink module.
Table 5. DEGs preserved in the modules.
| Module | DEGs |
|---|---|
| Black [26] | BAAT, BMP3, C16orf89, CFTR, CLDN2, CRTAC1, FIGF, HP, IGFALS, KCNK17, KLHDC7A, KNDC1,LRP2, MBL1P, NRGN, RASGRF1, RRAD, SCGB3A1, SCGB3A2, SCN1A, SCTR, SFTPD, SLC46A2, SLC5A9, SSTR1, WFDC2 |
| Pink [23] | EFCAB12, C1orf192, C22orf15, C5orf49,C6orf165, CCDC157, CCDC42B, CCDC65, DNAH6, DYDC2, DYNLRB2, EFCAB6, FAM166B, FAM183A, FAM92B, ROPN1L, SNTN, TEKT2, TEKT3, TMEM232, TSNAXIP1, WDR93, ZNF474 |
DEGs, differentially expressed genes.
Table 6. The function and pathway enrichment of DEGs of black module and pink module.
| ID | Terms | FDR | Genes |
|---|---|---|---|
| Black module-biological process | |||
| GO:0007586 | Digestion | 4.66E-05 | SSTR1, BAAT, SCTR |
| GO:0006811 | Ion transport | 0.00158761 | KCNK17, SCN1A, CFTR, SLC5A9 |
| GO:0055085 | Transmembrane transport | 0.00176614 | SCN1A, SLC46A2, CFTR, SLC5A9 |
| GO:0007165 | Signal transduction | 0.00987753 | RASGRF1, NRGN, IGFALS, RRAD |
| Black module-cellular component | |||
| GO:0005576 | Extracellular region | 3.96E-06 | C16orf89, SCGB3A2, HP, SFTPD, FIGF, WFDC2, SCGB3A1, IGFAL |
| GO:0005615 | Extracellular space | 3.72E-05 | SFTPD, FIGF, WFDC2, SCGB3A1,IGFALS,BMP3 |
| GO:0005886 | Plasma membrane | 0.000342 | RASGRF1, SSTR1, KCNK17, SCN1A, SCTR, SLC46A2, CLDN2, LRP2, RRAD |
| GO:0016021 | Integral to membrane | 0.013611 | KLHDC7A, KCNK17, SCN1A, SLC46A2, CLDN2, LRP2, CFTR, SLC5A9 |
| GO:0016020 | Membrane | 0.023071 | KLHDC7A, KCNK17, SCN1A, FIGF, LRP2, CFTR, SLC5A |
| Black-KEGG pathway | |||
| hsa04976 | Bile secretion | 1.91E-05 | CFTR, SCTR, BAAT |
| Pink-biological process | |||
| GO:0035085 | Cilium axoneme | 2.38E-06 | TEKT3, DNAH6, TEKT2 |
| GO:0005929 | Cilium | 1.34E-06 | SNTN, TEKT3, DNAH6, TEKT2 |
| GO:0005737 | Cytoplasm | 1.31E-05 | TEKT3, DNAH6, TEKT2, DYNLRB2 |
| GO:0005856 | Cytoskeleton | 1.31E-05 | TEKT3, DNAH6, TEKT2, DYNLRB2 |
| GO:0005874 | Microtubule | 1.31E-05 | TEKT3, DNAH6, TEKT2, DYNLRB2 |
FDR, false discovery rate; DEG, differentially expressed genes.
PPI network
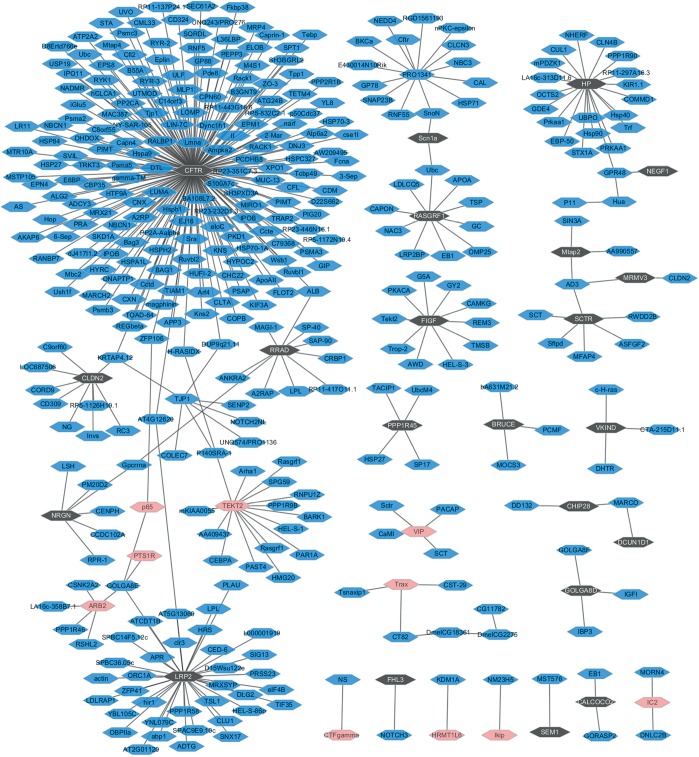
There are respective 26 and 23 DEGs related to lung SCC metastasis in the black module and the pink module. To obtain the interaction between the DEGs in the both of modules, PPI network was explored and visualize by Cytoscape. As Figure 2 shown, the network consisted of 396 nodes, 379 edges. In the PPIs network, the nodes with high degree are defined as hub protein. The most significant hub proteins were CFTR (degree =182), LRP2 (degree =37), HP (degree =20) and TEKT2 (degree =15). CFTR, LRP2, and HP were preserved in the black module; TEKT2 was preserved in the pink module.
Figure 2.
The constructed PPI networks of the DEGs in black module and the pink module. Pink nodes and black nodes represent DEGs in the pink module and black module, respectively. The blue nodes denote products of genes predicted to interact with the DEGs. The solid line means PPI correlation. PPI, protein-protein interaction; DEGs, differentially expressed genes.
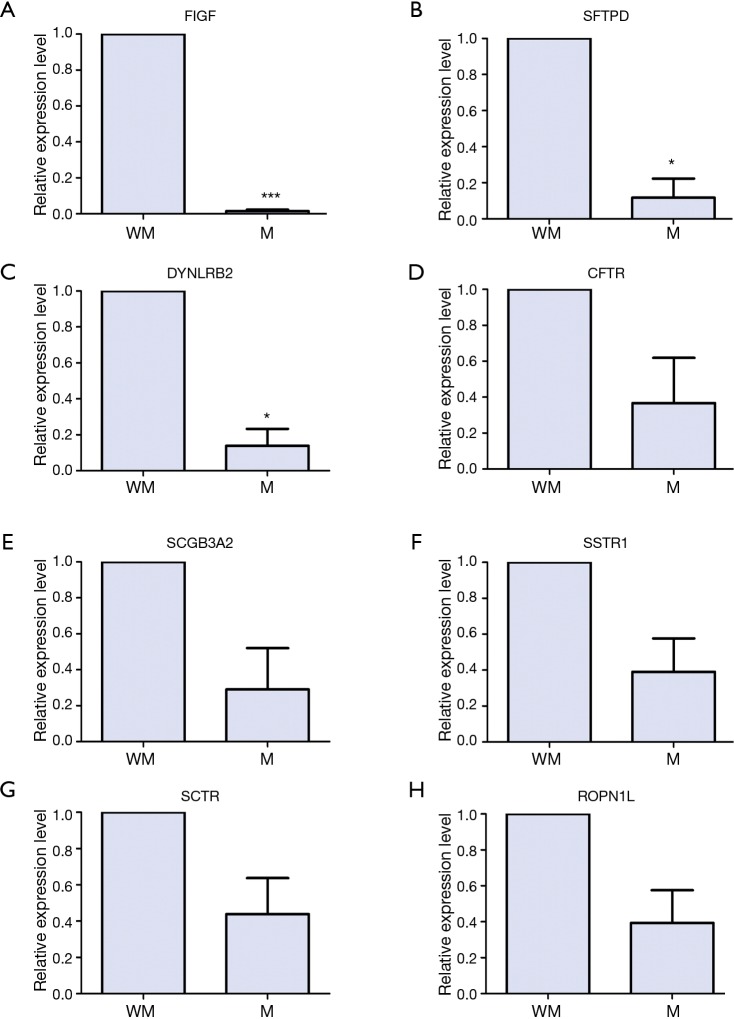
Verification of the expression level of DEGs through qRT-PCR
To verify our bioinformatics analyses, the expression level of DEGs were quantified by qRT-PCR in three primary tumor tissues of lung SCC patients with metastasis and three primary tumor tissues without metastasis. Seven DEGs including CFTR, FIGF, SSTR1, SFTPD, SCGB3A2, SCTR, DYNLRB2 were selected as candidate genes based on GO and KEGG annotation results from 49 genes in black and pink module, and literature review (22-26). One DEG, ROPN1L, was randomly selected as a candidate gene for qRT-PCR validation from 49 genes in black and pink module.
The biological roles of CFTR, FIGF, SSTR1, SFTPD and SCGB3A2 in lung cancer have been reported, but the biological roles of SCTR, DYNLRB2 an ROPN1L in lung SCC is unclear.
As shown in Figure 3A-C, FIGF (P<0.001), SFTPD (P<0.05), DYNLRB2 (P<0.05) were significantly down-regulated in the tumor samples of lung SCC with metastasis compared to those of lung SCC without metastasis; the expression levels of CFTR, SCGB3A2, SSTR1, SCTR and ROPN1L had no significance between lung SCC with metastasis and lung SCC without metastasis, but had the down-regulation tendency in lung SCC with metastasis (Figure 3D-H).
Figure 3.
The verification of mRNA expression level of DEGs between of lung SCC with and without metastasis in primary tumor tissues through qRT-PCR. (A) FIGF; (B) SFTPD; (C) DYNLRB2; (D) CFRT; (E) SCGB3A2; (F) SSTR1; (G) SCTR; (H) ROPN1L. M means patients with lung SCC metastasis; MW means patients without lung SCC metastasis. *, means P<0.05; and ***, means P<0.001. lung SCC, lung squamous cell carcinoma; DEGs, differentially expressed genes; qRT-PCR, quantitative real-time polymerase chain reactions.
Discussion
Two gene co-expression modules involved in the process of lung SCC metastasis were identified in our study. The black module and pink module contained 26 and 23 DEGs in lung SCC with metastasis compared to lung SCC without metastasis, respectively. The DEGs in the black module including CFTR, SCTR and BAAT were significantly enriched in the bile secretion pathway (hsa04976). Bile secretion is essential for digestion and absorption of fats and fat-soluble vitamins in the small intestine and the pathway is closely related to cholangiocarcinoma, gall bladder disease and familial cholestasis (27). Bile secretion pathway might be involved in the process of lung SCC metastasis.
The official name of CFTR is cystic fibrosis (CF) transmembrane conductance regulator, is a member of the ATP-binding cassette (ABC) transporter superfamily.
Mutations in this gene are associated with the autosomal recessive disorders CF, which leads to the abnormal transport of chloride and sodium across the epithelium, resulting in chronic lung obstruction, infection and inflammation. CF affects not only the physiological function of lungs, but also the pancreas, liver and intestines (22,28). In our study, CFTR was the hub protein in the PPI network, had the highest connectivity with 182 genes (Figure 2). It was down-regulated in lung SCC metastasis and its expression level was validated through qRT-PCR (Figure 3D), the result was accordance with the previous study (29). Several articles demonstrate that abnormal CFTR expression is related to the tumorigenesis and development of NSCLC. Low CFTR expression is significantly associated with advanced stage, lymph node metastasis and poor prognosis in NSCLC patients (22). The in vivo and in vitro experiments present knockdown of CFTR promotes epithelial-mesenchymal transition, invasion and migration of NSCLC cells; conversely, overexpression of CFTR suppresses cancer progression of NSCLC (22). Methylation of the CFTR gene is significantly greater in lung SCC than in lung adenocarcinomas and CFTR gene methylation is associated with significantly poorer survival in young patients, but not in elderly patients (30). Based on the above, low expression of CFTR might play pivotal roles in the process of lung SCC metastasis.
SCTR encodes secretin receptor, is a G protein-coupled receptor and belongs to the glucagon-VIP-secretin receptor family. It has been observed to be upregulated or down-regulated in several tumor types, and functions as promoting or suppressing the proliferation of tumor cells. SCTR was significantly underexpressed in primary pancreatic neuroendocrine tumors, nodal and liver metastases (31). Down-regulation of SCTR by promoter methylation promotes the cell proliferation and migration of breast cancer (32). In our study, the verification results of SCTR through qRT-PCR were accordance with our bioinformatics analysis. In addition to the bile secretion pathway, SCTR was significantly enriched in neuroactive ligand-receptor interaction and pancreatic secretion pathway (Table 3). SCTR had the connectivity with six genes in the PPI network (Figure 2). To our knowledge, this is the first reports presented SCTR was down-regulated in patients with lung SCC metastasis compared to those patients with lung SCC without metastasis. The biological function of SCTR in process of lung SCC needs to be further elucidated.
FIGF is also called VEGF-D or VEGFD. It encodes c-fos induced growth factor, a member of the platelet-derived growth factor/vascular endothelial growth factor family and is involved in angiogenesis, lymphangiogenesis and endothelial cell growth. VEGFD is a ligand for the VEGF receptor tyrosine kinases and activates it (33). The qRT-PCR shown FIGF was significantly down-regulated (P<0.001) in lung SCC metastasis compared to lung SCC without metastasis (Figure 3A). It was significantly enriched in cytokine-cytokine receptor interaction, mTOR signaling pathway and pathways in cancer. FIGF was a number of the black module and it had the connectivity with ten genes in the PPI network (Figure 2). In line with the previous article, FIGF is down-regulated in lung SCC (23). The mechanism of FIGF in lung SCC metastasis was unknown and further studies need to be investigated.
DYNLRB2 and ROPN1L were members of the pink module, encodes dynein light chain roadblock-type 2 and rhophilin associated tail protein 1 like, respectively.
ROPN1L variants were significantly associated with breast cancer risk in Korean women and Caucasian (34,35). The frequent amplification and copy number loss of DYNLRB2 is correlated with primary ductal carcinoma in situ (DCIS) and mixed DCIS, respectively (36). This is the first report displayed DYNLRB2 and ROPN1L were dysregulated in lung SCC, and might be related to the lung SCC metastasis.
SCGB3A2, SSTR1 and SFTPD were down-regulated in lung SCC metastasis compared to lung SCC without metastasis. SCGB3A2 encodes secretoglobin family 3A member 2. In lung cancer, SCGB3A2 were predominantly expressed in lung adenocarcinoma, compared with lung SCC and small cell carcinoma (24). It is a potentially useful marker for primary pulmonary tumors both in mice and humans (37). SSTR1 encodes somatostatin receptor 1, the expression level of SSTR1 mRNA was higher in both small cell lung cancer and lung SCC than in adenocarcinoma cell lines (26). SFTPD encodes surfactant protein D, is a lung-specific anti-inflammatory factor that antagonizes inflammation by inhibiting oxidative stress and stimulating innate immunity (38). In line with the previous article, SFTPD is down-regulated in NSCLC (25). The pathophysiology mechanism of dysregulated SCGB3A2, SSTR1 and SFTPD in lung SCC metastasis need further investigation.
In conclusion, we identified 482 DEGs in lung SCC without metastasis compared to lung SCC metastasis. Two gene co-expression modules including the black module and the pink module involved in the process of lung SCC metastasis were identified. Respective 26 and 23 dysregulated genes were enriched in the black module and the pink module. In the two of co-expression modules, CFTR, SCTR, FIGF might play key roles in the lung SCC metastasis. Our findings may contribute to the identification of early diagnosis biomarker for lung SCC metastasis and prognosis.
There are limitations in our study. Firstly, the biological roles of the key DEGs including SCTR and FIGF were unknown. In the future work, the in vivo and in vitro experiments were essential for exploring the function of genes mentioned above in the process of lung SCC metastasis. Secondly, additional studies with large cohorts of lung SCC with and without metastasis patients are needed to demonstrate the diagnostic value of identified genes as practical biomarkers.
Acknowledgements
Funding: This study was supported by Shandong Province Higher Educational Science and Technology Program (J11LF26).
Table S1. List of primers designed for the qRT-PCR verification.
| Primer | Primer sequence (5' to 3') |
|---|---|
| CFTR Forward | GATGGGGGCTGTGTCCTAAG |
| CFTR Reverse | GCATTGCTTCTATCCTGTGTTCA |
| FIGF Forward | ATCTAATCCAGCACCCCAAAAACT |
| FIGF Reverse | CTGGTATGAAAGGGGCATCTGTC |
| SCGB3A2 Forward | CTCTGGACAACATTCTTCCCTTTAT |
| SCGB3A2 Reverse | CCACCTCCGCTCTTTATCTTGA |
| SSTR1 Forward | GGGGCTATCTGCCTGTGCTAC |
| SSTR1 Reverse | CAAACACCATCACCACCATCAT |
| SCTR Forward | CCGTCCTCTACTGCTTCCTCAAC |
| SCTR Reverse | GGCTCTGCTCCAAGTGGCTG |
| SFTPD Forward | GCTACCTGGAAGCAGAAATGAA |
| SFTPD Reverse | AACAGAGCCATTGTCCCCTTT |
| DYNLRB2 Forward | CACCTGACAATGAAAGCCAAAAG |
| DYNLRB2 Reverse | TCACATGGATTCTGAATGACGAT |
| ROPN1L Forward | GTGCCTGCCGAAGGAAAA |
| ROPN1L Reverse | AAAACGTCTTGAAGGGGATGC |
| 18srRNA Forward | GTAACCCGTTGAACCCCATT |
| 18srRNA Reverse | CCATCCAATCGGTAGTAGCG |
qRT-PCR, quantitative real-time polymerase chain reaction.
Table S2. DEGs between lung SCC with and without metastasis.
| Gene | Gene ID | FC | FDR |
|---|---|---|---|
| Up-regulated genes | |||
| VGF | 7425 | 6.302272 | 1.90E-06 |
| VSIG10L | 147645 | 5.870895 | 1.28E-05 |
| MFAP5 | 8076 | 5.335057 | 0.000187 |
| HNF1A | 6927 | 5.184579 | 0.000382 |
| RPTOR | 57521 | 5.125631 | 0.000449 |
| F2 | 2147 | 5.062773 | 0.000535 |
| ATF5 | 22809 | 4.959337 | 0.000714 |
| GAL | 51083 | 4.944154 | 0.000714 |
| ITPKA | 3706 | 4.839489 | 0.001009 |
| MND1 | 84057 | 4.835881 | 0.001009 |
| TSPAN5 | 10098 | 4.772975 | 0.001176 |
| DRP2 | 1821 | 4.672904 | 0.001748 |
| DHCR7 | 1717 | 4.524834 | 0.003136 |
| C19orf76 | 199800 | 4.502532 | 0.003261 |
| KRT71 | 112802 | 4.393386 | 0.004713 |
| UPK2 | 7379 | 4.335088 | 0.00555 |
| SMARCD2 | 6603 | 4.324975 | 0.005592 |
| C4orf49 | 84709 | 4.297849 | 0.005983 |
| SRP68 | 6730 | 4.237439 | 0.007321 |
| TEX13B | 56156 | 4.221298 | 0.007491 |
| NDUFC1 | 4717 | 4.102 | 0.009792 |
| PRB3 | 5544 | 4.098234 | 0.009792 |
| PIWIL2 | 55124 | 4.084314 | 0.010104 |
| KRT18 | 3875 | 4.064585 | 0.010387 |
| C9orf70 | 84850 | 4.058666 | 0.010503 |
| TAC3 | 6866 | 4.049676 | 0.010714 |
| NPSR1 | 387129 | 4.036762 | 0.011201 |
| SLC9A2 | 6549 | 4.011459 | 0.011298 |
| GRIK1 | 2897 | 4.008088 | 0.011298 |
| ABCC2 | 1244 | 4.007931 | 0.011298 |
| C4BPB | 725 | 3.986718 | 0.011558 |
| TCHP | 84260 | 3.966038 | 0.011919 |
| KCNH6 | 81033 | 3.96427 | 0.011919 |
| TRIM54 | 57159 | 3.951173 | 0.012284 |
| PROCR | 10544 | 3.941815 | 0.012469 |
| NEB | 4703 | 3.92073 | 0.013004 |
| ZNF787 | 126208 | 3.920271 | 0.013004 |
| FOXH1 | 8928 | 3.920242 | 0.013004 |
| ZWINT | 11130 | 3.918314 | 0.013004 |
| GPR155 | 151556 | 3.916905 | 0.013004 |
| SYNPO2L | 79933 | 3.904368 | 0.01311 |
| FA2H | 79152 | 3.894921 | 0.01344 |
| NCAPG2 | 54892 | 3.873486 | 0.014273 |
| NBAS | 51594 | 3.86964 | 0.014273 |
| POLR3G | 10622 | 3.868808 | 0.014273 |
| COG1 | 9382 | 3.868597 | 0.014273 |
| PRR4 | 11272 | 3.856872 | 0.014627 |
| SFRS9 | 8683 | 3.839756 | 0.015322 |
| SCN3A | 6328 | 3.826201 | 0.015533 |
| ADI1 | 55256 | 3.826146 | 0.015533 |
| DDX54 | 79039 | 3.825855 | 0.015533 |
| CPS1 | 1373 | 3.80012 | 0.016528 |
| SAPS1 | 22870 | 3.77652 | 0.017456 |
| MAPT | 4137 | 3.766167 | 0.017892 |
| APOC3 | 345 | 3.76264 | 0.018047 |
| MIPOL1 | 145282 | 3.757985 | 0.018235 |
| HRC | 3270 | 3.752649 | 0.018276 |
| IL1RAPL1 | 11141 | 3.749319 | 0.018375 |
| PLK4 | 10733 | 3.747044 | 0.018375 |
| SMAGP | 57228 | 3.732997 | 0.018471 |
| DKK4 | 27121 | 3.730844 | 0.018471 |
| CADPS | 8618 | 3.723286 | 0.01859 |
| USP32 | 84669 | 3.722682 | 0.01859 |
| NPNT | 255743 | 3.711312 | 0.019352 |
| SYNGR4 | 23546 | 3.708512 | 0.019391 |
| PSMC5 | 5705 | 3.706852 | 0.019391 |
| USP7 | 7874 | 3.694849 | 0.020068 |
| NR2E1 | 7101 | 3.686376 | 0.020569 |
| ONECUT1 | 3175 | 3.683024 | 0.020731 |
| ASPSCR1 | 79058 | 3.669242 | 0.021581 |
| SAP30BP | 29115 | 3.667889 | 0.021597 |
| TESK1 | 7016 | 3.651205 | 0.022574 |
| PCDHGB4 | 8641 | 3.640394 | 0.022895 |
| RAD51C | 5889 | 3.63531 | 0.02311 |
| HES7 | 84667 | 3.632468 | 0.02311 |
| APOA2 | 336 | 3.627873 | 0.023268 |
| C19orf48 | 84798 | 3.626093 | 0.023293 |
| NADSYN1 | 55191 | 3.62274 | 0.023306 |
| NAA15 | 80155 | 3.62065 | 0.023398 |
| GALNT13 | 114805 | 3.608216 | 0.024172 |
| DCXR | 51181 | 3.606123 | 0.024193 |
| ING2 | 3622 | 3.605638 | 0.024193 |
| C9orf82 | 79886 | 3.59792 | 0.024535 |
| MED13 | 9969 | 3.596717 | 0.024552 |
| FAM104A | 84923 | 3.590437 | 0.024911 |
| PITPNC1 | 26207 | 3.589103 | 0.024911 |
| SMC6 | 79677 | 3.588884 | 0.024911 |
| CYP3A5 | 1577 | 3.581118 | 0.025177 |
| NTSR1 | 4923 | 3.579779 | 0.025198 |
| TRAP1 | 10131 | 3.57807 | 0.025198 |
| OR51E2 | 81285 | 3.575338 | 0.025228 |
| KPNA2 | 3838 | 3.570615 | 0.025532 |
| MELK | 9833 | 3.565422 | 0.025758 |
| RRM2 | 6241 | 3.539854 | 0.027316 |
| C4orf41 | 60684 | 3.539614 | 0.027316 |
| FUT6 | 2528 | 3.531912 | 0.028026 |
| HIST1H3H | 8357 | 3.525661 | 0.0283 |
| C4orf48 | 401115 | 3.524516 | 0.028325 |
| CELA1 | 1990 | 3.521018 | 0.028538 |
| COL25A1 | 84570 | 3.519272 | 0.028538 |
| TSPO2 | 222642 | 3.517829 | 0.028538 |
| MRPL38 | 64978 | 3.516225 | 0.028538 |
| DUSP13 | 51207 | 3.499272 | 0.029593 |
| CLTC | 1213 | 3.49865 | 0.029593 |
| GAR1 | 54433 | 3.489856 | 0.03009 |
| SLC25A10 | 1468 | 3.489392 | 0.03009 |
| ABCE1 | 6059 | 3.488184 | 0.03009 |
| CCDC124 | 115098 | 3.476216 | 0.031069 |
| XRCC6BP1 | 91419 | 3.467209 | 0.031602 |
| FDXR | 2232 | 3.465684 | 0.031616 |
| KLC2 | 64837 | 3.459955 | 0.032099 |
| C21orf45 | 54069 | 3.45586 | 0.032196 |
| ARHGDIA | 396 | 3.433486 | 0.034049 |
| HBQ1 | 3049 | 3.428947 | 0.034321 |
| CIT | 11113 | 3.419155 | 0.035096 |
| CENPE | 1062 | 3.409378 | 0.035835 |
| SNX31 | 169166 | 3.407213 | 0.036019 |
| MRPL21 | 219927 | 3.404929 | 0.036117 |
| DCTD | 1635 | 3.399858 | 0.03628 |
| KRTAP5-7 | 440050 | 3.399706 | 0.03628 |
| C17orf53 | 78995 | 3.398381 | 0.03628 |
| CLTA | 1211 | 3.377629 | 0.038227 |
| FOXK2 | 3607 | 3.368591 | 0.039039 |
| C14orf80 | 283643 | 3.361435 | 0.039747 |
| PWP2 | 5822 | 3.36065 | 0.039755 |
| FIBCD1 | 84929 | 3.358374 | 0.039889 |
| UBTF | 7343 | 3.356782 | 0.039896 |
| UCP1 | 7350 | 3.354004 | 0.040174 |
| LOC92659 | 92659 | 3.352846 | 0.040174 |
| FAM84B | 157638 | 3.342063 | 0.041043 |
| DDN | 23109 | 3.339528 | 0.041043 |
| MTL5 | 9633 | 3.336217 | 0.041112 |
| CST4 | 1472 | 3.335754 | 0.041112 |
| CENPO | 79172 | 3.33492 | 0.041132 |
| SLC5A5 | 6528 | 3.326312 | 0.04147 |
| RPS6KB1 | 6198 | 3.325852 | 0.04147 |
| H2AFB1 | 474382 | 3.320782 | 0.042023 |
| VWA5B2 | 90113 | 3.319972 | 0.042044 |
| C16orf75 | 116028 | 3.318308 | 0.042194 |
| LOC127841 | 127841 | 3.313993 | 0.042443 |
| LSM6 | 11157 | 3.312778 | 0.042527 |
| LMBR1 | 64327 | 3.30957 | 0.042815 |
| MUC6 | 4588 | 3.307915 | 0.042882 |
| PGP | 283871 | 3.306565 | 0.042882 |
| DDX4 | 54514 | 3.306498 | 0.042882 |
| VRK1 | 7443 | 3.303783 | 0.043098 |
| PCGF2 | 7703 | 3.302845 | 0.043105 |
| GINS2 | 51659 | 3.302439 | 0.043105 |
| RWDD4A | 201965 | 3.300758 | 0.043155 |
| HIST1H2BM | 8342 | 3.298546 | 0.043306 |
| C9orf140 | 89958 | 3.295731 | 0.043642 |
| C12orf43 | 64897 | 3.285002 | 0.044826 |
| ALDH1A1 | 216 | 3.283336 | 0.04499 |
| DHX8 | 1659 | 3.277351 | 0.045645 |
| UBE2O | 63893 | 3.273996 | 0.045794 |
| HSPBP1 | 23640 | 3.27391 | 0.045794 |
| NPPC | 4880 | 3.263528 | 0.047087 |
| THOC4 | 10189 | 3.258293 | 0.047551 |
| AKR7A3 | 22977 | 3.255861 | 0.047642 |
| GGNBP1 | 449520 | 3.253542 | 0.047824 |
| BCAT2 | 587 | 3.252302 | 0.047828 |
| LOC81691 | 81691 | 3.251006 | 0.047944 |
| TLK2 | 11011 | 3.247125 | 0.048489 |
| CLGN | 1047 | 3.24657 | 0.048489 |
| HSD17B1 | 3292 | 3.243008 | 0.04858 |
| CKM | 1158 | 3.237947 | 0.049187 |
| LONP1 | 9361 | 3.237664 | 0.049187 |
| GDF11 | 10220 | 3.236492 | 0.049227 |
| FGF3 | 2248 | 3.232167 | 0.049726 |
| Down-regulated genes | |||
| ZNF208 | 7757 | −6.99816 | 5.04E-08 |
| C8orf46 | 254778 | −6.37244 | 1.81E-06 |
| TNF | 7124 | −5.93751 | 1.28E-05 |
| GRIA1 | 2890 | −5.90534 | 1.28E-05 |
| LINGO4 | 339398 | −5.8607 | 1.28E-05 |
| S100A7A | 338324 | −5.42097 | 0.000144 |
| NDP | 4693 | −5.33403 | 0.000187 |
| IL22RA2 | 116379 | −5.12306 | 0.000449 |
| POU2F3 | 25833 | −5.09145 | 0.000493 |
| MIA | 8190 | −5.04813 | 0.000542 |
| FOXI2 | 399823 | −5.02927 | 0.000562 |
| PCDHGA4 | 56111 | −5.01861 | 0.000562 |
| SYCE1 | 93426 | −4.94241 | 0.000714 |
| FCER1A | 2205 | −4.88237 | 0.000926 |
| C7orf16 | 10842 | −4.82702 | 0.001009 |
| TRIM58 | 25893 | −4.82627 | 0.001009 |
| C2orf73 | 129852 | −4.82333 | 0.001009 |
| KLHDC7A | 127707 | −4.81194 | 0.001009 |
| SCG3 | 29106 | −4.81048 | 0.001009 |
| FAM169B | 283777 | −4.7216 | 0.001467 |
| LOC283731 | 283731 | −4.68577 | 0.001693 |
| CCDC42B | 387885 | −4.6414 | 0.001977 |
| FAIM2 | 23017 | −4.61988 | 0.002131 |
| SLC47A2 | 146802 | −4.56176 | 0.002738 |
| PCDHGA9 | 56107 | −4.52173 | 0.003136 |
| UPP2 | 151531 | −4.50633 | 0.003261 |
| PDC | 5132 | −4.48382 | 0.003474 |
| C1orf192 | 257177 | −4.4712 | 0.003598 |
| HTR3A | 3359 | −4.44092 | 0.004047 |
| HSD17B2 | 3294 | −4.42747 | 0.004129 |
| EMID1 | 129080 | −4.4268 | 0.004129 |
| S100B | 6285 | −4.38458 | 0.004789 |
| WBSCR17 | 64409 | −4.38066 | 0.004789 |
| DPYS | 1807 | −4.37219 | 0.004877 |
| CD1A | 909 | −4.36288 | 0.004988 |
| HP | 3240 | −4.32821 | 0.005592 |
| CDH20 | 28316 | −4.30398 | 0.005983 |
| TMPRSS5 | 80975 | −4.28326 | 0.006278 |
| NDN | 4692 | −4.27326 | 0.006453 |
| C5orf13 | 9315 | −4.26551 | 0.006567 |
| OLR1 | 4973 | −4.23143 | 0.007383 |
| MSTN | 2660 | −4.22817 | 0.007383 |
| C1orf186 | 440712 | −4.20012 | 0.008098 |
| GPR17 | 2840 | −4.18922 | 0.008366 |
| C9orf44 | 158314 | −4.1585 | 0.00943 |
| C4orf7 | 260436 | −4.1494 | 0.009666 |
| HEPACAM2 | 253012 | −4.14051 | 0.009791 |
| CRLF1 | 9244 | −4.13588 | 0.009791 |
| LOC284749 | 284749 | −4.12985 | 0.009791 |
| IGFN1 | 91156 | −4.1266 | 0.009791 |
| DYNLRB2 | 83657 | −4.12242 | 0.009791 |
| ART3 | 419 | −4.12232 | 0.009791 |
| MAGEE2 | 139599 | −4.12076 | 0.009791 |
| CCDC129 | 223075 | −4.11658 | 0.009791 |
| G6PC2 | 57818 | −4.11556 | 0.009791 |
| CORO2B | 10391 | −4.11263 | 0.009791 |
| RBM44 | 375316 | −4.11151 | 0.009791 |
| CNGA3 | 1261 | −4.10112 | 0.009792 |
| MBL1P | 8512 | −4.09773 | 0.009792 |
| HPD | 3242 | −4.09711 | 0.009792 |
| OR2W3 | 343171 | −4.08612 | 0.010104 |
| GRIK4 | 2900 | −4.07796 | 0.010114 |
| AGTR2 | 186 | −4.07697 | 0.010114 |
| FAM55D | 54827 | −4.07446 | 0.010114 |
| MOGAT2 | 80168 | −4.07341 | 0.010114 |
| PNMA5 | 114824 | −4.05686 | 0.010503 |
| CCDC141 | 285025 | −4.0289 | 0.011298 |
| ACOT12 | 134526 | −4.02808 | 0.011298 |
| WDR93 | 56964 | −4.02511 | 0.011298 |
| TPTE2 | 93492 | −4.02483 | 0.011298 |
| PCDHGA6 | 56109 | −4.01945 | 0.011298 |
| C13orf15 | 28984 | −4.01794 | 0.011298 |
| IL1B | 3553 | −4.01286 | 0.011298 |
| CTCFL | 140690 | −4.01208 | 0.011298 |
| SSTR1 | 6751 | −4.00644 | 0.011298 |
| HEPHL1 | 341208 | −3.99112 | 0.011558 |
| WFDC12 | 128488 | −3.9907 | 0.011558 |
| HTR3C | 170572 | −3.9881 | 0.011558 |
| PADI2 | 11240 | −3.98802 | 0.011558 |
| STAC2 | 342667 | −3.98702 | 0.011558 |
| ACSL4 | 2182 | −3.98589 | 0.011558 |
| CLDN2 | 9075 | −3.98299 | 0.011566 |
| RASD2 | 23551 | −3.98158 | 0.011566 |
| MYO1H | 283446 | −3.97564 | 0.011756 |
| GPIHBP1 | 338328 | −3.97226 | 0.011822 |
| TRPC2 | 7221 | −3.96666 | 0.011919 |
| CA4 | 762 | −3.961 | 0.011984 |
| FOXI1 | 2299 | −3.95747 | 0.012063 |
| CPB2 | 1361 | −3.94544 | 0.012448 |
| TEX9 | 374618 | −3.94412 | 0.012448 |
| SHC4 | 399694 | −3.93515 | 0.012719 |
| FAM123A | 219287 | −3.9236 | 0.013004 |
| PAGE5 | 90737 | −3.91727 | 0.013004 |
| PSAPL1 | 768239 | −3.91368 | 0.013026 |
| C1orf150 | 148823 | −3.91293 | 0.013026 |
| LOC646851 | 646851 | −3.91116 | 0.013026 |
| ROPN1L | 83853 | −3.90859 | 0.013031 |
| CRABP1 | 1381 | −3.90756 | 0.013031 |
| AMZ1 | 155185 | −3.90189 | 0.013151 |
| ADAMTS8 | 11095 | −3.88394 | 0.013963 |
| C1QL2 | 165257 | −3.88144 | 0.01401 |
| PLD5 | 200150 | −3.87838 | 0.014089 |
| TCTEX1D4 | 343521 | −3.86347 | 0.014479 |
| DRGX | 644168 | −3.8586 | 0.014627 |
| TMPRSS3 | 64699 | −3.85615 | 0.014627 |
| CYB561D1 | 284613 | −3.84443 | 0.015243 |
| CLEC4C | 170482 | −3.83844 | 0.015322 |
| FOLR3 | 2352 | −3.83843 | 0.015322 |
| ACRV1 | 56 | −3.83238 | 0.015533 |
| TMEM100 | 55273 | −3.83079 | 0.015533 |
| GPR128 | 84873 | −3.82961 | 0.015533 |
| FNDC5 | 252995 | −3.82395 | 0.015558 |
| CLDN23 | 137075 | −3.81787 | 0.015702 |
| DYDC2 | 84332 | −3.81611 | 0.015702 |
| KLK7 | 5650 | −3.81601 | 0.015702 |
| DRD2 | 1813 | −3.81539 | 0.015702 |
| SLC5A9 | 200010 | −3.81427 | 0.015702 |
| FAM166B | 730112 | −3.7975 | 0.016606 |
| CFP | 5199 | −3.79313 | 0.016803 |
| COL9A2 | 1298 | −3.78771 | 0.016969 |
| ALDH1A3 | 220 | −3.7876 | 0.016969 |
| SERPINA3 | 12 | −3.78639 | 0.016969 |
| RYR3 | 6263 | −3.78106 | 0.017238 |
| IQSEC3 | 440073 | −3.77321 | 0.017563 |
| HPR | 3250 | −3.7722 | 0.017563 |
| RIMS3 | 9783 | −3.75729 | 0.018235 |
| CYP2B6 | 1555 | −3.75556 | 0.018261 |
| RNF122 | 79845 | −3.75386 | 0.018276 |
| DNAH6 | 1768 | −3.74677 | 0.018375 |
| HAPLN4 | 404037 | −3.74597 | 0.018375 |
| DENND3 | 22898 | −3.74354 | 0.018391 |
| FAM183A | 440585 | −3.74312 | 0.018391 |
| TRPM5 | 29850 | −3.7399 | 0.018471 |
| SLC6A13 | 6540 | −3.73831 | 0.018471 |
| CHIT1 | 1118 | −3.73584 | 0.018471 |
| FIGF | 2277 | −3.73417 | 0.018471 |
| RXRG | 6258 | −3.73254 | 0.018471 |
| DBC1 | 1620 | −3.73111 | 0.018471 |
| MS4A8B | 83661 | −3.72991 | 0.018471 |
| TTTY2 | 60439 | −3.72925 | 0.018471 |
| RHO | 6010 | −3.72784 | 0.018478 |
| PTX3 | 5806 | −3.72667 | 0.018478 |
| CUX2 | 23316 | −3.70606 | 0.019391 |
| C22orf15 | 150248 | −3.70595 | 0.019391 |
| SPRR2G | 6706 | −3.70009 | 0.019751 |
| CHI3L1 | 1116 | −3.6862 | 0.020569 |
| TNFRSF19 | 55504 | −3.6782 | 0.021029 |
| CRTAC1 | 55118 | −3.67011 | 0.021581 |
| C1orf175 | 374977 | −3.66343 | 0.021827 |
| ACSBG1 | 23205 | −3.66287 | 0.021827 |
| KRT31 | 3881 | −3.65343 | 0.022543 |
| C6orf217 | 1E+08 | −3.6508 | 0.022574 |
| NEDD9 | 4739 | −3.64821 | 0.022701 |
| BST1 | 683 | −3.64606 | 0.022722 |
| C5orf49 | 134121 | −3.64571 | 0.022722 |
| TSNAXIP1 | 55815 | −3.64271 | 0.022888 |
| OCA2 | 4948 | −3.64101 | 0.022895 |
| ARHGAP6 | 395 | −3.63432 | 0.02311 |
| SCGB3A2 | 117156 | −3.63351 | 0.02311 |
| NANOS1 | 340719 | −3.63266 | 0.02311 |
| PCDHB18 | 54660 | −3.62948 | 0.023268 |
| CACNA1G | 8913 | −3.62744 | 0.023268 |
| DRD5 | 1816 | −3.62405 | 0.023306 |
| BMP3 | 651 | −3.62323 | 0.023306 |
| TRY6 | 154754 | −3.61309 | 0.023993 |
| CELF3 | 11189 | −3.60886 | 0.024172 |
| PSG8 | 440533 | −3.60801 | 0.024172 |
| ERVFRDE1 | 405754 | −3.60466 | 0.024193 |
| FBN3 | 84467 | −3.60111 | 0.02435 |
| PTCHD2 | 57540 | −3.60091 | 0.02435 |
| CHI3L2 | 1117 | −3.59413 | 0.0247 |
| PRY2 | 442862 | −3.58729 | 0.024968 |
| C16orf89 | 146556 | −3.58584 | 0.025011 |
| FAM151A | 338094 | −3.58328 | 0.02516 |
| PDK4 | 5166 | −3.58162 | 0.025177 |
| C3orf16 | 389161 | −3.57796 | 0.025198 |
| MMP3 | 4314 | −3.57631 | 0.025228 |
| PAK3 | 5063 | −3.57472 | 0.025228 |
| POLB | 5423 | −3.56716 | 0.025758 |
| BIRC2 | 329 | −3.56631 | 0.025758 |
| SCGB3A1 | 92304 | −3.56338 | 0.025865 |
| TSPAN2 | 10100 | −3.56094 | 0.026012 |
| C4A | 720 | −3.55936 | 0.026075 |
| COL29A1 | 256076 | −3.55698 | 0.02616 |
| LEP | 3952 | −3.55605 | 0.02616 |
| TCTEX1D1 | 200132 | −3.55567 | 0.02616 |
| TMEM217 | 221468 | −3.55251 | 0.026382 |
| TMEM232 | 642987 | −3.54522 | 0.027026 |
| CPAMD8 | 27151 | −3.54242 | 0.027217 |
| MMP1 | 4312 | −3.53001 | 0.028088 |
| LOC148824 | 148824 | −3.52948 | 0.028088 |
| SMYD1 | 150572 | −3.52739 | 0.028213 |
| PDGFA | 5154 | −3.51856 | 0.028538 |
| ADHFE1 | 137872 | −3.51763 | 0.028538 |
| DCLK1 | 9201 | −3.51695 | 0.028538 |
| KNDC1 | 85442 | −3.50966 | 0.028964 |
| C19orf59 | 199675 | −3.5085 | 0.028964 |
| CCR6 | 1235 | −3.50812 | 0.028964 |
| NRGN | 4900 | −3.5079 | 0.028964 |
| CCL17 | 6361 | −3.50786 | 0.028964 |
| SPOCK2 | 9806 | −3.50282 | 0.029391 |
| OBP2B | 29989 | −3.50221 | 0.029391 |
| SCN1A | 6323 | −3.49743 | 0.029631 |
| PDYN | 5173 | −3.49134 | 0.03009 |
| FABP7 | 2173 | −3.48896 | 0.03009 |
| LMOD3 | 56203 | −3.48822 | 0.03009 |
| SCTR | 6344 | −3.4856 | 0.030285 |
| SNTN | 132203 | −3.48335 | 0.030444 |
| SLC25A27 | 9481 | −3.47904 | 0.030841 |
| GNAO1 | 2775 | −3.47505 | 0.031107 |
| KCNK17 | 89822 | −3.47269 | 0.031284 |
| PCDHA9 | 9752 | −3.47128 | 0.031351 |
| LRP2 | 4036 | −3.46908 | 0.03151 |
| CCDC157 | 550631 | −3.46663 | 0.031602 |
| C21orf62 | 56245 | −3.46092 | 0.032082 |
| KAL1 | 3730 | −3.45759 | 0.032196 |
| RASGEF1B | 153020 | −3.45638 | 0.032196 |
| NPPA | 4878 | −3.45595 | 0.032196 |
| LCE3E | 353145 | −3.45363 | 0.032366 |
| GAS2 | 2620 | −3.45174 | 0.032496 |
| FAM92B | 339145 | −3.44844 | 0.032797 |
| TDRD6 | 221400 | −3.44545 | 0.033063 |
| MEGF11 | 84465 | −3.441 | 0.033388 |
| CX3CR1 | 1524 | −3.44075 | 0.033388 |
| CYP2D6 | 1565 | −3.44039 | 0.033388 |
| CLDN19 | 149461 | −3.43628 | 0.033799 |
| CLLU1 | 574028 | −3.42963 | 0.034321 |
| CAT | 847 | −3.42928 | 0.034321 |
| BOC | 91653 | −3.42237 | 0.03506 |
| IRX1 | 79192 | −3.4209 | 0.035093 |
| TMEM190 | 147744 | −3.42053 | 0.035093 |
| SLC46A2 | 57864 | −3.41846 | 0.035096 |
| RASGRF1 | 5923 | −3.41816 | 0.035096 |
| KANK4 | 163782 | −3.41106 | 0.035835 |
| KCNE1L | 23630 | −3.41057 | 0.035835 |
| BAAT | 570 | −3.41002 | 0.035835 |
| KIAA1644 | 85352 | −3.40608 | 0.036066 |
| OR4N4 | 283694 | −3.40208 | 0.03628 |
| PCDHA10 | 56139 | −3.40013 | 0.03628 |
| DNAH5 | 1767 | −3.39858 | 0.03628 |
| MT1F | 4494 | −3.39841 | 0.03628 |
| SH2D7 | 646892 | −3.39276 | 0.03693 |
| CFTR | 1080 | −3.3898 | 0.037229 |
| TAL1 | 6886 | −3.38671 | 0.037548 |
| IGFALS | 3483 | −3.38561 | 0.037597 |
| EFCAB12 | 90288 | −3.38423 | 0.037683 |
| CTSH | 1512 | −3.38248 | 0.037822 |
| ZNF98 | 148198 | −3.37939 | 0.038146 |
| CCDC65 | 85478 | −3.37732 | 0.038227 |
| C15orf37 | 283687 | −3.3738 | 0.038615 |
| CLEC3B | 7123 | −3.37133 | 0.03886 |
| RGS13 | 6003 | −3.36932 | 0.039039 |
| SFTPD | 6441 | −3.36378 | 0.03962 |
| TEKT3 | 64518 | −3.36159 | 0.039747 |
| KLHDC8B | 200942 | −3.35827 | 0.039889 |
| ROPN1 | 54763 | −3.35679 | 0.039896 |
| CRLF2 | 64109 | −3.35271 | 0.040174 |
| EFCAB6 | 64800 | −3.34864 | 0.040663 |
| RASL12 | 51285 | −3.34363 | 0.041043 |
| IL20 | 50604 | −3.3433 | 0.041043 |
| APOL4 | 80832 | −3.34189 | 0.041043 |
| CAMK1G | 57172 | −3.34104 | 0.041043 |
| DNAI1 | 27019 | −3.3407 | 0.041043 |
| COL14A1 | 7373 | −3.33932 | 0.041043 |
| CNTN4 | 152330 | −3.33921 | 0.041043 |
| PRSS1 | 5644 | −3.339 | 0.041043 |
| BMF | 90427 | −3.33803 | 0.041084 |
| TMEM151B | 441151 | −3.33632 | 0.041112 |
| LOC283050 | 283050 | −3.33424 | 0.041132 |
| KLF17 | 128209 | −3.3305 | 0.04147 |
| RRAD | 6236 | −3.32852 | 0.04147 |
| GP5 | 2814 | −3.32848 | 0.04147 |
| CLDN25 | 644672 | −3.32741 | 0.04147 |
| SOX11 | 6664 | −3.32658 | 0.04147 |
| WDR38 | 401551 | −3.3261 | 0.04147 |
| SRD5A2 | 6716 | −3.32582 | 0.04147 |
| C1orf228 | 339541 | −3.32106 | 0.042023 |
| NAV3 | 89795 | −3.31762 | 0.042197 |
| SPRR2B | 6701 | −3.31648 | 0.042268 |
| C14orf139 | 79686 | −3.31408 | 0.042443 |
| TKTL1 | 8277 | −3.31158 | 0.042608 |
| HAND1 | 9421 | −3.30775 | 0.042882 |
| RICTOR | 253260 | −3.30458 | 0.043076 |
| CACNG1 | 786 | −3.30069 | 0.043155 |
| RDH16 | 8608 | −3.30017 | 0.043155 |
| PVALB | 5816 | −3.2924 | 0.044062 |
| SEC14L3 | 266629 | −3.29154 | 0.044096 |
| HPCAL4 | 51440 | −3.28873 | 0.044437 |
| DIRC3 | 729582 | −3.2857 | 0.044817 |
| AGRP | 181 | −3.28238 | 0.045042 |
| FAM81B | 153643 | −3.27818 | 0.045613 |
| PLA2G2F | 64600 | −3.27614 | 0.04574 |
| CCDC30 | 728621 | −3.27408 | 0.045794 |
| PF4 | 5196 | −3.27303 | 0.045836 |
| CSDC2 | 27254 | −3.26912 | 0.046371 |
| RNF112 | 7732 | −3.26788 | 0.046472 |
| SLC17A8 | 246213 | −3.26127 | 0.04736 |
| PURG | 29942 | −3.25821 | 0.047551 |
| TG | 7038 | −3.25766 | 0.047551 |
| KIT | 3815 | −3.25764 | 0.047551 |
| DES | 1674 | −3.25617 | 0.047642 |
| TEKT2 | 27285 | −3.25453 | 0.047762 |
| ZNF474 | 133923 | −3.25294 | 0.047824 |
| CDX1 | 1044 | −3.24595 | 0.048491 |
| C6orf165 | 154313 | −3.24447 | 0.048565 |
| ATP2B3 | 492 | −3.24396 | 0.048565 |
| WFDC2 | 10406 | −3.2437 | 0.048565 |
| LPO | 4025 | −3.24181 | 0.048682 |
| WDR91 | 29062 | −3.23624 | 0.049227 |
| SGK223 | 157285 | −3.23513 | 0.049315 |
| AQP1 | 358 | −3.2312 | 0.049791 |
| TMEM114 | 283953 | −3.23056 | 0.049798 |
Lung SCC, lung squamous cell carcinoma; DEG, differentially expressed genes; FC, fold change; FDR, false discovery rate; ABC, ATP-binding cassette.
Ethical Statement: The study was approved by the ethics committee board of Linyi People’s Hospital (No. lyll2015N67) and written informed consent was obtained from all patients.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Lewis DR, Check DP, Caporaso NE, et al. US lung cancer trends by histologic type. Cancer 2014;120:2883-92. 10.1002/cncr.28749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braithwaite KL, Rabbitts PH. editors. Multi-step evolution of lung cancer. Amsterdam: Elsevier, 1999. [DOI] [PubMed] [Google Scholar]
- 3.Tiseo M, Gelsomino F, Alfieri R, et al. FGFR as potential target in the treatment of squamous non small cell lung cancer. Cancer Treat Rev 2015;41:527-39. 10.1016/j.ctrv.2015.04.011 [DOI] [PubMed] [Google Scholar]
- 4.Le Chevalier T, Scagliotti G, Natale R, et al. Efficacy of gemcitabine plus platinum chemotherapy compared with other platinum containing regimens in advanced non-small-cell lung cancer: a meta-analysis of survival outcomes. Lung Cancer 2005;47:69-80. 10.1016/j.lungcan.2004.10.014 [DOI] [PubMed] [Google Scholar]
- 5.Zhang C, Kuang M, Li M, et al. SMC4, which is essentially involved in lung development, is associated with lung adenocarcinoma progression. Sci Rep 2016;6:34508. 10.1038/srep34508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Zhou X, Fan Y, et al. WW45, a Gli1 binding protein, negatively regulated hedgehog signaling in lung cancer. Oncotarget 2016;7:68966-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang S, Zhou B, Wang L, et al. INO80 is required for oncogenic transcription and tumor growth in non-small cell lung cancer. Oncogene 2016. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maschietto M, Tahira AC, Puga R, et al. Co-expression network of neural-differentiation genes shows specific pattern in schizophrenia. BMC Med Genomics 2015;8:23. 10.1186/s12920-015-0098-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Y, Xing Y. Weighted gene co-expression network analysis of pneumocytes under exposure to a carcinogenic dose of chloroprene. Life Sci 2016;151:339-47. 10.1016/j.lfs.2016.02.074 [DOI] [PubMed] [Google Scholar]
- 10.Shi K, Bing ZT, Cao GQ, et al. Identify the signature genes for diagnose of uveal melanoma by weight gene co-expression network analysis. Int J Ophthalmol 2015;8:269-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reiner-Benaim A. FDR control by the BH procedure for two-sided correlated tests with implications to gene expression data analysis. Biom J 2007;49:107-26. 10.1002/bimj.200510313 [DOI] [PubMed] [Google Scholar]
- 13.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289-300. [Google Scholar]
- 14.Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol 2008;4:e1000117. 10.1371/journal.pcbi.1000117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saris CG, Horvath S, van Vught PWJ, et al. Weighted gene co-expression network analysis of the peripheral blood from Amyotrophic Lateral Sclerosis patients. BMC genomics 2009;10:405. 10.1186/1471-2164-10-405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bing Z, Yang G, Zhang Y, et al. Proteomic analysis of effects by x-rays and heavy ion in HeLa cells. Radiol Oncol 2014;48:142-54. 10.2478/raon-2013-0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 2008;24:719-20. 10.1093/bioinformatics/btm563 [DOI] [PubMed] [Google Scholar]
- 18.Carmona-Saez P, Chagoyen M, Tirado F, et al. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol 2007;8:R3. 10.1186/gb-2007-8-1-r3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res 2015;43:D470-8. 10.1093/nar/gku1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008;3:1101-8. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- 22.Darrah RJ, Mitchell AL, Campanaro CK, et al. Early pulmonary disease manifestations in cystic fibrosis mice. J Cyst Fibros 2016;15:736-44. 10.1016/j.jcf.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metodieva SN, Nikolova DN, Cherneva RV, et al. Expression analysis of angiogenesis-related genes in Bulgarian patients with early-stage non-small cell lung cancer. Tumori 2011;97:86-94. [DOI] [PubMed] [Google Scholar]
- 24.Tachihara-Yoshikawa M, Ishida T, Watanabe K, et al. Expression of secretoglobin3A2 (SCGB3A2) in primary pulmonary carcinomas. Fukushima J Med Sci 2008;54:61-72. 10.5387/fms.54.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Välk K, Vooder T, Kolde R, et al. Gene expression profiles of non-small cell lung cancer: survival prediction and new biomarkers. Oncology 2010;79:283-92. 10.1159/000322116 [DOI] [PubMed] [Google Scholar]
- 26.Kaemmerer D, Specht E, Sänger J, et al. Somatostatin receptors in bronchopulmonary neuroendocrine neoplasms: new diagnostic, prognostic, and therapeutic markers. J Clin Endocrinol Metab 2015;100:831-40. 10.1210/jc.2014-2699 [DOI] [PubMed] [Google Scholar]
- 27.Huang QX, Cui JY, Ma H, et al. Screening of potential biomarkers for cholangiocarcinoma by integrated analysis of microarray data sets. Cancer Gene Ther 2016;23:48-53. 10.1038/cgt.2015.66 [DOI] [PubMed] [Google Scholar]
- 28.Nyabam S, Wang Z, Thibault T, et al. A novel regulatory role for tissue transglutaminase in epithelial-mesenchymal transition in cystic fibrosis. Biochim Biophys Acta 2016;1863:2234-44. [DOI] [PubMed]
- 29.Li J, Zhang JT, Jiang X, et al. The cystic fibrosis transmembrane conductance regulator as a biomarker in non-small cell lung cancer. Int J Oncol 2015;46:2107-15. [DOI] [PubMed] [Google Scholar]
- 30.Son JW, Kim YJ, Cho HM, et al. Promoter hypermethylation of the CFTR gene and clinical/pathological features associated with non-small cell lung cancer. Respirology 2011;16:1203-9. 10.1111/j.1440-1843.2011.01994.x [DOI] [PubMed] [Google Scholar]
- 31.Carr JC, Sherman SK, Wang D, et al. Overexpression of membrane proteins in primary and metastatic gastrointestinal neuroendocrine tumors. Ann Surg Oncol 2013;20 Suppl 3:S739-46. 10.1245/s10434-013-3318-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang S, Kim B, Kang HS, et al. SCTR regulates cell cycle-related genes toward anti-proliferation in normal breast cells while having pro-proliferation activity in breast cancer cells. Int J Oncol 2015;47:1923-31. [DOI] [PubMed] [Google Scholar]
- 33.Laakkonen P, Waltari M, Holopainen T, et al. Vascular endothelial growth factor receptor 3 is involved in tumor angiogenesis and growth. Cancer Res 2007;67:593-9. 10.1158/0008-5472.CAN-06-3567 [DOI] [PubMed] [Google Scholar]
- 34.Sehrawat B, Sridharan M, Ghosh S, et al. Potential novel candidate polymorphisms identified in genome-wide association study for breast cancer susceptibility. Hum Genet 2011;130:529-37. 10.1007/s00439-011-0973-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim HC, Lee JY, Sung H, et al. A genome-wide association study identifies a breast cancer risk variant in ERBB4 at 2q34: results from the Seoul Breast Cancer Study. Breast Cancer Res 2012;14:R56. 10.1186/bcr3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao Y, Niu Y, Wang X, et al. Genetic changes at specific stages of breast cancer progression detected by comparative genomic hybridization. J Mol Med (Berl) 2009;87:145-52. 10.1007/s00109-008-0408-1 [DOI] [PubMed] [Google Scholar]
- 37.Kurotani R, Kumaki N, Naizhen X, et al. Secretoglobin 3A2/uteroglobin-related protein 1 is a novel marker for pulmonary carcinoma in mice and humans. Lung Cancer 2011;71:42-8. 10.1016/j.lungcan.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishii T, Hagiwara K, Ikeda S, et al. Association between genetic variations in surfactant protein d and emphysema, interstitial pneumonia, and lung cancer in a Japanese population. COPD 2012;9:409-16. 10.3109/15412555.2012.676110 [DOI] [PubMed] [Google Scholar]